OsFDML1, a rice homolog of Arabidopsis FACTOR OF DNA METHYLATION1, is directly targeted by OsMADS6 and regulates floral organ specification and meristem determination in rice.
Abstract
OsMADS6, an ancient AGAMOUS-LIKE6 (AGL6)-like gene, has essential functions in specifying floral organ and meristem identity in rice (Oryza sativa). However, how AGL6 genes control flower development remains largely unknown. In this study, we report that OsMADS6 directly targets FACTOR OF DNA METHYLATION LIKE 1 (OsFDML1), a rice homolog of the SUPPRESSOR OF GENE SILENCING3-like gene FACTOR OF DNA METHYLATION 1 (FDM1) from Arabidopsis (Arabidopsis thaliana). Arabidopsis FDM1 is involved in RNA-directed DNA methylation and OsFDML1 regulates flower development. The expression of OsFDML1 overlaps with that of OsMADS6 in the palea primordia and the ovule, and OsMADS6 directly promotes OsFDML1 expression through binding to regions containing putative CArG motifs within the OsFDML1 promoter during rice spikelet development. Consistent with the phenotypes of osmads6 mutants, the osfdml1 mutants showed floral defects, including altered palea identity with lemma-like shape containing no marginal region of palea, increased numbers of stigmas and fused carpels, and meristem indeterminacy. Moreover, transgenic plants overexpressing OsFDML1 displayed floral defects, such as abnormal paleae. Phylogenetic analysis showed that OsFDML1 homologs exist only in terrestrial plants. In addition, protein-protein interaction assays showed that OsFDML1 interacts with its close paralog OsFDML2, similar to the activity of OsFDML1 homologs in Arabidopsis. These results provide insight into how the ancient AGL6 gene regulates floral development.
Angiosperms exhibit diverse flower morphologies, which are determined by the precise control of floral organ initiation and formation (Zhang et al., 2013). Previously, extensive molecular and genetic investigations in eudicot species, such as Arabidopsis (Arabidopsis thaliana) and Antirrhinum majus, led to the classical ABC model, in which A genes define the sepals, A and B genes together specify the petals, B and C genes together determine the stamens, C gene defines the pistils, and A and C genes seem to be antagonistic to each other (Coen and Meyerowitz, 1991). A-function genes (APETALA1 [AP1], AP2, and OVULATA), B-function genes (AP3, PISTILLATA, DEFICIENS, and GLOBOSA), and C-function genes (AGAMOUS [AG] and PLENA) exhibit special expression patterns in Arabidopsis and Antirrhinum and are essential for flower development (Coen and Meyerowitz, 1991). The d-function gene SEEDSTICK specifies the ovule identity in Arabidopsis redundantly with AG, SHATTERPROOF1 (SHP1) and SHP2 (Pinyopich et al., 2003; Brambilla et al., 2007). Subsequently, a new class of MADS box genes, SEPALLATA1/2/3/4 (SEP1/2/3/4), were described as E-function genes that specify the identity of all four floral whorls (Pelaz et al., 2000, 2001a,b; Ditta et al., 2004). With these findings, in addition to the ABC model, the “quartet model” was developed, which is directly linked to floral organ specification in different complex combinations of the five types of homeotic transcription factors, i.e. A, B, C, D, and E (Theissen and Saedler, 2001).
Distinct from all eudicots and other monocots, in grass (Poaceae) species, the spikelet constitutes the basic unit of the inflorescence (Barker et al., 2001; Kellogg, 2001; Rudall et al., 2005; Zhang and Yuan, 2014). Each rice (Oryza sativa) spikelet contains two rudimentary glumes, two sterile lemmas and a fertile floret consisting of one lemma and one palea in whorl 1, two lodicules in whorl 2, six stamens in whorl 3, and one pistil in whorl 4 (Bommert et al., 2005; Itoh et al., 2005; Whipple et al., 2007; Yuan et al., 2009; Kellogg, 2015; Zhang et al., 2017).
Recent research in some grass species, such as rice and maize (Zea mays), suggests a modified ABCDE model (Kyozuka et al., 2000; Nagasawa et al., 2003; Prasad et al., 2005; Chen et al., 2006; Dreni et al., 2007; Ohmori et al., 2009; Thompson et al., 2009; Gao et al., 2010; Kobayashi et al., 2010; Li et al., 2010, 2011; Wang et al., 2010; Duan et al., 2012; Sang et al., 2012). Investigation of the expression pattern and function of these homeotic transcription factors in grasses and core eudicots suggests that the ABC model is partially applicable in regulating flower development, even though they have functional divergence (Kyozuka et al., 2000; Preston and Kellogg, 2006; Prusinkiewicz et al., 2007; Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Thompson and Hake, 2009; Zhang and Wilson, 2009; Li et al., 2011; Zhang et al., 2013; Zhang and Yuan, 2014). Briefly, the identities of whorl-1 organs in grasses are controversial, and the definition of A-function genes has not been well established yet.
The function of B genes seems highly conserved in both monocots and eudicots based on the finding of homeotic mutations of Silky1 in maize and SUPERWOMAN1 (SPW1) in rice, orthologous to the B gene AP3 in Arabidopsis. These mutations transform the lodicule and stamen into lemma/palea-like and carpel organs, respectively (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2007). The double mutants of two rice C genes, OsMADS3 and OsMADS58, display the conversion of carpel into a palea-like structure, suggesting that OsMADS3 and OsMADS58 redundantly regulate stamen and carpel identity and flower meristem determination, mimicking the function of the ortholog AG in Arabidopsis (Dreni et al., 2011). This clearly indicates that C function is conserved within flowering plants (Dreni et al., 2011; Dreni and Kater, 2014). Rice contains two D function genes, namely, OsMADS13 and OsMADS21 (Dreni et al., 2007; Li et al., 2011), and OsMADS13 specifies ovule identity and floral meristem determination, together with OsMADS3 and OsMADS58 (Dreni et al., 2011; Hu et al., 2015). Although OsMADS21 does not seem to be crucial for reproductive organ formation, its protein product retains the potential of ovule identity specification (Dreni et al., 2011).
Rice possesses at least five SEP-like genes, namely, OsMADS1/LEAFY HULL STERILE1 (LHS1), OsMADS5, OsMADS7, OsMADS8, and OsMADS34/PANICLE PHYTOMER2, and they have divergent functions during flower development (Malcomber and Kellogg, 2004, 2005; Zahn et al., 2005; Arora et al., 2007; Wu et al., 2018). OsMADS1 specifies the floral organ identity and meristem determination (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006; Hu et al., 2015). Transgenic plants in which the expression of OsMADS1, OsMADS5, OsMADS7, and OsMADS8 is simultaneously reduced show homeotic conversion of all floral organs, except the lemma, into leaf-like structures, mimicking the defects observed in sep1 sep2 sep3 sep4 quadruple mutants in Arabidopsis (Cui et al., 2010). OsMADS34 plays critical roles in rice inflorescence and spikelet development (Gao et al., 2010; Kobayashi et al., 2010). OsMADS32/CHIMERIC FLORAL ORGANS1 is a rice-specific MADS gene with critical roles in specifying floral organ identity by interacting with OsMADS2 and OsMADS4 (Sang et al., 2012; Wang et al., 2015).
AGL6 genes are ancient, found in both gymnosperms and angiosperms, and they are closely related to SEP- and AP1-like genes at the sequence level (Reinheimer and Kellogg, 2009; Li et al., 2010; Dreni and Zhang, 2016; Callens et al., 2018). Recent studies from monocots and core eudicots indicate that AGL6-like genes function in flower development (Reinheimer and Kellogg, 2009; Li et al., 2010; Dreni and Zhang, 2016). In grasses, AGL6-like genes have the ancient expression pattern in the ovule, but a diversified pattern in the palea (Reinheimer and Kellogg, 2009; Dreni and Zhang, 2016). Rice contains two AGL6-like genes: OsMADS17 plays minor and redundant roles, whereas OsMADS6 is a key regulator of flower development (Ohmori et al., 2009; Dreni and Zhang, 2016). OsMADS6 regulates flower development of four whorls in rice, similar to the function of SEP in Arabidopsis (Ohmori et al., 2009; Li et al., 2010; Dreni and Zhang, 2016). Allelic mutants of OsMADS6 exhibit obvious defects in floral organ identity specification and meristem determination, showing wider lemma-like palea (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010; Duan et al., 2012). In addition, their lodicules and stamens homeotically exhibit conversion into glume-like or mosaic structures, and ectopic carpels develop occasionally (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010; Duan et al., 2012). Moreover, OsMADS6 genetically interacts with other B, C, D, and E MADS genes in specifying floral organ identity and meristem fate during spikelet development (Li et al., 2010, 2011). OsMADS6 forms a protein complex with OsMADS1 (Moon et al., 1999), and osmads6 osmads1 (mfo1-2 lhs1-2; osmads1-z osmads6-1) display severe defects of floral meristem identity, which suggests that OsMADS6 and OsMADS1 together specify the state of rice flowers (Ohmori et al., 2009; Li et al., 2010). The specification of floral organ identity of the inner three whorls and meristem fate strongly depends on the additive effects and redundant functions of OsMADS6 with OsMADS16, OsMADS3, and OsMADS13 (Li et al., 2011). Interestingly, OsMADS6 may directly activate OsMADS58 by binding to a putative CArG box motif, and the double mutant osmads6-1 osmads58 exhibits higher numbers of stigmas and ectopic carpels/ovules, although it dramatically exhibits the phenotype of osmads6-1 in the outer three whorls (Li et al., 2011). These reports suggest that autoregulation exists between these MADS box genes. Undoubtedly, OsMADS6 plays key roles in rice spikelet specification by interacting with other homeotic MADS box genes. Thus, as an indispensable homeotic transcription factor in rice spikelet establishment, an underlying regulating network of OsMADS6 is expected.
In this study, we report that OsMADS6 directly activates the expression of one target gene OsFDML1, which is homologous to SUPPRESSOR OF GENE SILENCING3 (SGS3)-like genes in Arabidopsis. OsFDML1 belongs to the gene family found only in terrestrial plants and mainly expressed in inflorescence and spikelet organs. Mutants of OsFDML1 and ectopic expression of OsFDML1 caused abnormal flower development. This work defines a regulatory pathway of AGL6-like genes in regulating flower morphogenesis.
RESULTS
OsFDML1 Is a Direct Target of OsMADS6
To clarify the regulatory role of OsMADS6, we performed additional analyses from the microarray data of the osmads6-1 mutant at spikelet developmental stage Sp6 generated by Li et al. (2011). From the genes with changed expression in osmads6-1 (Li et al., 2011), we selected eight genes encoding transcription factors and kinases for further study (Supplemental Table S1). Reverse transcription quantitative PCR (RT-qPCR) analysis using 2- to 3-mm-long young inflorescences at stages of spikelet development Sp6 confirmed that seven genes were upregulated and one gene was downregulated in osmads6-1.
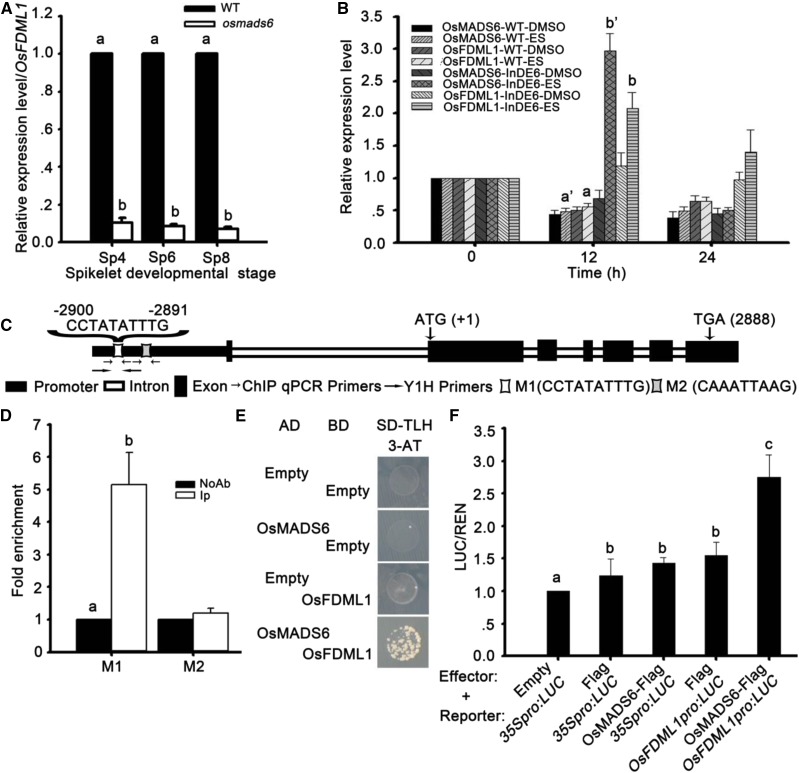
To test whether OsMADS6 directly regulates the expression of these eight genes, we cloned the coding sequence of OsMADS6 into the hormone-inducible expression vector LexA-VP16-ER (XVE), which employs the human estrogen receptor (hER) to confer β-estradiol inducible expression of a chimeric transcription factor (Zuo et al., 2006). Using the 0.5- to 1-cm root tips from 1-week-old young seedlings of transgenic plants containing OsMADS6 fused with XVE (henceforth referred to as InDE6), we observed that the expression of LOC_Os02g19130, encoding a putative transcription factor X1 gene, was rapidly increased after 12 h of treatment with 5 μmol/L β-estradiol (Fig. 1B). The expression level was similar to that of the expression of OsMADS6 in transgenic plants of InDE6 (Fig. 1B).
Figure 1.
OsMADS6 directly activates the expression of OsFDML1 in vivo during rice spikelet development. A, The expression of OsFDML1 decreased in osmads6-1 at spikelet developmental stages Sp4, Sp6, and Sp8. Spikelets at stages Sp4, Sp6, and Sp8 were collected from inflorescences of lengths 2, 2 to 3, and 4 to 7 mm, respectively. B, The expression of OsFDML1 increased with the induced expression of OsMADS6 in the transgenic plants InDE6. C, Diagram of OsFDML1 gene with promoter regions containing the putative CArG motif. D, ChIP-qPCR results showed OsMADS6 can directly bind to OsFDML1 promoter regions with putative CArG motif (M1) using 2- to 8-mm-long young inflorescences. The mock treatment included negative control treated without antibody and 200-bp OsFDML1 promoter region containing an unclassical CArG motif M2 close by to the motif M1. The fold enrichment was calculated relative to the negative control without antibody. E, OsMADS6 bound to the OsFDML1 promoter regions with putative CArG motif (M1) and activated the expression of nutritional reporter gene HIS3 by yeast one-hybrid assay. F, OsMADS6 triggered the expression of OsFDML1pro:LUC with integrating 3795-bp OsFDML1 genome sequence upstream of the ATG before LUC. All data are means ± sd (n ≥ 3). Letters a, b, c, a’, and b’ represent statistical significance, P < 0.05. IP, Immunoprecipitation with specific antibody against OsMADS6; InDE6, transgenic plants containing OsMADS6 fused with XVE; ES, β-estradiol.
Based on the decreased expression of this gene in osmads6-1 at stages Sp4, Sp6, and Sp8 during spikelet development (Fig. 1A), we expected that this gene (LOC_Os02g19130) would probably be a direct target of OsMADS6. Since LOC_Os02g19130 is homologous to the SGS3-like gene FACTOR OF DNA METHYLATION1/2/3/4/5 (FDM1/2/3/4/5) in Arabidopsis, we renamed it rice FACTOR OF DNA METHYLATION LIKE 1 (OsFDML1).
We then performed chromatin immunoprecipitation (ChIP)-qPCR analysis using 2- to 8-mm-long young inflorescences and purified rabbit polyclonal antibodies against OsMADS6 (Li et al., 2011). ChIP-qPCR showed specific enrichment of the 198-bp DNA fragment containing the putative CArG motif M1 (CCTATATTTG) within the OsFDML1 promoter, but not the 200-bp DNA fragment containing the unclassical CArG motif M2 (CAAATTAAG) close by to the motif M1 (Fig. 1, C and D). In addition, yeast one-hybrid results showed that OsMADS6 could bind to the fragment containing the putative CArG motif and activate the expression of the HIS3 nutritional reporter gene in the vector (Fig. 1E). Furthermore, compared with the empty vector and the modified one containing the tag Flag as negative controls, only the fused protein of OsMADS6-Flag triggered significantly increased expression of OsFDML1pro:LUC by coexpressing OsFDML1pro:LUC transiently with OsMADS6-Flag in Nicotiana benthamiana leaves (Fig. 1F). Taking these results together, we propose that OsMADS6 directly activates the expression of OsFDML1 by binding to the regions containing the putative CArG motif within the OsFDML1 promoter during rice spikelet development.
Expression and Phylogenetic Analysis of OsFDML1
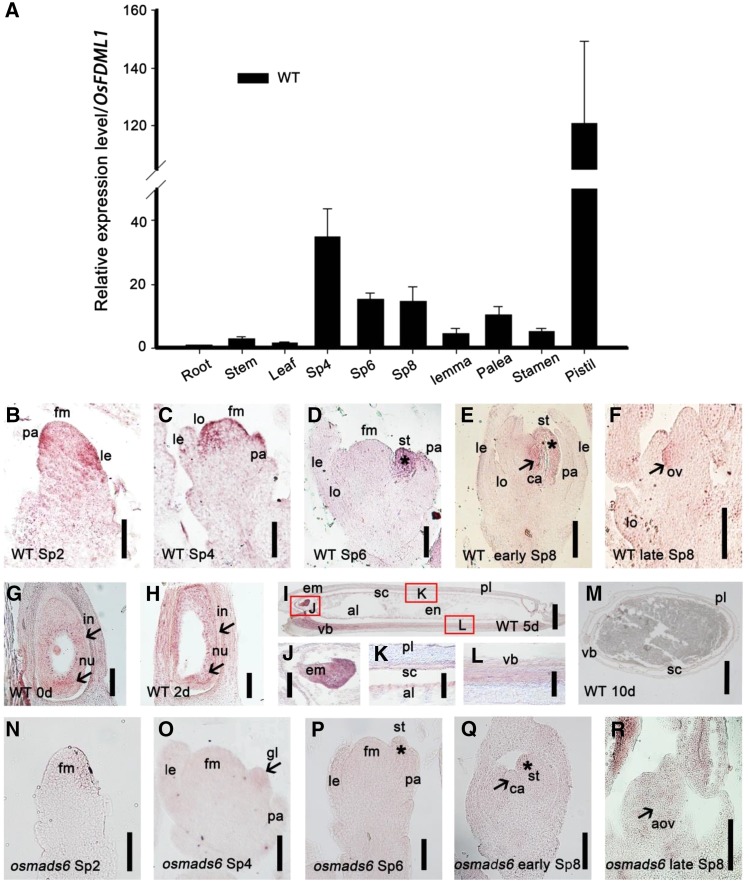
To address the function of OsFDML1, we performed RT-qPCR and showed that the expression of OsFDML1 mainly occurred in the spikelet and was strong in pistils but very weak in roots, stems, and leaves (Fig. 2A). In addition, our mRNA in situ hybridization results showed signals of OsFDML1 transcripts mainly at the tip of the palea in longitudinal sections (Fig. 2, B–D), and these signals disappeared very soon after the initiation of stamens (inflorescence length 4–7 mm; Fig. 2E). Furthermore, expression of OsFDML1 was seen in lemma primordia only at stage Sp2 (inflorescence length 1–1.5 mm; Fig. 2B). Surprisingly, we detected expression of OsFDML1 specifically in the stamen adjacent to the palea, but not in other stamens (Fig. 2, D and E). Moreover, expression of OsFDML1 was detected during the initiation of the carpel and ovule (Fig. 2, E and F). We detected OsFDML1 RNA signals in the nucellus and strong signals in the ovule, but relatively weak signals in the integument of the ovary (Fig. 2, G and H). The strongest signals were detected in 5-d-old young embryo and vascular bundle (Fig. 2, I, J, and L). With nucellus maturation, we detected relatively weak expression of OsFDML1 in the inner layer of the pericarp, young endosperm, and aleurone layer (Fig. 2, I and K), and no transcripts in the 10-d-old ovary (Fig. 2M). Moreover, expression of OsFDML1 was almost undetectable in osmads6-1 floral organs on mRNA in situ hybridization (Fig. 2, N–R), which is consistent with the regulatory role of OsMADS6 in activating the expression of OsFDML1. Overall, the expression of OsFDML1 mostly overlapped that of OsMADS6 in the floral organs, confirming that OsMADS6 is critical in modulating the expression of OsFDML1 during flower development.
Figure 2.
Expression pattern of OsFDML1 in the wild-type and osmads6-1 young spikelets. A, RT-qPCR analysis of the expression of OsFDML1 in wild-type root, stem, leaf, and spikelets at stages Sp4, Sp6, and early Sp8, and dissected lemma, palea, stamen, and pistil, respectively. Root was collected from young seedlings cultivated 10 d in water. Stem and leaf were collected when the plants changed to reproductive growth phase. Spikelets at stages Sp4, Sp6, and Sp8 were sampled from inflorescences of lengths ∼2, 2 to 3, and 4 to 7 mm, respectively. Lemma, palea, stamen, and pistil were dissected from the spikelets of length ∼6 mm. All data are means ± sd (n ≥ 3). B to F, In situ hybridization experiment showing the expression of OsFDML1 in wild-type young spikelets at stages Sp2, Sp4, Sp6, early Sp8, and late Sp8. Spikelets at stages Sp2, Sp4, Sp6, early Sp8, and late Sp8 were sampled from inflorescences of lengths ∼1 to 1.5, 2, 2 to 3, 4 to 7, and 10 to 20 mm, respectively. Asterisks indicate the palea side stamen in D and E. Arrowheads indicate the carpel primordia in E and the ovule in F. G to M, In situ hybridization experiment showing the expression of OsFDML1 in wild-type ovaries representing 0, 2, 5, and 10 d since pollination. J to L, partial enlargements of I. Arrowheads indicate the integument and nucellus in G and H, respectively. Red rectangles indicate the position of J to L in I. N to R, In situ hybridization experiment showing the expression of OsFDML1 in osmads6-1 spikelets at stages Sp2, Sp4, Sp6, early Sp8, and late Sp8, respectively. Asterisks indicate the palea side stamens in P and Q. Arrowheads indicate the glume-like structure of osmads6-1 in O, carpel primordia of osmads6-1 in Q, and the abnormal ovule of osmads6-1 in R. le, Lemma; pa, palea; gl, glume-like structure; lo, lodicules; st, stamen; fm, floral meristem; ca, carpel; ov, ovule; aov, abnormal ovule; nu, nucellus; in, integument; em, embryo; pl, pericarp; sc, seed capsule; al, aleurone; vb, vascular bundle. Bars = 50 μm in B, C, F, N, O, and R, 100 μm in D, E, J to L, P, and Q, and 500 μm in G to I and M.
To determine the evolutionary relationship of OsFDML1-like genes, we used the full-length protein sequence of OsFDML1 as a query to search the homologs of OsFDML1 in the proteome database (http://phytozome.jgi.doe.gov) and Congenie database (http://congenie.org). In total, we obtained 119 OsFDML1-like genes from 14 terrestrial plant species, including Physcomitrella patens, gymnosperms, monocots, and core eudicots, but no close relatives in green algae (Supplemental Table S2). In addition, most of the OsFDML1-like members contained the conserved XS (named after rice homolog X and Arabidopsis SGS3) and XH (rice gene X Homology) domains (Bateman, 2002), presented as tandem repeats with dramatic sequence variation. Therefore, phylogenetic analysis using the mostly conserved XH domain from P. patens moss; Pinus taeda (a gymnosperm); Amborella trichopoda (a sister species to all the extant angiosperms); Arabidopsis, Prunus persica, and Vitis vinifera Genoscope (core eudicots); rice and Brachypodium distachyon (commelinid monocots, Poaceae); and Ananas comosus and Musa acuminata (other commelinid monocots) from both the maximum likelihood and neighbor-joining approaches failed to obtain highly supported nodes. However, our sequence analysis suggested that these OsFDML1-like genes might have specific functions in terrestrial plants and undergo frequent tandem duplications and sequence variation as well as functional diversification. Further studies on the molecular mechanisms and generation of more knockout or knockdown mutants will clarify the global functions of this gene family.
The osfdml1 Mutants Exhibit Defects in Floral Organ Identity and Meristem Determination
To further clarify the function of OsFDML1, we identified two osfdml1 mutants: osfdml1-1 (PFG_1C-03064, Korea) containing a T-DNA inserted at the N terminus of the first exon, and osfdml1-2 (NE4801, Japan) containing a Tos17 retrotransposon inserted at the C terminus of the first exon (Supplemental Fig. S1A). RT-PCR results using mRNA extracted from the 0.5-cm-long inflorescence at spikelet developmental stage Sp8 revealed that expression of OsFDML1 was almost undetectable in osfdml1-1 (Supplemental Fig. S1B). The expression of OsFDML1 after Tos17 insertion was significantly reduced in osfdml1-2, whereas the transcript level of OsFDML1 before Tos17 insertion showed no significant change (Supplemental Fig. S1B). Moreover, RT-qPCR showed that the expression of OsFDML1 after Tos17 insertion was reduced to approximately half of that in osfdml1-2 (Supplemental Fig. S1C). Therefore, osfdml1-1 most likely represents a knockout mutant and osfdml1-2 is more likely to be a knockdown mutant. Both osfdml1 mutant alleles appeared recessive from the genetic analysis of osfdml1-1 and osfdml1-2 backcrossed with the wild-type Hwayoung and Nipponbare cultivars, respectively. In both cases, the F1 heterozygous plants showed normal spikelets but the segregated homozygous plants from F2 progenies displayed floral defects (Supplemental Fig. S2).
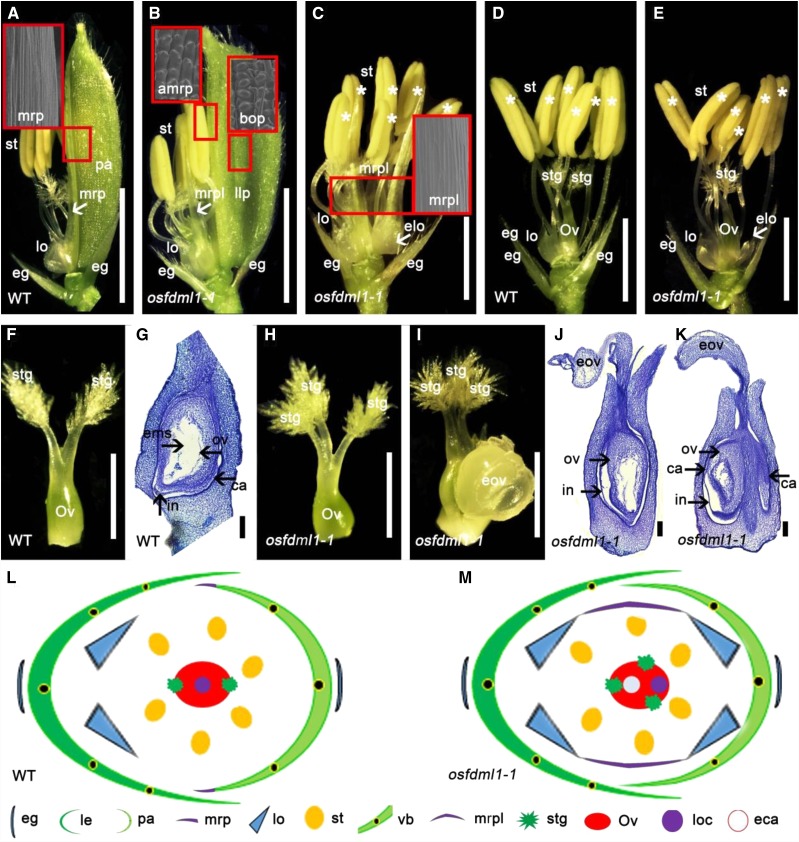
Both osfdml1 mutants displayed no visible differences during vegetative development compared with the wild type. Even though both mutants showed no obvious change in primary branch number, the numbers of secondary branches statistically decreased in the mutants (Supplemental Fig. S4, A–D). The number of secondary branches was ∼11.7 in osfdml1-1 compared with 14.3 in wild-type Hwayoung and 6.0 in osfdml1-2 compared with 8.3 in wild-type Nipponbare (Supplemental Fig. S4E). These values differ from that of osmads6, in which no defects of inflorescence branches were found. We observed that ∼33.3% of osfdml1-1 spikelets displayed defects in palea, lodicule, and pistil development (Supplemental Table S3). Compared with the wild-type plants, ∼3.4% of the spikelets of osfdml1-1 formed wider lemma-like paleae with abnormal marginal region of palea (MRP) tissues in whorl 1 and MRP-like structures and extra lodicules in whorl 2 (Fig. 3, A–C; Supplemental Table S3). Scanning electron microscopy revealed that the cell morphology in the margins of lemma-like palea was converted to that of the body region of the palea (BOP) (Fig. 3B). Normally, the MRP-like structures fused with lodicules (Fig. 3C). Approximately 9.2% of osfdml1-1 spikelets developed extra lodicules adjacent to the palea in whorl 2 (Fig. 3, D and E; Supplemental Table S3). In contrast to the wild-type pistil normally containing two stigmas, and one locule with one ovule covered by two layers of integuments and one embryo sac (Irish et al., 2003; Tanaka et al., 2014; Fig. 3, F and G), ∼20.7% of osfdml1-1 pistils developed three stigmas, of which ∼8.6% showed abnormal exposed ovules (Fig. 3, H–K; Supplemental Table S3). Occasionally, the defective pistils developed two carpels fused together (Fig. 3K). In addition, similar defects were observed in osfdml1-2 spikelets (Supplemental Fig. S3; Supplemental Table S4). These observations suggest that OsFDML1 can specify floral organ identity, including palea, lodicule, and pistil, and floral meristem determination in rice (Fig. 3, L and M). Given the similarity between osfdml1 and osmads6 in spikelet phenotypes, we proposed that OsFDML1 and OsMADS6 genetically function in the same pathway during rice spikelet development.
Figure 3.
Phenotypes of the wild-type and osfdml1-1 spikelets. A, Wild-type spikelet (genetic background Hwayoung) with dissected lemma. Arrowhead indicates the MRP structure. The red rectangles indicate the position of scanning electron microscopy and the cell identity of the MRP structure. B, osfdml1-1 spikelets developing a lemma-like palea in whorl 1 and MRP-like structures in whorl 2. Arrowhead indicates the MRP-like structure. The red rectangles indicate the positions of scanning electron microscopy and the cell identity of the BOP and abnormal MRP structures. C, osfdml1-1 spikelet with dissected lemma and the lemma-like palea showing extra lodicules and MRP-like structure in whorl 2. Arrowhead indicates the extra lodicules in the palea side. The red rectangles indicate the positions of scanning electron microscopy and the cell identity of the MRP-like structure. D, Wild-type spikelet with lodicules in lemma side, six stamens, and one pistil with two stigmas. Asterisks indicate the stamens. E, osfdml1-1 spikelets showing extra lodicules in palea side. Arrowhead indicates the extra lodicules in palea side. Asterisks indicate the stamens. F, Wild-type pistil with two stigmas. G, Vertical section of the wild-type ovary. Arrowheads indicate the carpel, integument, ovule, and embryo sac. H, osfdml1-1 pistil with three stigmas. I, osfdml1-1 pistil with three stigmas and an exposed ovule. J and K, Vertical section of the osfdml1-1 ovary. Occasionally two ovaries fused together (K). Arrowheads indicate the carpel, integument, and ovule. L and M, Diagrams of the wild-type and osfdml1-1 spikelets. eg, Empty glum; pa, palea; llp, lemma-like palea; st, stamen; lo, lodicule; elo, extra lodicules; mrp, marginal region of palea; mrpl, mrp-like structure; bop, body of palea; amrp, abnormal mrp structure; Ov, ovary; stg, stigma; ov, ovule; in, integument; ems, embryo sac; ca, carpel; eov, exposed ovule; vb, vascular bundle; loc, locule; eca, extra carpel. Bars = 2 mm in A and B, 1 mm in C to E, 500 μm in F, H, and I, and 100 μm in G, J, and K.
Ectopic Expression of OsFDML1 Causes Abnormal Floral Development
To further clarify the function of OsFDML1, we constructed OsFDML1-overexpressing transgenic (termed as OET6) plants driven by the constitutive CaMV 35S promoter. The expression of OsFDML1 increased significantly in the spikelets of OET6 lines, which displayed ∼25% of floral defects (Fig. 4, G and H). Compared with the wild-type palea, the OET6 developed a slightly wider palea with ectopic MRP tissues (Fig. 4, A–D). Paraffin section observation from OET6 lines showed that the ectopic MRP structures of OET6 developed an additional vascular bundle and additional BOP-like tissues composed of silicified cells, fibrous sclerenchyma, spongy parenchyma, and vacuolated inner surface; the MRP tissues showed a smooth epidermis lacking silicified thickening (Fig. 4, E and F). Moreover, scanning electron microscopy showed chimeric MRP/BOP-like mixed tissues extending into the MRP structures in OET6 palea (Fig. 4D). These findings suggest that OsFDML1 plays a key role in the establishment of palea identity, similar to that of OsMADS6.
Figure 4.
Phenotypes of OsFDML1 ectopic-expressed spikelets. A and B, Wild-type spikelet (A) with dissected lemma (B). Arrowhead indicates the MRP structure. The red rectangles indicate the positions of scanning electron microscopy and the cell identity of the BOP and MRP structures. C and D, Phenotype of OsFDML1 overexpressing spikelet (C) with ectopic wider MRP structure (D). Arrowhead indicates the ectopic MRP structure. The red rectangles indicate the position of scanning electron microscopy and the cell identity of the BOP, BOP-like, and MRP structures. E and F, Cross section of the wild-type MRP (E) and the ectopic wider MRP structure of OET6 (F). The vascular bundles are marked by red rectangles. Double-headed arrow indicates the additional BOP-like structure. G, Statistical analysis of abnormal flowers of the wild-type and OET6 inflorescences. All data are means ± sd (n ≥ 5). H, Relative expression level of OsFDML1 of the wild-type and OET6 spikelet. All data are means ± sd (n ≥ 3). Letters a and b represented statistical significance, P < 0.05. eg, Empty glume; le, lemma; pa, palea; mrp, marginal region of palea; emrp, ectopic mrp structure; bop, body of palea; bopl, bop-like structure; lo, lodicule; st, stamen; vb, vascular bundle; avb, additional vascular bundle. Bars = 2 mm in A to D and 100 μm in E and F.
OsFDML1 Forms Heterodimers with Its Close Paralog OsFDML2
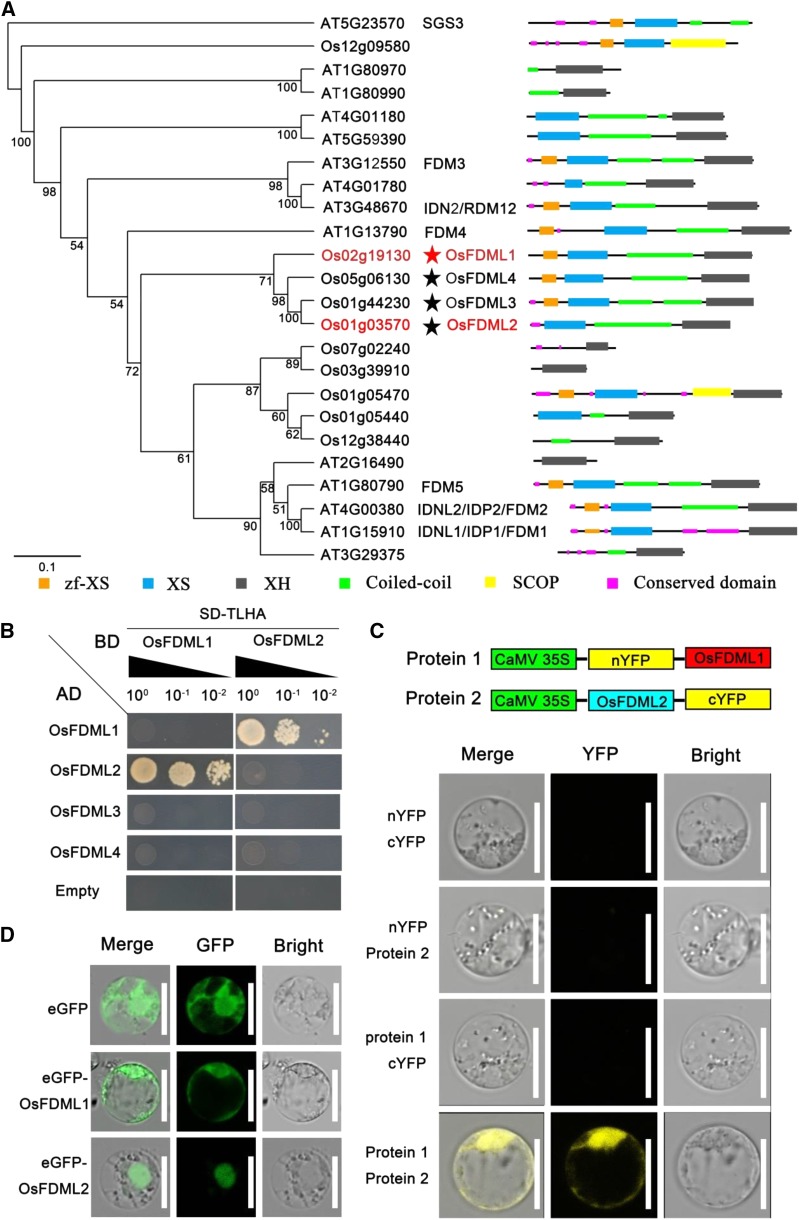
Sequence analysis showed that 10 members of OsFDML1-like genes were present in rice. Within the subgroup containing OsFDML1, the four paralogs (OsFDML1/2/3/4) exhibited high sequence similarity and contained the conserved domains of XS and XH (Fig. 5A). Given that the homolog in Arabidopsis INVOLVED IN DE NOVO 2/RNA-directed DNA methylation 12 (IDN2/RDM12) functions as homodimers or heterodimers with its paralogs IDN2-like 1 (IDN2L1)/IDN2 PARALOG 1 (IDP1)/FDM1, and IDN2L2/IDP2/FDM2 involved in the RdDM pathway in vivo (Ausin et al., 2012; Xie et al., 2012a,b; Zhang et al., 2012), we tested whether OsFDML1 could form protein complexes with its close paralogs in the same subgroup. Yeast two-hybrid analysis showed that only one paralog OsFDML2 could interact with OsFDML1 and that OsFDML1 could not form homodimers (Fig. 5B). To further confirm the interaction between OsFDML1 and OsFDML2, we performed a bimolecular fluorescence complementation (BiFC) assay in rice protoplasts and proved that OsFDML1 indeed interacted with its close paralog OsFDML2 in vivo (Fig. 5C).
Figure 5.
OsFDML1 functions as heterodimers with its close paralog OsFDML2. A, The neighbor-joining phylogenetic tree of OsFDML1 with homologs in rice and Arabidopsis. The bootstrap values (%) based on 100 replicates are marked at the branching points. The scale bar indicates the number of nucleotide substitutions per site. OsFDML1 and the interacting protein OsFDML2 are marked in red. The red star indicates OsFDML1. The black stars indicate the other three paralogs OsFDML2, OsFDML3, and OsFDML4 in rice belonging to the same subgroup. B, OsFDML1 interacts with its close paralog OsFDML2 by yeast two-hybrid assay. C, OsFDML1 interacts with OsFDML2 in vivo by BiFC assay in rice protoplast. D, OsFDML1 localizes in nucleus and cytoplasm, and OsFDML2 localizes in nucleus in rice protoplast. Bars = 10 μm in C and D.
In addition, we found that OsFDML2 is mainly expressed in the spikelet, particularly high in the pistil, which overlaps with OsFDML1 (Supplemental Fig. S6). Furthermore, both the fused proteins of eGFP-OsFDML1 and eGFP-OsFDML2 were shown to be localized specifically in the nucleus of rice protoplasts (Fig. 5D).
DISCUSSION
Rice OsMADS6 belongs to the ancient AGL6 gene family, found in both gymnosperms and angiosperms, which plays roles in flower development in monocots and core eudicots (Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Li et al., 2010; Dreni and Zhang, 2016). The rice genome contains two AGL6-like genes, OsMADS6 and OsMADS17, which are involved in flower development (Ohmori et al., 2009; Li et al., 2010; Dreni and Zhang, 2016). OsMADS6 is expressed in the floral meristem, palea primordia, lodicule, and carpel primordia, and with maturation of the spikelet, OsMADS6 transcripts are detectable in the ovule and integument (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010). Mutation of OsMADS6 caused severe defects in the spikelet, such as widened palea without MRP tissues, ectopic elongated lodicules and glume-like structures, and multiple pistils with multiple stigmas and independent or fused carpels, representing SEP-like function (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010; Duan et al., 2012; Dreni and Zhang, 2016). However, given that OsMADS6 genetically interacts with the floral homeotic genes SPW1, OsMADS3, OsMADS1, OsMADS7, and OsMADS8 and directly regulates OsMADS58 in specifying floral organ identity and meristem determination (Li et al., 2010, 2011), it remains largely unknown how AGL6 members regulate flower development.
In this study, we elucidated that OsFDML1 is a direct downstream target of OsMADS6, as evidenced by the down-regulation of OsFDML1 in osmads6 young spikelets and increased expression of OsFDML1 in transgenic plants when expression of OsMADS6 is ectopically induced (Fig. 1, A and B). In addition, we proved the direct binding by ChIP-PCR, yeast one-hybrid, and dual-LUC assays (Fig. 1, C–E). OsFDML1 was mainly expressed in the palea primordia, carpel primordia, and ovule (Fig. 2, C–F), overlapping with the expression of OsMADS6 (Ohmori et al., 2009; Li et al., 2010; Dreni and Zhang, 2016). The osfdml1 mutants exhibited lemma-shaped paleae without MRP tissues, increased number of stigmas, occasionally fused carpels, and indeterminate floral meristem, partially phenocopying the role of OsMADS6 in rice floral organ identity specification and meristem determination. The contribution of OsFDML1 to palea identity specification is demonstrated by the defective palea in osfdml1 with the lateral outgrowth of BOP in whorl 1 and MRP-like structures in whorl 2 (Fig. 3, B and C; Supplemental Fig. S3, B and C), and transgenic plants overexpressing OsFDML1 displayed abnormal palea phenotypes with lateral outgrowth of BOP (Fig. 4, D and F). Recent studies indicate that the palea may represent the fusion of three originally distinct organs that evolved into the two lateral MRP and the BOP (Reinheimer and Kellogg, 2009; Lombardo and Yoshida, 2015). Based on the disruption of MRP and BOP formation in osfdml1 mutants and OET6, we expect that some boundary control factors may determine the fusion of MRP and BOP tissues. For example, the rice mutant degenerated hull1, which contains the mutation of a lateral organ boundary domain gene (Li et al., 2008), developed extra MRP-like organs in inner whorls with degenerative lemma and palea (Li et al., 2008). RETARDED PALEA1 (REP1) encodes a CYCLOIDEA-like protein, which specifies palea identity and development (Yuan et al., 2009). It is important to determine the relationship of OsFDML1 with DH1 and REP1 in specifying palea identity. Consistent with the regulatory role of OsMADS6, osfdml1 developed abnormal pistils, including those with three stigmas and exposed ovules, which occasionally had two carpels fused together (Fig. 3, H–K). Similarly, osmads6 mutants produced multiple pistils with multiple stigmas and independent or fused carpels (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010). In maize, bearded-ear encoding the AGL6-like MADS box gene zea agamous3 exhibited protruding nucellus (Thompson et al., 2009).
The osfdml1 mutants have weaker floral defects than the osmads6 mutants because OsMADS6 may have multiple regulatory targets, which remain to be discovered. Alternatively, there is functional redundancy of OsFDML1 because rice has nine other homologs of OsFDML1 that contain the featured XS and/or XH domains (Chen and Bennetzen, 1996; Bateman, 2002). However, few of them have been functionally characterized due to lack of knockout or knockdown mutants. In contrast to rice osfdml1 mutants, the single mutation idn2/rdm12 and the sextuple mutations idn2-3 fdm1-1 idp2-1 fdm3-2 fdm4-2 fdm5-2 cause no obvious developmental defects, and they only result in minor changes in DNA methylation (Ausin et al., 2009; Zheng et al., 2010; Butt et al., 2014), suggesting strong redundancy of closely related proteins in Arabidopsis. These results indicate that members of this family may undergo a certain degree of functional diversification between monocots and eudicots during evolution. In Arabidopsis, the SGS3-like IDN2/RDM12, functioning as a homodimer and heterodimer in vivo with its two paralogs IDNL1/IDP1/FDM1 and IDNL2/IDP2/FDM2, potentially stabilizes the AGO4-delivered small interfering RNA to the correct chromatin sites, and is an important component in the RdDM pathway (Mourrain et al., 2000; Bateman, 2002; Ausin et al., 2012; Finke et al., 2012; Xie et al., 2012b; Zhang et al., 2012). This study revealed that rice OsFDML1 functions as a heterodimer by interacting with its close paralog OsFDML2 in both yeast two-hybrid and BiFC assays (Fig. 5, B and C), mimicking the homologs in Arabidopsis, which suggests that the genes responsible for the functions of this family might have been conserved in the evolution of flowering plants. Given that IDN2/RDM12 in Arabidopsis contributes to DNA methylation in the RdDM pathway, it would be interesting to test whether OsFDML1 regulates flower development via epigenetic modifications.
Moreover, in Arabidopsis, floral MADS box genes were suggested to regulate the expression of genes associated with chromatin structure remodeling (Yan et al., 2016). For example, Arabidopsis E-class protein SEP3 was identified to recruit SWITCH 2/SUC NONFERMENTING2 (SWI2/SNF2) chromatin-remodeling ATPases BRAHMA and SPLAYED to the regulatory loci of AP3 and AG, resulting in the induction of their expression by overcoming Polycomb repression (Smaczniak et al., 2012; Wu et al., 2012). In Arabidopsis, IDN2 dimers were shown to physically interact with SWI3B, a core subunit of SWI/SNF ATP-dependent chromatin-remodeling complex, which positioned nucleosomes and affected transcription (Zhu et al., 2013). Thus, our findings provide key insights into the regulatory network of AGL6-like genes and may illustrate a new link between genetics (AGL6) and epigenetics, adding to the current understanding of epigenetic mechanisms and chromatin structure in rice flower development.
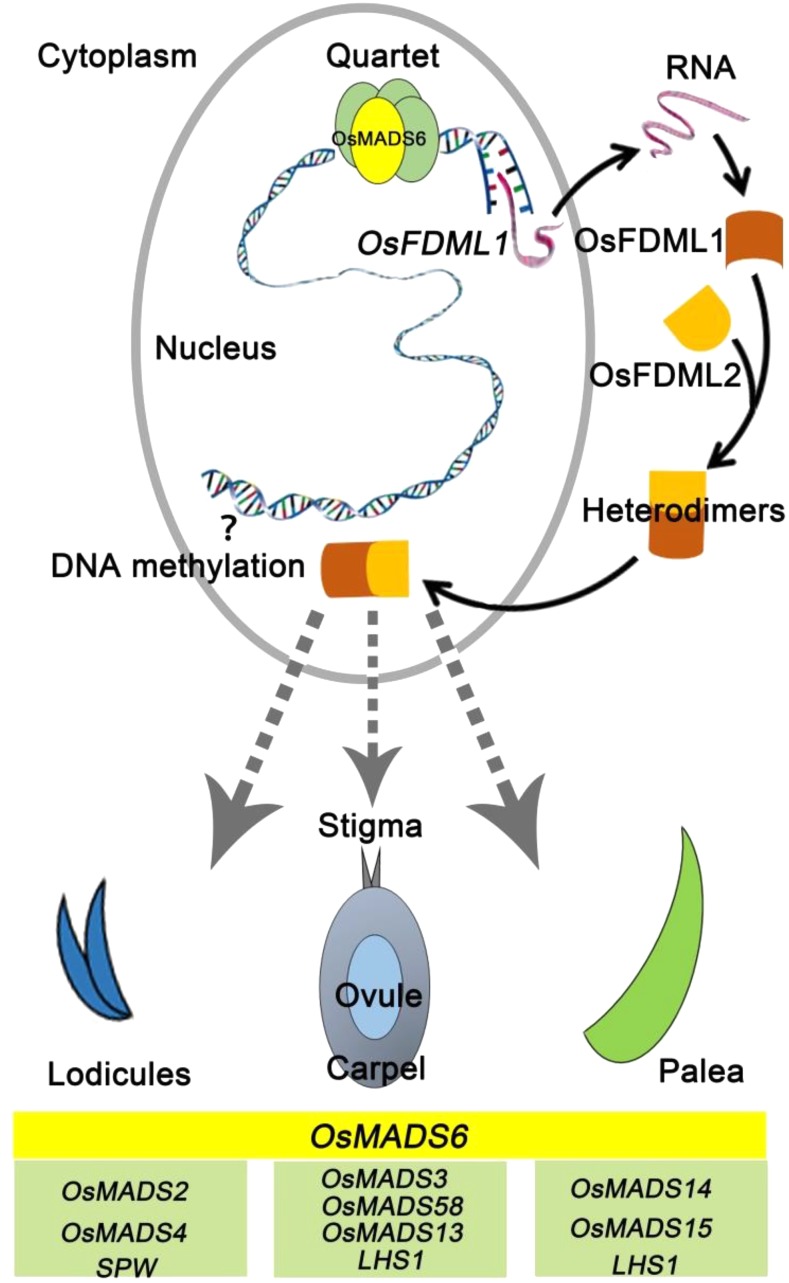
In summary, based on the observation of the spikelet phenotype of plants with altered expression of OsFDML1 and its expression pattern, as well as the heterodimers formed by OsFDML1 and its close paralog OsFDML2, we have presented a model in which the transcription factor OsMADS6 directly activates the expression of OsFDML1, and the protein OsFDML1 forms heterodimers with OsFDML2 to regulate rice flower development, probably via DNA methylation modification (Fig. 6).
Figure 6.
An updated model for the regulation of palea, lodicule, and pistil development by OsMADS6 via direct activation of OsFDML1. OsFDML1 functions as heterodimers with its close paralog OsFDML2.
MATERIALS AND METHODS
Plant Materials
The recessive mutants osfdml1-1 (PFG_1C-03064, ‘Hwayoung’) and osfdml1-2 (NE4801, ‘Nipponbare’) were obtained from the Rice Genome Resource Center, National Institute of Agrobiology Sciences (Japan) (Miyao et al., 2003) and Rice T-DNA Insertion Sequence Database (Korea), respectively. The mutant osmads6-1 (Li et al., 2010) with genetic background 9522 (Oryza sativa ssp. japonica) was a wild-type plant. The transgenic plants InDE6 were continuously grown and harvested for three planting seasons, and the T3 transgenic seeds were used for the OsMADS6-induced expression experiment. The OET6 transgenic plants were continuously grown and harvested for T2 transgenic plants and phenotype observation and floral tissue collection were performed. All rice plants were grown in the paddy field. The Nicotiana benthamiana plants were grown at 23°C under 16-h daylight conditions in a greenhouse. The phenotypes of osfdml1 and transgenic plants OET6 were sensitive to the environment, and all of the phenotypes reported in this study were observed twice successively in the paddy field under climatic conditions reported at www.tianqi.com and www.richurimo.51240.com (Supplemental Fig. S5).
ChIP-qPCR and Enrichment Analysis
Approximately 1 g of wild-type rice inflorescence 2 to 8 mm in length was cross-linked by 1% formaldehyde and sonicated four times for 10 s each, with a 30-s break after each round of sonication, using an Ultrasonic Crasher Noise Isolating Chamber (Scientz) at 100 W; this yielded chromatin DNA with a size of <500 bp. ChIP of the OsMADS6-DNA complexes followed the protocol of Li et al. (2011). The precipitated and recovered DNA was diluted with 100 μL of double-distilled water, and each reaction was performed in three biological replicates. Primer pairs FDML1LP1 + FDML1RP1 and FDML1LP2 + FDML1RP2 were used for qPCR (Supplemental Table S5). The reactions were performed using SYBR Green Master mix (Bio-Rad) on a real-time PCR instrument (Bio-Rad). The calculation of relative enrichment of the fragments with the putative CArG motifs followed the method suggested by Li et al. (2011).
RT-qPCR
We extracted total RNA from roots, stems, leaves, young inflorescences, and dissected lemmas, paleae, stamens, and pistils using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. About 1 μg total RNA was reverse transcribed with PrimeScript RT reagent Kit with gDNA Eraser (Takara). RT-qPCR was performed using SYBR Green Master Mix (Bio-Rad) on a real-time PCR instrument (Bio-Rad). Specific primers were designed and validated by melt curve and standard curve (R2 = 0.9996, E = 0.957 for FDML1F and FDML1R; R2 = 0.9998, E = 0.991 for FDML2F and FDML2R primer pairs) first before being applied to detect gene expression. We collected ∼30 1- to 2-cm-long roots from 15 seedlings, two 1-cm-long stems from two plants, one 2-cm-long leaves from one plant, 10 2-mm-long inflorescences, six 2- to 3-mm-long inflorescences, four 4- to 7-mm-long inflorescences, 15 dissected lemmas, 15n dissected paleae, 60 stamens, and 60 pistils for one biological replicate of different tissues. Data shown in this research were summarized from three biological replicates and three PCR replicates normalized to ACTIN expression.
OsMADS6-Induced Expression Experiment
We germinated ∼200 seeds of the wild-type and InDE6 T3 transgenic plants in water for ∼1 week. The young seedlings were divided into two equally sized groups and treated with 5 μm β-estradiol and the solvent DMSO, respectively. Thereafter, we collected 0.5- to 1-cm root tips from 10 young seedlings for each biological repeat at 0, 12, and 24 h. Each sample consisted of three biological replicates for total RNA extraction. The primers were FDML1F and FDML1R for OsFDML1 (Supplemental Table S5). RT-qPCR was performed using a real-time RCR instrument (Bio-Rad) with SYBR green Master mix (Bio-Rad). The relative expression levels of OsFDML1 and OsMADS6 were normalized relative to ACTIN expression.
Yeast One-Hybrid Assay
We cloned the full-length OsMADS6 cDNA to vector pGADT7 and the 102-bp DNA fragment containing the putative CArG motif to vector pHIS2, respectively, using the primer pair pHIS2F1F and pHIS2F1R (Supplemental Table S5). The yeast strain Y187 was then cotransformed and cultured at 30°C for 2 to 3 d on solid dropout media, SD-TL, with the empty vectors as negative controls. We chose three independent positive strains, incubated in liquid SD-TL media overnight and diluted to 1/10 in a gradient. Finally, we drew 5 μL of cotransformed host strains on the solid SD-TLH media with 20 nm 3-AT (Zhang et al., 2012).
Dual-Luciferase Assay
The LUC transactivation (dual-LUC) method was modified from the protocol described by Bennett et al. (1996). The effector OsMADS6-Flag was prepared by cloning the full length of OsMADS6 cDNA containing the primer pair pGM6F and pGM6R into the modified vector pGreenII-0000 with Flag driven by the CaMV 35S promoter (Supplemental Table S5). Both the empty vector and the modified vector with Flag were used as negative controls. The reporter OsFDML1pro:LUC was constructed by cloning the 3795-bp OsFDML1 genome sequence upstream of the ATG with primers pG0800F1F and pG0800F1R into the vector pGreenII-0800-LUC (Supplemental Table S5). The empty vector was used as a control.
The transformation of all the effectors and reporters followed the protocol of Li et al. (2014). Agrobacterium tumefaciens cultured overnight was collected and resuspended in Murashige and Skoog liquid medium to OD600 values of 0.6 to 0.8. The mixture with the effector and reporter strain at a ratio 4:1 was incubated for 3 h at room temperature with 10 mm MES and 200 μm acetosyringone and infiltrated into young N. benthamiana leaves, and the plants were kept for 48 h under weak light conditions. Four biological replicates were performed for each sample. The LUC/REN signals were calculated using dual-LUC assay solutions (Promega) on a luminometer (Promega).
Genotyping of osfdml1 Mutants and OsFDML1 Expression Level Test
Primer combinations for osfdml1-1 were LD82 + LD83 and LD82 + Tos7, and those for osfdml1-2 were LD55 + LD56 and LD56 + Osp79 (Supplemental Table S5). The precise positions of Tos17 and T-DNA were determined by PCR sequencing. To test the expression change of OsFDML1 in two osfdml1 mutants, we designed primers (i.e. F1, R1, F2, R2, and F3) for different sites according to the position of Tos17 and T-DNA (Supplemental Table S5).
Rice Transformation
For OsFDML1 overexpression, the full-length cDNA of OsFDML1 generated with primers pCAMF1F and pCAMF1R was cloned into vector pCAMBIA1301 driven by the CaMV 35S (Supplemental Table S5). We cultured the sheared young spikelets at 28°C under dark conditions for 12 to 15 d until the callus grew. The transformation of 35Spro:OsFDML1 was introduced to the wild-type callus by the A. tumefaciens-mediated method using hygromycin for selection.
Histochemical and Scanning Electron Microscopy Observation
All of the materials were fixed in formalin-acetic acid-alcohol solution with 3.7% formaldehyde at 4°C overnight, dehydrated in graded ethanol, infiltrated with Histo-Clear II (National Diagnostics), and embedded in paraffin (Sigma-Aldrich; the same products and procedure for in situ hybridization experiment). Sections of the tissues (8-μm thick) were prepared using a microtome (Leica), stained with 0.1% toluidine blue, and photographed under a bright-field microscope (Nikon). Scanning electron microscopy images were acquired using a scanning electron microscope (JEOL).
In Situ Hybridization
Tissues were fixed in formalin-acetic acid-alcohol solution with 3.7% formaldehyde at 4°C for 2 h and dehydrated in a series of different ethanol concentrations. Tissue sections (6- to 8-μm thick) were prepared following the same procedure used for histochemical observations.
The probes were amplified with primers F1F + F1T7R (henceforth referred to as “antisense probe”) and F1T7F + F1R (henceforth referred to as “sense probe”), respectively. F1T7F and F1T7R were designed by adding 20-bp T7 promoter sequences TAATACGACTCACTATAGGG to the 5ʹ end of the primers (Supplemental Table S5). The antisense and sense probes were labeled with digoxigenin using an in vitro transcription kit (Roche) with T7 polymerase. The procedures for in situ hybridization and digoxigenin signal detection followed the protocol described by Dreni et al. (2007).
Yeast Two-Hybrid Assay
To perform the yeast two-hybrid analysis, we first cloned the full-length cDNAs of OsFDML1, OsFDML2, OsFDML3, and OsFDML4 into vectors pGADT7 and pGBKT7, respectively. Subsequently, we cotransformed the corresponding pGADT7 and pGBKT7 constructs into the yeast strain AH109 and cultured the yeast on solid dropout media SD-TL at 30°C for 2 to 3 d. The positive strains were incubated in liquid SD-TL media overnight and collected by centrifugation, and the serial decimal dilution was used for spot assay on dropout media SD-TLHA (the main procedures followed the yeast protocol handbook from Clontech). The GAL4-AD-OsFDML1-fused protein and GAL4-BD-OsFDML2-fused protein activated the expression of the reporter gene HIS3, which was confirmed by the results from GAL4-AD-OsFDML2-fused protein and GAL4-BD-OsFDML1 expression. Primers for these constructs are listed in Supplemental Table S5.
Subcellular Localization of OsFDML1 in Protoplast
The full-length cDNAs of OsFDML1 and OsFDML2, amplified with the primer combinations pA7F1F + pA7F1R and pA7F2F + pA7F2R, respectively, were cloned into vector pA7 encoding GFP driven by CaMV 35S (Supplemental Table S5). The recombinant plasmid expressing eGFP-OsFDML1 and eGFP-OsFDML2 fused proteins was introduced into rice protoplasts following the protocol described by Bart et al. (2006). The images were visualized using a confocal microscope (Leica).
BiFC Assay in Protoplast
The full-length cDNAs of OsFDML1 and OsFDML2 were cloned into vectors pSAT-cEYFP-c1 and pSAT-nEYFP-c1, respectively. All the steps for the BiFC assay followed the protocol of Zhang et al. (2011).
Phylogenetic Analysis
The 119 homologs were obtained using the full-length protein sequence of OsFDML1 as the query to search in the Phytozome database (http://phytozome.jgi.doe.gov) and Congenie database (http://congenie.org). The 24 protein sequences from Arabidopsis and rice were multiple-aligned by MAFFT (http://mafft.cbrc.jp/alignment/server) and kept the conserved XS-XH domain as determined by Genedoc (Nicholas et al., 1997). The neighbor-joining phylogenetic tree was constructed following a method described previously (Pelucchi et al., 2002). The bootstrap values (%) based on 100 replicates are marked at the branching points.
Statistical Analysis
Statistical works were run with SigmaPlot 12.0. All data were expressed as means ± sd (n ≥ 3). Statistical significance level of P < 0.05 between different sample groups was tested using ANOVA.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project and the Arabidopsis Information Resource data libraries under the following accession numbers: OsFDML1 (Os02g0293300), OsFDML2 (Os01g0126600), OsFDML3 (Os01g0633200), OsFDML4 (Os05g0153200), SGS3 (AT5G23570), IDN2/RDM12 (AT3G48670), FDM1 (AT1G15910), FDM2 (AT4G00380), FDM3 (AT3G12550), FDM4 (AT1G13790), and FDM5 (AT1G80790). Accession numbers for the sequences used in the phylogenetic analysis are listed on the tree.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of two allelic osfdml1 mutants.
Supplemental Figure S2. Spikelet phenotypes of heterozygous and homozygous osfdml1-1 and osfdml1-2 mutants.
Supplemental Figure S3. Phenotypes of wild-type and osfdml1-2 spikelets.
Supplemental Figure S4. The phenotype and statistical analysis of the primary branch and secondary branch of the mutant inflorescences.
Supplemental Figure S5. The climatic conditions of mutant growth in the field in 2017.
Supplemental Figure S6. The expression pattern of OsFDML2 in the wild type.
Supplemental Table S1. List of genes putatively encoding transcription factors and kinases with the changed expression in osmads6-1 from microarray analysis by Li et al. (2011).
Supplemental Table S2. Homologs of OsFDML1 in 14 species of terrestrial plants.
Supplemental Table S3. Number of floral organs in whorls 1, 2, 3, and 4 of osfdml1-1.
Supplemental Table S4. Number of floral organs in whorls 1, 2, 3, and 4 of osfdml1-2.
Supplemental Table S5. Sequence information of primers.
Acknowledgments
We thank Dr. Ludovico Dreni for assisting in data analysis; Zhijing Luo, Mingjiao Chen, and Zibo Chen for rice materials planting; Yun Hu for providing OsMADS6 rabbit polyclonal antibody and for InDE6 transgenic plant construction; and Changsong Yin and Jie Wang for their excellent assistance on in situ hybridization.
Footnotes
This research was supported by the National Key Technologies Research and Development Program of China, Ministry of Science and Technology (grant nos. 2016YFD 0100804 and 2016YFE0101000); by the National Natural Science Foundation of China (31230051); by the H2020-MSCA-RISE-2015 project ExpoSEED (691109); by the Innovative Research Team, Ministry of Education; by the 111 Project (grant no. B14016); by the Australian Research Council (DP170103352); and by Australia-China Science and Research Fund Joint Research Centre grant ACSRF48187.
Articles can be viewed without a subscription.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Mockler TC, Chory J, Jacobsen SE (2009) IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol 16: 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Greenberg MV, Simanshu DK, Hale CJ, Vashisht AA, Simon SA, Lee TF, Feng S, Española SD, Meyers BC, et al. (2012) INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 109: 8374–8381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker NP, Clark LG, Davis JI, Duvall MR, Guala GF, Hsiao C, Kellogg EA, Linder HP, et al. (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. (2002) The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46: 69–78 [DOI] [PubMed] [Google Scholar]
- Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater MM, Colombo L (2007) Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H, Graner S, Luschnig C (2014) Expression analysis of Arabidopsis XH/XS-domain proteins indicates overlapping and distinct functions for members of this gene family. J Exp Bot 65: 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA (2018) Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot [DOI] [PubMed] [Google Scholar]
- Chen M, Bennetzen JL (1996) Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol Biol 32: 999–1001 [DOI] [PubMed] [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH (2006) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Kater MM (2014) MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol 201: 717–732 [DOI] [PubMed] [Google Scholar]
- Dreni L, Zhang D (2016) Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. J Exp Bot 67: 1625–1638 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM (2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Xing Z, Diao Z, Xu W, Li S, Du X, Wu G, Wang C, Lan T, Meng Z, et al. (2012) Characterization of Osmads6-5, a null allele, reveals that OsMADS6 is a critical regulator for early flower development in rice (Oryza sativa L.). Plant Mol Biol 80: 429–442 [DOI] [PubMed] [Google Scholar]
- Finke A, Kuhlmann M, Mette MF (2012) IDN2 has a role downstream of siRNA formation in RNA-directed DNA methylation. Epigenetics 7: 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liang W, Yin C, Yang X, Ping B, Li A, Jia R, Chen M, Luo Z, Cai Q, et al. (2015) Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant 8: 1366–1384 [DOI] [PubMed] [Google Scholar]
- Irish EE, Szymkowiak EJ, Garrels K (2003) The wandering carpel mutation of Zea mays (Gramineae) causes misorientation and loss of zygomorphy in flowers and two-seeded kernels. Am J Bot 90: 551–560 [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. (2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. (2015) Inflorescence Structure. In: Flowering Plants, Monocots: Poaceae. The Families and Genera of Vascular Plants 13: 25–38 [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K (2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Li A, Zhang Y, Wu X, Tang W, Wu R, Dai Z, Liu G, Zhang H, Wu C, Chen G, Pan X (2008) DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol Biol 66: 491–502 [DOI] [PubMed] [Google Scholar]
- Li G, Liang W, Zhang X, Ren H, Hu J, Bennett MJ, Zhang D (2014) Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc Natl Acad Sci USA 111: 10377–10382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D (2011) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Yoshida H (2015) Interpreting lemma and palea homologies: a point of view from rice floral mutants. Front Plant Sci 6: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YH, Kang HG, Jung JY, Jeon JS, Sung SK, An G (1999) Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol 120: 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DWI (1997) Gene-Doc: analysis and visualization of genetic variation. Embnew News 4: 14 [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001b) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF (2001a) Conversion of leaves into petals in Arabidopsis. Curr Biol 11: 182–184 [DOI] [PubMed] [Google Scholar]
- Pelucchi N, Fornara F, Favalli C, Masiero S, Lago C, Pè ME, Colombo L, Kater MM (2002) Comparative analysis of rice MADS-box genes expressed during flower development. Sex Plant Reprod 15: 113–122 [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2006) Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007) Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Reinheimer R, Kellogg EA (2009) Evolution of AGL6-like MADS box genes in grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell 21: 2591–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ, Stuppy W, Cunniff J, Kellogg EA, Briggs BG (2005) Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am J Bot 92: 1432–1443 [DOI] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, Ren D, Fang L, Wang N, Zhao F, Ling Y, Yang Z, Liu Y, He G (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol 160: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Hirano HY (2014) Flower development in rice. Adv Bot Res 72: 221–262 [Google Scholar]
- Theissen G, Saedler H (2001) Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Hake S (2009) Translational biology: from Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S (2009) bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang L, Cai Q, Hu Y, Jin Z, Zhao X, Fan W, Huang Q, Luo Z, Chen M, Zhang D, Yuan Z (2015) OsMADS32 interacts with PI-like proteins and regulates rice flower development. J Integr Plant Biol 57: 504–513 [DOI] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Zanis MJ, Kellogg EA, Schmidt RJ (2007) Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc Natl Acad Sci USA 104: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Liang W, Zhu W, Chen M, Ferrándiz C, Burton RA, Dreni L, Zhang D (2018) Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol 176: 1646–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Ren G, Zhang C, Yu B (2012a) The DNA- and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J 72: 491–500 [DOI] [PubMed] [Google Scholar]
- Xie M, Ren G, Costa-Nunes P, Pontes O, Yu B (2012b) A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res 40: 4422–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Chen D, Kaufmann K (2016) Molecular mechanisms of floral organ specification by MADS domain proteins. Curr Opin Plant Biol 29: 154–162 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB (2009) RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 1–12 [Google Scholar]
- Zhang D, Yuan Z (2014) Molecular control of grass inflorescence development. Annu Rev Plant Biol 65: 553–578 [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Ning YQ, Zhang SW, Chen Q, Shao CR, Guo YW, Zhou JX, Li L, Chen S, He XJ (2012) IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet 8: e1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yuan Z, An G, Dreni L, Hu J, Kater MM (2013) Panicle development. Rice Genom Genet 5: 279–295 [Google Scholar]
- Zhang J, Nallamilli BR, Mujahid H, Peng Z (2010) OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J 64: 604–617 [DOI] [PubMed] [Google Scholar]
- Zhang T, Li Y, Ma L, Sang X, Ling Y, Wang Y, Yu P, Zhuang H, Huang J, Wang N, et al. (2017) LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc Natl Acad Sci USA 114: 9984–9989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xing Y, He XJ, Li W, Hu Y, Yadav SK, Oh J, Zhu JK (2010) An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J 62: 92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT (2013) A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 49: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Hare PD, Chua NH (2006) Applications of chemical-inducible expression systems in functional genomics and biotechnology. Methods Mol Biol 323: 329–342 [DOI] [PubMed] [Google Scholar]