A method for concurrent quantification of a large number of metabolites representing the metabolic flux of seven major classes of plant hormones provides a simple and sensitive tool for phytohormone studies.
Abstract
Phytohormones are physiologically important small molecules that play essential roles in intricate signaling networks that regulate diverse processes in plants. We present a method for the simultaneous targeted profiling of 101 phytohormone-related analytes from minute amounts of fresh plant material (less than 20 mg). Rapid and nonselective extraction, fast one-step sample purification, and extremely sensitive ultra-high-performance liquid chromatography-tandem mass spectrometry enable concurrent quantification of the main phytohormone classes: cytokinins, auxins, brassinosteroids, gibberellins, jasmonates, salicylates, and abscisates. We validated this hormonomic approach in salt-stressed and control Arabidopsis (Arabidopsis thaliana) seedlings, quantifying a total of 43 endogenous compounds in both root and shoot samples. Subsequent multivariate statistical data processing and cross-validation with transcriptomic data highlighted the main hormone metabolites involved in plant adaptation to salt stress.
During the last decade, the techniques used in metabolomic analyses have advanced tremendously. In plant science, the most widely used methods are based on separation by liquid chromatography (LC) or gas chromatography combined with tandem mass spectrometric detection (MS/MS). The main advantages of these combinations are high sensitivity and versatility. To enhance signals of trace compounds, such as the plant hormones (phytohormones) considered here, it is essential to reduce the influence of abundant interfering compounds present in plant matrices by rigorous purification of extracts before the instrumental analysis (Du et al., 2012). The sample preparation steps usually include solid-phase extraction (SPE) with general-purpose sorbents or more selective immunosorbents that specifically target the selected compounds (Pencík et al., 2009; Turečková et al., 2009; Oklestkova et al., 2017; Plačková et al., 2017). Many analytical methods (particularly the immunological methods) have been described for the determination of a single compound or a specific class of phytohormones (Du et al., 2012; Tarkowská et al., 2014). However, there is growing interest in methods capable of simultaneously analyzing phytohormones of several classes together with their precursors and metabolites, for the following reasons.
Phytohormones are naturally occurring signaling molecules that play key roles in the regulation of plant physiology, development, and adaptation to environmental stimuli. Generally, their concentrations in plant tissues are extremely low (fmol to pmol g−1 fresh weight). They are also exceptionally diverse compounds with wide ranges of physicochemical properties and are divided into several structural classes: cytokinins (CKs) and 2-methylthiocytokinins (2MeSCKs), auxins (AXs), ethylene, gibberellins (GAs), abscisic acid and its metabolic products (hereafter referred to as abscisates [ABAs]), brassinosteroids (BRs), jasmonates (JAs), salicylic acid (SA), and strigolactones (Davies, 2010; Zwanenburg et al., 2016). Their biological activities depend on their availability, which is controlled by their biosynthetic and metabolic rates, cellular and subcellular localization, transport, and responses of the signal perception and transduction pathways (Davies, 2010). Modulations at any of these levels can directly affect myriad physiological processes. Although certain phytohormones are usually related to specific biological functions or responses, there is increasing evidence that plant hormone signaling involves complex interactions (cross talk) among all the pathways involved (Vanstraelen and Benková, 2012). Indeed, this is hardly surprising, as plants in natural environments may have to cope simultaneously with (for example) salt, water, and temperature stresses, pathogen attack, competition, and a need to complete certain physiological processes within environmentally dictated time frames. Thus, plants’ physiological regulation involves challenging coordination of the biosynthesis, transport, and metabolism of multiple hormones, their highly interacting signal transduction pathways, transcription factors, and responsive genes.
Clearly, a convenient method to simultaneously quantify as wide a range as possible of plant signaling molecules of all known classes would greatly facilitate the investigation of hormone functions and networks. Thus, several plant hormone-profiling techniques have been published, and the number of covered compounds is increasing (Chiwocha et al., 2003; Pan et al., 2008; Kojima et al., 2009; Farrow and Emery, 2012; Cao et al., 2016; Wang et al., 2017). The most extensive analysis of primary and secondary metabolites published to date included 53 plant hormone-related compounds (Schäfer et al., 2016), and a more focused analysis of plant growth substances covered 54 compounds (Cai et al., 2016). However, there is scope for further extension. An ideal method should provide both a qualitative overview and precise quantitative information for all covered compounds. It also requires appropriate sample preparation and high instrumental performance (in terms of both robustness and sensitivity), due to the low concentrations of phytohormones (relative to those of primary and secondary metabolites) and wide ranges of chemical structure and stability.
Here, we present a methodology with these features, designed to afford rapid, sensitive, and simultaneous LC-MS/MS-based profiling of 101 CKs, AXs, GAs, BRs, ABAs, JAs, and SA. The analytes include bioactive forms of the hormones, their precursors, and metabolites to acquire quantitative snapshots of the physiological status of sampled tissues (Supplemental Table S1). The protocol for isolating all 101 compounds combines rapid, one-step, nonselective extraction from milligram amounts of plant tissues (less than 20 mg fresh weight) followed by their LC separation and extremely sensitive MS-based quantification. To assess the practical utility of this hormonomic approach, the method was applied to characterize phytohormone profiles in root and shoot tissues of control and salt-stressed Arabidopsis (Arabidopsis thaliana) seedlings. Our results highlight the value of such analysis, which (together with multivariate data analysis and cross-validation with transcriptomic data) revealed the seedlings’ hormonal responses to salinity stress, one of the major factors limiting crop production (Munns and Tester, 2008), and differences in the responses of their roots and shoots.
RESULTS
One-Step Extraction of Distinct Phytohormone Classes
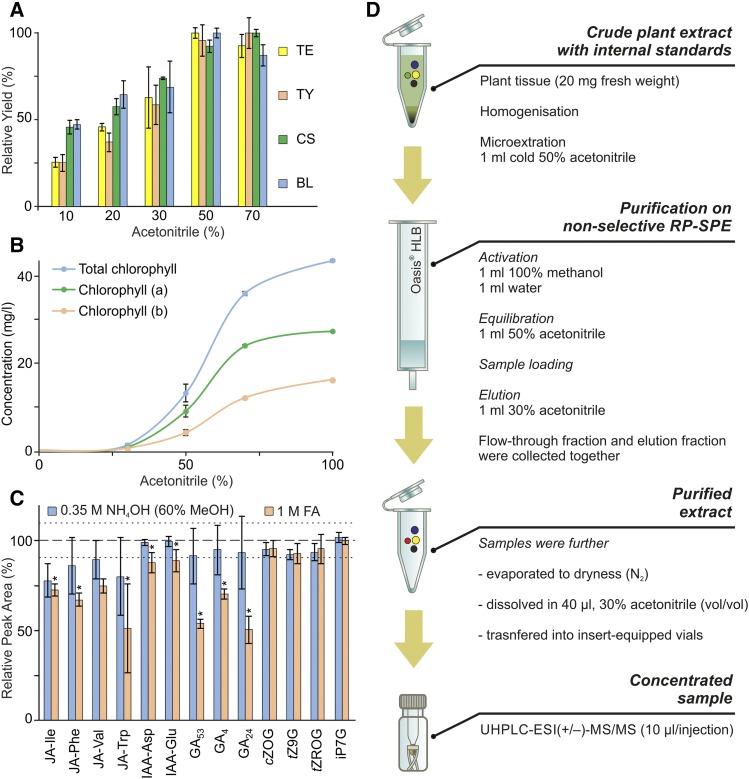
In an attempt to effectively extract targeted compounds and minimize the risk of their decomposition by elevated temperatures and enzymatic degradation, the samples had to be processed at the lowest temperature reliably above freezing (less than 4°C; Ljung et al., 2010). Plant tissue was first homogenized and extracted with a suitable solvent in which the phytohormones are soluble and chemically stable. Ice-cold acetonitrile (ACN) was chosen here as the extraction solvent in accordance with several previously published studies dealing with analyses of hydrophobic phytohormones, such as diterpenoid GAs (Urbanová et al., 2013) and triterpenoid BRs (Tarkowská et al., 2016). We tested aqueous water:ACN mixtures with ACN contents ranging from 0% to 100% (v/v), focusing on the solubility of the most hydrophobic compounds included in our study (Fig. 1). Furthermore, to quantify the impairment of the final LC-MS/MS analysis by signal suppression, contents of the most abundant interfering plant pigments, chlorophyll a and chlorophyll b, were determined. The solubility of all investigated BRs (calculated as a percentage of the maximal signal intensity) reached, on average, 95% in solvents with 50% or greater (v/v) ACN (Fig. 1A); however, the concentration of interfering plant pigments also increased rapidly with increases in ACN (Fig. 1B). Thus, ice-cold 50% ACN was selected as the optimal extraction solvent, providing the best balance between signal intensity for selected phytohormones and chlorophyll coextraction.
Figure 1.
Optimization of sample preparation. A, Solubility of selected BRs (TE, teasterone; TY, typhasterol; CS, castasterone; BL, brassinolide), the most hydrophobic phytohormone class included in our study, in extraction solvents with varied ACN contents. Relative yield (ratio, in percentage, of average peak area to maximal average peak area per extraction solvent) is shown (error bars represent sd; Supplemental Table S10). B, Amounts of chlorophyll extracted (mg L−1) using extraction solvents with the indicated ACN concentrations (error bars represent sd; Supplemental Table S11). C, pH-dependent stability (relative peak area, in percentage) of selected compounds (JA-Ile, jasmonoyl-l-Ile; JA-Phe, jasmonoyl-l-Phe; JA-Val, jasmonoyl-l-Val; JA-Trp, jasmonoyl-l-Trp; IAA-Asp, indole-3-acetyl-l-Asp; IAA-Glu, indole-3-acetyl-l-Glu; cZOG, cis-zeatin-O-glucoside; tZ9G, trans-zeatin-9-glucoside; tZROG, trans-zeatin riboside-O-glucoside; iP7G, N6-isopentenyladenine-7-glucoside) dissolved in 0.35 m NH4OH in 60% methanol (MeOH) and 1 m formic acid (FA), based on peak areas relative to peak areas of compounds dissolved in control solvent (50% ACN). The dashed line represents the average peak area, and the dotted lines represent the average of sd values for compounds dissolved in the control solvent; asterisks indicate significant changes compared with the control: *, P < 0.05, by Student’s t test (Supplemental Table S12). D, Scheme of sample microextraction and purification prior to UHPLC-ESI-MS/MS analysis. RP, Reverse phase.
It was also essential to consider the chemical stability of the wide spectrum of targeted analytes during sample preparation. For instance, GAs are pH sensitive and should only be exposed to solvents with pH 2.5 to 8.5 (Urbanová et al., 2013). To test the pH sensitivity of our analytes, selected metabolites (Fig. 1C) were dissolved in different aqueous solutions of 1 m formic acid (pH < 3), 0.35 m NH4OH in 60% (v/v) MeOH (pH > 12), or 50% ACN solution (as a control), solutions that often are used in sample preparation of CKs and other phytohormones during ion-exchange SPE (Dobrev and Kamínek, 2002; Kojima et al., 2009; Záveská Drábková et al., 2015; Schäfer et al., 2016). A mixture with known amounts of each compound (0.4 pmol of CKs and JAs with 4 pmol of AXs and GAs) was incubated in each solution for 15 min at 4°C. After evaporation under a gentle stream of nitrogen, samples were dissolved in 30% ACN and analyzed by LC-MS/MS. The peak area of each compound relative to the corresponding peak’s area in control samples was then calculated (Fig. 1C). In the case of the NH4OH solvent, levels of most of the tested compounds remained close to those found in control samples, but in the acidic extraction solvent, the recoveries of GAs, JAs, and indole-3-acetic acid (IAA) amino acid conjugates were significantly lower compared with control samples. To preserve the levels of all targeted compounds and limit their possible structural changes and hydrolysis during sample preparation, cold extraction using 50% aqueous ACN with no additives was used in all subsequent phytohormone experiments (see scheme in Fig. 1D).
Reduction of a Complex Plant Matrix by a Highly Efficient Purification Step
In the simultaneous analysis of phytohormones, several multistep SPE methods combining the use of either silica-based RP sorbents with long 18-carbon alkyl chains (C18) or polymer-based RP materials with ion-exchange properties (mixed-mode sorbents) have proven efficacy for purifying samples and enrichment of the targeted analyte fraction (Kojima et al., 2009; Balcke et al., 2012; Floková et al., 2014; Záveská Drábková et al., 2015; Cao et al., 2016; Schäfer et al., 2016). These approaches often work with solvents of various pH values, but (as mentioned above) the pH sensitivity of some of our analytes limited the use of acidic conditions during sample preparation. Moreover, sample preparation utilizing multiple-step SPEs is very time consuming and often includes several evaporation steps, which significantly reduce the effectiveness of sample preparation protocols, especially for highly volatile compounds such as methyl jasmonate or methyl salicylate (Floková et al., 2014). To avoid these problems, we used 50% ACN without any additives as both the sample extraction and SPE loading solution (Fig. 1D), thus eliminating one evaporation step, reducing the effects of the plant matrix, and minimizing losses caused by enzymatic degradation and pH-dependent hydrolysis. To remove coextracted plant pigments with maximum efficiency while maintaining high analyte recovery, we utilized RP polymer-based SPE Oasis HLB columns, which are packed with a hydrophilic-lipophilic-balanced (HLB) water-wettable sorbent. Recoveries following this purification step were studied using extracts from 20 mg fresh weight of Arabidopsis plant material supplemented with authentic phytohormone standards before and after SPE purification steps (Caban et al., 2012; see “Materials and Methods”). The HLB sorbent was used to retain possible interfering compounds, while targeted compounds passed through the SPE column sorbent in the loading step (the flow-through fraction) or were eluted subsequently with 30% (v/v) ACN. These fractions were pooled, and average total extraction yields per class ranged from 87% to 97%, except for BRs, which averaged 52% (Table I; Supplemental Table S2). Samples were evaporated further under a gentle stream of gaseous nitrogen and then dissolved in 40 µL of 30% ACN prior to LC-MS/MS analysis (Fig. 2).
Table I. Overview of average recovery, minimal and maximal limits of detection (LOD), and average method precision and analytical accuracy (absolute value) for each phytohormone class.
For detailed information, see Supplemental Table S2.
| Phytohormone Class | No. of Compounds | Minimal/Maximal LOD | Spiked Contents | Average Recovery | Average Method Precision | Average Method Accuracy |
|---|---|---|---|---|---|---|
| fmol | pmol | % | % RSD | % bias | ||
| CKs | 41 | 0.005/0.5 | 0.5–5 | 87.0 | 3.30 | 7.49 |
| AXs | 15 | 0.05/10 | 1–50 | 91.5 | 4.31 | 6.93 |
| JAs | 11 | 0.1/50 | 10 | 95.3 | 5.70 | 5.20 |
| ABAs | 5 | 1.0/10 | 10 | 95.9 | 1.06 | 5.91 |
| GAs | 14 | 0.25/25 | 5 | 89.2 | 3.00 | 5.54 |
| SA | 1 | 25 | 100 | 97.3 | 4.05 | 8.68 |
| BRs | 14 | 2.5/25 | 10 | 52.5 | 7.80 | 5.26 |
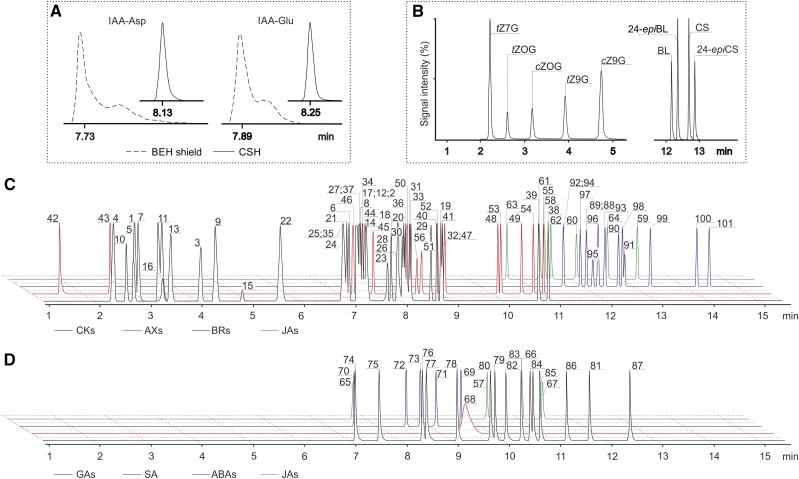
Figure 2.
Optimization of baseline chromatographic separation. A, Peak shape of IAA-Asp and IAA-Glu separated on Acquity UPLC CSH (solid lines) and Acquity UPLC BEH shield (dashed lines) columns. B, Isomer separation of N/O-glucoside forms of cis-/trans-zeatins (left) and brassinolide (BL/24-epiBL) and castasterone (CS/24-epiCS) isobars (right) using an Acquity UPLC CSH column. Compound abbreviations are as in Figure 1C. C and D, Overlay of UHPLC-ESI-MS/MS separation of targeted compounds in ESI(+) mode (C) and ESI(–) mode (D). Phytohormones are numbered according to Supplemental Table S1.
Profiling of 101 Phytohormone-Related Compounds by Ultrafast LC-MS/MS
One of the main inherent difficulties in profiling more than 100 plant hormones (Supplemental Table S1) is that many of the compounds have similar core structures, including isomers with the same MS fragmentation patterns (e.g. cis- and trans-zeatin, topolin isomers, brassinolide and 24-epibrassinolide [24-epiBL], and castasterone and 24-epicastasterone). To optimize baseline separation, we tested two RP ultra-HPLC (UHPLC) columns packed with charged-surface hybrid (CSH) and ethylene-bridged hybrid (BEH) polymer-based sub-2-μm sorbents. In good agreement with previously reported results (Urbanová et al., 2013; Floková et al., 2014), the CSH column provided better peak shapes and peak-to-peak resolution of the above-mentioned isomeric compounds than the BEH column (Fig. 2A). The composition of the mobile phase and the use of different mobile phase additives strongly influenced the separation, peak shape, and analyte ionization. Cao et al. (2016) found that increasing the concentration of formic acid in the range from 0.05% to 0.2% impaired CK separation. Confirming this trend, we investigated the separation of CKs using 0.001%, 0.01%, and 0.1% formic acid. However, the optimal baseline separation of CK isomers was achieved on the CSH column using isocratic elution with 0.01% formic acid in both mobile phase solutions (water and ACN; Fig. 2B).
Retention times of the 101 targeted compounds were further determined by separate injections and compared with those of 74 stable isotope-labeled standards (Supplemental Table S3). Each nonlabeled and stable isotope-labeled couple coeluted with the same or almost identical retention time, although deuterated analogs usually eluted slightly earlier than corresponding authentic standards due to the chromatographic isotope effect (Pratt, 1986). Under our chromatographic conditions, the monitored compounds were eluted from 1.28 to 14.47 min with good reproducibility of retention times below 2% (except 3.4% for the most polar compound, tryptamine; Supplemental Table S4). Not all compounds were separated to baseline (Fig. 2), but their determination in multiple reaction monitoring (MRM) mode allowed precise detection due to specific precursors to product ion transitions in the fragmentation of co-eluting compounds (Supplemental Table S3). Appropriate precursor ions ([M+H]+ or [M–H]–) and the most abundant product ions for each compound were carefully selected, partly for this purpose. The metabolites of CKs, 2MeSCKs, AXs, and BRs, and some JA precursors and its amino acid conjugates, were determined in positive electrospray ionization [ESI(+)] mode, which provided good agreement with previously published data (Tarkowski et al., 2010; Novák et al., 2012; Svačinová et al., 2012; Floková et al., 2014; Tarkowská et al., 2016). All other phytohormones (including ABAs, GAs, SA, and JAs) were determined in ESI(–) mode (Turečková et al., 2009; Urbanová et al., 2013; Floková et al., 2014). Finally, the collision energy and cone voltage were optimized to maximize signal intensities. The optimized MS conditions are listed in Supplemental Table S3. Under these parameters, LODs for 101 targeted plant hormones and their metabolites ranged from 0.005 fmol for 2MeSCK ribosides to 50 fmol for 12-hydroxy-jasmonic acid. The LOD and limit of quantification (LOQ) were experimentally determined and defined as 3 and 10 times the noise level, respectively (for details, see Table I; Supplemental Table S2).
The proposed method was initially designed for simultaneous analysis of positively and negatively charged ions, utilizing both MRM modes (polarity switching) in single analytical run. However, the ESI(–) mode is known to be generally less sensitive than the positive mode, and polarity switching for simultaneous analysis of 101 phytohormones caused further 4- to 12-fold reductions in signal intensity of negatively charged compounds (Supplemental Fig. S1). Instrument performance declined due to duty cycle problems and the few milliseconds required for signal recovery after every such switch. Moreover, the acquisition rate in MRM modes also is determined by the dwell time, which is the amount of time spent collecting a data point at a set transition or peak before switching to the next value in the MRM method (O’Mahony et al., 2013). Therefore, to improve the sensitivity of MS-based detection, samples were analyzed in two separate runs under the same chromatographic conditions, with the flow rate set to 0.5 mL min−1 and the column temperature to 50°C. Under these conditions, UHPLC-ESI-MS/MS analysis of the targeted compounds in one sample takes 32 min in total (Fig. 2, C and D). To further increase the duty cycle, and thus potentially the signal intensity, the 17- and 15-min chromatographic separations of the targeted analytes in ESI(+) and ESI(–) modes, respectively, were both divided into nine MRM scan segments.
Calibration curves constructed after repeatedly injecting standard solutions revealed a broad linear concentration range for most compounds, spanning at least 3 orders of magnitude with R2 ≥ 0.993 (Supplemental Table S3). The method sensitivity and linearity were found to be comparable to those reported by authors using MS/MS for simultaneous phytohormone analysis (Kojima et al., 2009; Balcke et al., 2012; Floková et al., 2014; Záveská Drábková et al., 2015; Cai et al., 2016; Schäfer et al., 2016; Delatorre et al., 2017).
Method Validation
Using the standard isotope dilution method, concentrations of all the analytes were calculated as ratios of nonlabeled compounds to labeled internal standards (IS) or closely eluting stable isotope-labeled tracers (Supplemental Table S3). To validate the UHPLC-MS/MS method, spiked Arabidopsis seedling extracts were analyzed, and the endogenous levels were subtracted from the amounts of nonlabeled standards added (see “Materials and Methods”). Finally, the calculated concentrations of each analyte were compared with the known amounts added to samples and are presented as method accuracy, ranging on average from 5.2% bias (JAs) to 8.68% bias (SA). Method precision was calculated as the relative sd (RSD) of analyte concentrations determined in three replicates. The precision ranged, on average, from 1.06% RSD (ABAs) to 7.8% RSD (BRs; Table I; for details, see Supplemental Table S2). Furthermore, the reproducibility test of the LC-MS/MS method also was performed by reinjection of standard mixtures prepared in low, medium, and high concentrations of targeted compounds in a range of 4 orders of magnitude (Supplemental Table S4). Solutions kept in the autosampler at 4°C were analyzed in triplicate on three consecutive days. For three concentration levels of the 101 targeted compounds, the RSD values of analyte responses were below 15% (Supplemental Table S4). Moreover, the intraday and interday precisions of the analytical method were quantified by evaluating the closeness of a set of analytical results obtained for a series of replicate samples and were expressed in terms of the RSD for those measurements. For endogenous phytohormones determined in 20 mg fresh weight of 10-d-old Arabidopsis seedlings, the intraday and interday precisions ranged from 0.6% to 10.5% and from 1.3% to 17.3%, respectively (Supplemental Table S5). The overall validation parameters of the developed method demonstrate its reliability and utility for simultaneous quantification of multiple classes of phytohormones in minute amounts of plant material.
Phytohormone Quantification in Plants under Salinity Stress
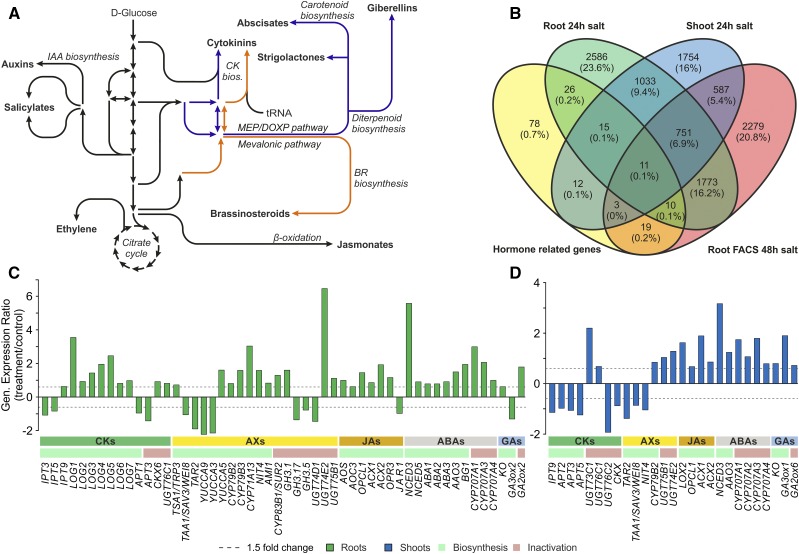
To assess the applicability of the newly developed, targeted metabolomics approach, we used it in a comparison of hormone-related transcript and metabolite levels in samples of root and shoot tissues (less than 20 mg fresh weight) of stressed Arabidopsis plants and controls. Published microarray data sets were screened to identify a stimulus that affects genes involved in most phytohormone metabolic pathways (Fig. 3; Supplemental Table S6). This bioinformatics analysis was performed in an unsupervised manner (without checking for up- or down-regulation). Salinity stress was identified as an appropriate condition, which is likely to cause major alterations in the metabolite levels of most hormones and corresponding changes in transcript profiles (Fig. 3B).
Figure 3.
Alterations in expression levels of plant hormone-related genes induced by salinity stress in Arabidopsis. A, Simplified scheme of plant hormone biosynthesis pathways. B, Venn diagram showing numbers, percentages, and overlaps of hormone-related genes (yellow oval; Supplemental Table S6) and transcripts with affected expression levels (1.5-fold or greater change, P < 0.01) in response to salt stress experiments as described in “Materials and Methods” (genes in green and blue ovals correspond to Arabidopsis root and shoot samples, respectively, from the Genevestigator Experiment ID: AT-00120, and genes in the red oval correspond to sorted root cell-specific protoplasts from Genevestigator Experiment ID: AT-00656). FACS, Fluorescence-activated cell sorting. C and D, Changes in hormone-related gene expression levels in shoot (C) and root (D) tissues under salinity stress (log ratio treated/control, 1.5-fold or greater change, P < 0.01; Genevestigator Experiment ID: AT-00120; Supplemental Tables S8 and S9).
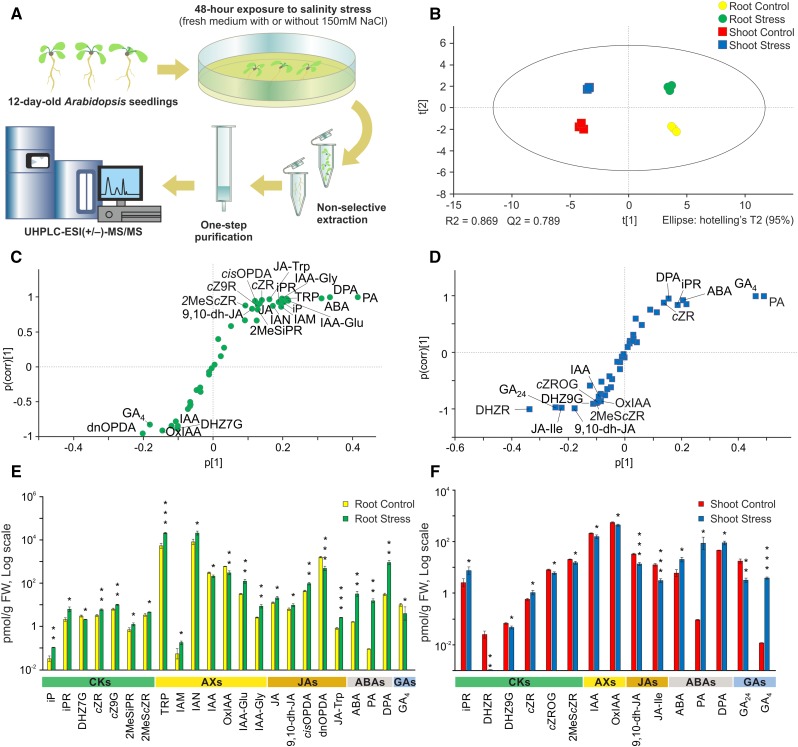
Therefore, we conducted a salinity stress experiment where we gently transferred 12-d-old Arabidopsis seedlings to new media with and without 150 mm NaCl for an additional 48 h. Shoots and roots were harvested separately, and their hormonomic profiles were examined (Supplemental Table S7). To show peak shapes, peak retentions compared with appropriate IS, and signal intensities of endogenously determined phytohormones, we present representative chromatograms of treated Arabidopsis root samples (Supplemental Figs. S2 and S3). Moreover, the univariate scatterplots of endogenous phytohormone levels provide information about the distribution of phytohormone levels in salt-stressed and control Arabidopsis seedlings under our experimental conditions (Supplemental Fig. S4). In total, 45 endogenous compounds out of the 101 phytohormone-related analytes were detected in both root and shoot samples. Two BRs (24-epiBL and 28-norcastasterone) were identified, but their levels were below the LOQ. Thus, they were omitted in subsequent statistical data analysis. According to Student’s t test, the levels of 23 of 43 determined compounds differed significantly between samples of roots of salt-stressed and control seedlings (Fig. 4E), and the levels of 15 compounds differed between their respective shoots (Fig. 4F). In addition, multivariate statistical analysis revealed clear separation between the hormone profiles of root and shoot samples and between the profiles of control and salt-stressed seedlings (Fig. 4B). Orthogonal projections to latent structures discriminant analysis-based S-plots revealed compounds that were strongly affected by salinity stress and, thus, were primarily responsible for the latter separation (Fig. 4, C and D).
Figure 4.
Comparison of phytohormones in Arabidopsis roots and shoots exposed to salinity stress and controls. A, Schematic illustration of the salinity stress experiment. B, Principal component analysis (PCA) score plot showing the separation and grouping of samples (roots and shoots of control and salt-stressed plants) according to the composition and abundance of determined compounds. C and D, Orthogonal projections to latent structures discriminant analysis (OPLS-DA) S-plots showing the variables responsible for separating roots of control and salt-stressed plants (C) and their shoots (D), combining covariance (p[1], the farther the distance from zero, the higher the contribution to the difference between two groups) and correlation (p(corr)[1], the farther the distance from zero, the higher the reliability). E and F, Levels of targeted compounds in roots (E) and shoots (F) in pmol g−1 fresh weight (FW), logarithmic scale. Asterisks indicate significant differences between salt-stressed and control plants: *, P < 0.05; **, P < 0.01; and ***, P < 0.001, by Student’s t test; error bars represent sd. Compound abbreviations are listed in Supplemental Table S1.
The hormonomic results were further cross-validated by comparing the transcriptomic data (Fig. 3, C and D; Supplemental Tables S8 and S9) and hormonal profiles (Fig. 4, E and F). ABA and JA, often referred to as plant stress hormones, have been shown previously to promote salt tolerance (Ryu and Cho, 2015). Accordingly, salt stress was associated with increases in the levels of ABA, its oxidation products phaseic acid and dihydrophaseic acid, and JA in roots, together with the up-regulation of JA biosynthesis and ABA biosynthesis and oxidation genes. A similar correspondence was found between GA metabolite and transcript profiles and the differential responses of the active GA4 to salt stress in shoots and roots. GA biosynthesis and inactivation genes (KO and GA2ox, encoding ent-kaurene oxidase and gibberellin 2-oxidase, respectively) were induced in both tissues under salt stress. However, GA3ox, catalyzing the last biosynthetic step of bioactive GAs, was induced only in shoots, in accordance with an observed increase in GA4 concentration in this tissue. By contrast, AX and CK metabolite outputs showed a more dynamic balance that could not be readily linked to changes in the expression profiles of genes involved in their biosynthesis and metabolism under salinity stress. These findings suggest that physiological responses to stimuli such as salinity are not controlled solely by a single active form of a hormone (or even single active forms of several hormones) but by the combined activities and ratios of multiple hormones, metabolites, and (hence) genes. Our hormonomic analysis and the cross-validation with transcriptomic data highlight the value of such profiling methods, which provide potent tools to assess not only transcript levels but also the levels of corresponding metabolites in samples, thereby obtaining global views of hormonal responses and interactions.
DISCUSSION
Recent technical advances in analytical methods have helped to detect more hormone metabolites (precursors, catabolites, and conjugates) in one sample and, thus, to obtain information about the overall pattern of the hormone metabolome (Novák et al., 2017). We present here a plant hormonomics technique involving a nonselective extraction and SPE purification followed by high-throughput UHPLC-ESI-MS/MS separation and analysis for profiling 101 phytohormones and their metabolites in a single plant sample (Supplemental Table S1). Several challenges were addressed during its development and should be considered in any attempts to establish such techniques. First, the use of an appropriate extraction solvent is crucial to minimize possible enzymatic degradation, reduce levels of interfering substances, and efficiently extract the analytes from plant tissues (Hoyerová et al., 2006). This poses a dilemma because increasing concentrations of organic solvents, such as MeOH, ethanol, ACN, isopropanol, and chloroform (singly or in various combinations), generally increases the extraction efficiency of both the analytes and interfering substances (e.g. pigments, proteins, phenolics, or lipids). Such ballast compounds increase background noise, detection limits, risks of analytes co-eluting with other substances, and fouling of the instruments (Tarkowská et al., 2014). Thus, at least one purification step before final MS-based analysis is desirable, or even essential, for high-throughput analyses (Nováková and Vlčková, 2009). The risk of enzymatic or chemical breakdown of the analyte also can be minimized by performing the extraction at low temperatures (Ljung et al., 2010); however, the effect of this on the extraction and determination of phytohormones was not assessed here. Therefore, our protocol includes the extraction of samples with an optimized solvent (ice-cold 50% aqueous ACN) and purification of the extracts by one-step, nonselective, RP SPE (Fig. 1). ACN-based extraction is frequently used in nontargeted metabolomic analysis (Hyötyläinen, 2013). The relatively high polarity of ACN may be less suited for nonpolar substances, although the ACN aqueous solution has been used as an extraction solvent in methods dedicated to analyses of GAs and BRs (Urbanová et al., 2013; Tarkowská et al., 2016). Moreover, to prevent pH-dependent hydrolysis and/or other structural changes that may occur during extraction (Novák et al., 2012), we avoided use of acidified solvents during sample extraction and purification. Based on the results of our method validation, ice-cold ACN-based extraction combined with the stable isotope dilution method enables efficient control of hormone losses during sample extraction and homogenization. After applying 74 IS added before homogenization, the phytohormone levels of spiked Arabidopsis samples were recovered in the range 85.3% to 114.4% (Supplemental Table S2). To produce adequate and precise results, therefore, the IS-based correction is effective for different hormone-metabolizing activities. In addition, rapid extraction and purification methods also can minimize a risk of analyte degradation or readsorption onto the matrix (Albaseer et al., 2010).
LC was selected partly because most known phytohormones are nonvolatile compounds that require chemical derivatization for gas chromatography- (but not LC-) MS-based multitargeted profiling (Müller et al., 2002; Birkemeyer et al., 2003). The increasing availability of chromatographic columns with diverse physicochemical properties also has significantly improved the versatility of LC techniques. Furthermore, the rapid development of UHPLC, using columns with sub-2-μm particles, has greatly improved the separation, resolution, sensitivity, and overall speed of LC-based analytical methods (Nováková, 2013). Thus, LC is now the most robust, convenient, and widely utilized technique for simultaneous phytohormone analysis (Kojima et al., 2009; Müller and Munné-Bosch, 2011; Balcke et al., 2012; Floková et al., 2014; Záveská Drábková et al., 2015; Cai et al., 2016; Cao et al., 2016; Schäfer et al., 2016; Delatorre et al., 2017; Wang et al., 2017). Furthermore, compound determination based on specific MRM with a combination of known retention times of the targeted compounds and its stable isotopically labeled IS provide a reliable tool for the compensation of losses during extraction or the matrix effect and are used widely as such throughout LC-MS/MS plant hormone analyses (Tarkowská et al., 2014). Combining one-step, nonselective SPE with optimized UHPLC-MS/MS, including the use of a 1.7-µm particle size mix-mode hybrid C18 column, enabled sensitive and selective quantification of 101 underivatized phytohormones and related compounds in two independent 17-min ([M+H]+) and 15-min ([M+H]−) chromatographic runs without positive/negative polarity switching (Fig. 2).
As mentioned above, the LC-MS/MS method was originally established for simultaneous analysis operating in both modes; however, analysis of plant hormones requires high sensitivity due to the very low amounts of some hormones in plant tissues. The application of separate polarity modes indeed provided us with higher signal intensity, up to 12-fold in ESI(–) (Supplemental Fig. S1). This may be due to the use of a longer dwell time and final reduction of the overall cycle time for each MRM scan (O’Mahony et al., 2013). In our method, the dwell times in ESI(–) modes of polarity switch- and non-switch-based approaches were in the range of 8 to 20 ms and 20 to 100 ms, respectively. Dwell time also represents a compromise between the signal-to-noise (S/N) ratio and the number of data points actually needed to define a chromatographic peak (Mastovská and Lehotay, 2003). Increasing the number of data points improves the S/N ratio but also increases the scan cycle time, resulting in the reduction of MS signal (Hird et al., 2014). Furthermore, time sectoring is another important factor when setting up LC-MS/MS methods (O’Mahony et al., 2013). Splitting of the chromatographic run into more scan windows also could help to increase the sensitivity. Under our chromatographic conditions, the MRM channels were time sectored to increase the cycle time for each analyte and acquire at least 15 data points across a chromatographic peak to ensure reliable integration. Finally, nine time scan segments were used for analysis of the same samples in positive and negative ion modes compared with 12 scan windows in the method utilizing polarity switching in a single analytical run.
Given the very low amounts of some hormones in plants, the risk of false positive is extremely high, and one should ensure unambiguous detection of the analytes of interest without any interference by other matrix compounds. The following quality criteria were used to ensure the correct identification and quantification of the targeted compounds: (1) the retention times should match those of the standard compounds below 2% RSD; (2) the intensity ratio of the selected MRMs should be within ±15% of that observed for the standard compounds; and (3) the S/N ratios should be greater than 3:1. We previously showed the positive effect of small amounts of plant tissue on IAA recovery using silica (C8) or polymer-based (HLB) SPE columns (Novák et al., 2012). This result suggests a reduction in the matrix effect depending on the increasing amount of fresh weight. Our general purification method is able to separate a broad range of phytohormone metabolites from small amounts of tissue (20 mg fresh weight). However, not all targeted compounds are present in every plant tissue or plant species, even if their actual levels could still be below the LOD of the method presented. As shown in Table I and Supplemental Tables S2 to S5, the method combining microextraction and purification prior to UHPLC-ESI-MS/MS analysis shows good performance with respect to all the validation parameters tested. The measured coefficients were mostly within 15% of the nominal values, meaning that the method’s reproducibility, repeatability, accuracy, and precision are consistent with the typical validation parameters and the requirements of bioanalytical methods (Nováková, 2013).
The optimized method was applied to the phytohormone profiling of Arabidopsis plants grown under salt stress and control conditions. In total, 43 phytohormones and their metabolites in single plant samples were quantified simultaneously (Supplemental Table S7; Supplemental Fig. S4). Univariate and multivariate analyses of the acquired data revealed significant differences in profiles of these compounds between roots and shoots and between controls and salt-stressed plants (Fig. 4). The analysis also revealed compounds primarily responsible for the significant differences in profiles (Fig. 4, E and F), which mostly corresponded well with transcriptomic data (Fig. 3, C and D). The cross-validation of the hormonomic results with salt stress-induced changes in gene expression thus highlights the potential of this technique in unraveling the network of plant hormone-signaling cascades.
In summary, our method for phytohormone profiling provides a simple, sensitive, and powerful tool for phytohormonal studies. The validation experiments, in conjunction with the demonstration of the hormonomic method’s accuracy and precision, confirm its reliability and utility for routine quantifications of phytohormones in minute amounts of plant tissue. LC-MS/MS-based methods were already used to quantify phytohormone classes in carefully collected material, such as plant organs and specific organ parts (e.g. root apex; Plačková et al., 2017), isolated cells (Petersson et al., 2009; Jin et al., 2013; Pencík et al., 2013; Antoniadi et al., 2015), or even organelles (Ranocha et al., 2013; Jiskrová et al., 2016). In the future, such material also could be used in the hormonomic analysis. Thus, the combination of precise sample preparation (using, for example, FACS or laser microdissection; Immanen et al., 2016) with sensitive analysis also will yield more precise information about the localization of determined compounds and their function.
MATERIALS AND METHODS
Chemicals and Materials
Authentic standards and their isotopically labeled counterparts are listed in Supplemental Table S1). CKs, AXs, GAs, JAs, SA, ABA, phaseic acid, BRs, and their corresponding isotopically labeled analogs were purchased from Olchemim and Chemiclones; dihydrophaseic acid, neophaseic acid, 7-hydroxy-abscisic acid, and their corresponding isotopically labeled analogs were obtained from the compound library of the Laboratory of Growth Regulators (Turečková et al., 2009). Formic acid, ACN (hypergrade for LC-MS), and MeOH (hypergrade for LC-MS) were purchased from Merck. Deionized (Milli-Q) water was obtained using a Simplicity 185 water system (Millipore) and used to prepare all aqueous solutions.
Plant Material and Salinity Stress Experiments
Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) was used for method validation and the salt stress experiments. Seedlings were grown vertically in petri dishes in standard Murashige and Skoog medium in a growth chamber under long-day conditions at a light intensity of 100 µE m−2 s−1 (16-h-light, 24°C/8-h-dark, 18°C cycle) for 10 and 12 d. On day 12, plants assigned to the salt stress treatment were transferred to new medium supplemented with 150 mm NaCl (8.77 g L−1) prior to autoclaving, and seedlings were grown vertically for an additional 48 h. Control seedlings were grown in the same way and transferred during day 12 to standard Murashige and Skoog medium. On day 14, seedlings of both sets were harvested, and shoots and roots were separated, weighed into microtubes, immediately frozen in liquid nitrogen, and stored at −80°C until extraction and analysis.
Solubility Experiment
The solubility of selected BRs (Supplemental Table S10) was tested by adding 40-µL portions of aqueous ACN, with concentrations ranging from 10% to 70% (v/v), to mixtures containing 50 pmol of each of the compounds in solid state. The samples were thoroughly mixed by sonication for 5 min at 4°C in a Transsonic T310a laboratory ultrasonicator with an ice block-filled bathtub (Elma), then filtered using modified nylon 0.2-µm Centrifugal Filters (VWR International). Portions (20 µL) of the filtrates were transferred to new insert-equipped vials and analyzed by UHPLC-ESI-MS/MS (2 µL per injection). Finally, average peak areas of each compound extracted in each solvent and relative yields (percentages of average peak areas in each solvent relative to those obtained using the most effective tested solvent) were calculated.
Chlorophyll Extraction
Chlorophyll a and chlorophyll b were extracted from 100-mg (fresh weight) samples of Arabidopsis seedlings using 1 mL of aqueous ACN at concentrations ranging from 10% to 100% (v/v; n = 4, extraction solvent). After extraction and removal of solid particles by centrifugation (36,670 g, 4°C, and 10 min), the supernatants were transferred to new Eppendorf tubes and evaporated to dryness under a gentle stream of gaseous nitrogen using a TurboVap LV evaporation system (Caliper Life Sciences). Before chlorophyll determination, the pellets were dissolved in acetone (reagent grade, Lach-ner). The light absorbance of the resulting suspensions was measured at 663.2-, 646.8-, and 750-nm wavelengths using an Infinite 200 PRO spectrophotometer (Tecan), and the sample’s chlorophyll contents were calculated according to Lichtenthaler (1987); Supplemental Table S11).
Stability Experiment
Portions of solutions containing known amounts of selected analytes (0.4 pmol of CKs and JAs with 4 pmol of AXs and GAs per sample [n = 3]; Supplemental Table S12) were transferred to new vials and evaporated to dryness under a gentle stream of gaseous nitrogen using a TurboVap LV evaporation system (Caliper Life Sciences). The compounds were then dissolved in 1 mL of aqueous solutions of 1 m formic acid (pH < 3), 0.35 m NH4OH (60% MeOH [v/v], pH > 12), or 50% ACN using ultrasound (5 min, 4°C; Transsonic T310a laboratory ultrasonicator [Elma]). After incubation for 15 min at 4°C, samples were filtered using modified nylon 0.2-µm Centrifugal Filters (VWR International). Filtered samples were evaporated to dryness as described above, dissolved in 40 µL of 30% ACN, and subjected to UHPLC-MS/MS analysis (10 µL per injection). Relative peak areas (percentage) of the compounds were calculated as ratios to respective peak areas obtained from analyses of reference samples in 50% ACN (Fig. 1C). However, the extent of enzymatic activity in plant extracts obtained using the proposed sample preparation protocol was not investigated.
Sample Extraction
For the quantification of targeted plant hormones and related compounds, 20-mg (fresh weight) portions of separately harvested roots and shoots were weighed into 2-mL plastic microtubes (Eppendorf) and frozen in liquid nitrogen. To minimize the risk of false-positive detections, possible background levels of analytes were subtracted from measured sample values. Therefore, in the blank controls, 1 mL of extraction buffer containing a mixture of stable isotopically labeled IS also was purified. Before extraction, three 3-mm ceria-stabilized zirconium oxide beads (Next Advance) were added to each sample. A mixture of stable, isotopically labeled IS was added to validate the method and enable the precise quantification of endogenous levels of targeted compounds. The amounts of IS ranged from 0.4 to 50 pmol per sample (precise amounts are listed in Supplemental Table S3). Frozen plant material was extracted in 1 mL of ice-cold 50% aqueous (v/v) ACN using an MM 301vibration mill (Retsch) operating at a frequency of 27 Hz for 5 min. Samples were then sonicated for 3 min at 4°C using a Transsonic T310 ultrasonicator with an ice block-filled bathtub (Elma) and subsequently extracted using a Stuart SB3 benchtop laboratory rotator (Bibby Scientific) for 30 min at 4°C. After centrifugation (10 min, 36,670 g, and 4°C; Beckman Avanti 30), the supernatant was transferred to clean plastic microtubes. Samples were further purified according to the scheme shown in Figure 1D.
Sample Purification
All samples were purified using Oasis HLB RP, polymer-based SPE cartridges (1 cc per 30 mg), obtained from Waters, that had been washed with 1 mL of 100% MeOH and 1 mL of deionized water, then equilibrated with 50% aqueous (v/v) ACN. After loading a sample (supernatant obtained following the procedure described above), the flow-through fraction was collected in a glass tube (Fisherbrand). The cartridge was then rinsed with 1 mL of 30% (v/v) ACN, and this fraction was collected in the same glass tube as the flow-through fraction. After this single-step SPE, the samples were evaporated to dryness under a gentle stream of nitrogen using a TurboVap LV evaporation system (Caliper Life Sciences) and stored at −20°C until analysis. For UHPLC-ESI-MS/MS analysis, the samples were dissolved in 40 µL of 30% ACN (v/v) and transferred to insert-equipped vials, then 20-µL portions of each sample were injected (in two 10-µL injections) into the UHPLC-ESI-MS/MS system.
UHPLC-ESI-MS/MS Conditions
Targeted compounds were analyzed using an Acquity UPLC I-Class System equipped with a Binary Solvent Manager, a Sample Manager with Flow-Through Needle, and an Acquity UPLC CSH C18 RP column (150 × 2.1 mm, particle size of 1.7 µm) coupled to a triple quadrupole mass spectrometer (Xevo TQ-S MS), all from Waters. The mobile UPLC phase consisted of binary gradients of ACN with 0.01% (v/v) formic acid (A) and 0.01% (v/v) aqueous formic acid (B), flowing at 0.5 mL min−1, which depended on the ESI mode, as described below. MassLynx software (version 4.1; Waters) was used to control the instrument and to acquire and process the MS data.
Separation of Compounds Detected in ESI(+) Mode
Analytes were initially eluted isocratically with 5% (v/v) A for 5 min, then the proportion of A was increased linearly to 80% over the following 10 min. After this, the column was washed with 100% A and then reequilibrated under the initial conditions for 2 min. The column temperature was held at 50°C.
Separation of Compounds Detected in ESI(−) Mode
The mobile phase was the same until the A:B ratio reached 65:35 (v/v) in the 13th min. Then, the column was washed with 100% A and reequilibrated under the initial conditions for 2 min.
During analytical runs in both ESI modes, the UHPLC eluate was switched to waste until acquisition of the first targeted compound and back to waste after elution of the last compound to minimize impairment of the MS system’s sensitivity by ballast compounds. During the acquisition of analytes, the eluate was introduced into the electrospray ion source of the MS/MS analyzer operating under the following conditions: source/desolvation temperature, 125°C/600°C; cone/desolvation gas flow, 150/1,000 L h−1; capillary voltage, 2.1 kV for ESI(+) and 1.5 kV for ESI(–); cone voltage, 10 to 40 V; collision energy, 12 to 30 eV; collision gas flow (argon), 0.21 mL min−1. The analyzed compounds and appropriate IS were quantified in MRM mode using optimized MS conditions (Supplemental Table S3). The interscan and interchannel delays were set to 3 ms when switching between successive MRM channels, and 20 ms was required for interchannel delay when switching from positive to negative ionization mode between successive channels. The dwell times ranged from 8 to 100 ms to provide at least 15 data points across each chromatographic peak using automatic mode. The MRM transitions were recorded over each chromatographic run in nine targeted scan windows to maximize the MS signal intensity for each compound. For ESI(+) runs, these windows were as follows: 1 to 5.3, 5.31 to 7.65, 7.3 to 8.4, 8.35 to 8.85, 8.86 to 10, 10.1 to 10.95, 11.3 to 11.85, 11.5 to 12.7, and 12.6 to 15 min. For ESI(–) runs, they were as follows: 6.3 to 7.3, 7.3 to 8.25, 8.2 to 8.7, 8.7 to 9.3, 9 to 9.9, 9.8 to 10.85, 10.9 to 11.3, 11.3 to 12, and 12 to 13 min.
Method Validation
UHPLC-ESI-MS/MS calibration curves were constructed using serially diluted phytohormone standards, listed in Supplemental Table S3, and the labeled IS (added in known concentrations). The LOD and LOQ were defined as S/N ratios of 3:1 and 10:1, respectively.
To evaluate losses of analytes during the purification process and validate the method, three sets of samples were each prepared in triplicate and analyzed by the UHPLC-ESI-MS/MS system. In the first set, 20 mg fresh weight of Arabidopsis seedlings was extracted in ice-cold 50% ACN spiked with known amounts of stable isotope-labeled IS (at levels listed in Supplemental Table S2) and subsequently purified by the SPE protocol (Fig. 1D). The second set consisted of identical plant tissue extracts spiked with a mixture of authentic and stable isotope-labeled IS (at levels listed in Supplemental Table S2) before SPE. The third set consisted of 20-mg (fresh weight) portions of Arabidopsis plant tissue extracted and spiked by adding nonlabeled standards, in varied concentrations, directly to purified eluates after the SPE step.
Analyte recovery (percentage; Caban et al., 2012) following the purification process was calculated as the ratio of the mean peak area of a nonlabeled analyte spiked before SPE (set 2) to the mean peak area of the same analyte spiked after SPE purification (set 3) multiplied by 100.
Concentrations of plant hormones were quantified using the standard isotope dilution method (Rittenberg and Foster, 1940). Concentrations of nonlabeled, targeted compounds added to samples from sample set 2 were calculated after subtracting their determined endogenous levels (average values for each compound obtained from analyses of sample set 1). Finally, determined analyte concentrations were compared with known theoretical amounts of appropriate standards added to samples and presented as method accuracy (expressed as percentage bias). The method precision for each analyte was calculated as the RSD (percentage) of its determined concentration in three replicates of samples of set 2.
To test the reproducibility and repeatability of the method, two experiments were carried out. First, standard mixtures containing known concentrations of the IS (medium) and three concentration levels (low/medium/high) of the targeted compounds were prepared as follows: CKs and 2MeSCKs, 0.001/0.05/1; AXs, 0.01/0.5/10; JAs [ESI(+)], 0.0025/0.05/2.5; JAs and ABAs [ESI(–)], 0.05/1/50; SA, 0.1/5/100; GAs, 0.005/0.1/5; and BRs, 0.05/0.05/10 pmol per injection. Three replicates of each of these analyte levels were injected on three consecutive days, and all samples were kept at 4°C throughout the experiment. The stability of the retention times for all compounds was expressed as percentage RSD. The reproducibility of the IS-normalized response for each analyte also was calculated. In the second experiment, 10-d-old Arabidopsis seedlings (20 mg fresh weight) were spiked with known concentrations of isotope-labeled IS listed in Supplemental Table S2, and then endogenous phytohormones were isolated using the developed method (Fig. 1D). All samples were analyzed as an independent batch in five replicates on three separate days. The intraday and interday calculations were based on the IS-normalized response of endogenous compounds. The results are expressed as percentage RSD of the measurements.
Genevestigator Analysis
The meta-analytical approach of Genevestigator (Hruz et al., 2008) has proven value for designing new experiments and validating existing results (Saito et al., 2008). Therefore, the software was initially used (in an unsupervised manner) to identify a single stress condition that alters the expression of genes involved in the biosynthesis of most hormones and their metabolism pathways (Supplemental Table S6). Salt stress was identified as the most appropriate condition by screening using the Differential Expression tool (Genevestigator Experiment ID: AT-00656), and its suitability was confirmed by screening shifts in the expression of hormone-related genes in one more salt stress experiment (Genevestigator Experiment ID: AT-00120). For representation of these results (Fig. 3B), a Venn diagram was constructed using Venny 2.1 software (Oliveros, 2007-2015). The input consisted of hormone-related genes listed in Supplemental Table S6 and Arabidopsis genes that exhibited altered expression (up- or down-regulation in a greater than 1.5-fold change, P < 0.01) in response to salt stress after a 24-h treatment in Arabidopsis roots and shoots (Genevestigator Experiment ID: AT-00120) and after a 48-h treatment in root cell-specific protoplasts (Genevestigator Experiment ID: AT-00656) isolated through FACS. To compare the data on hormonomic and transcriptomic shifts under salt stress, the Perturbations tool was used. The genes listed in Supplemental Table S6 that were significantly up- or down-regulated (fold change ≥ 1.5, P < 0.01) in response to salinity stress were identified. Log values of changes in their expression were then extracted, and their activities in hormone pathways were noted to check the consistency between their responses and the changes we detected in the corresponding hormone metabolites (Supplemental Tables S8 and S9).
The Genevestigator interface is a JAVA applet running in the user’s browser. Information about the individual tools and the respective statistical analysis of the data is provided on the Genevestigator Web site (www.genevestigator.com).
Statistical Analysis (Univariate and Multivariate Statistics)
Before multivariate statistical analysis, the missing values of the targeted compounds found in some plant tissue samples under the LOD were imputed with two-thirds of their respective LODs listed in Supplemental Table S2 (Martín-Fernández et al., 2003). Compounds for which more than 50% of values were missing were removed from the data set. Multivariate analysis was performed using SIMCA software (version 14; Umetrics). Unsupervised PCA and supervised OPLS-DA were applied to log-transformed and Pareto-scaled data. PCA was used to obtain a general overview of the data structure, and OPLS-DA derived S-plots to identify compounds responsible for the separation of roots and shoots and samples from control and salinity-stressed plants. Differences in the levels of each determined metabolite between these groups also were evaluated using Student’s t test at P < 0.05, P < 0.01, and P < 0.001.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under the accession numbers listed in Supplemental Table S6.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of signal intensities of indicated analytes of various phytohormone classes.
Supplemental Figure S2. Representative MRM chromatograms of endogenous CKs and AXs in 20 mg fresh weight of Arabidopsis roots 48 h after the salt stress treatment.
Supplemental Figure S3. Representative MRM chromatograms of endogenous JAs, ABAs, SA, and GAs in 20 mg fresh weight of Arabidopsis roots 48 h after the salt stress treatment.
Supplemental Figure S4. Scatterplots of phytohormone distributions in root and shoot samples of control and salt-stressed Arabidopsis plants.
Supplemental Table S1. List of targeted compounds.
Supplemental Table S2. Method validation data.
Supplemental Table S3. Optimized UHPLC-MS/MS parameters.
Supplemental Table S4. Reproducibility of the LC-MS/MS method.
Supplemental Table S5. Intraday and interday precision of the method.
Supplemental Table S6. Phytohormone-related genes used in the experimental design process.
Supplemental Table S7. Determined levels of plant hormones in root and shoot samples of salt-stressed and control Arabidopsis plants.
Supplemental Table S8. Genes showing shifts in expression levels under salt stress in roots.
Supplemental Table S9. Genes showing shifts in expression levels under salt stress in shoots.
Supplemental Table S10. Solubility of BRs.
Supplemental Table S11. Determination of chlorophylls.
Supplemental Table S12. Tests of pH stability.
Acknowledgments
We thank the Swedish Metabolomics Centre for the use of instrumentation and Sees-editing for careful revision of the article. We also thank Eva Hrdličková Hirnerová for technical support and sample preparation during the plant hormone analyses.
Footnotes
This work was funded by the Internal Grant Agency of Palacký University (project no. IGA_PrF_2018_023), the Ministry of Education, Youth, and Sports of the Czech Republic (National Program for Sustainability I, grant no. LO1204), and the Czech Science Foundation (grant no. GA17-06613S). Support was also provided by the Swedish Governmental Agency for Innovation Systems (Vinnova) and the Swedish Research Council to K.L. and I.A.
Articles can be viewed without a subscription.
References
- Albaseer SS, Rao RN, Swamy YV, Mukkanti K (2010) An overview of sample preparation and extraction of synthetic pyrethroids from water, sediment and soil. J Chromatogr A 1217: 5537–5554 [DOI] [PubMed] [Google Scholar]
- Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, Novák O (2015) Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 27: 1955–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcke GU, Handrick V, Bergau N, Fichtner M, Henning A, Stellmach H, Tissier A, Hause B, Frolov A (2012) An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkemeyer C, Kolasa A, Kopka J (2003) Comprehensive chemical derivatization for gas chromatography-mass spectrometry-based multi-targeted profiling of the major phytohormones. J Chromatogr A 993: 89–102 [DOI] [PubMed] [Google Scholar]
- Caban M, Migowska N, Stepnowski P, Kwiatkowski M, Kumirska J (2012) Matrix effects and recovery calculations in analyses of pharmaceuticals based on the determination of β-blockers and β-agonists in environmental samples. J Chromatogr A 1258: 117–127 [DOI] [PubMed] [Google Scholar]
- Cai WJ, Ye TT, Wang Q, Cai BD, Feng YQ (2016) A rapid approach to investigate spatiotemporal distribution of phytohormones in rice. Plant Methods 12: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZY, Sun LH, Mou RX, Zhang LP, Lin XY, Zhu ZW, Chen MX (2016) Profiling of phytohormones and their major metabolites in rice using binary solid-phase extraction and liquid chromatography-triple quadrupole mass spectrometry. J Chromatogr A 1451: 67–74 [DOI] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35: 405–417 [DOI] [PubMed] [Google Scholar]
- Davies PJ. (2010) Plant Hormones: Biosynthesis, Signal Transduction, Action! Ed 3 Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Delatorre C, Rodríguez A, Rodríguez L, Majada JP, Ordás RJ, Feito I (2017) Hormonal profiling: development of a simple method to extract and quantify phytohormones in complex matrices by UHPLC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci 1040: 239–249 [DOI] [PubMed] [Google Scholar]
- Dobrev PI, Kamínek M (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Du F, Ruan G, Liu H (2012) Analytical methods for tracing plant hormones. Anal Bioanal Chem 403: 55–74 [DOI] [PubMed] [Google Scholar]
- Farrow SC, Emery RN (2012) Concurrent profiling of indole-3-acetic acid, abscisic acid, and cytokinins and structurally related purines by high-performance-liquid-chromatography tandem electrospray mass spectrometry. Plant Methods 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floková K, Tarkowská D, Miersch O, Strnad M, Wasternack C, Novák O (2014) UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 105: 147–157 [DOI] [PubMed] [Google Scholar]
- Hird SJ, Lau BPY, Schuhmacher R, Krska R (2014) Liquid chromatography-mass spectrometry for the determination of chemical contaminants in food. Trends Anal Chem 59: 59–72 [Google Scholar]
- Hoyerová K, Gaudinová A, Malbeck J, Dobrev PI, Kocábek T, Solcová B, Trávnícková A, Kamínek M (2006) Efficiency of different methods of extraction and purification of cytokinins. Phytochemistry 67: 1151–1159 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyötyläinen T. (2013) Sample collection, storage and preparation. In Hyotylainen T, Wiedmer SK, eds, Chromatographic Methods in Metabolomics. Royal Society of Chemistry, London, pp 11–42 [Google Scholar]
- Immanen J, Nieminen K, Smolander OP, Kojima M, Alonso Serra J, Koskinen P, Zhang J, Elo A, Mähönen AP, Street N, et al. (2016) Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr Biol 26: 1990–1997 [DOI] [PubMed] [Google Scholar]
- Jin X, Wang RS, Zhu M, Jeon BW, Albert R, Chen S, Assmann SM (2013) Abscisic acid-responsive guard cell metabolomes of Arabidopsis wild-type and gpa1 G-protein mutants. Plant Cell 25: 4789–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiskrová E, Novák O, Pospíšilová H, Holubová K, Karády M, Galuszka P, Robert S, Frébort I (2016) Extra- and intracellular distribution of cytokinins in the leaves of monocots and dicots. N Biotechnol 33: 735–742 [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Ljung K, Sandberg G, Moritz T (2010) Methods of plant hormone analysis. In Davies PJ, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Ed 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 717–740 [Google Scholar]
- Martín-Fernández JA, Barceló-Vidal C, Pawlowsky-Glahn V (2003) Dealing with zeros and missing values in compositional data sets using nonparametric imputation. Math Geol 35: 253–278 [Google Scholar]
- Mastovská K, Lehotay SJ (2003) Practical approaches to fast gas chromatography-mass spectrometry. J Chromatogr A 1000: 153–180 [DOI] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S (2011) Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Düchting P, Weiler EW (2002) A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216: 44–56 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- Novák O, Napier R, Ljung K (2017) Zooming in on plant hormone analysis: tissue- and cell-specific approaches. Annu Rev Plant Biol 68: 323–348 [DOI] [PubMed] [Google Scholar]
- Nováková L. (2013) Challenges in the development of bioanalytical liquid chromatography-mass spectrometry method with emphasis on fast analysis. J Chromatogr A 1292: 25–37 [DOI] [PubMed] [Google Scholar]
- Nováková L, Vlčková H (2009) A review of current trends and advances in modern bio-analytical methods: chromatography and sample preparation. Anal Chim Acta 656: 8–35 [DOI] [PubMed] [Google Scholar]
- O’Mahony J, Clarke L, Whelan M, O’Kennedy R, Lehotay SJ, Danaher M (2013) The use of ultra-high pressure liquid chromatography with tandem mass spectrometric detection in the analysis of agrochemical residues and mycotoxins in food: challenges and applications. J Chromatogr A 1292: 83–95 [DOI] [PubMed] [Google Scholar]
- Oklestkova J, Tarkowská D, Eyer L, Elbert T, Marek A, Smržová Z, Novák O, Fránek M, Zhabinskii VN, Strnad M (2017) Immunoaffinity chromatography combined with tandem mass spectrometry: A new tool for the selective capture and analysis of brassinosteroid plant hormones. Talanta 170: 432–440 [DOI] [PubMed] [Google Scholar]
- Oliveros JC. (2007-2015) Venny: an interactive tool for comparing lists with Venn’s diagrams.http://bioinfogp.cnb.csic.es/tools/venny/index.html
- Pan X, Welti R, Wang X (2008) Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 1773–1781 [DOI] [PubMed] [Google Scholar]
- Pencík A, Rolcík J, Novák O, Magnus V, Barták P, Buchtík R, Salopek-Sondi B, Strnad M (2009) Isolation of novel indole-3-acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655 [DOI] [PubMed] [Google Scholar]
- Pencík A, Simonovik B, Petersson SV, Henyková E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zazímalová E, et al. (2013) Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 25: 3858–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plačková L, Oklestkova J, Pospíšková K, Poláková K, Buček J, Stýskala J, Zatloukal M, Šafařík I, Zbořil R, Strnad M, et al. (2017) Microscale magnetic microparticle-based immunopurification of cytokinins from Arabidopsis root apex. Plant J 89: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Pratt JJ. (1986) Isotope dilution analysis using chromatographic separation of isotopic forms of the compound to be measured. Ann Clin Biochem 23: 251–276 [DOI] [PubMed] [Google Scholar]
- Ranocha P, Dima O, Nagy R, Felten J, Corratgé-Faillie C, Novák O, Morreel K, Lacombe B, Martinez Y, Pfrunder S, et al. (2013) Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun 4: 2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg D, Foster GL (1940) A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. J Biol Chem 133: 737–744 [Google Scholar]
- Ryu H, Cho Y (2015) Plant hormones in salt stress tolerance. J Plant Biol 58: 147–155 [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics: ‘majority report by precogs.’ Trends Plant Sci 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Schäfer M, Brütting C, Baldwin IT, Kallenbach M (2016) High-throughput quantification of more than 100 primary- and secondary-metabolites, and phytohormones by a single solid-phase extraction based sample preparation with analysis by UHPLC-HESI-MS/MS. Plant Methods 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svačinová J, Novák O, Plačková L, Lenobel R, Holík J, Strnad M, Doležal K (2012) A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowská D, Novák O, Floková K, Tarkowski P, Turečková V, Grúz J, Rolčík J, Strnad M (2014) Quo vadis plant hormone analysis? Planta 240: 55–76 [DOI] [PubMed] [Google Scholar]
- Tarkowská D, Novák O, Oklestkova J, Strnad M (2016) The determination of 22 natural brassinosteroids in a minute sample of plant tissue by UHPLC-ESI-MS/MS. Anal Bioanal Chem 408: 6799–6812 [DOI] [PubMed] [Google Scholar]
- Tarkowski P, Václavíková K, Novák O, Pertry I, Hanuš J, Whenham R, Vereecke D, Šebela M, Strnad M (2010) Analysis of 2-methylthio-derivatives of isoprenoid cytokinins by liquid chromatography-tandem mass spectrometry. Anal Chim Acta 680: 86–91 [DOI] [PubMed] [Google Scholar]
- Turečková V, Novák O, Strnad M (2009) Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80: 390–399 [DOI] [PubMed] [Google Scholar]
- Urbanová T, Tarkowská D, Novák O, Hedden P, Strnad M (2013) Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 112: 85–94 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28: 463–487 [DOI] [PubMed] [Google Scholar]
- Wang Q, Cai WJ, Yu L, Ding J, Feng YQ (2017) Comprehensive profiling of phytohormones in honey by sequential liquid-liquid extraction coupled with liquid chromatography-mass spectrometry. J Agric Food Chem 65: 575–585 [DOI] [PubMed] [Google Scholar]
- Záveská Drábková L, Dobrev PI, Motyka V (2015) Phytohormone profiling across the Bryophytes. PLoS ONE 10: e0125411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanenburg B, Pospíšil T, Ćavar Zeljković S (2016) Strigolactones: new plant hormones in action. Planta 243: 1311–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]