Rapid reanimation of a photosynthetic bacterium following nitrogen starvation is facilitated by anticipation and requires two parallel routes of glycogen catabolism and a particular glycogen phosphorylase paralog.
Abstract
Many organisms survive stressful conditions via entry into a dormant state that can be rapidly exited when the stressor disappears; this ability provides a strong selective advantage. In the cyanobacterium Synechocystis sp. PCC 6803, the exit from nitrogen chlorosis takes less than 48 h and is enabled by the impressive metabolic flexibility of these cyanobacteria, which pass through heterotrophic and mixotrophic phases before reentering photoautotrophic growth. Switching between these states requires delicate coordination of carbohydrate oxidation, CO2 fixation, and photosynthesis. Here, we investigated the contribution of the different carbon catabolic routes by assessing mutants of these pathways during nitrogen chlorosis and resuscitation. The addition of nitrate to nitrogen-starved cells rapidly starts the awakening program. Metabolism switches from maintenance metabolism, characterized by residual photosynthesis and low cellular ATP levels, to an initial heterotrophic phase, characterized by respiration and an immediate increase in ATP levels. This respiration relies on glycogen breakdown catalyzed by the glycogen phosphorylase GlgP2. In the following transient mixotrophic phase, photosynthesis and CO2 fixation restart and glycogen is consumed. During the mixotrophic phase, parallel operation of the oxidative pentose phosphate cycle and the Entner-Doudoroff pathway is required for resuscitation to proceed; the glycolytic route via the Embden-Meyerhof-Parnas pathway has minor importance. Our data suggest that, during resuscitation, only the Entner-Doudoroff and oxidative pentose phosphate pathways supply the metabolic intermediates necessary for the anabolic reactions required to reconstitute a vegetative cell. Intriguingly, the key enzymes for glycogen catabolism are already expressed during the preceding chlorotic phase, in apparent preparation for rapid resuscitation.
Cyanobacteria are oxygenic photoautotrophic organisms adapted to a wide range of environments (Stanier and Cohen-Bazire, 1977). Nitrogen shortage represents one of the most common growth limitations in terrestrial and marine ecosystems (Vitousek and Howarth, 1991). Nondiazotrophic cyanobacteria, including Synechocystis sp. PCC 6803 (hereafter Synechocystis), respond to the lack of a usable nitrogen source by undergoing a process called chlorosis (Allen and Smith, 1969; Luque and Forchhammer, 2008). This adaptation mechanism is characterized by the degradation of photosynthetic pigments, which causes cells to turn from a blue-green to a yellow color. During chlorosis, the cells divide one more time and then enter cell cycle arrest, where they degrade the bulk of cellular proteins and the photosynthetic apparatus, leaving only residual photosynthetic activity. Additionally, they tune down their metabolism by minimizing energy-consuming reactions, such as protein synthesis and anabolic processes. These molecular adaptations lead the cells into a dormant state that allows them to survive for a long period of time (Görl et al., 1998; Sauer et al., 2001). The energy produced by the residual photosynthetic activity seems to be sufficient to keep cells alive, since they consume almost no ATP. Furthermore, chlorotic cells rapidly accumulate reserve polymers, including glycogen and polyhydroxybutyrate (PHB; Sauer et al., 2001; Schlebusch and Forchhammer, 2010).
Synechocystis is capable of rapidly recovering from chlorosis when a usable nitrogen source becomes available (Klotz et al., 2016). The resuscitation process occurs in two major phases. First, the energy necessary to reinstall central cellular processes is rapidly provided by carbohydrate oxidation and respiration. Second, phase 2 starts approximately 16 h after the addition of nitrate and is characterized by reconstitution of the photosynthetic apparatus; after 48 h, cells have turned green again and photosynthesis resumes. We have demonstrated previously that glycogen is the major reserve polymer required for resuscitation. Although cells also accumulate PHB during nitrogen starvation, no significant decrease in PHB content is observed during the first phase of resuscitation, and a PHB-free mutant is able to recover from chlorosis as efficiently as the wild type (Klotz et al., 2016). By contrast, mutants deficient in glycogen synthesis present a nonbleaching phenotype and die during prolonged nitrogen starvation, which implies that glycogen synthesis is necessary for chlorosis (Gründel et al., 2012). Upon nitrogen depletion, cells accumulate glycogen up to 60% of the cell’s dry weight, and this glycogen is used as a substrate for respiration during resuscitation (Klotz and Forchhammer, 2017).
Glycogen is a biopolymer composed of α-d-glucosyl units connected by α-1,4 linkages and branched through α-1,6 linkages, which account for approximately 7% to 10% of the total linkages, and are organized in a specific way (Shearer and Graham, 2002). The synthesis and degradation of glycogen granules involve several enzymes with specific activities (Preiss, 1984). Among them, glycogen phosphorylase catalyzes the phosphate-dependent splitting of the α-1,4 linkage, thereby releasing Glc-1-P. Synechocystis harbors two homologous glycogen phosphorylase genes, glgP1 and glgP2 (corresponding to sll1356 and slr1367). Fu and Xu (2006) showed the different physiological roles of these two enzymes: GlgP1 seems to be important during heat stress, whereas GlgP2 provides the main glycolytic activity during day/night cycles. This functional divergence suggests that these two enzymes also may play different roles during resuscitation from nitrogen starvation-induced chlorosis.
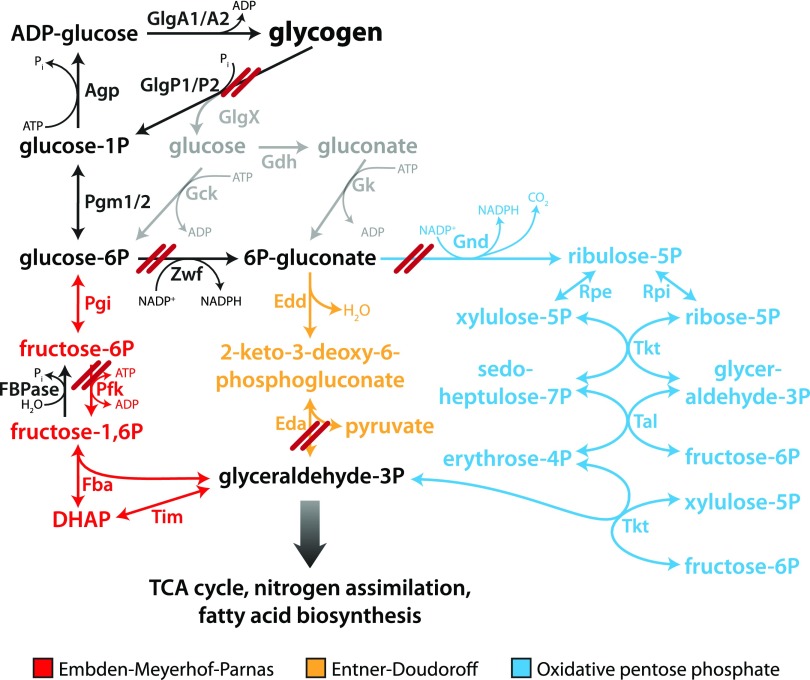
The Glc-1-P molecule released by glycogen phosphorylase is converted to Glc-6-P by phosphoglucomutase (Pgm) and then can be channeled into different glycolytic routes. Synechocystis also has two homologs of Pgm, Pgm1 (sll0726) and Pgm2 (slr1334), although Pgm1 is responsible for 97% of the activity (Liu et al., 2013). Conventionally, cyanobacteria were known to oxidize Glc-6-P via the Embden-Meyerhof-Parnas (EMP) pathway (glycolysis) and the oxidative pentose phosphate (OPP) pathway. However, Synechocystis was recently discovered to also possess the Entner-Doudoroff (ED) glycolytic pathway (Chen et al., 2016). Therefore, there are three possible degradation routes for Glc-6-P in Synechocystis (Fig. 1). The role of these different pathways in the process of resuscitation was hitherto unclear.
Figure 1.
Depiction of glycogen catabolism in Synechocystis. Deletion of specific genes is indicated as a double red line. These deletions resulted in a total of nine mutants that were analyzed in this study: ΔglgP1, ΔglgP2, ΔglgP1/2, ΔpfkB1/2, Δzwf, Δzwf/pfkB1/2, Δgnd, Δeda, and Δgnd/eda. TCA, Tricarboxylic acid.
Here, we investigated the importance of different routes of glycogen degradation for energy metabolism and their regulation during nitrogen starvation and resuscitation. We employed various deletion mutants to identify the crucial pathways in glycogen catabolism during resuscitation. Our results demonstrate that the two glycogen phosphorylase paralogs have different functions and that the ED and OPP pathways play a major role in resuscitation. The fact that the key enzymes for glycogen degradation are already expressed during nitrogen starvation demonstrates that Synechocystis anticipates the awakening process and is prepared for rapid glycogen degradation, once nitrogen is available. To maintain this machinery in a quiescent state unless it is needed requires a sophisticated, yet unknown, mechanism of regulation.
RESULTS
Energy Metabolism during Nitrogen Starvation and Resuscitation in Synechocystis
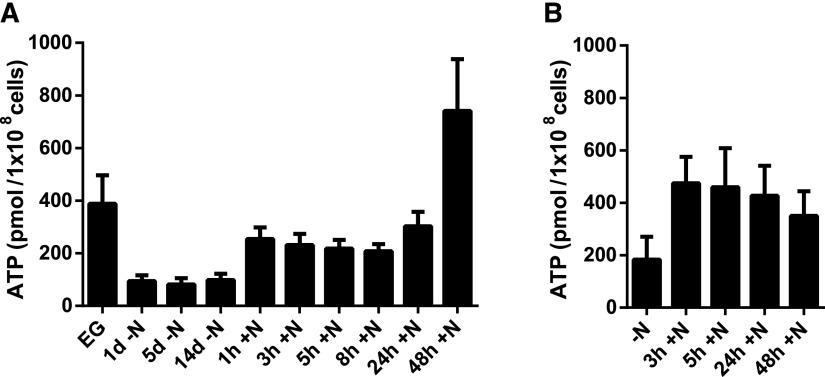
In order to better understand energy metabolism during nitrogen starvation and resuscitation, we measured the ATP content of Synechocystis during these phases. As shown in Figure 2A, after nitrogen depletion, the ATP level dropped from ∼400 to ∼100 nm 108 cells−1 mL−1 and stayed constant at this level while the cells were in dormancy. After providing the cells with nitrate, the ATP level almost immediately increased to ∼250 nm 108 cells−1 mL−1, a level that was intermediate between that in exponentially growing cells and that during nitrogen starvation. The ATP content then stayed constant during the first phase of resuscitation until about 24 h. Thereupon, concomitant with the onset of photosynthetic electron transport, the ATP levels increased strongly and, at 48 h, reached a value 2 times higher than in exponentially growing cells (greater than 700 nm 108 cells−1 mL−1). At this time point, photosynthetic activity is completely restored, although cells have not yet resumed cell division (Klotz et al., 2016). These results are consistent with the metabolic adaptations of Synechocystis during nitrogen starvation and resuscitation (Klotz et al., 2016).
Figure 2.
Determination of ATP content during exponential growth (EG), nitrogen starvation (−N), and resuscitation (+N) of the wild type (A) and ΔglgP1/2 (B). The ATP content was normalized to 1 × 108 cells. At least three biological replicates were measured; error bars represent the sd.
GlgP2 Is the Crucial Enzyme for Glycogen Degradation during Resuscitation
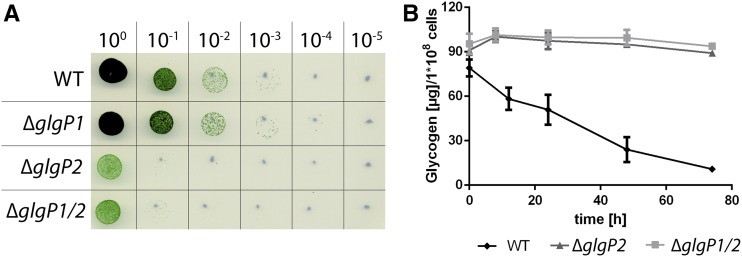
To understand the importance of glycogen degradation during resuscitation, mutants deficient in the two homologous glycogen phosphorylases (glgP1 and glgP2) were generated. The two genes were replaced by antibiotic resistance cassettes, resulting in a total of three strains: ΔglgP1, ΔglgP2, and the double mutant ΔglgP1/2. We starved these three phosphorylase-deficient mutants for 1 month and analyzed their ability to recover from nitrogen starvation on BG11 solid agar plates in comparison with the Synechocystis wild type (Fig. 3A). ΔglgP1 presented no phenotype, since it was able to recover with the same efficiency as the wild type. However, neither ΔglgP2 nor ΔglgP1/2 could efficiently recover from nitrogen starvation. We measured the ability of ΔglgP2 and ΔglgP1/2 to degrade glycogen by measuring the glycogen content of these mutants during resuscitation (Fig. 3B). Our results showed that neither of the two mutants degraded a significant amount of glycogen after nitrate addition. These findings indicate that glycogen degradation is essential for recovery from nitrogen chlorosis and that GlgP2 is the major glycogen-degrading enzyme during resuscitation.
Figure 3.
Characterization of glycogen phosphorylase-deficient mutants (ΔglgP1, ΔglgP2, and ΔglgP1/2). A, Spot assay on solid BG11 agar of the phosphorylase-deficient mutants to test resuscitation from long-term chlorosis. Dilutions are indicated on the top row. B, Determination of the glycogen content during resuscitation from 1-month chlorosis of ΔglgP2 and ΔglgP1/2. The glycogen content was normalized to 1 × 108 cells. At least three biological replicates were measured. Error bars represent the sd. WT, Wild type.
Glycogen Degradation Is Necessary for Turning on Respiration during Resuscitation
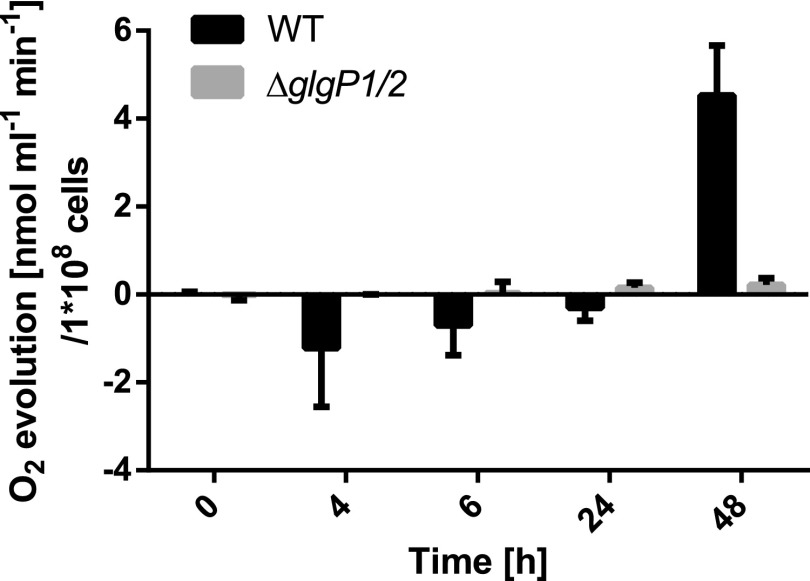
Degradation of glycogen is necessary for successful resuscitation, but it is not known how glycogen degradation affects the onset of respiration and the switch to heterotrophic metabolism upon the addition of nitrate. To address this, we further characterized the ΔglgP1/2 mutant by measuring its oxygen evolution during resuscitation. Figure 4 shows a comparison of how oxygen is consumed/produced in the wild type and ΔglgP1/2. While the wild type turned on respiration soon after nitrate addition, no significant oxygen consumption was observed for ΔglgP1/2 in the first 24 h of recovery. Rather, this mutant exhibited a tiny amount of oxygen evolution after 6 h of resuscitation, while the wild type was still respiring. In the wild type, residual photosynthesis is completely suppressed after the addition of nitrate (Klotz et al., 2016), suggesting that ATP is obtained from respiration. Suppression of photosynthesis during illumination might be due to the induction of respiration, resembling an inverse Kok effect. The Kok effect describes the phenomenon that, during exponential growth, photosynthesis inhibits respiration (Healey and Myers, 1971), the opposite of which seems to occur during Synechocystis resuscitation. In the ΔglgP1/2 mutant, since respiration is not turned on, residual photosynthesis may continue after nitrate addition. To test this hypothesis, we used pulse-amplitude modulation (PAM) fluorometry to determine the PSII activity of the ΔglgP1/2 mutant during resuscitation (Supplemental Fig. S1). In fact, instead of being suppressed, the PAM-measured PSII activity increased slightly during the first hours of resuscitation, confirming that ΔglgP1/2, unable to degrade glycogen, does not turn on respiration but, instead, continues its residual photosynthesis.
Figure 4.
Oxygen evolution of ΔglgP1/2 and the wild type (WT) during recovery. At least three biological replicates were measured; error bars represent the sd.
To investigate how this residual photosynthesis in the ΔglgP1/2 strain affects its energy metabolism, we measured the ATP levels during resuscitation (Fig. 2B). Strikingly, as in the wild type, the ATP concentration increased after the addition of nitrate and stayed constant during the first few hours of resuscitation. However, in contrast to the wild type, wherein a further increase in ATP levels was observed after 24 h, the ATP levels in the mutant decreased at the later time points, consistent with its inability to recover from chlorosis. Conversely, in the dark, only a small increase in ATP levels was observed in the ΔglgP1/2 mutant (Supplemental Fig. S2).
Functionality of the ED and OPP Pathways Plays a Key Role during Resuscitation
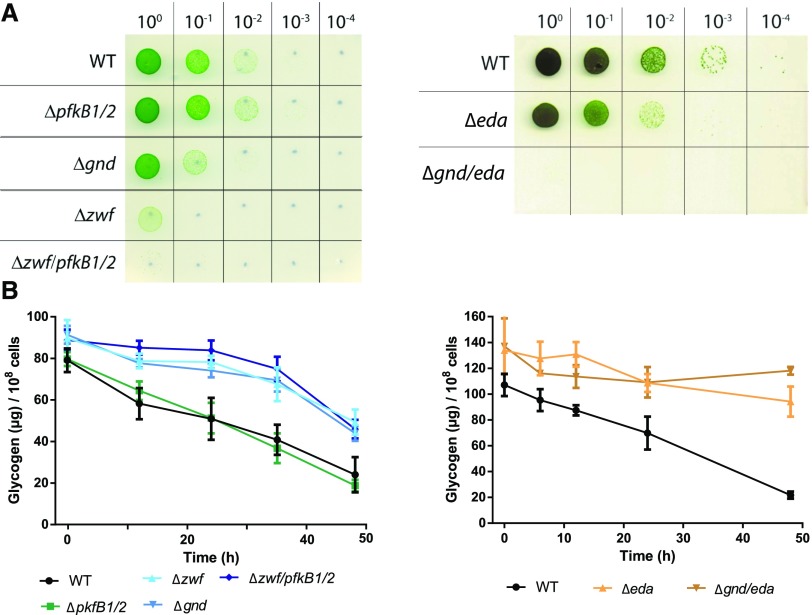
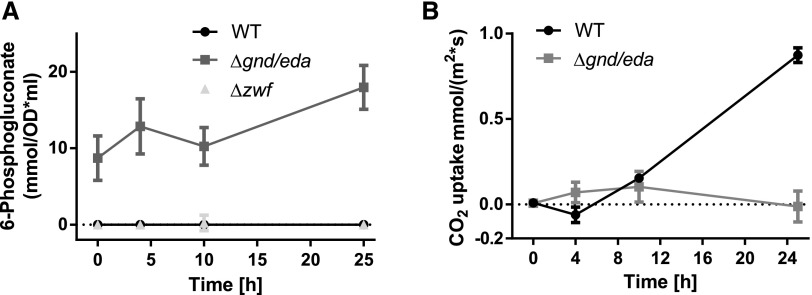
To determine the importance of the different glycolytic routes during resuscitation, mutants of the key enzymes of these pathways were analyzed. Phosphofructokinase (Pfk), unique to the EMP pathway, catalyzes the phosphorylation of Fru-6-P to Fru-1,6-bisphosphate. Synechocystis harbors two pfk paralogs, pfkB1 and pfkB2. Glc-6-P dehydrogenase (Zwf) converts Glc-6-P to 6-phosphogluconate, which can be further metabolized via the ED or OPP pathway. 6-Phosphogluconate dehydrogenase (Gnd) is unique to the OPP pathway, catalyzing the conversion of 6-phosphogluconate to ribulose-5-phosphate. The key enzyme unique to the ED pathway is 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase (Eda), which converts KDPG into glyceraldehyde-3-phosphate and pyruvate. The following mutants were analyzed: single mutants Δeda, Δgnd, and Δzwf, the double mutants ΔpfkB1/2 and Δgnd/eda, and the triple mutant Δzwf/pfkB1/2. A depiction of the blocked pathways is shown in Figure 1. The mutants were starved for 1 month and then resuscitated on solid BG11 agar plates in parallel with the wild type (Fig. 5A). If the possible bypass via Glc dehydrogenase (Gdh) and gluconate kinase (Gk; indicated in gray in Fig. 1) can be neglected, the deletion of zwf and pfkB1/2 should block all glycogen catabolic pathways. In fact, the respective mutant Δzwf/pfkB1/2 could not recover from nitrogen starvation. This result confirmed that Glc, which arises as a minor by-product from the hydrolytic activity of the debranching enzyme (GlgX), is not efficiently metabolized via Gdh and Gk. In contrast to the zwf mutant, no substantial difference between the recovery of ΔpfkB1/2 and the wild type could be observed, indicating that the EMP pathway does not play a role in resuscitation. Deletion of the ED pathway alone in Δeda resulted in poorer recovery compared with the wild type, whereas when the OPP pathway alone was interrupted in Δgnd, the effect was even more pronounced. Interruption of both the ED and OPP pathways in Δzwf resulted in a mutant that recovered poorly, demonstrating that the EMP pathway could only very inefficiently compensate for the loss of the ED and OPP pathways (compare Δzwf/pfkB1/2 and Δzwf). Strikingly, the double mutant Δgnd/eda could not recover at all. Since, in both mutants, neither the OPP nor the ED pathway is functioning, we expected no difference in recovery between Δzwf and Δgnd/eda. Thus, there must be another reason why Δzwf recovered, albeit inefficiently, whereas Δgnd/eda did not recover at all. We hypothesized that 6P-gluconate might accumulate in Δgnd/eda but not in Δzwf, thereby possibly inhibiting CO2 fixation as soon as photosynthesis starts. 6P-gluconate is known to bind to Rubisco with the potential to inhibit CO2 fixation. Measurement of 6P-gluconate levels under chlorotic conditions confirmed that this metabolite heavily accumulated in Δgnd/eda, whereas it was barely detectable in either Δzwf or wild-type cells (Fig. 6A). In agreement with this, CO2 fixation was severely impaired in Δgnd/eda, whereas the wild type first evolved CO2 as a consequence of respiration and only then turned on CO2 fixation with the onset of photosynthesis (Fig. 6B). Taken together, these results indicate that the EMP pathway does not play an important role in resuscitation, whereas the OPP and ED pathways are the main glycogen-degrading routes during this process. As the Δeda mutant showed a slightly better recovery than the Δgnd mutants, it appears that the OPP pathway plays a more crucial role than the ED pathway during resuscitation. Furthermore, initiation of CO2 fixation via the Calvin-Benson cycle following the respiratory phase is essential for a successful recovery.
Figure 5.
Characterization of glycolytic mutants ΔpfkB1/2, Δzwf, Δzwf/pfkB1/2, Δgnd, Δeda, and Δgnd/eda. A, Spot assay on solid BG11 agar of glycolytic mutants to test resuscitation from long-term chlorosis. Dilutions are indicated in the top row. B, Determination of the glycogen content of glycolytic mutants during resuscitation from 1-month chlorosis of the same mutants. The glycogen content was normalized to 1 × 108 cells. At least three biological replicates were measured; error bars represent the sd. WT, Wild type.
Figure 6.
Intracellular 6-phosphogluconate content and CO2 uptake of mutants blocked in the ED and OPP pathways (Δgnd/eda and Δzwf). A, Intracellular 6-phosphogluconate content during resuscitation from chlorosis of the wild type (WT), Δgnd/eda, and Δzwf. Slightly negative values were set to 0. B, CO2 uptake of the wild type and Δgnd/eda during resuscitation. At least three biological replicates were measured; error bars represent the sd.
To reveal how the various mutations affect glycogen consumption, we measured the glycogen content of all six glycolytic mutants during resuscitation. As shown in Figure 5B, the initiation of resuscitation caused a linear degradation of glycogen in the wild type as well as in ΔpfkB1/2. However, Δzwf, Δzwf/pfkB1/2, Δgnd, Δeda, and Δgnd/eda seemed to accumulate more glycogen than the wild type and showed a much slower degradation of this polymer. Degradation of a portion of the glycogen (20%–40%) in these mutants is possible because they have a functional GlgP2, but degradation of hexose phosphates is totally or partially blocked, and this inhibits further glycogen degradation. To prove this, we measured the levels of Glc-6-P and Fru-6-P in the wild type and Δzwf/pfkB1/2, which is blocked in all hexose phosphate degradation routes. Already during nitrogen starvation, both hexose phosphates accumulated in Δzwf/pfkB1/2 when compared with the wild type. After the onset of resuscitation, the levels of both sugars dropped below the detection limit in the wild type, indicating their fast consumption. By contrast, in the Δzwf/pfkB1/2 triple mutant, the level of Glc-6-P continued to increase and that of Fru-6-P is maintained at high levels, confirming the inability of the mutant to metabolize these molecules (Supplemental Fig. S3). These results show that glycogen breakdown is activated by nitrate addition independent of the start of the glycolytic reactions, explaining the degradation of glycogen in the various glycolytic mutants.
Synechocystis Is Prepared for Glycogen Degradation during Nitrogen Starvation
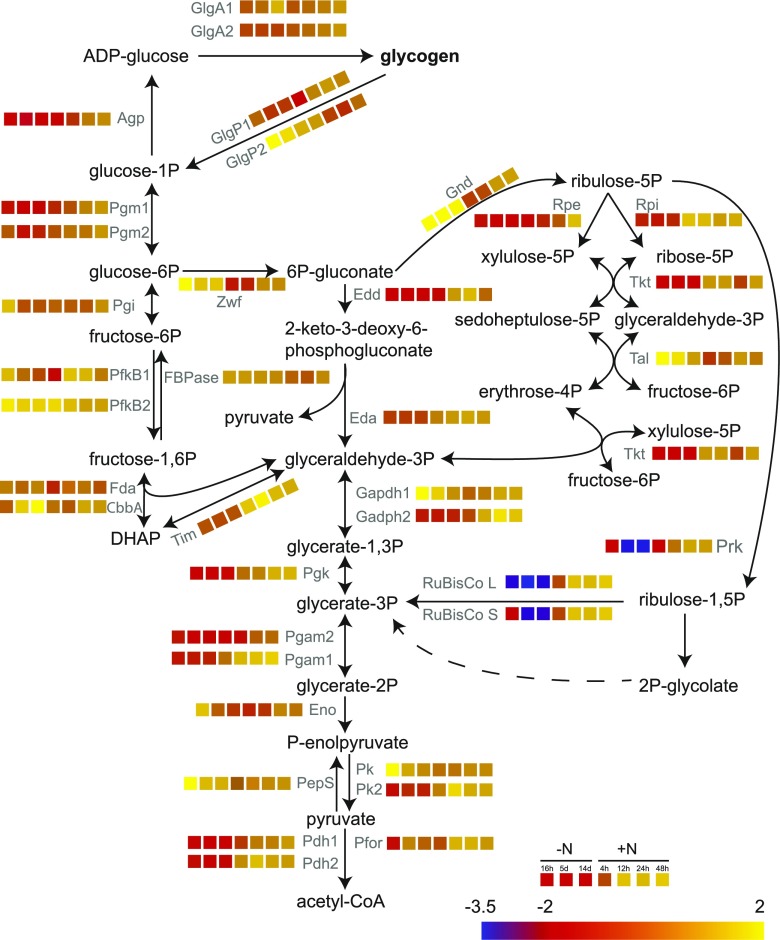
Previously, we performed a transcriptomic analysis of cells undergoing chlorosis and resuscitation (Klotz et al., 2016). The transcriptional regulation of the genes involved in glycogen catabolism is shown in Figure 7. The experiments described above led us to conclude that GlgP2 is the major glycogen-degrading enzyme during resuscitation, while GlgP1 does not play a relevant role here. In accord, our transcriptomic data set revealed that glgP1 is not subjected to strong regulation, whereas glgP2 is strongly up-regulated during nitrogen starvation but repressed when resuscitation is induced. Intriguingly, the genes for the key enzymes for the OPP and ED pathways, zwf and gnd, which play a major role in recovery from chlorosis, showed a similar regulation to glgP2: they were up-regulated during nitrogen starvation and down-regulated during resuscitation. A quantitative proteomic analysis of chlorotic and recovering cells (Spaet et al., 2018) revealed that protein levels of GlgP2, Zwf, and Gnd were indeed higher during nitrogen starvation than in exponentially growing cells (Supplemental Fig. S4), confirming that the glgP2, zwf, and gnd genes are not only transcribed but also translated into proteins during chlorosis. This shows that the cells produce the glycogen-degrading enzymes together with the glycogen granules, so that degradation can start as soon as nitrogen is available again. During resuscitation, glgP2, zwf, and gnd transcripts decrease while the protein levels are maintained, indicating that these enzymes are not turned over as long as they are active. Of the glycogen-degrading enzymes, the major phosphoglucomutase Pgm1 is the only one that is regulated in a different way. The pgm1 gene is repressed during chlorosis and turned on during resuscitation, and the same trend is observed at the protein level. Moreover, Pgm1 is strongly subjected to posttranslational modification via Ser phosphorylation at two sites (Spaet et al., 2018), indicating that it might play a key role in the control of glycogen degradation upon nitrate addition.
Figure 7.
Transcriptional regulation of genes needed for glycogen catabolism during nitrogen starvation (16 h, 5 d, and 14 d) and resuscitation (4, 12, 24, and 48 h). Analysis was performed using a high-resolution microarray (Agilent; format, 8 × 60 K; slide layout = IS-62976-8-V2) as described by Klotz et al. (2016). Relative transcript abundance normalized to exponential growth is shown as log2 and encoded in a color code from blue (2−3.5) to yellow (22).
To further reveal the activation of the glycogen-degrading enzymes in chlorotic cells, we measured the oxygen-exchange rate after dark incubation in the wild type and the Δglgp1/2 mutant (Supplemental Fig. S5). In the light, chlorotic wild-type cells neither consume nor produce oxygen; however, upon transfer to darkness, a low but clearly measurable consumption of oxygen could be observed. By contrast, Δglgp1/2 was unable to start respiration in the dark, like due to its inability to initiate respiration upon nitrate addition (see above). These results suggest that the equipment of chlorotic cells with the glycogen-degrading enzymes serves a double role: it allows rapid resuscitation when nitrate becomes available again and it enables cells to use glycogen for survival in the dark.
DISCUSSION
Synechocystis is able to survive long-term periods of nitrogen starvation and to rapidly resuscitate after the readdition of a usable nitrogen source. Glycogen, which quickly accumulates during nitrogen starvation and plays a major role in the transition to chlorosis, is rapidly mobilized upon the onset of resuscitation (Klotz et al., 2016). In this study, we investigated the importance of glycogen degradation for revival and the implication of the different glycogen catabolic pathways during resuscitation.
When Synechocystis is exposed to nitrogen starvation, cells tune down their metabolism and remain alive with minimal photosynthetic activity (Klotz et al., 2016). The metabolic adaptations during chlorosis reduce both energy consumption and generation, reaching a metabolic equilibrium that allows them to survive for a long time under starvation conditions. In agreement with the assumption that, even in the dormant state, a minimal amount of energy is necessary to keep cells alive (Sauer et al., 2001), ATP levels were shown to be maintained at a level that corresponds to one-fourth of the value at exponential growth (Fig. 2A). We propose that this residual level is adjusted to keep dormant cells alive. Chlorotic cells consume only very low amounts of ATP, since they perform almost no anabolic reactions or protein synthesis. As such, ATP regeneration should be tuned down correspondingly. In agreement, our previous transcriptomic data showed that genes encoding the F-ATPase machinery were strongly repressed in long-term chlorotic cells (Klotz et al., 2016). When resuscitation was initiated by the addition of nitrate, the first genes that were induced comprised all components for the translational apparatus, for nitrogen assimilation, and the F0-ATPase subunits. This implies an increased need for ATP due to the onset of anabolism and protein synthesis, which must be matched with an increased capacity for ATP regeneration by reinstallation of the F-ATPase machinery (Klotz et al., 2016). In line with this scenario, the measurement of ATP levels revealed a sudden increase of ATP upon the addition of nitrate. This first increase reaches a plateau, lying between the levels in dormant and vegetative growing cells. This plateau indicates that the increased need for ATP is perfectly balanced by the increased capacity to regenerate ATP. Only after 24 h of the resuscitation process does the concentration of ATP start to increase further, reaching almost double the value of normal growing cells after 48 h. At this point, cells have restored the powerful energy-generating photosynthetic machinery, but they have not yet consumed all reserve polymers and have not yet entered exponential growth, which gives rise to a transient surplus of energy.
This study confirmed our previous assumption that glycogen degradation is pivotal for resuscitation. Characterization of the individual glycogen phosphorylase mutants (ΔglgP1 and ΔglgP2) revealed that only GlgP2 plays a crucial role for glycogen breakdown during recovery from chlorosis. GlgP2 also is known to be responsible for glycogen degradation during periods of darkness (Fu and Xu, 2006). By contrast, GlgP1 has only been shown to play a crucial role during heat stress conditions (Fu and Xu, 2006), and any further physiological function remains elusive. Both ΔglgP2 and the ΔglgP1/2 double mutant were unable to degrade glycogen during resuscitation (Fig. 3B), confirming that no other pathway can degrade glycogen. Furthermore, we showed that glycogen degradation is essential for the initiation of respiration upon nitrate addition, since no significant oxygen consumption is observed in the ΔglgP1/2 double mutant (Fig. 4). In the wild type, respiration during the first hours of resuscitation leads to a complete inhibition of photosynthesis (Klotz et al., 2016). The ΔglgP1/2 double mutant, however, does not respire and, therefore, photosynthesis is not shut down. Rather, it seems that these mutant cells attempt to awake their metabolism when nitrate is added and use the remaining photosynthetic machinery to produce ATP (Fig. 2B). However, due to the lack of metabolic intermediates provided by glycogen catabolism, resuscitation is abortive. Nevertheless, the initial increase in ATP in these mutants suggests that the ATP levels are actively regulated by an unknown metabolic trigger, which responds to the addition of nitrate.
Glycogen phosphorylase catalyzes the excision of a Glc-1-P molecule by breaking the α-1,4 linkage. This Glc-1-P can be further catabolized via three different glycolytic routes, namely, the EMP, ED, and OPP pathways. Our findings indicate that the ED and OPP pathways are the major glycogen-degrading routes during resuscitation, since the mutants blocked in either or both pathways (Δzwf, Δgnd, Δeda, and Δgnd/eda) are either retarded or inhibited in their ability to recover from chlorosis and degrade glycogen. The lack of phenotype of the ΔpfkB1/2 mutant indicates that the EMP pathway does not play a crucial role in this process (Fig. 5). However, a minor contribution of the EMP pathway in glycogen breakdown might not be specific to the resuscitation process. During resuscitation, a first heterotrophic phase (0–24 h +N), in which glycogen is degraded and fuels respiration, is followed by a gradual increase in photosynthetic activity, while glycogen degradation continues (16–48 h; Klotz et al., 2016). It appears that CO2 fixation is activated as soon as photosynthesis is initiated. The concomitant use of glycogen as a carbon source with photosynthetic energy generation and CO2 fixation resembles mixotrophic conditions. Finally, cells reenter fully autotrophic conditions under which they again rely only on photosynthesis. The restoration of transcription and translation takes place early in resuscitation (Klotz et al., 2016) and requires precursors for nucleic and amino acids. As long as the Calvin-Benson cycle is not running, ribose components for the synthesis of RNA and erythrose-4-phosphate for the synthesis of aromatic compounds can be provided exclusively via the OPP pathway. In line with this, deleting genes that participate in the OPP pathway has the severest impact on the cell’s ability to recover from chlorosis (Δgnd, Δzwf, ΔzwfpfkB1/B2, and Δgnd/eda). Following the first respiratory phase, mixotrophic conditions are created when glycogen degradation continues while photosynthesis starts. During this phase, the Calvin-Benson cycle, which in many steps is a reversal of the OPP pathway, is activated. The transcriptomic data (Klotz et al., 2016) show that genes encoding enzymes of the Calvin-Benson cycle (e.g. Prk, RubiscoL, RubiscoS, Pgk, Gapdh2, Tkt, Rpi, and Rpe) are already up-regulated in the early phase of resuscitation (4 h +N). Under mixotrophic conditions, the ED pathway is probably the most physiologically important (Chen et al., 2016). The ED pathway does not share reactions and intermediates with the Calvin-Benson cycle; this allows concomitant glycogen breakdown via the ED pathway and operation of the reductive pentose phosphate cycle. The ED pathway might thus be especially important in the mixotrophic phase of resuscitation. Furthermore, this pathway provides a shortcut for the delivery of pyruvate to the tricarboxylic acid cycle. In line with this, the impact of deletion of the ED pathway on resuscitation is less severe than deletion of the OPP pathway. Remarkably, deletion of both gnd and eda completely abrogated resuscitation, whereas deletion of zwf resulted in a mutant that was still able to recover poorly (Fig. 5A). In line with this, we detected an overaccumulation of 6P-gluconate in Δgnd/eda but not in Δzwf (Fig. 6A). 6P-gluconate is known to compete with ribulose-1,5-bisphosphate for binding to Rubisco (Badger and Lorimer, 1981). It follows that CO2 fixation is impaired in Δgnd/eda in addition to the lack of the OPP and ED pathways (Fig. 6B). The limited recovery of Δzwf is thus probably enabled by a minor consumption of Glc-6-P via the EMP pathway in the early phase followed by CO2 fixation as soon as photosynthesis starts (compare Δzwf and Δzwf/pfkB1/2 in Fig. 5A). However, in Δgnd/eda, the consumption of Glc-6-P via the EMP pathway is not followed by CO2 fixation and, accordingly, resuscitation is not successful. These results convincingly show that the awakening of dormant cyanobacteria from chlorosis requires glycogen breakdown in the early phase followed by CO2 fixation.
A transcriptomic analysis of the chlorosis and resuscitation processes revealed that Synechocystis anticipates and prepares for glycogen degradation during nitrogen starvation (Klotz et al., 2016). Transcription of glgP2, zwf, and gnd, which we found to be the main enzymes in glycogen catabolism during resuscitation, was up-regulated during nitrogen starvation and turned down during resuscitation (Fig. 7). Moreover, quantitative proteomic analyses (Spaet et al., 2018) showed that the GlgP2, Zwf, and Gnd protein levels also increased during chlorosis and are maintained after the addition of nitrate. This indicates that the cells anticipate the resuscitation process and produce the proteins necessary to degrade glycogen while they undergo chlorosis, which allows an immediate response when a nitrogen source is available. Furthermore, chlorotic cells are able to rapidly turn on respiration in the dark (Supplemental Fig. S5), which substantiates that GlgP2 is present and active during chlorosis. These findings raised the question of how glycogen mobilization is initiated, since most of the enzymes involved in the first steps of glycogen catabolism (GlgP2, Zwf, and Gnd) are present throughout the course of chlorosis and resuscitation, yet glycogen degradation only starts after the addition of nitrate. This requires a mechanism that prevents unintended degradation of glycogen during chlorosis by GlgP2. Direct regulation of GlgP2 activity is conceivable; however, posttranslational control of glycogen phosphorylase activity in cyanobacteria has not been demonstrated so far. Another key enzyme that might control glycogen degradation is Pgm1. This enzyme has been described as a target of thioredoxin regulation in Synechocystis (Lindahl and Florencio, 2003) and showed conspicuous protein Ser phosphorylation dynamics during chlorosis and resuscitation (Spaet et al., 2018). Whatever the mechanism involved, glycogen mobilization must be activated upon the onset of nitrogen assimilation as well as after transfer to darkness. How these signals are perceived by the cells and transmitted to activate the necessary enzymes requires further investigation. These studies will lead to insights into how cyanobacteria coordinate metabolic transitions, which is poorly understood. The awakening of dormant Synechocystis cells from chlorosis offers a unique model system in which to study the delicate interplay of carbohydrate oxidation, CO2 fixation, and photosynthesis.
MATERIALS AND METHODS
Cultivation of Escherichia coli
Escherichia coli was grown in Luria-Bertani medium at 37°C (Bertani, 1951). For growth on solid medium, 1.5% (w/v) agar-agar was added to the regular Luria-Bertani medium. Strains containing plasmids have been propagated with the appropriate concentration of antibiotics.
Cyanobacterial Cultivation Conditions
All Synechocystis sp. PCC 6803 strains used in this study were grown in BG11 supplemented with 5 mm NaHCO3, as described previously (Rippka et al., 1979). A list of the strains used is provided in Supplemental Table S1. Two kinds of wild-type strains, Glc sensitive and Glc tolerant, were used; both strains responded equally during nitrogen starvation and resuscitation. Cultivation was performed with continuous illumination (40–50 μmol photons m−2 s−1) and shaking (130–140 rpm) at 27°C. Induction of nitrogen starvation and resuscitation was induced as described previously (Schlebusch and Forchhammer, 2010; Klotz et al., 2016). If mutants or strains containing antibiotic markers were used, the precultures were propagated with the appropriate concentration of antibiotics. Biological replicates were inoculated with the same precultures but propagated, nitrogen starved, and resuscitated independently in different flasks under identical conditions.
Isothermal, Single-Reaction DNA Assembly (Gibson Cloning)
Cloning was performed as described by Gibson et al. (2009) using primers containing sequences of the specific vector backbones (Supplemental Table S2). pUC19 (New England Biolabs) was used for the generation of cyanobacterial mutants.
ATP Determination
One-milliliter aliquots of bacterial cultures were taken and immediately frozen in liquid nitrogen. ATP was extracted by boiling and freezing samples three times consecutively (boiling at 100°C and freezing in liquid nitrogen) and spinning them down at 25,000g for 1 min at 4°C. ATP in the supernatant was quantified with the ATP Determination Kit (Molecular Probes; A22066) following the manufacturer’s protocol. A total of 450 µL of a reaction mix containing 25 mm Tricine buffer, pH 7.8, 5 mm MgSO4, 0.1 mm EDTA, 0.1 mm sodium azide, 1 mm DTT, 0.5 mm d-luciferin, and 1.25 µg mL−1 firefly luciferase was mixed with 50 µL of the samples, and the luminescence was read in a luminometer (Sirius Luminometer; Berthold Detection Systems). An ATP standard curve was generated and used to calculate ATP content in the collected samples.
Spot Assay
Serial dilutions of chlorotic cultures were prepared (100, 10−1, 10−2, 10−3, 10−4, and 10−5), starting with an OD750 of 1.5 µL of these dilutions, dropped on solid BG11 agar plates, and cultivated at 50 μmol photons m−2 s−1 and 27°C for 5 to 7 d.
Glycogen Determination
Glycogen determination was performed as described previously (Gründel et al., 2012) with modifications described by Klotz et al. (2015).
Oxygen Evolution Measurement
Oxygen evolution was measured in vivo using a Clark-type oxygen electrode (Hansatech DW1). Light was provided from a high-intensity white light source (Hansatech L2). Oxygen evolution of 2 mL of recovering cultures at an OD750 of 0.5 was measured at room temperature and 50 μmol photons m−2 s−1.
PAM
PSII activity was analyzed in vivo with a WATER-PAM chlorophyll fluorometer (Walz). All samples were dark adapted for 5 min before measurement. The maximal PSII quantum yield was determined with the saturation pulse method (Schreiber et al., 1995). Cultures were diluted 1:20 in BG11 medium before the measurements in a final volume of 2 mL.
Quantification Assay for 6P-Gluconate
A total of 100 µL of cells with an OD750 of 50 was pelleted, resuspended in 1 mL of 0.2 m HCl, and incubated at 95°C for 15 min. The solution was centrifuged (10 min at 18,000g at room temperature), and the supernatant was transferred to a new cup and neutralized with 1 mL of 1 m Tris-HCl (pH 8). The solution was divided into two parts (2 × 900 µL) for a positive and a blank sample. A total of 90 µL of 11 mm NADP+ solution was added to all samples as well as 10 µL of 5 units mL−1 Gnd (Megazyme) solution to all positive samples and 10 µL of water to all blank samples. Absorption at 340 nm was measured, and blank sample absorption values were subtracted from positive sample absorption values. The 6-phosphogluconate concentration was then calculated by using a standard curve.
Quantification Assay for Glc-6-P
A total of 100 μL of cells with an OD750 of 50 was pelleted, resuspended in 1 mL of 0.2 m HCl, and incubated at 95°C for 15 min. The solution was centrifuged (10 min at 18,000g at room temperature), and the supernatant was transferred to a new cup and neutralized with 1 mL of 1 m Tris-HCl (pH 8). The solution was divided into two parts (2 × 900 μL) for a positive and a blank sample. A total of 90 μL of 11 mm NADP+ solution was added to all samples as well as 10 μL of 5 units mL−1 Glc-6-P dehydrogenase (Sigma-Aldrich) solution to all positive samples and 10 μL of water to all blank samples. Absorption at 340 nm was measured, and blank sample absorption values were subtracted from positive absorption values. Glc-6-P concentration was then calculated by using a standard curve.
Quantification Assay for Fru-6-P
A total of 100 μL of cells with an OD750 of 50 was pelleted, resuspended in 1 mL of 0.2 m HCl, and incubated at 95°C for 15 min. The solution was centrifuged (10 min at 18,000g at room temperature), and the supernatant was transferred to a new cup and neutralized with 1 mL of 1 m Tris-HCl (pH 8). The solution was divided into two parts (2 × 900 μL) for a positive and a blank sample. A total of 90 μL of 11 mm NADP+ solution containing 5 units mL−1 Glc-6-P dehydrogenase (Sigma-Aldrich) was added to all samples as well as 10 μL of 5 units mL−1 phosphoglucose isomerase (Boehringer Mannheim) solution to all positive samples and 10 μL of water to all blank samples. Absorption at 340 nm was measured, and blank sample absorption values were subtracted from positive absorption values. Fru-6-P concentration was then calculated by using a standard curve.
CO2 Gas-Exchange Measurement
BG11 medium devoid of bicarbonate was mixed with agar, heated, and poured onto cutoff lids of 5-mL centrifugation cups. After solidification of the agar, 100 µL of cells with an OD750 of 50 was added and dried in front of a ventilator while being exposed to 50 µE of light emitted by a fluorescent tube. Two such lids were loaded simultaneously into the cuvette of a GFS-3000 gas-exchange measuring system from Walz containing an atmosphere of 400 µL L−1 CO2 and 60% humidity at 28°C. The cuvette was exposed to 50 µE of light emitted by a fluorescent tube. After a short adaptation period, CO2-exchange rates were monitored for 90 s, while measurements were taken every 15 s. The six measurements were averaged. In order to correct for diffusion of CO2 into or out of the agar, the CO2 exchange of cell-free agar lids was measured and subtracted from sample values.
Accession Numbers
Sequence data from this article can be found in the UniProt database under accession numbers P73511 (GlgP1), P73546 (GlgP2), P74643 (Pgm1), P73411 (Zwf), P72830 (PfkB1) Q55988 (PfkB2), P52208 (Gnd), and Q55872 (Eda). The accession number of the microarray data cited in this article is GEO:GSE83363.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PSII quantum yield determined by PAM fluorometry of Synechocystis wild type and ΔglgP1/2 during recovery from chlorosis.
Supplemental Figure S2. ATP content during nitrogen starvation and resuscitation of ΔglgP1/2 in the absence of light.
Supplemental Figure S3. Glc-6-P and Fru-6-P levels in Synechocystis wild type and Δzwf/pfkB1/2 during nitrogen starvation and resuscitation.
Supplemental Figure S4. Expression ratios of transcripts and proteins of the main enzymes involved in glycogen degradation during resuscitation.
Supplemental Figure S5. Oxygen-exchange rate of chlorotic ΔglgP1/2 and wild-type cells in the light and after 3 min of incubation in the dark.
Supplemental Table S1. List of strains used.
Supplemental Table S2. List of primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Lars Nichelmann for assistance with the CO2 fixation measurements and Selina Schwarzbach for involvement in ATP measurements.
Footnotes
This work was supported by the German Research Council (Deutsche Forschungsgemeinschaft, DFG) GRK 1708 “Molecular Principles of Bacterial Survival Strategies”, the DFG SFB 766 (A11), grant GU 1522/1-1, and by BMBF (FP3 09).
References
- Allen MM, Smith AJ (1969) Nitrogen chlorosis in blue-green algae. Arch Mikrobiol 69: 114–120 [DOI] [PubMed] [Google Scholar]
- Badger MR, Lorimer GH (1981) Interaction of sugar phosphates with the catalytic site of ribulose-1,5-bisphosphate carboxylase. Biochemistry 20: 2219–2225 [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schreiber K, Appel J, Makowka A, Fähnrich B, Roettger M, Hajirezaei MR, Sönnichsen FD, Schönheit P, Martin WF, et al. (2016) The Entner-Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc Natl Acad Sci USA 113: 5441–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Xu X (2006) The functional divergence of two glgP homologues in Synechocystis sp. PCC 6803. FEMS Microbiol Lett 260: 201–209 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Görl M, Sauer J, Baier T, Forchhammer K (1998) Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology 144: 2449–2458 [DOI] [PubMed] [Google Scholar]
- Gründel M, Scheunemann R, Lockau W, Zilliges Y (2012) Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158: 3032–3043 [DOI] [PubMed] [Google Scholar]
- Healey FP, Myers J (1971) The Kok effect in Chlamydomonas reinhardi. Plant Physiol 47: 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz A, Forchhammer K (2017) Glycogen, a major player for bacterial survival and awakening from dormancy. Future Microbiol 12: 101–104 [DOI] [PubMed] [Google Scholar]
- Klotz A, Reinhold E, Doello S, Forchhammer K (2015) Nitrogen starvation acclimation in Synechococcus elongatus: redox-control and the role of nitrate reduction as an electron sink. Life (Basel) 5: 888–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz A, Georg J, Bučinská L, Watanabe S, Reimann V, Januszewski W, Sobotka R, Jendrossek D, Hess WR, Forchhammer K (2016) Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr Biol 26: 2862–2872 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Florencio FJ (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci USA 100: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hu HH, Gao H, Xu XD (2013) Role of two phosphohexomutase genes in glycogen synthesis in Synechocystis sp. PCC6803. Chin Sci Bull 58: 4616–4621 [Google Scholar]
- Luque I, Forchhammer K (2008) Nitrogen assimilation and C/N balance sensing. In Herrero A, Flores E, eds, The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press, Norfolk, UK, pp 335–382 [Google Scholar]
- Preiss J. (1984) Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol 38: 419–458 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61 [Google Scholar]
- Sauer J, Schreiber U, Schmid R, Völker U, Forchhammer K (2001) Nitrogen starvation-induced chlorosis in Synechococcus PCC 7942: low-level photosynthesis as a mechanism of long-term survival. Plant Physiol 126: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch M, Forchhammer K (2010) Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 76: 6101–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Endo T, Mi H, Asada K (1995) Quenching analysis of chlorophyll fluorescence by the saturation pulse method: particular aspects relating to the study of eukaryotic algae and cyanobacteria. Plant Cell Physiol 36: 873–882 [Google Scholar]
- Shearer J, Graham TE (2002) New perspectives on the storage and organization of muscle glycogen. Can J Appl Physiol 27: 179–203 [DOI] [PubMed] [Google Scholar]
- Spaet P, Klotz A, Sascha R, Boris M, Karl F (2018) Chlorosis as a developmental program in cyanobacteria: the proteomic fundament for survival and awakening. BioRxiv https://doi.org/10.1101/325761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Cohen-Bazire G (1977) Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol 31: 225–274 [DOI] [PubMed] [Google Scholar]
- Vitousek PM, Howarth RW (1991) Nitrogen limitation on land and in the sea: how can it occur. Biogeochemistry 13: 87–115 [Google Scholar]