Abstract
DNA hybridization onto DNA-functionalized nanoparticle surfaces (e.g., in the form of a spherical nucleic acid (SNA)) is known to be enhanced relative to hybridization free in solution. Surprisingly, via isothermal titration calorimetry, we reveal that this enhancement is enthalpically, as opposed to entropically, dominated by ~20 kcal/mol. Coarse-grained molecular dynamics simulations suggest that the observed enthalpic enhancement results from structurally confining the DNA on the nanoparticle surface and preventing it from adopting enthalpically unfavorable conformations like those observed in the solution case. The idea that structural confinement leads to the formation of energetically more stable duplexes is evaluated by decreasing the degree of confinement a duplex experiences on the nanoparticle surface. Both experiment and simulation confirm that when the surface-bound duplex is less confined, i.e., at lower DNA surface density or at greater distance from the nanoparticle surface, its enthalpy of formation approaches the less favorable enthalpy of duplex formation for the linear strand in solution. This work provides insight into one of the most important and enabling properties of SNAs and will inform the design of materials that rely on the thermodynamics of hybridization onto DNA-functionalized surfaces, including diagnostic probes and therapeutic agents.
Spherical nucleic acids (SNAs) are a class of structures typically made by arranging linear nucleic acids at high density around a nanoparticle core.1,2 SNAs have become important entities in the development of medical diagnostic probes,3,4 intracellular small-molecule detection agents, RNA tracking agents,5–8 and building blocks for colloidal crystal engineering.9–14 Their unique properties, which are highly differentiated from linear structures, make them very attractive for such uses. One of these properties is a higher affinity constant for complementary nucleic acids. Depending on the sequence, SNAs can bind complements orders of magnitude more tightly than linear forms of the same sequence.15–17 Despite the importance of this enhanced binding for many of the SNA applications, its origin remains unknown.
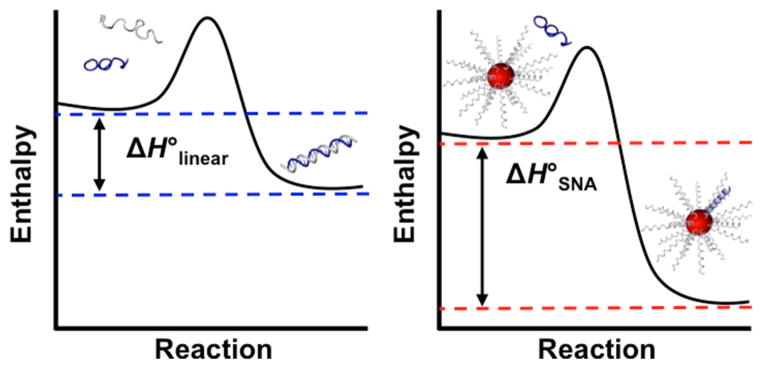
Given the restricted nature and preorientation of the short strands that define SNAs, enhancement of hybridization could be attributed to entropic contributions. However, here we demonstrate that complement binding on SNAs carries a higher entropic penalty than binding in the linear form.15 We show that the binding enhancement is instead enthalpically driven (Scheme 1) and explain its thermodynamic origin. We use temperature-dependent fluorescence melting studies, isothermal titration calorimetry (ITC), and coarse-grained molecular dynamics (MD) simulations to determine the entropy and enthalpy of hybridization for linear DNA binding as well as binding of a complement to an SNA. Via a combination of experiment and simulation, we explain that structural confinement on a nanoparticle surface prevents DNA from adopting unfavorable binding conformations that ultimately account for the observed enthalpically dominated binding enhancement on SNAs.
Scheme 1.
Comparison of Complementary DNA Hybridization to Either Linear DNA (left) or Spherical Nucleic Acids (SNAs, right) to Elucidate the Thermodynamic Origin of Binding Enhancement Observed on SNAs
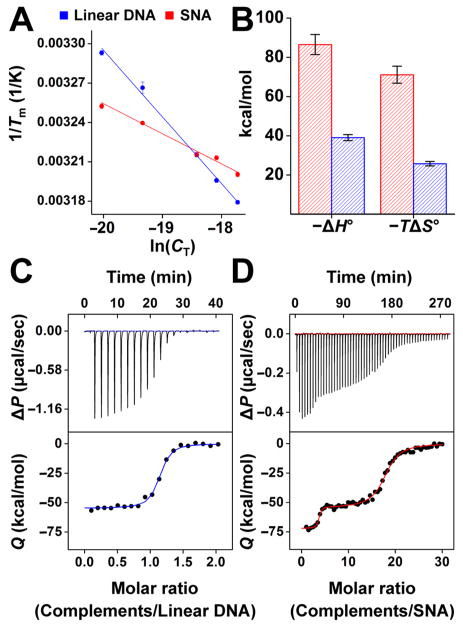
To determine the entropies and enthalpies of binding, we performed concentration-dependent fluorescence hybridization experiments (Figure 1A,B).18 We studied, under identical conditions, a linear 12-mer DNA system and 5.9 nm gold nanoparticle SNAs functionalized with ~46 DNA strands of the same sequence. Both systems were prepared in a 1:1 stoichiometry of either SNA or linear DNA to a complementary strand, and a DNA helix-coil transition temperature was measured over a range of concentrations (Figures 1A, S1, and S2). The concentration dependence of the helix-coil transition temperature reflects the thermodynamics of hybridization through the van ’t Hoff relationship19,20
Figure 1.
(A) van ’t Hoff plots from which thermodynamic constants are extracted for binding between complementary linear strands and either linear 12-mer DNA (blue) or SNAs (red). (B) Comparison of enthalpic gain and entropic cost derived from the van’t Hoff plots. (C) Isothermal titration calorimetry of 12-mer DNA duplex hybridization free in solution and (D) 12-mer DNA duplex hybridization on SNAs functionalized with ~46 strands per particle. Upper panel: differential heating power ΔP vs time. Lower panel: integrated heats of reaction Q vs molar ratio.
where Tm is the transition temperature, CT is the combined concentration of SNA (or linear DNA) and complement, R is the gas constant, and ΔH° and ΔS° are the enthalpy and entropy of hybridization, respectively. Importantly, this analysis treats the SNA as a single molecular entity and concentrations are adjusted to ensure 1:1 binding of complementary strand to SNA (see SI for details). This is the case for many of the SNA’s uses as probes for high-sensitivity detection or as antisense gene-regulation agents, where target concentration is relatively low with respect to probe.
The less steep slope for the SNA system reveals that the binding enthalpy on SNAs, ΔH° = −91.3 ± 5.5 kcal/mol, is far more favorable than for linear DNA, ΔH° = −40.6 ± 2.4 kcal/mol (Figure 1A,B and Table S1). Remarkably, the SNA system also exhibits a higher entropic loss upon hybridization, with TΔS° = −75.8 ± 5.3 kcal/mol at 298 K vs TΔS° = −27.2 ± 2.3 kcal/mol for linear DNA. Since the increased enthalpic gain in the SNA system more than compensates for the larger entropic cost, the free energy of hybridization is lower for SNAs than for linear DNA (ΔG°SNA = −15.5 ± 0.2 kcal/mol vs ΔG°linear = −13.4 ± 0.1 kcal/mol), and the association constant is correspondingly higher, KeqSNA = (2.3 ± 0.8) × 1011 M−1 vs Keqlinear = (6.8 ± 1.1) × 10sM−1. Whereas the enhanced binding confirms prior observations,15–17 the larger entropic penalty for SNA binding and the increased enthalpic gain are puzzling given the conformational constraints of the nanoparticle-bound DNA, and the view that the dense packing of DNA on the SNA is likely to result in destabilizing steric and electrostatic interactions.21–25 Indeed, we have observed such thermodynamic trends before,15 but refrained from commenting on their origin because of the lack of a suitable explanation.
The van ’t Hoff analysis assumes that DNA hybridization proceeds in the dilute limit in a two-state manner and that the enthalpy of this process is independent of temperature.19 Since these assumptions have been shown to significantly affect van ’t Hoff-derived enthalpies of linear DNA hybridization,26–28 we sought to corroborate our findings with a model-independent technique. Specifically, to confirm the larger enthalpy of hybridization for SNAs, we performed ITC experiments on the same systems.29,30 The ITC curve shapes (Figures 1C,D and S3–S5) indicate that DNA hybridization on the SNA differs significantly from hybridization free in solution. The linear-DNA system shows a sigmoidal binding isotherm with an inflection point at a molar ratio of 1, reflecting 1:1 binding stoichiometry (Figure 1C). In contrast, the SNA system exhibits double-sigmoidal behavior with inflection points at molar ratios of 4 and 15 strands per particle (Figure 1D). This shape implies that SNAs exhibit a type of negative cooperativity, where binding of the first four strands is enthalpically more favorable than subsequent hybridization events. Such negative cooperativity is consistent with prior observations.16
The ITC curves also directly yield the hybridization enthalpies from the released heat Q, showing an enthalpy gain that is 20.7 ± 2.2 kcal/mol higher for binding on SNAs (Figure 1C,D and Table S2). The qualitative agreement between these data and the fluorescence data suggests that the relative entropic and enthalpic contributions determined from the van ’t Hoff analysis are qualitatively reliable, despite the assumptions of the model. Discrepancies in the absolute values of hybridization enthalpies derived from calorimetry and the van ’t Hoff analysis have been previously observed in linear DNA systems.31,32 Differences can be explained by deviations from two-state behavior19 and changes in heat capacity associated with DNA melting.26–28,32 Additionally, ITC experiments are conducted at much higher concentrations than van ’t Hoff experiments and are therefore more susceptible to excluded-volume effects.33,34 We suspect that all of these factors play a role in the systems under study and, if considered in the van ’t Hoff analysis, may lead to better agreement with calorimetric values.
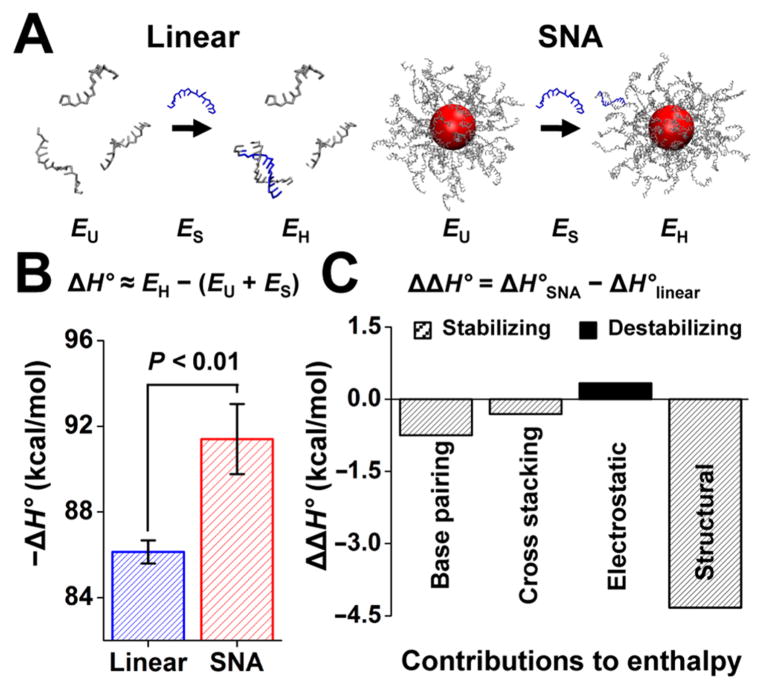
With strong experimental evidence to support that binding to the SNA is enthalpically more favorable than binding to linear DNA, we turned to coarse-grained MD simulations to understand the origin of this enhancement. Simulations of the hybridized and the unhybridized state were performed for both the linear and the SNA systems using the 3SPN.2 model (see SI for computational details).35–37 This model separates the DNA into three sites per nucleotide, one each for the phosphate, sugar, and base, and has been parametrized to reproduce correct structural, thermodynamic, mechanical, and kinetic properties of DNA. Assuming incompressibility, we computed the enthalpy of duplex formation as38
where EH is the internal energy of the system with a hybridized duplex, EU is the internal energy of the unhybridized strands, and ES is the energy of a single complementary DNA strand in solution (Figure 2A). The simulations confirmed the experimentally observed trend for the enthalpy of hybridization, with an enhancement of ~5.3 kcal/mol associated with hybridization on the SNA relative to free in solution (Figure 2B). To achieve high statistical accuracy, the simulations were performed with implicit ions, using the Debye-Hüsckel approximation, but we confirmed that the same trends are obtained when using explicit salt and counterions (details in SI). Owing to the lack of explicit solvent, ΔH° differs quantitatively from the experimental values. However, the simulations make it possible to separate the inter-and intramolecular contributions. To identify the primary origin of the observed enthalpic enhancement, we broke down the hybridization enthalpies of the linear duplex and the SNA duplex into (i) interstrand base-pairing, (ii) cross-stacking, (iii) electrostatic, and (iv) intrastrand structural energies (Figure 2C and Table S3). For each of these contributions, we defined ΔΔH as the difference between the enthalpy of hybridization on the SNA (3HSNA) and the enthalpy of hybridization for linear DNA (ΔHlinear). We found that the main contribution to the enhancement of ΔH° on the SNA was the change in structural energy, which comprises covalent-bond, angle, dihedral, and base-stacking energies, and was most pronounced for the T10-linker region of 10 thymine bases that connects the duplex to the nanoparticle surface (Figure S7). Since experimentally the presence of the T10 linker did not affect the enthalpy of hybridization for the linear DNA (Figure S4), we conclude that the SNA architecture must give rise to the change ΔΔH in the structural hybridization energy. Confinement due to surface attachment and molecular crowding prevents hybridized DNA on the nanoparticle from adopting energetically unfavorable conformations that cause distortions in the bond angles, dihedrals, and intrastrand base stacking away from the minimum-energy conformation, as would occur in the unconfined linear DNA case.
Figure 2.
(A) Coarse-grained MD simulations of 12-mer DNA before (EU) and after (EH) duplex formation free in solution and on an SNA functionalized with 46 strands. (B) Comparison of simulation-derived enthalpies of hybridization for a duplex formed free in solution and one formed on an SNA. (C) Breakdown of contributions to the enthalpy of hybridization.
It has been observed that molecular crowding or excluded-volume effects increase local DNA concentration and as a result stabilize duplex formation.33,34 Yet, excluded volume also restricts the degrees of freedom of hybridizing molecules and biases the DNA toward more enthalpically stable conformations. This effect is reminiscent of the stability observed for locked nucleic acid (LNA) hybridization. LNA is a synthetic RNA analog for which the ribose moiety is structurally constrained by a 2' oxygen to 4' carbon methylene bridge.39 Incorporation of LNA bases into DNA oligomers has led to a demonstrated enhancement of the thermodynamic stability of duplexes.40 This effect is enthalpically dominated, as shown by calorimetry41 and is thought to result from the conformational restriction of base-stacking and hydrogen bonding interactions.42,43
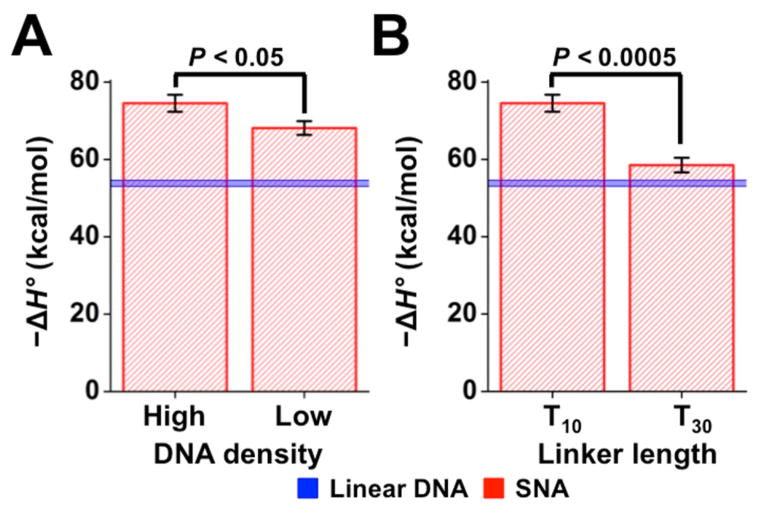
To test if DNA confinement on the surface of the nanoparticles indeed resulted in an enhanced enthalpy of hybridization, as suggested by the MD simulations, we performed ITC on two SNA systems with less confined DNA. We hypothesized that if conformational restriction of duplexes on the SNA resulted in an enhanced enthalpy of hybridization, then less confined SNA duplexes should have a less favorable enthalpy of hybridization. First, we decreased the DNA surface density by functionalizing nanoparticles with only 30 DNA strands, to obtain a density 33% lower than that of the original SNAs. In support of our hypothesis, the hybridization enthalpy of the first four DNA strands on these low-density SNAs was 6.4 ± 2.7 kcal/mol less favorable (Figures 3A and S6A). To further explore the degree to which confinement on SNAs could be tuned, we tested an SNA architecture with an even lower degree of confinement. We moved the duplex-forming region of the DNA further away from the nanoparticle surface by replacing the T10-linker region with a linker region composed of 30 thymine bases (T30). For this design, we maintained the high DNA density of ~46 strands per particle. The increased distance from the nanoparticle surface caused a striking decrease in the enthalpy of duplex formation of 16.0 ± 2.8 kcal/mol (Figures 3B and S6B). The effect was so strong that nearly all enhancement of the enthalpy disappeared, with the enthalpy of hybridization on the T30-SNA nearly identical to the enthalpy of hybridization of linear DNA. The combined density and linker data were corroborated by simulation (Figure S8) and demonstrated that the enthalpy of complementary DNA hybridization onto SNAs can be tuned by as much as 20 kcal/mol simply by varying the degree of confinement of a surface-bound strand.
Figure 3.
Enthalpy of hybridization onto SNAs for the first four complementary strands as a function of (A) DNA surface density and (B) linker length. Linear DNA hybridization enthalpy (blue) is provided for comparison.
Structural confinement also helps explain the considerable entropic cost of complement hybridization on an SNA. In simulations, we found that upon hybridization single-stranded DNA surrounding the duplex on the surface became structurally more ordered (Figure S9). While this effect was minor on a per-strand basis, collectively these contributions significantly reduced the ensemble degrees of freedom. This entropic cost counteracts hybridization of the first DNA strand on an SNA but has been shown to reduce the entropic cost for subsequent hybridization events.16
In conclusion, we have demonstrated that, relative to linear DNA, the enthalpy of complement hybridization is more favorable on spherical nucleic acids and results in an enhanced free energy of binding. While one could make intuitive arguments that the observed binding enhancement on SNAs is entropically driven, experimental and computational data show that it is an enthalpically driven process. This new insight can inform future engineering of DNA-functionalized surfaces. The surface architecture of SNAs can be modified to increase or decrease the enthalpic contributions to hybridization and consequently influence therapeutically and diagnostically relevant association constants.
Supplementary Material
Acknowledgments
This material is based upon work supported by the Air Force Office of Scientific Research under awards FA9550-17-1-0348 and FA9550-14-1-0003; the U.S. Department of Commerce, National Institute of Standards and Technology under award 70NANB14H012 as part of the Center for Hierarchical Materials Design (CHiMaD); the National Science Foundation award DMR-1610796; and the Northwestern University Keck Biophysics Facility and a Cancer Center Support Grant (NCI CA060553). L.K.F. gratefully acknowledges the National Science Foundation for a graduate research fellowship and the Center for Computation and Theory of Soft Materials at Northwestern University for a graduate research fellowship. We thank Dr. Kevin Metcalf for insightful comments that greatly improved the manuscript.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b03459.
Complete experimental and computational details, supporting figures and tables, and additional discussion (PDF)
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Cutler JI, Auyeung E, Mirkin CA. J Am Chem Soc. 2012;134(3):1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 3.Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, Mirkin CA. Anal Chem. 2012;84(9):4153–4160. doi: 10.1021/ac3004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289(5485):1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 5.Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA. Proc Natl Acad Sci U S A. 2015;112(31):9591–9595. doi: 10.1073/pnas.1510581112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, Mirkin CA. Anal Chem. 2012;84(4):2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. ACS Nano. 2009;3(8):2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA. Science. 2011;334(6053):204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]
- 10.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451(7178):549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, Tagawa M, Xin HL, Wang T, Emamy H, Li H, Yager KG, Starr FW, Tkachenko AV, Gang O. Science. 2016;351(6273):582–586. doi: 10.1126/science.aad2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Veracoechea FJ, Mladek BM, Tkachenko AV, Frenkel D. Phys Rev Lett. 2011;107(4):45902. doi: 10.1103/PhysRevLett.107.045902. [DOI] [PubMed] [Google Scholar]
- 13.Angioletti-Uberti S, Mognetti BM, Frenkel D. Nat Mater. 2012;11(6):518–522. doi: 10.1038/nmat3314. [DOI] [PubMed] [Google Scholar]
- 14.Feng L, Dreyfus D, Sha R, Seeman NC, Chaikin PM. Adv Mater. 2013;25(20):2779–2783. doi: 10.1002/adma.201204864. [DOI] [PubMed] [Google Scholar]
- 15.Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2005;127(37):12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 16.Randeria PS, Jones MR, Kohlstedt KL, Banga RJ, Olvera de la Cruz M, Schatz GC, Mirkin CA. J Am Chem Soc. 2015;137(10):3486–3489. doi: 10.1021/jacs.5b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt TJ, Knotts TA. J Chem Phys. 2011;134(20):205105. doi: 10.1063/1.3592557. [DOI] [PubMed] [Google Scholar]
- 18.Morrison LE, Stols LM. Biochemistry. 1993;32(12):3095–3104. doi: 10.1021/bi00063a022. [DOI] [PubMed] [Google Scholar]
- 19.Marky LA, Breslauer KJ. Biopolymers. 1987;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 20.Mergny JL, Lacroix L. Oligonucleotides. 2003;13(6):515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Craig SL. J Am Chem Soc. 2005;127(38):13227–13231. doi: 10.1021/ja052352h. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Wang W, Ge J, Zhao XS. Nucleic Acids Res. 2009;37(11):3756–3765. doi: 10.1093/nar/gkp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irving D, Gong P, Levicky R. J Phys Chem B. 2010;114(22):7631–7640. doi: 10.1021/jp100860z. [DOI] [PubMed] [Google Scholar]
- 24.Wong IY, Melosh NA. Nano Lett. 2009;9(10):3521–3526. doi: 10.1021/nl901710n. [DOI] [PubMed] [Google Scholar]
- 25.Wong IY, Melosh NA. Biophys J. 2010;98(12):2954–2963. doi: 10.1016/j.bpj.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holbrook JA, Capp MW, Saecker RM, Record MT. Biochemistry. 1999;38(26):8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 27.Wu P, Nakano S, Sugimoto N. Eur J Biochem. 2002;269(12):2821–2830. doi: 10.1046/j.1432-1033.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 28.Mikulecky PJ, Feig AL. Biochemistry. 2006;45(2):604–616. doi: 10.1021/bi0517178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freire E, Mayorga OL, Straume M. Anal Chem. 1990;62(18):950A–959A. [Google Scholar]
- 30.Jelesarov I, Bosshard HR. J Mol Recognit. 1999;12(1):3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 31.Riccelli PV, Vallone PM, Kashin I, Faldasz BD, Lane MJ, Benight AS. Biochemistry. 1999;38(34):11197–11208. doi: 10.1021/bi9904407. [DOI] [PubMed] [Google Scholar]
- 32.Rouzina I, Bloomfield VA. Biophys J. 1999;77(6):3242–3251. doi: 10.1016/S0006-3495(99)77155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT. Proc Natl Acad Sci U S A. 2011;108(31):12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia F, Lu X, Tan X, Wang D, Cao X, Zhang K. Angew Chem, Int Ed. 2017;56(5):1239–1243. doi: 10.1002/anie.201610753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knotts TA, Rathore N, Schwartz DC, de Pablo JJ. J Chem Phys. 2007;126(8):84901. doi: 10.1063/1.2431804. [DOI] [PubMed] [Google Scholar]
- 36.Sambriski EJ, Schwartz DC, de Pablo JJ. Biophys J. 2009;96(5):1675–1690. doi: 10.1016/j.bpj.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinckley DM, Freeman GS, Whitmer JK, de Pablo JJ. J Chem Phys. 2013;139(14):144903. doi: 10.1063/1.4822042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomzov AA, Vorobjev YN, Pyshnyi DV. J Phys Chem B. 2015;119(49):15221–15234. doi: 10.1021/acs.jpcb.5b09645. [DOI] [PubMed] [Google Scholar]
- 39.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54(14):3607–3630. [Google Scholar]
- 40.Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. J Am Chem Soc. 1998;120(50):13252–13253. [Google Scholar]
- 41.Kaur H, Wengel J, Maiti S. Biochemistry. 2008;47(4):1218–1227. doi: 10.1021/bi700996z. [DOI] [PubMed] [Google Scholar]
- 42.Kierzek E, Pasternak A, Pasternak K, Gdaniec Z, Yildirim I, Turner DH, Kierzek R. Biochemistry. 2009;48(20):4377–4387. doi: 10.1021/bi9002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owczarzy R, You Y, Groth CL, Tataurov AV. Biochemistry. 2011;50(43):9352–9367. doi: 10.1021/bi200904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.