Abstract
Inspired by our previous efforts on the modifications of diarylpyrimidines as HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTI) and reported crystallography study, novel diarylnicotinamide derivatives were designed with a “triazole tail” occupying the entrance channel in the NNRTI binding pocket of the reverse transcriptase to afford additional interactions. The newly designed compounds were then synthesized and evaluated for their anti-HIV activities in MT-4 cells. All the compounds showed excellent to good activity against wild-type HIV-1 strain with EC50 of 0.02–1.77 μM. Evaluations of selected compounds against more drug-resistant strains showed these compounds had advantage of inhibiting E138K mutant virus which is a key drug-resistant mutant to the new generation of NNRTIs. Among this series, propionitrile (3b2, EC50(IIIB) = 0.020 μM, EC50(E138K) = 0.015 μM, CC50 = 40.15 μM), pyrrolidin-1-ylmethanone (3b8, EC50(IIIB) = 0.020 μM, EC50(E138K) = 0.014 μM, CC50 = 58.09 μM) and morpholinomethanone (3b9, EC50(IIIB) = 0.020 μM, EC50(E138K) = 0.027 μM, CC50 =180.90 μM) derivatives are the three most promising compounds which are equally potent to the marketed drug Etravirine against E138K mutant strain but with much lower cytotoxicity. Furthermore, detailed SAR, inhibitory activity against RT and docking study of the representative compounds are also discussed.
Keywords: HIV-1, NNRTIs, Triazole, Diarylnicotinamide, Entrance channel, Drug design
1. Introduction
Human immunodeficiency virus (HIV) is the pathogenic factor for acquired immune deficiency syndrome (AIDS) which severely harms human health. From the WHO most updated report: by the end of 2016, there were still 36.7 million people living with HIV globally [1]. Among them, 1.8 million became newly infected in 2016. And in the same year, 1.0 million people died from AIDS related illnesses. Fortunately, due to the antiretroviral treatment, especially the application of highly active antiretroviral therapy (HAART) which combines multiple drugs that act on different viral targets, AIDS-related deaths have fallen by 39% since the peak in 2000.
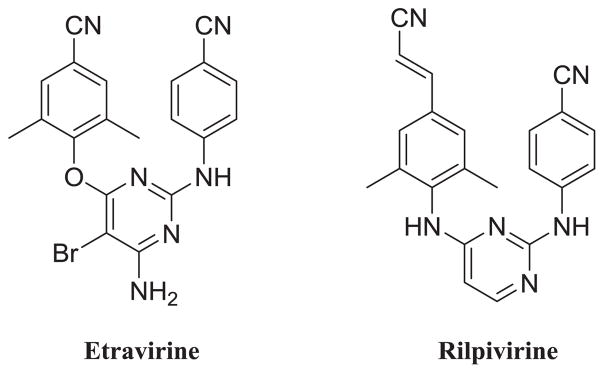
Non-nucleoside reverse transcriptase inhibitors (NNRTIs) are essential components of HARRT for their excellent potencies and low toxicities. With superior activity against wild-type (WT) and mutant HIV-1 strains, diarylpyrimidine (DAPY) derivatives, as the second generation NNRTIs, have attracted considerable attentions over the past few years. FDA approved drugs Etravirine (TMC125, ETR) and Rilpivirine (TMC278, RPV) are the two representative compounds of this series (Fig. 1). However, the unsatisfactory pharmacokinetic properties of these DAPY derivatives [2–4] and adverse effects caused by them [5–7] demands efforts to identify more NNRTI agents with higher potency and better drug-likeness [8,9]. More importantly, the low genetic barrier of viral mutations led to the rapid emergence of drug resistance. From the study in the lab of Steegen, only one-third of patients retained full susceptibility to Etravirine (36.5%) and Rilpivirine (27.3%) [10]. Among these mutations, E138K in RT is not only the most common NNRTI resistance associated mutation found in patients who failed RPV-containing regimens [11] but also the earliest mutation for ETR resistance in all subtypes in vitro. Besides, E138K mutation also confers phenotypic resistance to Nevirapine (NVP) and Efavirenz (EFV) [12,13]. Therefore, the broad cross-resistance against nearly all of the approved NNRTIs make the development of E138K must be avoided whenever possible [14].
Fig. 1.
U.S. FDA approved DAPY NNRTIs.
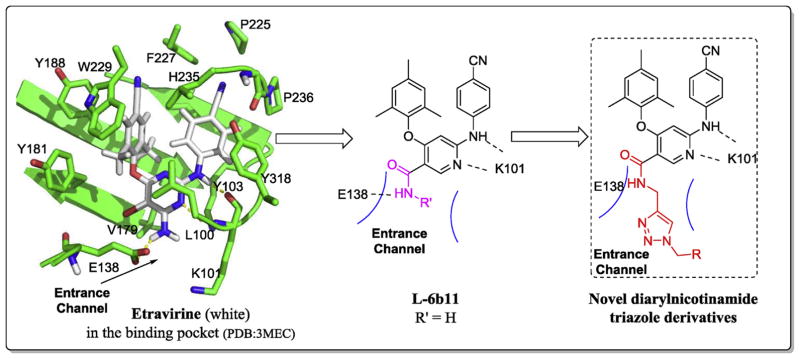
The binding mode of ETR with the NNRTI binding pocket (NNIBP) in RT is well depicted by the crystallography studies [15] (Fig. 2): the central pyrimidine ring and the NH linker connecting to its 2-position form two important hydrogen bonds with the main chain of K101; two aromatic wings of the molecule fit into two adjacent grooves composed mainly of lipophilic residues; additionally, the NH2 and Br groups on the central pyrimidine of ETR point to an entrance channel opening to the solvent surface [16,17] which formed by amino acids L100, K101, E138 and V179.
Fig. 2.
The design of a novel series of diarylnicotinamide 1,4-disubstituted 1,2,3-triazole derivatives.
Great efforts from our group [18–24] focused on the modifications of the central ring of DAPY series trying to discover optimal moieties that can occupy the tolerant space of the entrance channel. Among these different scaffolds, nicotinamide core which can form new hydrogen bond with E138 endowed the molecules not only excellent activities (e.g. L-6b11, EC50 (IIIB) = 0.027 μM, SI > 12,518) [20] but also a port to connect more favorable groups. Thus, we designed a novel series of nicotinamide derivatives based on the L-6b11 template which is the best compound from our last series, hoping to acquire better potency especially against the E138K strain. We kept the core and two phenyl wings as original while linked various substituted benzyl to explore the chemical space of the entrance channel or other groups (mainly hydrophilic groups) by suitable linker. Owing to the importance and realizability of 1,2,3-triazole compounds, the triazolewas used as a linker in the structure to afford potential additional interactions with the surrounding residues [25] (Fig. 2). Unique properties of 1,2,3-triazole such as rigidity and stability in vivo, hydrogen bonding capability, and dipole moment are considered as decisive factors for their improved biological activity. For example, some 1,2,3-triazoles synthesized in our group showed a broad spectrum of antiviral activities [25–28]. Besides, the application of “click chemistry” make it very easy to form the triazole ring.
The newly designed compounds were then synthesized, evaluated for their anti-HIV activities against WT (IIIB and ROD) and multiple mutant strains. The representative compounds were also tested for their ability to inhibit the RT of HIV-1. Herein, the result of evaluation, SAR and modeling analysis will be discussed in this article.
2. Results and discussion
2.1. Chemistry
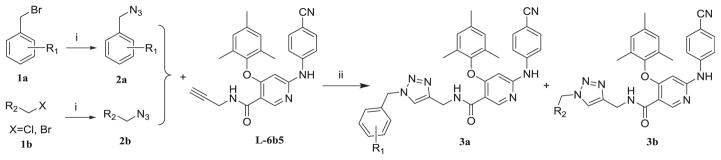
The straightforward synthetic route of target diarylnicotinamide 1,4-disubstituted 1,2,3-triazole derivatives is depicted in Scheme 1. For the preparation of the designed series, substituted haloalkanes (1a or 1b) were used as the starting materials which reacted with sodium azide by nucleophilic substitution to form the appropriate azides (2a or 2b). Then the desired compounds were generated by copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) “click chemistry” reaction between the azides and 6-((4-cyanophenyl) amino)-4-(mesityloxy)-N-(prop-2-yn-1-yl)nicotinamide (L-6b5) which was obtained as one of the active compounds in our previous series [20]. These synthesized compounds were characterized by physicochemical and spectral means and the MS, 1H NMR, 13C NMR spectral data were found in agreement with the assigned molecular structures.
Scheme 1.
The synthetic route of diarylnicotinamide 1,4-disubstituted 1,2,3-triazole derivatives.
Reagents and conditions
i: NaN3 ii: CuSO4, VcNa, THF-H2O, 65 °C.
2.2. Anti-HIV activity evaluation
The newly synthesized diarylnicotinamide triazole derivatives were evaluated for their activities against WT HIV-1 strain (IIIB), most common K103 N + Y181C double mutant HIV-1 strain (RES056) and HIV-2 strain (ROD) in MT-4 cells using the MTT method [29,30]. And the inhibition assay for a panel of more mutant strains L100I, K103 N, E138K, Y181C, Y188L and F227L + V106A were conducted as well for selected compounds. Nevirapine (NVP), Delavirdine (DLV), Efavirenz (EFV), Etravirine and Zidovudine (azidothymidine, AZT) were used as reference drugs. The cytotoxicity of these compounds was determined in parallel. The results, expressed as EC50, CC50, SI and RF are illustrated in Tables 1–4.
Table 1.
Activity against IIIB and RES056 HIV-1 strains, cytotoxicity and SI of the title compounds in MT-4 cells.
| ||||||
|---|---|---|---|---|---|---|
| No. | R | EC50 (μM)a
|
CC50 (μM)b | SIc
|
||
| IIIB | RES056 | IIIB | RES056 | |||
| 3a1 | 2-Me | 0.077 ± 0.051 | >164.2 | ≥164.2 | ≥2132 | < or X1d |
| 3a2 | 2-F | 0.095 ± 0.023 | >222.5 | >222.5 | >2348 | X1 |
| 3a3 | 2-Cl | 0.104 ± 0.022 | >188.5 | ≥188.5 | ≥1807 | < or X1 |
| 3a4 | 2-CN | 0.039 ± 0.007 | >219.8 | >219.8 | >5589 | X1 |
| 3a5 | 2-NO2 | 0.057 ± 0.019 | >212.3 | >212.3 | >3724 | X1 |
| 3a6 | 3-Me | 0.106 ± 0.046 | >113.8 | 113.84 ± 27.32 | 1074 | <1 |
| 3a7 | 3-F | 0.079 ± 0.009 | >204.7 | ≥204.7 | ≥2608 | < or X1 |
| 3a8 | 3-Cl | 0.104 ± 0.054 | >187.4 | 187.4 | 1799 | <1 |
| 3a9 | 3-CN | 0.025 ± 0.008 | >129.6 | ≥129.6 | ≥5213 | < or X1 |
| 3a10 | 3-NO2 | 0.037 ± 0.014 | >156.3 | ≥156.3 | ≥4220 | < or X1 |
| 3a11 | 4-Me | 0.167 ± 0.031 | >165.1 | 165.1 | 990 | <1 |
| 3a12 | 4-F | 0.074 ± 0.003 | >153.1 | ≥153.1 | ≥2056 | < or X1 |
| 3a13 | 4-Cl | 0.194 ± 0.087 | >120.0 | ≥120.0 | ≥620 | < or X1 |
| 3a14 | 4-NO2 | 0.040 ± 0.015 | >139.6 | ≥139.6 | ≥3498 | < or X1 |
| 3b1 |
|
0.037 ± 0.007 | 5.23 ± 2.93 | 25.24 ± 5.03 | 678 | 5 |
| 3b2 |
|
0.020 ± 0.005 | 2.31 ± 0.57 | 40.15 ± 16.62 | 2044 | 17 |
| 3b3 |
|
0.029 ± 0.016 | 2.21 ± 1.77 | 19.37 ± 11.42 | 670 | 9 |
| 3b4 |
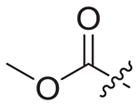
|
1.77 ± 0.23 | >13.8 | 13.8 ± 7.03 | 8 | <1 |
| 3b5 |
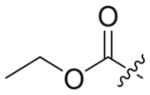
|
0.653 ± 0.31 | >9.53 | 9.53 | 15 | <1 |
| 3b6 |
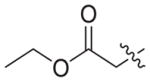
|
0.847 ± 0.30 | >21.47 | 21.47 ± 1.38 | 25 | <1 |
| 3b7 |
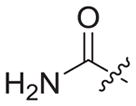
|
0.172 ± 0.007 | 5.33 ± 1.21 | 24.01 ± 2.71 | 139 | 5 |
| 3b8 |
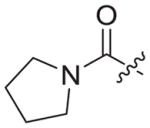
|
0.020 ± 0.005 | 1.43 ± 0.31 | 58.09 ± 34.60 | 2897 | 41 |
| 3b9 |
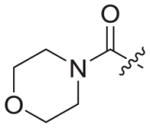
|
0.020 ± 0.009 | 15.70 ± 7.16 | 180.9 ± 36.74 | 9279 | 12 |
| L-6b5e | – | 0.029 ± 0.005 | 6.10 ± 0.54 | 72.96 ± 26.85 | 2471 | 12 |
| NVP | – | 0.26 | >15.02 | >15.02 | >58 | – |
| DLV | – | 0.038 | – | 36.8 | 733 | – |
| EFV | – | 0.005 | 0.31 ± 0.16f | >6.34 | >1212 | – |
| ETR | – | 0.004 | 0.017 ± 0.001f | 2.46 | 733 | – |
| AZT | – | 0.007 | 0.011 ± 0.005f | >7.48 | >1053 | – |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method.
CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method.
SI: selectivity index, the ratio of CC50/EC50.
X1: stands for ≥1 or <1.
L-6b5: the activity data were quoted from our published paper [20].
Table 4.
Selectivity against a panel of mutant HIV-1 stains of selected compounds in MT-4 cells.
| No. | SIa
|
|||||
|---|---|---|---|---|---|---|
| L100I | K103 N | E138K | Y181C | Y188L | F227L + V106A | |
| 3b1 | 13 | 62 | 651 | 109 | 8 | 32 |
| 3b2 | 38 | 158 | 2631 | 450 | 18 | 121 |
| 3b3 | 19 | 73 | 756 | 123 | 6 | 33 |
| 3b7 | 7 | 23 | 159 | 29 | 5 | 13 |
| 3b8 | 60 | 214 | 4189 | 686 | 39 | 245 |
| 3b9 | 30 | 473 | 6800 | 761 | 25 | 297 |
| NVP | >18 | >2 | >64 | >2 | >2 | >2 |
| DLV | 14 | 12 | 715 | 15 | 44 | 16 |
| EFV | >62 | >52 | >1235 | >928 | >19 | >30 |
| ETR | 370 | 773 | 179 | 159 | 111 | 143 |
| AZT | >1444 | >901 | >581 | >1058 | >704 | >1544 |
SI: selectivity index, the ratio of CC50/EC50.
All the 23 compounds exhibited good anti-HIV-1 (WT) activity with sub-micromolar EC50 ranging from 0.02 to 0.85 μM, except for compound 3b4 with EC50 of 1.77 μM (Table 1). Regarding the substituted benzyl derivatives, i.e. 3a1–3a14, they all had EC50 values below 200 nM. Among several different groups on the benzyl ring, the cyano- and nitro-with higher polarity apparently showed their advantage in potency. It also indicated that the 3-position was more favorable for these two polar groups than 4-or 2-positions, as 3a9 (3-CN, EC50 = 25 nM) was more potent than 3a4 (2-CN, EC50 = 39 nM) while 3a10 (3-NO2, EC50 = 37 nM) was better than 3a14 (4-NO2, EC50 = 40 nM) and 3a5 (2-NO2, EC50 = 57 nM). Fluorine had a secondary effect on the potency, furthermore 3a12 (4-F, EC50 =74 nM) was slightly better than 3a7 (3-F, EC50 =79 nM) and 3a2 (2-F, EC50 =104 nM). Additionally, the compounds with hydrophobic Me or Cl were less potent and especially for the para-substituted ones. This sub-series compounds were not cytotoxic with CC50 higher than 110 μM and most of them had SI higher than 1000. Unfortunately, none of these substituted benzyl derivatives showed potency against the RES056 strain.
We also tried various tails attached to the triazole, like ketone, esters or amides instead of the benzyl group. The esters (3b4, 3b5, 3b6) displayed EC50 from 0.65 to 1.77 μM with relatively higher cytotoxicity (9.53–21.5 μM), while 3b7 with an acetamide had better potency (EC50 =172 nM). The rest of compounds all exhibited very good potency against the WT HIV-1 virus with EC50 below 37 nM. More importantly, some of them (3b1, 3b2, 3b3, 3b7, 3b8 and 3b9) showed 50% of inhibition against double-mutant stain RES056 at low micromolar concentration. 3b2, 3b8 and 3b9 containing a Cyanomethyl, 2-oxo-2-(pyrrolidin-1-yl)ethyl or 2-morpholino-2-oxoethyl, respectively, are the three most active compounds with the same EC50 against WT HIV-1 at 20 nM, which were more potent than the control drugs NVP (EC50 = 0.26 μM) and DLV (EC50 = 0.26 μM). Notably, 3b8 had the best SI (= 41) against RES056 strain whereas 3b9 had the best SI (= 9279) against HIV IIIB strain.
Regarding the cytotoxicity of these compounds, it is apparent that 3a sub-series with substituted benzyl were much less toxic with all the CC50 values higher than 100 μM. Whereas, the 3b subseries had CC50 values ranging from 9.53 to 180.9 μM, and most of them had CC50 below 40 μM. From the detailed information, there seemed to be a rough trend that the cyclic (bulkier) group helped to reduce the cytotoxicity: 3b1–7 < 3b8 < 3b9 ≈3a series (in CC50).
With the comparison of L-6b5 (EC50 = 0.029 μM, CC50 =72.96 μM), we could conclude that longer and diverse groups connecting to the amide of the nicotinamide are tolerant for this region. That may because these groups were supposed to stay at the interface between the enzyme and the solvent. And some groups are helpful to make the molecules slightly more active. That may indicate these “tail tip” fragments indeed have subtle interactions with the amino acid residues nearby. And as expected, none of 23 compounds inhibited the HIV-2 strain (Data is not showed in the article) showing that they all act as classical NNRTIs.
Based on the results of the first round of screening, six compounds 3b1–3b3, 3b7–3b9 were selected for testing against six more different HIV-1 mutant strains due to their good potencies against both WT and RES056 strains. (Table 2). All six compounds showed same sensitivity order against the six mutant viruses as follows: Y188L (least sensitive to these compounds) < L100I < F227L + V106A < K103 N < Y181L < E138K (most sensitive to these compounds). For example, these molecules all had single digit micromolar EC50 values against Y188L strain; for K103 N mutated virus, six compounds exhibited EC50 ranging from 0.25 to 1.03 μM; all tested compounds except for 3b7 with amide showed EC50 under 40 nM against E138K strain which is better than the control drug NVP and DLV and comparable to ETR and AZT. Regarding the resistance fold (RF, ratio of EC50 against mutant strain/EC50 against WT strain), compounds 3b2, 3b3, 3b7, 3b8 showed the values for E138K less than 1 while ETR had the RF of 3.5 (Table 3). That means these compounds inhibited the E138K mutant strain specifically with a better EC50 than against the WT virus. These findings implicated a same inhibitory mechanism of these analogues, especially verified the introduction of a longer triazole tail may help to bind tighter to the RT containing E138K. Among them, compounds 3b2 and 3b8 which contain N atom as hydrogen bond acceptor have the best activity against E138K strain. Besides, all six triazole analogues had lower EC50 values than both NVP and DLV against K103 N, Y181C, F227L + V106A, meanwhile they have better potency against Y188L than NVP.
Table 2.
Activity against a panel of mutant HIV-1 stains of selected compounds in MT-4 cells.
| No. | EC50 (μM)a
|
|||||
|---|---|---|---|---|---|---|
| L100I | K103 N | E138K | Y181C | Y188L | F227L + V106A | |
| 3b1 | 1.95 ± 0.45 | 0.41 ± 0.15 | 0.039 ± 0.001 | 0.23 ± 0.030 | 3.17 ± 0 | 0.78 ± 0.053 |
| 3b2 | 1.06 ± 0.22 | 0.25 ± 0.028 | 0.015 ± 0.003 | 0.089 ± 0.004 | 2.24 ± 0.070 | 0.33 ± 0.21 |
| 3b3 | 1.00 ± 0.16 | 0.26 ± 0.072 | 0.026 ± 0.001 | 0.16 ± 0.014 | 3.20 ± 0.67 | 0.59 ± 0.006 |
| 3b7 | 3.52 ± 0.37 | 1.03 ± 0.026 | 0.15 ± 0.005 | 0.83 ± 0.033 | 4.46 ± 0.12 | 1.80 ± 0.89 |
| 3b8 | 0.96 ± 0.12 | 0.27 ± 0.006 | 0.014 ± 0.001 | 0.085 ± 0.020 | 1.48 ± 0.081 | 0.24 ± 0.008 |
| 3b9 | 6.13 ± 0.17 | 0.38 ± 0.048 | 0.027 ± 0.001 | 0.24 ± 0.085 | 7.16 ± 0.46 | 0.61 ± 0.007 |
| NVP | 0.83 ± 0.32 | 6.78 ± 0.08 | 0.23 ± 0.05 | 6.21 ± 0.08 | 9.61 ± 1.86 | 7.17 ± 1.86 |
| DLV | 2.54 ± 0.31 | 3.00 ± 0 | 0.051 ± 0.006 | 2.46 ± 0.54 | 0.84 ± 0.01 | 2.27 ± 0.14 |
| EFV | 0.10 ± 0.01 | 0.12 ± 0.004 | 0.005 ± 0.001 | 0.007 ± 0.001 | 0.34 ± 0.01 | 0.21 ± 0.008 |
| ETR | 0.007 ± 0.002 | 0.003 ± 0 | 0.014 ± 0.002 | 0.016 ± 0 | 0.011 ± 0.002 | 0.005 ± 0 |
| AZT | 0.005 ± 0 | 0.008 ± 0 | 0.013 ± 0.004 | 0.007 ± 0.001 | 0.011 ± 0.002 | 0.005 ± 0 |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method.
Table 3.
Resistance folds for a panel of mutant HIV-1 stains of selected compounds in MT-4 cells.
| No. | Resistance Foldsa
|
|||||
|---|---|---|---|---|---|---|
| L100I | K103 | N E138K | Y181C | Y188L | F227L + V106A | |
| 3b1 | 52.7 | 11.1 | 1.1 | 6.2 | 85.7 | 21.1 |
| 3b2 | 53.0 | 12.5 | 0.8 | 4.5 | 112.0 | 16.5 |
| 3b3 | 34.5 | 9.0 | 0.9 | 5.5 | 110.3 | 20.3 |
| 3b7 | 20.5 | 6.0 | 0.9 | 4.8 | 25.9 | 10.5 |
| 3b8 | 48.0 | 13.5 | 0.7 | 4.3 | 74.0 | 12.0 |
| 3b9 | 306.5 | 19.0 | 1.4 | 12.0 | 358.0 | 30.5 |
| NVP | 3.2 | 26.1 | 0.9 | 23.9 | 37.0 | 27.6 |
| DLV | 66.8 | 78.9 | 1.3 | 64.7 | 22.1 | 59.7 |
| EFV | 20.0 | 24.0 | 1.0 | 1.4 | 68.0 | 42.0 |
| ETR | 1.7 | 0.8 | 3.5 | 4.0 | 2.8 | 1.2 |
| AZT | 0.7 | 1.2 | 1.9 | 1.0 | 1.6 | 0.7 |
Resistance Fold: RF, ratio of EC50 against mutant strain/EC50 against WT strain.
Generally, in terms of the potency against all six mutants, the inhibitory ability order of tested compounds was similar as that against WT virus, i.e. 3b2 ≈ 3b8 > 3b3 > 3b1 ≈ 3b9 >3b7, only with few exceptions to certain mutant strains. 3b2 and 3b8 which showed very similar potency were the best two compounds against these mutants. They showed better potencies than the first generation NNRTIs NVP and DLV against most of the mutants and were equally potent with ETR and AZT against E138K strain. But since the two compounds and 3b9 had better cytotoxic properties, they showed much higher selectivity (3b2: SI = 2631, 3b8: SI = 4189, 3b9: SI = 6800 against E138K; 3b2: SI = 450, 3b8: SI = 686, 3b9: SI = 761 against Y181C) than ETR (SI = 179 against E138K and SI = 159 against Y181C) when against E138K and Y181C mutant virus strains (Table 4).
To further verify the above results, two compounds 3b8 and 3b9 were selected for their excellent potency and low cytotoxicity along with RPV as a control to test their potency against wild-type HIV-1 (NL4-3) replication in TZM-bl cell lines. Their anti-HIV potency (EC50, as measured by a luciferase gene expression assay [32]) and cytotoxicity (CC50) as well as selective index (SI) data are summarized in Table 5. The data for 3b8 and 3b9 were consistent between the two independent assays.
Table 5.
Activity against NL4-3 HIV-1 strain, cytotoxicity and SI of compounds 3b8, 3b9 and RPV in TZM-bl cells.
EC50: Concentration of compound that causes 50% inhibition of viral infection, presented as the mean ± standard deviation (SD) and determined in at least triplicate against HIV-1 in TZM-bl cells.
CC50: Concentrations that cause cytotoxicity to 50% of cells; CytoTox-Glo cytotoxicity assays (Promega) were used, and values were averaged from two independent tests.
SI: selectivity index, the ratio of CC50/EC50.
2.3. Inhibition of HIV-1 RT
With the aim to further confirm the target of the newly designed diarylnicotinamide 1,4-disubstituted 1,2,3-triazole derivatives, the two most promising compounds 3b8 and 3b9 were also tested in enzymatic assays against HIV-1 RT [33,34]. ETR was used as a reference drug in this assay. As shown in Table 6, compound 3b8 and 3b9 exhibited moderate inhibitory activity of the enzyme with IC50 value of 2.70 and 1.57 μM, respectively, which are about one to three time less potent than that of ETR (0.75 μM). To a certain extent, the result reflected their activity against the WT HIV-1 virus in cell-based assays, and also manifested that these derivatives represented by compound 3b8 and 3b9 targeted HIV-1 RT, thus acting as genuine NNRTIs.
Table 6.
Inhibitory activity of compound 3b8, 3b9 and ETR against HIV-1 RT.
| Compd. | 3b8 | 3b9 | ETR |
|---|---|---|---|
| IC50 (μM)a | 2.70 | 1.57 | 0.75 |
50% inhibitory concentration of tested compounds required to inhibit biotin deoxyuridine triphosphate (biotin-dUTP) incorporation into the HIV-1 RT by 50%.
2.4. Molecular modeling analysis
To better explain the SAR and find the potential binding modes of this series of molecules, the two representative compounds 3b8 and 3b9 were docked into the NNRTIs binding pocket in WT and E138K mutated RT using the Surflex-Dock of SYBYL-X 2.0. The proposed binding modes of these compounds to the NNIBP are shown in Fig. 3 & Fig. 4.
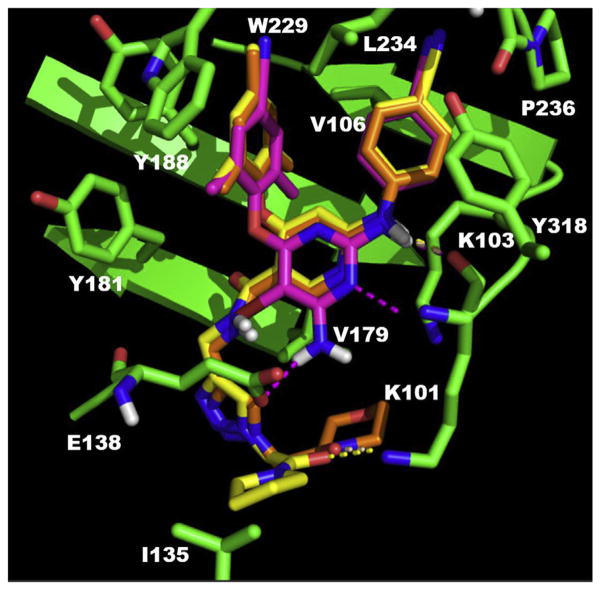
Fig. 3.
Predicted binding mode of compound 3b8 (yellow), 3b9 (orange) in the allosteric site of HIV-1 WT RT (PDB code: 3MEC) with the comparison with the original ligand of this crystal structure ETR (pink). The docking results are shown by PyMOL. hydrogen bond interactions are indicated by dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
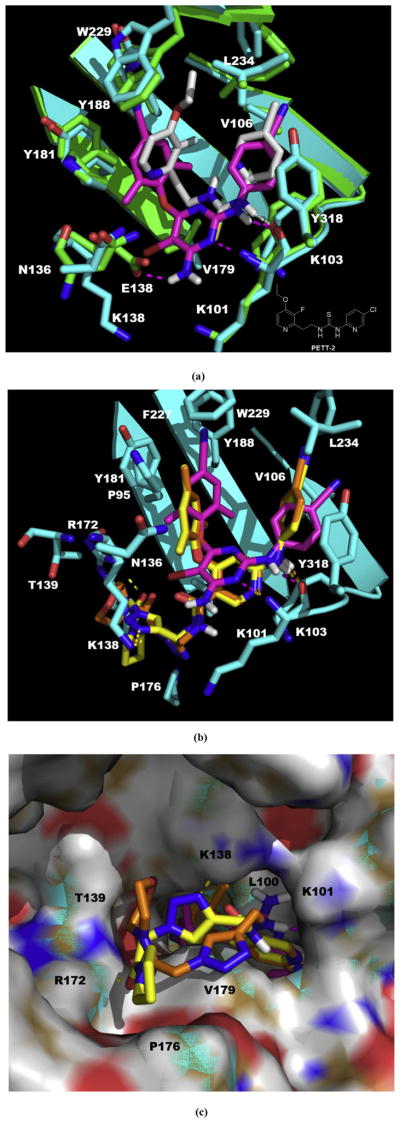
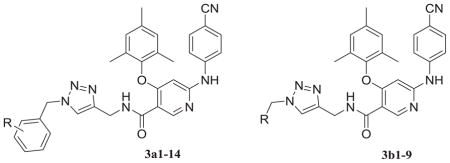
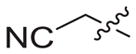
Fig. 4.
(a) The structure of PETT-2, superimposition of docked ETR (pink) to the PETT-2 (gray) extracted from PDB 2HNZ and the overlap of WT (green, from PDB 3MEC) and E138K mutant (cyan, from PDB 2HNZ) RT. (b) The proposed binding mode of 3b8 (yellow), 3b9 (orange) and ETR (pink) with E138K RT (cyan). (c) The proposed binding mode of 3b8 (yellow), 3b9 (orange) and ETR (pink) with E138K RT (cyan): view from the entrance channel. The docking results are shown by PyMOL. hydrogen bond interactions are indicated by dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In the binding mode with WT RT (Fig. 3), the main structures of 3b8, 3b9 and ETR, the original ligand of 3MEC, overlapped very well, and the three compounds shared a common “U” shape binding mode [35–37] with NNIBP in reverse transcriptase of WT HIV-1 virus: i) the left substituted phenol of these structures pointed to a hydrophobic pocket composed by V108, K181, K188, F227 and W229, with a π-π stacking with residue K188; ii) the right 4-cyanoaniline interacted with another pocket which contains residues V106, L234, H235, P236 and Y318; iii) the NH between the right aniline and the central pyridine core in three structures all had a robust hydrogen bond with the key residue K101 in RT. The differences between two novel compounds and ETR in the binding mode were mainly the parts pointing to the entrance channel. Particularly, the amino group on the pyrimidine of ETR formed a hydrogen bond with the carboxyl of residue E138; while the triazoles of two new structures pointing to the solvent-exposed channel oriented the connected amides to form a hydrogen bond with the side chain of K101. Furthermore, the pyrrolidine of 3b8 and morpholine of 3b9 at the far end exposed to the solvent and approached to hydrophobic I135 and V179, respectively. It is possible that the amphiphilic property of these two moieties are beneficial to increase the binding affinity to RT.
Since there was no available crystal complex of E138K RT with ETR, we obtained the RT harboring E138K from the crystal complex with PETT-2 (PDB: 2HNZ). The ligand PETT-2 of 2HNZ is from PETT series which was obtained by simplify the structure of NNRTI tetrahydroimidazobenzodiazepinthione derivatives through bond disconnection [38]. PETT-2 showed excellent potency against HIV-1 RT (IC50 of 5 nM) [39], and shared a very similar binding mode of NNIBP with ETR (Fig. 4a. Note: The 3D structure of PETT-2 in the NNIBP was extracted directly from the PDB complex 2HNZ, which bearing a methyl on the pyridine connected to the thiourea instead of a Cl atom of the correct structure but should not confer a big effect of whole binding mode). The extracted ligand PETT-2 (gray) overlapped well with the upper part of ETR (pink) which was docked into the E138K mutant RT (cyan): two pyridine rings just occupied the position where the two phenyl wings of ETR were; the thiourea also could form a hydrogen bond with carboxyl of K101. Comparison of the mutated RT (E138K, from PDB 2HNZ, cyan) with WT RT (from PDB 3MEC, green) showed that most parts of the two structures superposed on each other with a perturbation in the positions of some of sidechains and only bearing one large change at the mutation site (Fig. 4a), which consistent with the description in the original study [40]: TheWT E138 and mutated K138 residues overlapped as far as the CB carbon atom, with the rest of the side chain of K103 swung away from the binding pocket by a rotation giving a different orientation of amino and carboxyl groups; besides, there was a movement in the K101 side chain which involved a rotation about the CD–CE bond resulting in the amino group movement toward further away from the NNRTI pocket. Above findings indicated the feasibility of using the 2HNZ in our modeling study.
Then we docked 3b8 (yellow) and 3b9 (orange) into this mutated RT with ETR (pink) as a reference (Fig. 4b and c). There were some obvious adjustments of the main structure of two target structures. It seemed like there was a rotation of the whole parts deep in the pocket compared to ETR, but all the key interactions remained including the hydrophobic interactions with the two sub-pocket and the double hydrogen bonds with backbone of K101 (Fig. 4b). For the parts pointing to the entrance channel, ETR lost a hydrogen bond with E138 due to the mutation, while 3b8 and 3b9 kept the interactions with the residues located at the interface with solvent which may involve K101, K138, T139, R172 and P176 (Fig. 4c). Especially, for 3b8, the most potent compounds against E138K strain, there was two additional hydrogen bonds with K138 and R172, respectively. These findings may explain the reason that this series had good potency against the key drug resistant mutant to the new generation HIV-1 NNRTIs E138K and confirmed our rational design as effective strategy to overcome the drug-resistant issues [37].
3. Conclusion
In general, a series of novel diarylnicotinamide triazole analogues were rationally designed based on structure-guided approach, synthesized and evaluated for their bioactivities against HIV-1 (IIIB, K103 N + Y181C, L100I, K103 N, E138K, Y181C, Y188L and F227L + V106A) and HIV-2 (ROD) in MT-4 cells. Several promising compounds (3b2, 3b8, 3b9) were discovered with very good potency against WT and several mutant strains. Noteworthily, regarding the inhibitory activity against E138K, the major drug resistant mutant to the new generation HIV-1 NNRTIs, 3b8 and 3b9 with triazole linker targeting the entrance channel of NNRTIs in RT are equally potent to marketed drug ETR but with much lower cytotoxicity. The inhibitory activity of representative compounds confirmed their target is HIV-1 RT. More valuable information from the SAR and molecular modeling study should be helpful to develop potential preclinical candidates for necessary back-up drugs of the second generation NNRTIs. Ongoing study in our lab will be reported in due course.
4. Experimental section
4.1. Chemistry
All melting points were determined on a micromelting point apparatus and are uncorrected. 1H NMR and 13C NMR spectra were recorded on a Bruker AV-400 spectrometer using DMSO-d6 as solvent and tetramethylsilane (TMS) as internal standard. Chemical shifts are reported in parts per million (δ), and signals are expressed as s (singlet), d (doublet), t (triplet), q (quartet) or m (multiplet). Mass spectra were taken on a LC Autosampler Device: Standard G1313A instrument. TLC was performed on Silica Gel GF254 for TLC (Merck) and spots were visualized by iodine vapors or by irradiation with UV light (254 nm). Flash column chromatography was performed on column packed with Silica Gel 60 (200–300 mesh). Solvents were reagent grade and, when necessary, were purified and dried by standard methods. Concentration of the reaction solutions involved the use of rotary evaporator at reduced pressure.
4.1.1. General procedure for the synthesis of substituted (azidomethyl)benzene (2a) and other nonaromatic substituted azidomethane (2b)
4.1.1.1. Method A
To a flask, appropriate benzyl bromide (1a, 0.50 g) and NaN3 (1.5 eq) was added and then dissolved in 6mL DMF: H2O (5:1) as solvent. The mixture was stirred under oil bath at 50 °C for 12 h. After the reaction, the solvent was removed by rotary evaporation, and 20 mL of water was added. The solution was then extracted by ethyl acetate for three times. The organic phase was combined, dried and concentrated to yield the crude which was used for the next step without further purification. This method was applied in the preparation of 2a1–2a14.
4.1.1.2. Method B
To a flask, appropriate halide (1b2–1b4 and 1b7, 0.50 g) and NaN3 (2–5 eq) (plus 0.1 eq of tetrabutylammonium, when the halide was bromoacetamide) was added and then dissolved in 5–10 mL DMF as solvent. The mixture was stirred under oil bath at 70–80 °C for 12 h. After the reaction, the solvent was removed by rotary evaporation, and 15 mL of water was added. The solution was then extracted by ethyl acetate for three times. The organic phase was combined, dried and concentrated to yield the crude product which was used for the next step without further purification. This method was applied in the preparation of 2b2–2b4 and 2b7.
4.1.1.3. Method C
To a flask, appropriate halide (1b5 and 1b6, 0.70 g) was dissolved in 15mL of acetone and then NaN3 (3–5 eq) (plus 0.1 eq of KI, when the halide was 3-Bromo-1-propanol) and 5mL of H2O was added. The mixture was stirred bath at 50 °C for 12 h. After the reaction, the solvent was removed by rotary evaporation, and 15 mL of water was added. The solution was then extracted by ethyl acetate for three times. The organic phase was combined, dried and concentrated to yield the crude product which was used for the next step without further purification. This method was applied in the preparation of 2b5 and 2b6.
4.1.1.4. Method D
To a flask containing 10 mL anhydrous methylene chloride, bromoacetyl bromide (1b8) or chloroacetyl chloride (1b9) (1eq) was added and the mixture was stirred at −30 °C for 15min. Then morpholine (1.5 eq, for 2b8) or tetrahydropyrrole (2 eq, for 2b9) was added dropwise. After 5 h of stirring at rt, the solution was washed by saturated NH4Cl aq, saturated Na2CO3 aq, H2O and brine successively. The organic phase was dried by Na2SO4, filtered, and concentrated. To the residue, 5–10 mL DMF and NaN3 (1.8 eq) was added. The mixture was stirred under oil bath at 70–80 °C for 12 h. After the reaction, the solvent was removed by rotary evaporation, and 15 mL of water was added. The solution was then extracted by ethyl acetate for three times. The organic phase was combined, dried and concentrated to yield the crude product which was used for the next step without further purification. This method was applied in the preparation of 2b8 and 2b9.
4.1.2. General procedure for the synthesis of N-((1-(substituted benzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a) and N-((1-substituted-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy) nicotinamide (3b)
L-6b5 (0.15 g, 0.37 mmol), appropriate azide (3 eq), CuSO4 (0.0091 g, 0.036 mmol), and VcNa (0.0217 g, 0.11 mmol) was added to the flask together with the 10 mL of THF:H2O (1:1) as solvent. After the mixture was stirred at 65 °C for 12 h, 10 mL of H2O was added to the flask. Then it was extracted by ethyl acetate, concentrated and then the product (3a and 3b) was obtained by column chromatography.
4.1.2.1. N-((1-(2-methylbenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4(mesityloxy)nicotinamide (3a1)
2-Methylbenzyl bromide (1a1)was used as starting material. White solid, yield: 92.0%, m. p.: 220–223 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.70 (s, 1H, pyridine-H), 8.66 (t, 1H, J = 5.6 Hz, NH), 7.88 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.8 Hz, Ph-H), 7.68 (d, 2H, J = 8.7 Hz, Ph-H), 7.25–7.07 (m, 4H, Ph-H), 7.05 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.58 (s, 2H, CH2), 4.59 (d, 2H, J = 5.5 Hz, CH2), 2.30 (s, 3H, CH3), 2.23 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.60, 162.69, 158.62, 152.17, 147.27, 145.89, 145.66, 136.77, 135.85, 134.63, 133.54 (2 × C, Ph), 130.87, 130.57, 130.26, 129.14, 128.74, 126.65, 123.21 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.21, 102.47, 94.41 (pyridine-C3), 51.38 (CH2), 35.44 (CH2), 20.85 (CH3), 19.04 (CH3), 16.13 (2 × CH3). ESI-MS: m/z 558.5 [M+H]+, 580.5 [M+Na]+. C33H31N7O2 (557.25).
4.1.2.2. N-((1-(2-fluorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a2)
2-Fluorobenzyl bromide (1a2) was used as starting material. White solid, yield: 80.0%, m. p.: 180–183 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.69 (s, 1H, NH), 8.69 (s, 1H, pyridine-H), 7.97 (s, 1H, triazole-H), 7.84 (d, 2H, J = 7.9 Hz, Ph-H), 7.68 (d, 2H, J = 8.0 Hz, Ph-H), 7.41–7.40 (m, 1H), 7.32–7.30 (m, 1H), 7.25–7.19 (m, 2H), 7.05 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.63 (s, 2H, CH2), 4.62 (s, 2H, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.61, 162.70, 160.55 (d, J = 245 Hz), 158.64, 152.21, 147.28, 146.00, 145.66, 135.85, 133.56 (2 × C, Ph), 131.21, 131.16, 130.59 (2 × C, Ph), 130.26 (2 × C, Ph), 125.26 (d, J = 3.3 Hz), 123.47 (triazole-CH), 123.32, 120.05, 118.45 (2 × C, Ph), 116.06 (d, J = 20.7 Hz), 112.18, 102.47, 94.41 (pyridine-C3), 47.29 (CH2), 35.43 (CH2), 20.86 (CH3), 16.14 (2 × CH3). ESI-MS: m/z 562.4 [M+H]+, 584.4 [M+Na]+. C32H28FN7O2 (561.23).
4.1.2.3. N-((1-(2-chlorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a3)
2-Chlorobenzyl bromide (1a3) was used as starting material. White solid, yield: 90.0%, m. p.: 188–190 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.71 (s, 1H, NH), 7.97 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.6 Hz, Ph-H), 7.69 (d, 2H, J = 8.6 Hz, Ph-H), 7.50 (d, 1H, J = 7.5 Hz, Ph-H), 7.38 (t, 1H, J = 6.9 Hz, Ph-H), 7.32 (t, 1H, J = 7.1 Hz, Ph-H), 7.20 (d, 1H, J = 6.7 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.68 (s, 2H, CH2), 4.61 (d, 2H, J = 4.6 Hz, CH2), 2.31 (s, 3H, CH3), 2.04 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.70, 158.64, 152.20, 147.29, 145.93, 145.66, 135.85, 133.82, 133.55 (2 × C, Ph), 133.08, 130.91, 130.66, 130.58, 130.27 (2 × C, Ph), 130.07, 128.13, 123.55 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.20, 102.47, 94.41 (pyridine-C3), 50.99 (CH2), 35.44 (CH2), 20.85 (CH3), 16.15 (2 × CH3). ESI-MS: m/z 578.5 [M+H]+, 600.4 [M+Na]+. C32H28ClN7O2 (577.20).
4.1.2.4. N-((1-(2-cyanobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a4)
2-Cyanobenzyl bromide (1a4) was used as starting material. White solid, yield: 81.8%, m. p.: 161–163 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.70 (s, 1H, pyridine-H), 8.70 (s, 1H, NH), 8.06 (s, 1H, triazole-H), 7.90 (d, 1H, J = 7.2 Hz, Ph-H), 7.85 (d, 2H, J = 8.5 Hz, Ph-H), 7.68 (d, 3H, J = 8.2 Hz, Ph-H), 7.55 (t, 1H, J = 7.1 Hz, Ph-H), 7.34 (d, 1H, J = 7.5 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.79 (s, 2H, CH2), 4.60 (d, 2H, J = 4.7 Hz, CH2), 2.30 (s, 3H, CH3), 2.04 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.64, 162.71, 158.64, 152.22, 147.29, 146.12, 145.66, 139.39, 135.85, 134.22, 133.80, 133.56 (2 × C, Ph), 130.59, 130.26 (2 × C, Ph), 129.82, 129.64, 123.75 (triazole-CH), 120.05 (CN), 118.45 (2 × C, Ph), 117.44, 112.17, 111.69, 102.47, 94.40 (pyridine-C3), 51.41 (CH2), 35.43 (CH2), 20.86 (CH3), 16.16 (2 × CH3). ESI-MS: m/z 569.5 [M+H]+, 591.5 [M+Na]+. C33H28N8O2 (568.23).
4.1.2.5. N-((1-(2-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a5)
2-Nitrobenzyl bromide (1a5) was used as starting material. White solid, yield: 83.7%, m. p.: 149–153 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.70 (s, 1H, NH), 8.70 (s, 1H, pyridine-H), 8.12 (d, 1H, J = 7.8 Hz, Ph-H), 8.03 (s, 1H, triazole-H), 7.84 (d, 2H, J = 8.2 Hz, Ph-H), 7.68 (d, 3H, J = 8.2 Hz, Ph-H), 7.64–7.60 (m, 1H), 7.05–7.01 (m, 3H), 5.94 (s, 2H, CH2), 5.92 (s, 1H, pyridine-H), 4.62 (d, 2H, J = 4.3 Hz, CH2), 2.30 (s, 3H, CH3), 2.04 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.64, 162.71, 158.64, 152.20, 148.05, 147.29, 146.12, 145.65, 135.85, 134.70, 133.55 (2 × C, Ph), 131.49, 130.58, 130.39, 130.27 (2 × C, Ph), 130.04, 125.48, 124.11 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.18, 102.47, 94.41 (pyridine-C3), 50.27 (CH2), 35.46 (CH2), 20.85 (CH3), 16.14(2 × CH3). ESI-MS: m/z 589.5 [M+H]+, 611.4 [M+Na]+. C32H28N8O4 (588.22).
4.1.2.6. N-((1-(3-methylbenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a6)
3-Methylbenzyl bromide (1a6)was used as starting material. White solid, yield: 88.3%, m. p.: 253–255 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.68 (s, 1H, NH), 7.98 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.4 Hz, Ph-H), 7.68 (d, 2H, J = 8.4 Hz, Ph-H), 7.22 (t, 1H, J = 7.0 Hz, Ph-H), 7.14–7.05 (m, 5H), 5.93 (s, 1H, pyridine-H), 5.52 (s, 2H, CH2), 4.59 (d, 2H, J = 4.6 Hz, CH2), 2.30 (s, 3H, CH3), 2.26 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.61, 162.70, 158.64, 152.23, 147.28, 145.99, 145.66, 138.40, 136.48, 135.85, 133.55 (2 × C, Ph), 130.58 (2 × C, Ph), 130.26 (2 × C, Ph), 129.19, 129.08, 128.99, 125.54, 123.17 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.18, 102.47, 94.41 (pyridine-C3), 53.22 (CH2), 35.46 (CH2), 21.37 (CH3), 20.85 (CH3), 16.14 (2 × CH3). ESI-MS: m/z 558.6 [M+H]+, 580.5 [M+Na]+. C33H31N7O2 (577.25).
4.1.2.7. N-((1-(3-fluorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a7)
3-Fluorobenzyl bromide (1a7) was used as starting material. White solid, yield 80.4%, m. p.: 222–223 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.69 (s, 1H, NH), 8.05 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.3 Hz, Ph-H), 7.68 (d, 2H, J = 8.3 Hz, Ph-H), 7.40–7.39 (m, 1H, Ph-H), 7.18–7.12 (m, 3H, Ph-H), 7.05 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.60 (s, 2H, CH2), 4.60 (d, 2H, J = 3.8 Hz, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.70 162.60 (d, J = 243 Hz), 158.64, 152.23, 147.28, 146.14, 145.66, 139.31 (d, J = 7.4 Hz), 135.85 (PhC-CH3), 133.55 (2 × C, Ph), 131.25 (J = 8.4 Hz), 130.57 (2 × C, Ph), 130.25 (2 × C, Ph), 124.46 (d, J = 2.7 Hz), 123.42 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 115.42 (d, J = 18.6 Hz), 115.21 (d, J = 19.8 Hz), 112.18, 102.47, 94.41 (pyridine-C3), 52.51 (CH2), 35.45 (CH2), 20.85 (CH3), 16.13 (2 × CH3). ESI-MS: m/z 562.5 [M+H]+, 584.5 [M+Na]+. C32H28FN7O2 (561.23).
4.1.2.8. N-((1-(3-chlorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a8)
3-Chlorobenzyl bromide (1a8) was used as starting material. White solid, yield: 88.7%, m. p.: 243–244 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.68 (s, 1H, NH), 8.05 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.4 Hz, Ph-H), 7.68 (d, 2H, J = 8.4 Hz, Ph-H), 7.38 (s, 3H, Ph-H), 7.26 (s, 1H, Ph-H), 7.04 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.60 (s, 2H, CH2), 4.60 (d, 2H, J = 4.4 Hz, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.71, 158.64, 152.25, 147.28, 146.16, 145.66, 139.01, 135.85, 133.73, 133.55 (2 × C, Ph), 131.11, 130.58, 130.26, 128.56, 128.28, 127.13, 123.43 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.16, 102.48, 94.41 (pyridine-C3), 52.42 (CH2), 35.45 (CH2), 20.85 (CH3), 16.14(2 × CH3). ESI-MS: m/z 578.4 [M+H]+, 600.4 [M+Na]+ C32H28ClN7O2 (577.20).
4.1.2.9. N-((1-(3-cyanobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a9)
3-Cyanobenzyl bromide (1a9) was used as starting material. White solid, yield: 84.2%, m. p.: 203–205 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.69 (s, 1H, NH), 8.08 (s, 1H, triazole-H), 7.86–7.79 (m, 4H), 7.68 (d, 2H, J = 8.3 Hz, Ph-H), 7.63–7.57 (m, 2H), 7.04 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.66 (s, 2H, CH2), 4.60 (d, 2H, J = 4.4 Hz, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.63, 162.70, 158.64, 152.24, 147.28, 146.23, 145.65, 138.19, 135.85, 133.55 (2 × C, Ph), 133.39, 132.42, 132.06, 130.57, 130.51, 130.26, 123.51 (triazole-CH), 120.05,118.88, 118.45 (2 × C, Ph),112.15, 102.48, 94.41 (pyridine-C3), 52.24 (CH2), 35.45 (CH2), 20.85 (CH3), 16.14 (2 × CH3). ESI-MS: m/z 569.5 [M+H]+, 591.5 [M+Na]+. C33H28N8O2 (568.23).
4.1.2.10. N-((1-(3-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a10)
3-Nitrobenzyl bromide (1a10) was used as starting material. White solid, yield: 83.7%, m. p.: 221–223 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.71 (s, 1H, NH), 8.21–8.18 (m, 2H), 8.12 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.6 Hz, Ph-H), 7.76 (d, 1H, J = 7.3 Hz, Ph-H), 7.69–7.64 (m, 3H), 7.03 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.76 (s, 2H, CH2), 4.60 (d, 2H, J = 4.8 Hz, CH2), 2.30 (s, 3H, CH3), 2.02 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.65, 162.70, 158.64, 152.22, 148.33, 147.27, 146.24, 145.65, 138.74, 135.85, 135.18, 133.55 (2 × C, Ph), 130.83, 130.56, 130.24, 123.59, 123.56, 123.23, 120.05, 118.45 (2 × C, Ph), 112.17, 102.48, 94.41 (pyridine-C3), 52.15 (CH2), 35.46 (CH2), 20.84 (CH3), 16.12 (2 × CH3). ESI-MS: m/z 589.5 [M+H]+, 611.4 [M+Na]+. C32H28N8O4 (588.22).
4.1.2.11. N-((1-(4-methylbenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a11)
4-Methylbenzyl bromide (1a11) was used as starting material. White solid, yield: 88.3%, m. p.: 245–246 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.69 (s, 1H, pyridine-H), 8.65 (s, 1H, NH), 7.94 (s, 1H, triazole-H), 7.84 (d, 2H, J = 8.4 Hz, Ph-H), 7.68 (d, 2H, J = 8.5 Hz, Ph-H), 7.18 (d, 2H, J = 7.1 Hz, Ph-H), 7.13 (d, 2H, J = 7.4 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.50 (s, 2H, CH2), 4.57 (d, 2H, J = 5.6 Hz, CH2), 2.30 (s, 3H, CH3), 2.27 (s, 3H, CH3), 2.02 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.60, 162.68, 158.62, 152.17, 147.28, 145.97, 145.66, 137.88, 135.84, 133.56 (2 × C, Ph), 130.59 (2 × C, Ph), 130.26 (2 × C, Ph), 129.70 (2 × C), 128.44 (2 × C), 123.06 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.23, 102.47, 94.41 (pyridine-C3), 53.02 (CH2), 35.45 (CH2), 21.15 (CH3), 20.86 (CH3), 16.15(2 × CH3). ESI-MS: m/z 558.6 [M+H]+, 580.5 [M+Na]+. C33H31N7O2 (577.25).
4.1.2.12. N-((1-(4-fluorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a12)
4-Fluorobenzyl bromide (1a12) was used as starting material. White solid, yield: 87.7%, m. p.: 226–228 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.70 (s, 1H, pyridine-H), 8.67 (s, 1H, NH), 8.00 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.5 Hz, Ph-H), 7.68 (d, 2H, J = 8.4 Hz, Ph-H), 7.36 (t, 2H, J = 5.8 Hz, Ph-H), 7.17 (t, 2H, J = 8.5 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.56 (s, 2H, CH2), 4.58 (d, 2H, J = 4.7 Hz, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.69,162.33 (d, J = 243 Hz), 158.63, 152.19, 147.28, 146.07, 145.66, 135.85, 133.55 (2 × C, Ph), 132.86 (d, J = 2.8 Hz), 130.72 (d, J = 8.4 Hz), 130.58, 130.26 (2 × C, Ph), 123.16 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 116.00 (d, J = 21.4 Hz), 112.21, 102.47, 94.41 (pyridine-C3), 52.40 (CH2), 35.45 (CH2), 20.84 (CH3), 16.14(2 × CH3). ESI-MS: m/z 562.5 [M+H]+, 584.4 [M+Na]+. C32H28FN7O2 (561.23).
4.1.2.13. N-((1-(4-chlorobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a13)
4-Chlorobenzyl bromide (1a13) was used as starting material. White solid, yield: 89.9%, m. p.: 232–234 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.67 (t, 1H, J = 5.6 Hz, NH), 8.02 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.8 Hz, Ph-H), 7.68 (d, 2H, J = 8.8 Hz, Ph-H), 7.40 (d, 2H, J = 8.4 Hz, Ph-H), 7.31 (d, 2H, J = 8.5 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.58 (s, 2H, CH2), 4.59 (d, 2H, J = 5.6 Hz, CH2), 3.35 (s, 3H, CH3), 2.31 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.68, 158.63, 152.19, 147.27, 146.10, 145.65, 135.85, 135.60, 133.54 (2 × C, Ph), 133.29, 130.57, 130.32 (2 × C, Ph), 130.25, 129.16 (2 × C, Ph), 123.31 (triazole-CH), 120.05, 118.44 (2 × C, Ph), 112.21, 102.47, 94.41 (pyridine-C3), 52.39 (CH2), 35.44 (CH2), 20.85 (CH3), 16.13 (2 × CH3). ESI-MS: m/z 578.5 [M+H]+, 600.5 [M+Na]+. C32H28ClN7O2 (577.20).
4.1.2.14. N-((1-(4-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3a14)
4-Nitrobenzyl bromide (1a14) was used as starting material. White solid, yield: 87.2%, m. p.: 269–271 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.70 (s, 1H, pyridine-H), 8.70 (s, 1H, NH), 8.20 (d, 2H, J = 8.4 Hz, Ph-H), 8.10 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.4 Hz, Ph-H), 7.68 (d, 2H, J = 8.5 Hz, Ph-H), 7.51 (d, 2H, J = 8.2 Hz, Ph-H), 7.03 (s, 2H, Ph-H), 5.92 (s, 1H, pyridine-H), 5.77 (s, 2H, CH2), 4.61 (d, 2H, J = 4.3 Hz, CH2), 2.30 (s, 3H, CH3), 2.03 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.67, 162.68, 158.63, 152.15, 147.68, 147.28, 146.24, 145.66, 144.05, 135.86, 133.55 (2 × C, Ph), 130.56, 130.24, 129.46 (2 × C, Ph), 124.31 (2 × C, Ph), 123.75 (triazole-CH), 120.05,118.44 (2 × C, Ph),112.26, 102.47, 94.43 (pyridine-C3), 52.30 (CH2), 35.45 (CH2), 20.83 (CH3), 16.15 (2 × CH3). ESI-MS: m/z 589.4 [M+H]+, 611.4 [M+Na]+. C32H28N8O4 (588.22).
4.1.2.15. 6-((4-cyanophenyl)amino)-N-((1-(3-hydroxypropyl)-1H-1,2,3-triazol-4-yl)methyl)-4-(mesityloxy)nicotinamide (3b1)
3-Bromo-propan-1-ol (1b1) was used as starting material. White solid, yield: 80.2%, m. p.: 198–199 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.67 (s, 1H, NH), 7.96 (s, 1H, triazole-H), 7.86 (d, 2H, J = 8.5 Hz, Ph-H), 7.68 (d, 2H, J = 8.6 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.94 (s, 1H, pyridine-H), 4.65 (s, 1H, O-H), 4.59 (d, 2H, J = 4.8 Hz, CH2), 4.39 (t, 2H, J = 6.8 Hz, CH2), 3.39–3.34 (m, 2H, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3), 1.94–1.91 (m, 2H, CH2). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.62, 162.72, 158.64, 152.23, 147.29, 145.67, 145.47, 135.87, 133.55 (2 × C, Ph), 130.60, 130.28 (2 × C, Ph), 123.16 (triazole-CH), 120.05, 118.45, 112.22, 102.47, 94.43 (pyridine-C3), 57.93 (CH2), 47.01 (CH2), 35.47 (CH2), 33.48 (CH2), 20.85 (CH3), 16.16 (2 × CH3). ESI-MS: m/z 512.7 [M+H]+, 534.5 [M+Na]+. C28H29N7O3 (511.23).
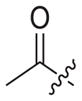
4.1.2.16. N-((1-(2-cyanoethyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3b2)
3-Bromoethyleyanide (1b2) was used as starting material. White solid, yield: 89.1%, m. p.: 144–146 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.72 (s, 1H, NH), 8.07 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.3 Hz, Ph-H), 7.68 (d, 2H, J = 8.3 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 4.64–4.60 (m, 4H, 2 × CH2), 3.16 (t, 2H, J = 5.6 Hz, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 163.65, 162.75, 158.67, 152.31, 147.31, 145.97, 145.66, 135.86, 133.55 (2 × C, Ph), 130.63, 130.27 (2 × C, Ph), 123.44 (triazole-CH), 120.05, 118.58, 118.46 (2 × C, Ph), 112.10, 102.48, 94.41 (pyridine-C3), 45.40 (CH2), 35.44 (CH2), 20.85 (CH3), 18.95 (CH2), 16.19(2 × CH3). ESI-MS: m/z 507.6 [M+H]+, 529.5 [M+Na]+. C28H26N8O2 (506.22).
4.1.2.17. 6-((4-cyanophenyl)amino)-4-(mesityloxy)-N-((1-(2-oxopropyl)-1H-1,2,3-triazol-4-yl)methyl)nicotinamide (3b3)
Bromoacetone (1b3) was used as starting material. White solid, yield: 72.5%, m. p.: 246–250 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.72 (s, 1H, NH), 7.86 (s, 1H, triazole-H), 7.85 (d, 2H, J = 7.9 Hz, Ph-H), 7.68 (d, 2H, J = 8.3 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.94 (s, 1H, pyridine-H), 5.44 (s, 2H, CH2), 4.62 (d, 2H, J = 4.2 Hz, CH2), 2.30 (s, 3H, CH3), 2.18 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 201.32 (C=O), 163.63, 162.74, 158.67, 152.31, 147.30, 145.66, 145.56, 135.86, 133.55 (2 × C, Ph), 130.62, 130.27 (2 × C, Ph), 124.58 (triazole-CH), 120.05, 118.46 (2 × C, Ph), 112.09, 102.48, 94.41 (pyridine-C3), 58.61 (CH2), 35.47 (CH2), 27.46 (CH3), 20.85 (CH3), 16.18 (2 × CH3). ESIMS: m/z 510.6 [M+H]+, 532.5 [M+Na]+. C28H27N7O3 (509.22).
4.1.2.18. Methyl 2-(4-((6-((4-cyanophenyl)amino)-4-(mesityloxy) nicotinamido)methyl)-1H-1,2,3-triazol-1-yl)acetate (3b4)
Azidoacetic acid methyl ester (2b4) was used as starting material. White solid, yield: 88.5%, m. p.: 234–236 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.76 (s, 1H, NH), 8.73 (s, 1H, NH), 8.73 (s, 1H, pyridine-H), 8.00 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.6 Hz, Ph-H), 7.68 (d, 2H, J = 8.6 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.94 (s, 1H, pyridine-H), 5.39 (s, 2H, CH2), 4.62 (d, 2H, J = 5.4 Hz, CH2), 3.70 (s, 3H, CH3), 2.51 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 168.21, 163.65, 162.74, 158.68, 152.32, 147.30, 145.76, 145.65, 135.86, 133.55 (2 × C, Ph), 130.62 (2 × C, Ph), 130.27 (2 × C, Ph), 124.68 (triazole-CH), 120.05, 118.46 (2 × C, Ph), 112.07, 102.48, 94.40 (pyridine-C3), 52.92 (CH2), 50.67 (CH3), 35.43 (CH2), 20.85 (CH3), 16.18 (2 × CH3). ESI-MS: m/z 526.5 [M+H]+, 548.5 [M+Na]+. C28H27N7O4 (525.21).
4.1.2.19. Ethyl 2-(4-((6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamido) methyl)-1H-1,2,3-triazol-1-yl)acetate (3b5)
Ethyl bromoacetate (1b5) was used as starting material. White solid, yield: 76.1%, m. p.: 177–179 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.72 (s, 1H, NH), 7.99 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.4 Hz, Ph-H), 7.68 (d, 2H, J = 8.4 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.94 (s, 1H, pyridine-H), 5.36 (s, 2H, CH2), 4.62 (d, 2H, J = 4.4 Hz, CH2), 4.16 (q, 2H, J = 7.0 Hz, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3), 1.21 (t, 3H, J = 7.0 Hz, CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 167.71 (C=O), 163.64, 162.75, 158.68, 152.32, 147.31, 145.73, 145.66, 135.86, 133.55 (2 × C, Ph), 130.62 (2 × C, Ph), 130.27 (2 × C, Ph), 124.69 (triazole-CH), 120.05, 118.47 (2 × C, Ph), 112.08, 102.49, 94.41 (pyridine-C3), 61.87 (CH2), 50.78 (CH2), 35.44 (CH2), 20.85 (CH3), 16.19 (2 × CH3), 14.43 (CH3). ESI-MS: m/z 540.5 [M+H]+, 562.4 [M+Na]+. C29H29N7O4 (539.23).
4.1.2.20. Ethyl 3-(4-((6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamido) methyl)-1H-1,2,3-triazol-1-yl)propanoate (3b6)
Ethyl 3-bromopropionate (1b6) was used as starting material. White solid, yield: 81.6%, m. p.: 149–151 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.75 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.67 (s, 1H, NH), 8.00 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.5 Hz, Ph-H), 7.68 (d, 2H, J = 8.5 Hz, Ph-H), 7.05 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.60 (s, 2H, CH2), 4.58–4.55 (m, 4H, 2 × CH2), 4.03 (q, 2H, J = 7.0 Hz, CH2), 2.93 (t, 2H, J = 6.2 Hz, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3), 1.12 (t, 3H, J = 7.0 Hz, CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 170.74 (C=O), 163.60, 162.72, 158.65, 152.28, 147.29, 145.66, 145.54, 135.86, 133.55 (2 × C, Ph), 130.61 (2 × C, Ph), 130.27 (2 × C, Ph), 123.37 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.12, 102.48, 94.40 (pyridine-C3), 60.72 (CH2), 45.64 (CH2), 35.43 (CH2), 34.52 (CH2), 20.85 (CH3), 16.17 (2 × CH3), 14.41 (CH3). ESI-MS: m/z 554.5 [M+H]+, 576.4 [M+Na]+. C30H31N7O4 (553.24).
4.1.2.21. N-((1-(2-amino-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl)-6-((4-cyanophenyl)amino)-4-(mesityloxy)nicotinamide (3b7)
Bromoacetamide (1b7) was used as starting material. White solid, yield: 72.4%, m. p.: 227–229 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.76 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.72 (s, 1H, NH), 7.92 (s, 1H, triazole-H), 7.85 (d, 2H, J = 8.3 Hz, Ph-H), 7.69 (s, 1H), 7.68 (d, 2H, J = 8.6 Hz, Ph-H), 7.34 (s, 1H), 7.06 (s, 2H, Ph-H), 5.94 (s, 1H, pyridine-H), 5.04 (s, 2H, CH2), 4.61 (d, 2H, J = 4.1 Hz, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 167.80 (C=O), 163.60, 162.74, 158.66, 152.32, 147.29, 145.66, 145.28, 135.86, 133.55 (2 × C, Ph), 130.63, 130.27 (2 × C, Ph), 124.70 (triazole-CH), 120.05, 118.45 (2 × C, Ph), 112.09, 102.46, 94.40 (pyridine-C3), 51.85 (CH2), 35.47 (CH2), 20.85 (CH3), 16.20 (2 × CH3). ESI-MS: m/z 511.7 [M+H]+, 533.5 [M+Na]+. C27H26N8O3 (510.21).
4.1.2.22. 6-((4-cyanophenyl)amino)-4-(mesityloxy)-N-((1-(2-oxo-2-(pyrrolidin-1-yl)ethyl)-1H-1,2,3-triazol-4-yl)methyl)nicotinamide (3b8)
Chloroacetyl chloride (1b8) was used as starting material. White solid, yield: 87.2%, m. p.: 135–138 °C. 1H NMR (400 MHz, DMSO-d6DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.72 (s, 1H, pyridine-H), 8.70 (s, 1H, NH), 7.86 (s, 1H, triazole-H), 7.85 (d, 2H, J = 9.9 Hz, Ph-H), 7.68 (d, 2H, J = 8.2 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.31 (s, 2H, CH2), 4.61 (d, 2H, J = 4.4 Hz, CH2), 3.50 (t, 2H, J = 6.0 Hz, CH2), 3.31 (t, 2H, J = 5.8 Hz, CH2), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 1.80–1.76 (m, 4H). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 164.24, 163.61, 162.74, 158.66, 152.30, 147.30, 145.66, 145.26, 135.86, 133.55 (2 × C, Ph), 130.63, 130.28, 124.73 (triazole-CH), 120.05, 118.46 (2 × C, Ph), 112.12, 102.47, 94.41 (pyridine-C3), 67.50, 51.66 (CH2), 35.50, 24.18, 20.85 (CH3), 16.20 (2 × CH3). ESI-MS: m/z 565.4 [M+H]+, 587.5 [M+Na]+. C31H32N8O3 (564.26).
4.1.2.23. 6-((4-cyanophenyl)amino)-4-(mesityloxy)-N-((1-(2-morpholino-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl)nicotinamide (3b9)
Bromoacetyl bromide (1b9) was used as starting material. White solid, yield: 76.6%, m. p.: 164–167 °C. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 9.74 (s, 1H, NH), 8.71 (s, 1H, pyridine-H), 8.69 (s, 1H, NH), 7.86 (s, 1H, triazole-H), 7.84 (d, 2H, J = 9.8 Hz, Ph-H), 7.68 (d, 2H, J = 8.5 Hz, Ph-H), 7.06 (s, 2H, Ph-H), 5.93 (s, 1H, pyridine-H), 5.43 (s, 2H, CH2), 4.60 (d, 2H, J = 4.5 Hz, CH2), 3.63–3.43 (m, 8H), 2.30 (s, 3H, CH3), 2.07 (s, 6H, 2 × CH3). 13C NMR (100 MHz, DMSO-d6, ppm) δ: 164.99 (C=O), 163.61, 162.74, 158.65, 152.28, 147.30, 145.66, 145.31, 135.86, 133.56 (2 × C, Ph), 130.63, 130.28, 124.88 (triazole-CH), 118.45 (2 × C, Ph), 112.12, 102.47, 94.41 (pyridine-C3), 66.41(morpholine-CH2), 66.33(morpholine-CH2), 50.95 (CH2), 45.23 (morpholine-CH2), 42.37(morpholine-CH2), 35.49 (CH2), 20.86 (CH3), 16.20 (2 × CH3). ESI-MS: m/z 581.6 [M+H]+, 603.6 [M+Na]+. C31H32N8O4 (580.25).
4.2. In vitro anti-HIV assay
The methodology of the anti-HIV assay used in this project has been described in detail previously [24,29,30]. HIV-1 (IIIB, K103 N/Y181C (RES056) [41,42], F227L/V106A, L100I, K103 N, E138K, Y181C, and Y188L) or HIV-2 (ROD) stock (50 μL at 100–300 CCID50) (50% cell culture infectious dose) or culture medium was added to either the infected or mock-infected wells of the microtiter tray. Mock-infected cells were used to evaluate the effect of test compound on uninfected cells in order to assess the cytotoxicity of the tested compound. Exponentially growing MT-4 cells (Derived from a 50-year old Japanese male with generalised lymphadenopathy and heptosplenomegaly by co-culture of his peripheral leukocytes with male umbilical cord lymphocytes.) were resuspended and transferred to the microtiter tray wells. Five days after infection, the viability of mock- and HIV-infected cells was examined spectrophotometrically by the MTT assay.
The MTT assay is based on the reduction of yellow colored 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Acros Organics, Geel, Belgium) by mitochondrial dehydrogenase of metabolically active cells to a blue-purple formazan that can be measured spectrophotometrically [24].
Inhibitory activity of compounds against HIV-1 Infection of TZM-bl Cells was measured as a reduction in the level of luciferase gene expression after a single round of virus infection of the cells similar to that described previously [43]. Briefly, TZM-bl cells were infected with 800 TCID50 of virus (NL4-3) in the presence of compounds at various concentrations. Two days after infection, the culture medium was removed from each well, and 100 μL of Bright Glo reagent (Promega, San Luis Obispo, CA) was added to the cells to measure luminescence using a Victor 2 luminometer. The effective concentration (EC50) against HIV-1 strains was defined as the concentration that caused a 50% decrease in luciferase activity (relative light units) compared to that of virus control wells.
4.3. HIV-1 RT inhibition assay
The inhibition assay of HIV-1 RTwt was implemented by utilizing the template/primer hybrid poly (A) × oligo (dT)15, digoxigenin- and biotin-labeled nucleotides, an antibody to digoxigenin which conjugated to peroxidase (anti-DIG-POD), and the peroxidase substrate ABTS. The incorporation quantities of the digoxigenin- and biotin-labeled dUTP into DNA represented the activity of HIV-1 RT. The HIV-RT inhibition assay was performed by using an RT assay kit (Roche), and the procedure was reported in detail previously [19]. IC50 values corresponded to the concentration of the tested compounds required to inhibit biotin-dUTP incorporation by HIV-1 RT by 50%.
4.4. Molecular modeling
The two representative compounds 3b8 and 3b9 were docked into the NNRTIs binding pocket in WT (PDB code: 3MEC, crystal complex with ETR [44]) and E138K mutated (PDB code: 2HNZ, crystal complex with PETT-2 as a phenethylthiazolylthiourea derivative [40]) RT using the Surflex-Dock of SYBYL-X 2.0. Structures of 3b8 and 3b9 was drawn, optimized and docked into the published three-dimensional crystal structures of WT RT (complexes with ETR, PDB code: 3MEC, retrieved from the Protein Data Bank) by means of surflex-docking module of Sybyl-2.0. Default parameters were used as described in the Sybyl-2.0 manual unless otherwise specified. Top scoring poses were shown by PyMOL version 1.7.0 (http://www.pymol.org/), in overlap with the bound ligand (ETR) in the binding site of RT. The secondary structure of RT is shown in cartoons, and only the key residues for interactions with the inhibitors were shown in sticks and labeled. The potential hydrogenbonds were presented by dashed lines. More detailed method is described previously [19].
Acknowledgments
Financial support from the National Natural Science Foundation of China (NSFC Nos. 81273354, 81573347), Key Project of NSFC for International Cooperation (Nos. 81420108027), Young Scholars Program of Shandong University (YSPSDU, YSPSDU, No. 2016WLJH32), Key Research and Development Project of Shandong Province (No. 2017CXGC1401), KU Leuven (GOA 10/014) and NIH grant AI033066 from the National Institute of Allergy and Infectious Disease (awarded to K.H. L.) is gratefully acknowledged.
Footnotes
Notes
The authors declare no conflict of interest.
References
- 1.HIV/AIDS Fact Sheet. http://www.who.int/mediacentre/factsheets/fs360/en/
- 2.Aouri M, Barcelo C, Guidi M, Rotger M, Cavassini M, Hizrel C, Buclin T, Decosterd LA, Csajka C, Swiss HIVCS. Population pharmacokinetics and pharmacogenetics analysis of rilpivirine in HIV-1-infected individuals. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00899-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatri A, Dutta S, Dunbar M, Podsadecki T, Trinh R, Awni W, Menon R. Evaluation of drug-drug interactions between direct-acting anti-hepatitis C virus combination regimens and the HIV-1 antiretroviral agents raltegravir, tenofovir, emtricitabine, efavirenz, and rilpivirine. Antimicrob Agents Chemother. 2016;60:2965–2971. doi: 10.1128/AAC.02605-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson A, McGowan I. Long-acting rilpivirine for HIV prevention. Curr Opin HIV AIDS. 2015;10:253–257. doi: 10.1097/COH.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 5.Schrijvers R. Etravirine for the treatment of HIV/AIDS. Expet Opin Pharmacother. 2013;14:1087–1096. doi: 10.1517/14656566.2013.787411. [DOI] [PubMed] [Google Scholar]
- 6.Schrijvers R. Etravirine for the treatment of HIV/AIDS. Expet Opin Pharmacother. 2013;14:1087–1096. doi: 10.1517/14656566.2013.787411. [DOI] [PubMed] [Google Scholar]
- 7.Rilpivirine First-line treatment of HIV infection: efavirenz is better documented. Prescrire Int. 2012;21:262–265. [PubMed] [Google Scholar]
- 8.Song Y, Fang Z, Zhan P, Liu X. Recent advances in the discovery and development of novel HIV-1 NNRTI platforms (Part II): 2009–2013 update. Curr Med Chem. 2014;21:329–355. doi: 10.2174/09298673113206660298. [DOI] [PubMed] [Google Scholar]
- 9.Zhan P, Pannecouque C, De Clercq E, Liu X. Anti-HIV drug discovery and development: current innovations and future trends. J Med Chem. 2016;59:2849–2878. doi: 10.1021/acs.jmedchem.5b00497. [DOI] [PubMed] [Google Scholar]
- 10.Steegen K, Bronze M, Papathanasopoulos MA, van Zyl G, Goedhals D, Variava E, MacLeod W, Sanne I, Stevens WS, Carmona S. HIV-1 antiretroviral drug resistance patterns in patients failing NNRTI-based treatment: results from a national survey in South Africa. J Antimicrob Chemother. 2017;72:210–219. doi: 10.1093/jac/dkw358. [DOI] [PubMed] [Google Scholar]
- 11.Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr. 2012;59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 12.Asahchop EL, Oliveira M, Wainberg MA, Brenner BG, Moisi D, Toni T, Tremblay CL. Characterization of the E138K resistance mutation in HIV-1 reverse transcriptase conferring susceptibility to etravirine in B and non-B HIV-1 subtypes. Antimicrob Agents Chemother. 2011;55:600–607. doi: 10.1128/AAC.01192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradshaw D, Mandalia S, Nelson M. How common is the non-nucleoside reverse transcriptase inhibitor mutation E138K in clinical practice? J Infect. 2011;63:172–173. doi: 10.1016/j.jinf.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Wainberg MA. Combination therapies, effectiveness, and adherence in patients with HIV infection: clinical utility of a single tablet of emtricitabine, rilpivirine, and tenofovir. HIV AIDS (Auckl) 2013;5:41–49. doi: 10.2147/HIV.S32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J Med Chem. 2010;53:4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]
- 16.Ragno R, Coluccia A, La Regina G, De Martino G, Piscitelli F, Lavecchia A, Novellino E, Bergamini A, Ciaprini C, Sinistro A, Maga G, Crespan E, Artico M, Silvestri R. Design, molecular modeling, synthesis, and anti-HIV-1 activity of new indolyl aryl sulfones. Novel derivatives of the indole-2-carboxamide. J Med Chem. 2006;49:3172–3184. doi: 10.1021/jm0512490. [DOI] [PubMed] [Google Scholar]
- 17.Ragno R, Artico M, De Martino G, La Regina G, Coluccia A, Di Pasquali A, Silvestri R. Docking and 3-d QSAR studies on indolyl aryl sulfones. Binding mode Exploration at the HIV-1 reverse transcriptase non-nucleoside binding site and design of highly active N-(2-Hydroxyethyl)carboxamide and N-(2-Hydroxyethyl)carbohydrazide derivatives. J Med Chem. 2005;48:213–223. doi: 10.1021/jm040854k. [DOI] [PubMed] [Google Scholar]
- 18.Tian Y, Du D, Rai D, Wang L, Liu H, Zhan P, De Clercq E, Pannecouque C, Liu X. Fused heterocyclic compounds bearing bridgehead nitrogen as potent HIV-1 NNRTIs. Part 1: design, synthesis and biological evaluation of novel 5,7-disubstituted pyrazolo[1,5-a]pyrimidine derivatives. Bioorg Med Chem. 2014;22:2052–2059. doi: 10.1016/j.bmc.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Tian Y, Chen W, Liu H, Zhan P, Li D, Liu H, De Clercq E, Pannecouque C, Liu X. Fused heterocycles bearing bridgehead nitrogen as potent HIV-1 NNRTIs. Part 2: discovery of novel [1,2,4]Triazolo[1,5-a]pyrimidines using a structure-guided core-refining approach. Eur J Med Chem. 2014;85:293–303. doi: 10.1016/j.ejmech.2014.07.104. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Chen W, Zhan P, De Clercq E, Pannecouque C, Liu X. Design, synthesis and anti-HIV evaluation of novel diarylnicotinamide derivatives (DANAs) targeting the entrance channel of the NNRTI binding pocket through structure-guided molecular hybridization. Eur J Med Chem. 2014;87:52–62. doi: 10.1016/j.ejmech.2014.09.054. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Zhan P, Daelemans D, Yang J, Huang B, De Clercq E, Pannecouque C, Liu X. Structural optimization of pyridine-type DAPY derivatives to exploit the tolerant regions of the NNRTI binding pocket. Eur J Med Chem. 2016;121:352–363. doi: 10.1016/j.ejmech.2016.05.054. [DOI] [PubMed] [Google Scholar]
- 22.Meng Q, Chen X, Kang D, Huang B, Li W, Zhan P, Daelemans D, De Clercq E, Pannecouque C, Liu X. Design, synthesis and evaluation of novel HIV-1 NNRTIs with dual structural conformations targeting the entrance channel of the NNRTI binding pocket. Eur J Med Chem. 2016;115:53–62. doi: 10.1016/j.ejmech.2016.02.068. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Chen W, Kang D, Lu X, Li X, Liu Z, Huang B, Daelemans D, Pannecouque C, De Clercq E, Zhan P, Liu X. Design, synthesis and anti-HIV evaluation of novel diarylpyridine derivatives targeting the entrance channel of NNRTI binding pocket. Eur J Med Chem. 2016;109:294–304. doi: 10.1016/j.ejmech.2015.11.039. [DOI] [PubMed] [Google Scholar]
- 24.Kang D, Fang Z, Li Z, Huang B, Zhang H, Lu X, Xu H, Zhou Z, Ding X, Daelemans D, De Clercq E, Pannecouque C, Zhan P, Liu X. Design, synthesis, and evaluation of thiophene[3,2-d]pyrimidine derivatives as HIV-1 non-nucleoside reverse transcriptase inhibitors with significantly improved drug resistance profiles. J Med Chem. 2016;59:7991–8007. doi: 10.1021/acs.jmedchem.6b00738. [DOI] [PubMed] [Google Scholar]
- 25.Fang Z, Kang D, Zhang L, Huang B, Liu H, Pannecouque C, De Clercq E, Zhan P, Liu X. Synthesis and biological evaluation of a series of 2-((1-substituted-1H-1,2,3-triazol-4-yl)methylthio)-6-(naphthalen-1-ylmethyl)pyri midin-4(3H)-one as potential HIV-1 inhibitors. Chem Biol Drug Des. 2015;86:614–618. doi: 10.1111/cbdd.12524. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Huang B, Liu X, Zhan P. Discovery of bioactive molecules from CuAAC click-chemistry-based combinatorial libraries. Drug Discov Today. 2016;21:118–132. doi: 10.1016/j.drudis.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Gao P, Sun L, Zhou J, Li X, Zhan P, Liu X. Discovery of novel anti-HIV agents via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) click chemistry-based approach. Expet Opin Drug Discov. 2016;11:857–871. doi: 10.1080/17460441.2016.1210125. [DOI] [PubMed] [Google Scholar]
- 28.Kang D, Zhang H, Zhou Z, Huang B, Naesens L, Zhan P, Liu X. First discovery of novel 3-hydroxy-quinazoline-2,4(1H,3H)-diones as specific anti-vaccinia and adenovirus agents via ‘privileged scaffold’ refining approach. Bioorg Med Chem Lett. 2016;26:5182–5186. doi: 10.1016/j.bmcl.2016.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 30.Pannecouque C, Daelemans D, De Clercq E. Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later. Nat Protoc. 2008;3:427–434. doi: 10.1038/nprot.2007.517. [DOI] [PubMed] [Google Scholar]
- 31.Kang D, Fang Z, Huang B, Lu X, Zhang H, Xu H, Huo Z, Zhou Z, Yu Z, Meng Q, Wu G, Ding X, Tian Y, Daelemans D, De Clercq E, Pannecouque C, Zhan P, Liu X. Structure-based optimization of thiophene[3,2-d]pyrimidine derivatives as potent HIV-1 non-nucleoside reverse transcriptase inhibitors with improved potency against resistance-associated variants. J Med Chem. 2017;60:4424–4443. doi: 10.1021/acs.jmedchem.7b00332. [DOI] [PubMed] [Google Scholar]
- 32.Liu N, Wei L, Huang L, Yu F, Zheng W, Qin B, Zhu DQ, Morris-Natschke SL, Jiang S, Chen CH, Lee KH, Xie L. Novel HIV-1 non-nucleoside reverse transcriptase inhibitor agents: optimization of diarylanilines with high potency against wild-type and rilpivirine-resistant E138K mutant virus. J Med Chem. 2016;59:3689–3704. doi: 10.1021/acs.jmedchem.5b01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chander S, Ashok P, Singh A, Murugesan S. De-novo design, synthesis and evaluation of novel 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline derivatives as HIV-1 reverse transcriptase inhibitors. Chem Cent J. 2015;9:33. doi: 10.1186/s13065-015-0111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki K, Craddock BP, Okamoto N, Kano T, Steigbigel RT. Poly A-linked colorimetric microtiter plate assay for HIV reverse transcriptase. J Virol Meth. 1993;44:189–198. doi: 10.1016/0166-0934(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Zhan P, Li D, De Clercq E, Liu X. Recent advances in DAPYs and related analogues as HIV-1 NNRTIs. Curr Med Chem. 2011;18:359–376. doi: 10.2174/092986711794839142. [DOI] [PubMed] [Google Scholar]
- 36.Li D, Zhan P, De Clercq E, Liu X. Strategies for the design of HIV-1 non-nucleoside reverse transcriptase inhibitors: lessons from the development of seven representative paradigms. J Med Chem. 2012;55:3595–3613. doi: 10.1021/jm200990c. [DOI] [PubMed] [Google Scholar]
- 37.Zhan P, Chen X, Li D, Fang Z, De Clercq E, Liu X. HIV-1 NNRTIs: structural diversity, pharmacophore similarity, and implications for drug design. Med Res Rev. 2013;33(Suppl 1):E1–E72. doi: 10.1002/med.20241. [DOI] [PubMed] [Google Scholar]
- 38.Ahgren C, Backro K, Bell FW, Cantrell AS, Clemens M, Colacino JM, Deeter JB, Engelhardt JA, Hogberg M, Jaskunas SR, et al. The PETT series, a new class of potent nonnucleoside inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:1329–1335. doi: 10.1128/aac.39.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren J, Diprose J, Warren J, Esnouf RM, Bird LE, Ikemizu S, Slater M, Milton J, Balzarini J, Stuart DI, Stammers DK. Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses. J Biol Chem. 2000;275:5633–5639. doi: 10.1074/jbc.275.8.5633. [DOI] [PubMed] [Google Scholar]
- 40.Ren J, Nichols CE, Stamp A, Chamberlain PP, Ferris R, Weaver KL, Short SA, Stammers DK. Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138. FEBS J. 2006;273:3850–3860. doi: 10.1111/j.1742-4658.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- 41.Torti C, Pozniak A, Nelson M, Hertogs K, Gazzard BG. Distribution of K103N and/or Y181C HIV-1 mutations by exposure to zidovudine and non-nucleoside reverse transcriptase inhibitors. J Antimicrob Chemother. 2001;48:113–116. doi: 10.1093/jac/48.1.113. [DOI] [PubMed] [Google Scholar]
- 42.Tantillo C, Ding J, Jacobo-Molina A, Nanni RG, Boyer PL, Hughes SH, Pauwels R, Andries K, Janssen PA, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 43.Dang Z, Lai W, Qian K, Ho P, Lee KH, Chen CH, Huang L. Betulinic acid derivatives as human immunodeficiency virus type 2 (HIV-2) inhibitors. J Med Chem. 2009;52:7887–7891. doi: 10.1021/jm9004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J Med Chem. 2010;53:4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]