Abstract
Objective
Obesity and type 2 diabetes (T2D) arise from the interplay between genetic, epigenetic, and environmental factors. The aim of this study was to combine bioinformatics and functional studies to identify miRNAs that contribute to obesity and T2D.
Methods
A computational framework (miR-QTL-Scan) was applied by combining QTL, miRNA prediction, and transcriptomics in order to enhance the power for the discovery of miRNAs as regulative elements. Expression of several miRNAs was analyzed in human adipose tissue and blood cells and miR-31 was manipulated in a human fat cell line.
Results
In 17 partially overlapping QTL for obesity and T2D 170 miRNAs were identified. Four miRNAs (miR-15b, miR-30b, miR-31, miR-744) were recognized in gWAT (gonadal white adipose tissue) and six (miR-491, miR-455, miR-423-5p, miR-132-3p, miR-365-3p, miR-30b) in BAT (brown adipose tissue). To provide direct functional evidence for the achievement of the miR-QTL-Scan, miR-31 located in the obesity QTL Nob6 was experimentally analyzed. Its expression was higher in gWAT of obese and diabetic mice and humans than of lean controls. Accordingly, 10 potential target genes involved in insulin signaling and adipogenesis were suppressed. Manipulation of miR-31 in human SGBS adipocytes affected the expression of GLUT4, PPARγ, IRS1, and ACACA. In human peripheral blood mononuclear cells (PBMC) miR-15b levels were correlated to baseline blood glucose concentrations and might be an indicator for diabetes.
Conclusion
Thus, miR-QTL-Scan allowed the identification of novel miRNAs relevant for obesity and T2D.
Keywords: QTL, Computational biology, Insulin signalling, miR-31, Adipogenesis, Type 2 diabetes
Highlights
-
•
A computational biology approach identified ten miRNAs relevant for obesity/T2D.
-
•
miR-31 is upregulated in adipose tissue of obese and diabetic subjects.
-
•
miR-31 targets genes of the insulin signaling pathway and adipogenesis.
1. Introduction
The excessive accumulation of body fat, obesity is a lifestyle driven and genetically heritable disorder, as well as a major risk factor for secondary diseases like type 2 diabetes (T2D) [1]. A classical and powerful tool for the investigation of the genetic architecture of these polygenic diseases is genome-wide association studies (GWAS) [2]. However, the challenge of GWAS is the requirement of a large number of participants due to the heterogeneity of the human genome and the influence of environmental factors [3], [4]. Another approach for the identification of disease-related genes is to perform linkage studies with mouse inbred strains that differ in their susceptibility for the disease leading to the detection of quantitative trait loci (QTL) harboring genetic variants [5]. The subsequent breeding and characterization of recombinant congenic lines is needed for the identification of gene variants by positional cloning [6].
Nevertheless, not only variants within genes but also single nucleotide polymorphisms (SNP), insertions and deletions (indels) in promoter or enhancer regions could be responsible for a QTL. To identify these alterations, the concept of expression QTL (eQTL) has been established, which links a genetic variant with the expression of a specific gene [7]. However, different studies have already described the existence of genes related to polygenic diseases, which are located outside of QTL and eQTL regions [8]. Those genes must be regulated by trans elements like transcription factors or non-coding RNAs such as miRNAs that are located within a QTL.
miRNAs are small non-coding RNAs with a length of 19–24 nucleotides which are known to bind to the 3′UTR region of specific mRNAs leading to an altered expression or translation of the corresponding target gene(s) [9]. Most miRNAs have hundreds of targets involved in different signaling cascades. Thus, they are potent regulators and suitable candidates in polygenic diseases like obesity or T2D [10], [11]. For example, it was demonstrated that miR-107 and miR-103 play an important role in insulin sensitivity [12] and that miR-375 is involved in the regulation of alpha- and beta-cell mass in pancreatic islets [13] and may be a novel pharmacological target for the treatment of diabetes [14].
For the discovery of genes involved in obesity and T2D, we generated a backcross population derived from obese diabetes-susceptible New Zealand Obese (NZO) and lean diabetes-resistant C57BL/6 (B6) mice and performed a genome-wide linkage study for approximately 20 metabolic traits. Overall, we detected 17 partially overlapping loci on 11 chromosomes and were able to identify genetic variants in Ifi202b [15] and Lefty1 [16] on chromosome 1 and Zfp69 [17] on chromosome 4. The aim of the current study was to develop a comprehensive and quantitative in silico approach (miR-QTL-Scan), which allows the identification of miRNAs within QTL that mediate the differential expression of targets in metabolically relevant tissues and to verify their role in humans. To provide direct functional evidence for the achievement of the bioinformatics approach one candidate miRNA, miR-31 was investigated in human and mouse tissues and manipulated in vitro.
2. Materials and methods
2.1. Genome-wide linkage study
In a collaboration within the DZD network, a large linkage study of diabetes-susceptible NZO and diabetes-resistant B6 mice was performed to identify genes related to metabolic diseases. In the current study we took advantage of this linkage study (data not published) and used the identified loci of more than 300 N2 backcross male mice. The linkage analysis (1-QTL scan) was performed with the R-package R/qtl version - 1.40–8 using the EM-algorithm, 1,000 permutations (non parametric), and 141 SNPs leading to a median distance of 17.4 Mbp. The threshold for a significant logarithm of the odds score (LOD) was set to ≥3.0 as described by Lander et al. for N2 backcross populations [18]. The 2-QTL analysis was performed under the same settings.
2.2. Array-based transcriptomics of parental mice
Total RNA was isolated from murine tissue using TRIzol™ reagent (Invitrogen, Germany). From parental NZO and B6 mice (6 weeks of age), samples of gonadal white adipose tissue (gWAT), brown adipose tissue (BAT), and skeletal muscle were aliquoted for RNA extraction. The procedure was carried out according to manufacturer's instructions. RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Germany), and the manufacturer's instructions were followed to measure RNA integrity (RIN). Samples with RIN values >8 were subsequently selected for microarray analysis. Microarray analysis of NZO and B6 (n = 4/strain) was performed by OakLabs GmbH (Germany) using a SurePrint G3 Mouse GE 8 × 60 k chip (Agilent Technologies). Before analyzing the fold change and calculating p-values, the samples were quantile normalized using the “preprocessCore” R-package, version 1.36. P-values of transcriptomics were calculated using a two-tailed Students t-test with a threshold of p ≤ 0.05 with the “stats” R-package version 3.1.1. The threshold for a significant expression difference between groups was a log2 fold change of |0.7|. The results of the array analysis are available via accession ID: GSE111142 on GEO.
2.3. Identification of putative regulatory elements of miRNAs
Putative regulatory elements located 50 kb–4 kb upstream of miRNA loci were mapped by using the histone modification publicly available in ENCODE. Enhancers were characterized by genomic regions highly enriched with H3K27ac and H3K4me1, promoters retained H3K27ac and include H3K4me3.
2.4. Use of the online tool https://146.107.176.32/miR-QTL-Scan/
The tool can be used by several web browsers (e.g. Firefox, Chrome, InternetExplorer). Please press “continue loading this website” and follow the description provided online and in Fig. S1.
2.5. miRNA isolation and quantitative PCR (qPCR) in mice
Small RNA was isolated from 30 mg gWAT and 5 mg liver using the PureLink miRNA Kit (Invitrogen) according to the manufacturer's protocol. miR-31 (000185, Thermo Fisher Scientific, Germany), miR-31* (002495, Thermo Fisher Scientific), and snoRNA202 (001232, Thermo Fisher Scientific) were reverse transcribed individually using 30 ng for gWAT and 150 ng miRNA for liver with the TaqMan miRNA Reverse Transcription Kit according to vendor's instructions (Thermo Fisher Scientific). All qPCRs were performed with the Roche LightCycler® 480/384 (Roche, Switzerland). Relative expression of miRNA levels was evaluated using snoRNA202 as endogenous control.
2.6. SGBS cell culture
All cell culture experiments were performed in at least three independent experiments. Simpson-Golabi-Behmel syndrome (SGBS) cells were cultured as described [19]. To induce adipogenic differentiation, cells were washed once with PBS and serum-free DMEM-F12 supplemented with 10 μg/ml transferrin, 20 nM insulin, 100 nM cortisol, 200 pM T3, 25 nM dexamethasone, 250 μM IBMX, and 2 μM rosiglitazone was added [20]. After 4 days, medium was changed and serum-free DMEM-F12 supplemented with 10 μg/ml transferrin, 20 nM insulin, 100 nM cortisol, and 200 pM T3 was added. The differentiation rate was determined by counting the number of lipid-laden, differentiated adipocytes (defined by five clearly visible lipid droplets) and undifferentiated cells. Three microscopic fields were counted per well using a net micrometer. The triglyceride content was measured using Triglyceride Reagent, Free Glycerol Reagent, and a Glycerol Standard (Sigma–Aldrich, Germany) according to the manufacturer's instructions.
2.7. miRNA mimic transfection in SGBS cells
Transfection was performed as previously described [21]. In brief, SGBS cells were transfected with 20 nM miR-31-5p mimic (Syn-hsa-miR-31-5p, Qiagen, Germany; 5′AGGCAAGAUGCUGGCAUAGCU) or non-target control (AllStars Negative Control siRNA, Qiagen) and 0.66 μl/cm2 Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Lipofectamine/RNA complexes were added drop wise to the cells without a change of media one day after seeding the cells. Adipogenic differentiation was induced two days later as described above.
2.8. RNA isolation in SGBS cells
Total RNA and miRNA isolation was performed with the Direct-zol RNA mini Prep Kit (Zymo Research, Germany) on day 0 and day 14 of adipogenic differentiation according to manufacturer's instructions.
2.9. Reverse transcription and quantitative PCR (qPCR) in SGBS cells
For miRNA quantification, miRNA was reverse transcribed using the miScript II RT Kit (Qiagen) and analyzed by qPCR using the miScript SYBR Green PCR Kit and the miScript primer assay for hsa-miR-31*_1 (Qiagen). Results were normalized to SNORD68_11 (sno68) (Qiagen). To investigate mRNA expression, total RNA was reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen). qPCRs were performed with the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Germany). Results were normalized to HPRT. All qPCR experiments were performed with a Bio-Rad CFX Connect System (Bio-Rad). Primer sequences are available upon request.
2.10. Human participants and whole blood collection
A random sample of 89 German individuals was recruited as study population from the ongoing Tübingen Family Study (TÜF) [22]. The TÜF study currently comprises >3000 non-related individuals at increased risk for T2D, i.e. non-diabetic subjects with family history of T2D, BMI ≥ 27 kg/m2, impaired fasting glycaemia (IGT), and/or previous gestational diabetes. All TÜF participants undergo assessment of medical history, smoking status and alcohol consumption habits, physical examination, routine blood tests, and oral glucose tolerance tests (oGTTs). The study population assessed here consisted of individuals with complete oGTT and documented absence of medication known to influence glucose tolerance, insulin sensitivity or insulin secretion. The study adhered to the Declaration of Helsinki, and each participant signed a written informed consent. The Ethical Committee of the Medical Faculty of the University of Tübingen approved the study protocol. Subjects were grouped into normal glucose tolerant subjects with BMI < 27 (NGT.BMI < 27, n = 25) or BMI>30 (NGT.BMI>30, n = 16) and subjects with impaired glucose tolerance with BMI < 27 (IGT.BMI < 27, n = 20) or BMI > 30 (IGT.BMI > 30, n = 28). A standardized 75-g oGTT was performed following a 10-h overnight fast. For the determination of plasma glucose, venous blood samples were drawn at baseline and at time-points 30, 60, 90, and 120 min of the oGTT.
2.11. miRNA in human adipose tissue and in human PBMC
miRNAs were isolated from visceral and subcutaneous adipose tissue with the miRNeasy Mini Kit (Qiagen). For miRNA analysis in human PBMC, whole blood was collected in PAXgene Blood RNA Tubes (PreanalytiX) after overnight fasting. Total RNA was isolated using the Paxgene miRNA kit (PreanalytiX) according to manufacturer's specifications. cDNA synthesis was performed by using the miScript II RT Kit (Qiagen). qPCRs were performed with the miScript SYBR Green PCR Kit (Qiagen) and measured with the 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) or a LightCycler 450 (Roche). QPCR measurements were conducted in duplicates. For relative expression levels of all miRNAs, hsa-miR-92a-3p was used as control.
2.12. Statistics
Plotting of all data was performed by using GraphPad Prism, version 7.0 (USA), if not mentioned otherwise. p-values were calculated by two-tailed Student's t-test or by one-way ANOVA in the case of comparing more than two groups. qPCR expression data were evaluated by the 2−ΔCT method [23]. Circos plots were created with R-package RCircos version 1.2.0 [24]. The two-dimensional genome scan was generated with the R-package R/qtl version - 1.40–8. Statistics for target prediction was performed by a 4-field-chi-square-test. The data were divided into four groups: Putative targets/non-putative target genes and differentially expressed/not differentially expressed genes. The maximum number of genes within the mouse genome was set to 30,000. Statistical analyses of array results are described in 2.2.
2.13. Gene enrichment in pathway analysis
The pathway enrichment analysis of the predicted miRNA target genes was done for KEGG pathways and GO-Terms using DAVID, version 6.7 [25]. The cutoff enrichment score was set above 3 and p-value below 0.1.
3. Results
3.1. miR-QTL-scan, a framework for the discovery of miRNA within quantitative trait loci
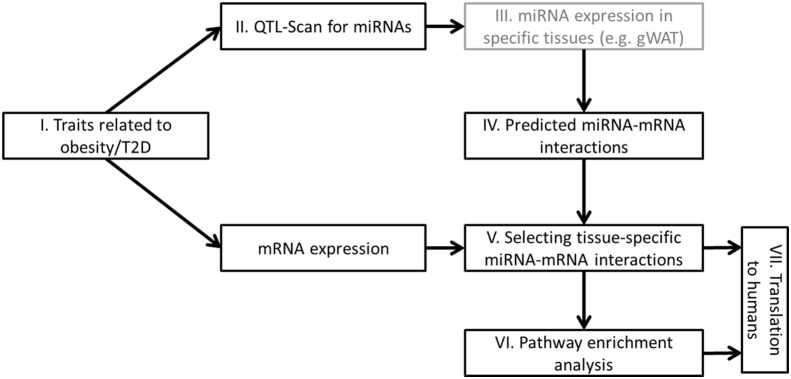
In order to identify miRNAs within QTL that negatively regulate the expression of considerable genes and their participation in obesity and/or T2D, miR-QTL-Scan, an extensive framework was developed that uses different information layers (Figure 1).
Figure 1.
Workflow of the miR-QTL-Scan for the identification of miRNAs and the putative targets genes in obesity- and diabetes-related QTL. Numbers refer to working steps. Step III is optional and used to identify tissue-specific miRNAs. gWAT: gonadal white adipose tissue.
The linkage study of the NZO and B6 cross identified 17 partially overlapping QTL on 11 chromosomes. Ten partially overlapping loci on chromosomes 1, 3, 4, 11, 13, 14, 15, and 17 were linked to traits of obesity (body weight week 6 and week 14, fat mass, adipose tissue weight, triglycerides, and free fatty acids) and seven on chromosomes 1, 4, 9, 11, 13, 18, and 19 to parameters related to T2D (blood glucose, plasma and total pancreatic insulin, and liver triglycerides). For the identification of miRNAs that are located in the peak regions of these QTL, we used MiRBase [27] and found 138 miRNAs to be located in the obesity-related QTL and 111 in the diabetes-related QTL; 79 were detected in both.
3.2. Development of a target prediction tool
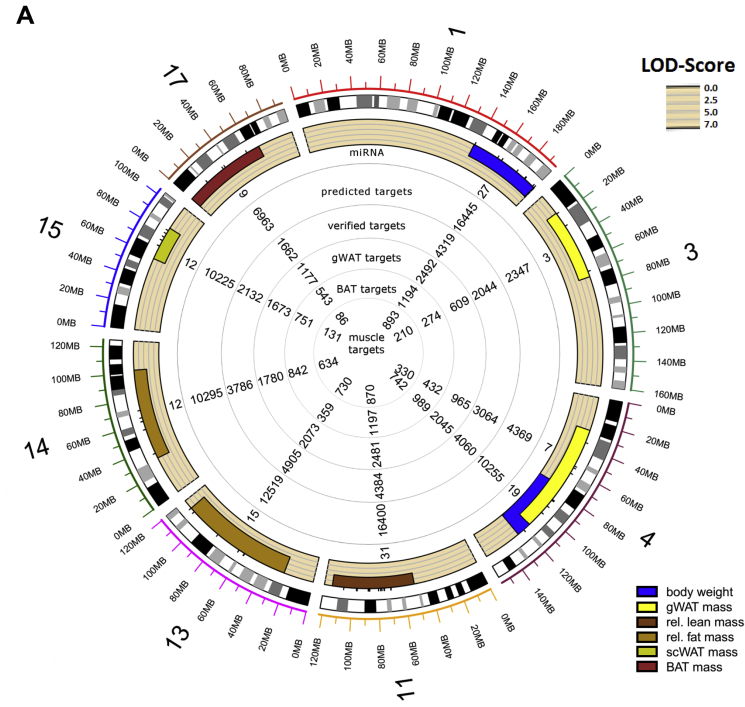
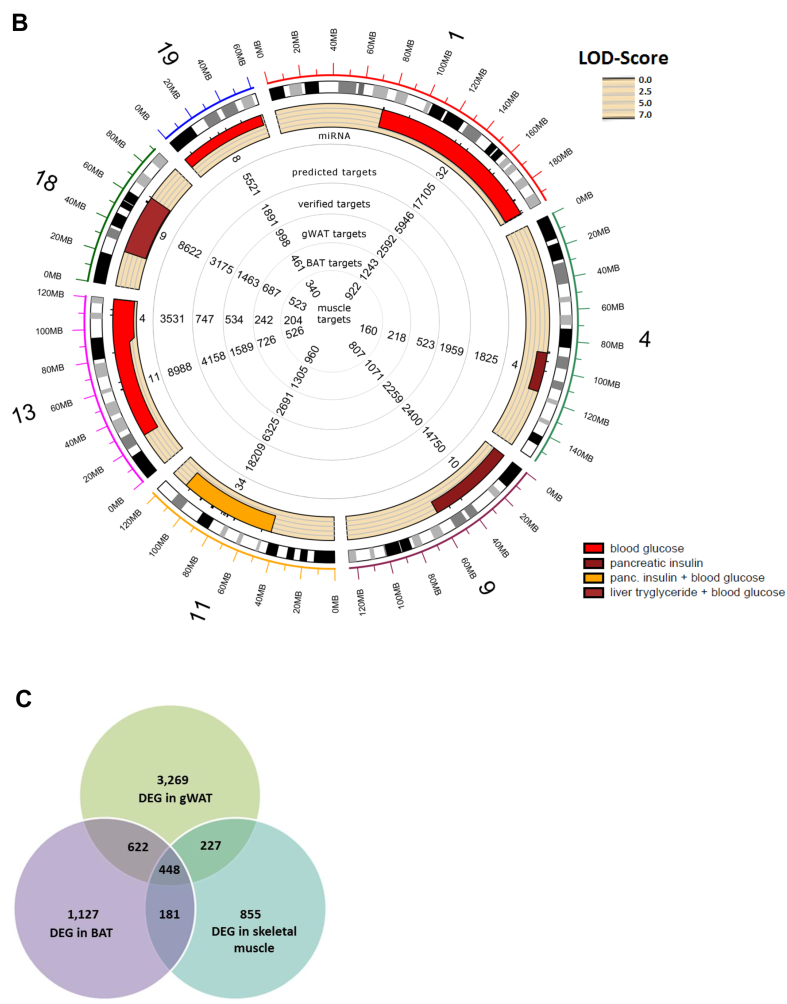
In order to predict reliable targets for the miRNAs that are located in the QTL (Figure 1 IV and V), five prediction tools, DIANA-microT [28], miRDB [29], TarPmiR [30], TargetScan7.1 [31], and RNA22 [32] were applied on the first level. An optimal selection of miRNA-specific targets was given when at least three tools showed overlapping results. As shown in Figure 2A and B, each QTL contains 3 to 34 miRNAs which theoretically target between 1,825 and 18,209 transcripts.
Figure 2.
Overview of miRNAs located in QTL for obesity and T2D and differently expressed target genes. Circos plot of miRNAs located in QTL for obesity (A) and T2D (B), predicted targets, targets in experimental databases, and putative differentially expressed targets in gWAT, BAT, and muscle. (C) Number of differentially expressed genes (DEG) in BAT, WAT, and skeletal muscle of B6 and NZO mice. gWAT: gonadal white adipose tissue; scWAT: subcutaneous white adipose tissue; BAT: brown adipose tissue; DEG: differently expressed genes.
The second level of target prediction (Figure 1 IV.) was based on three databases (DIANA-TarBase, miRecords, and miRTarBase [33], [34], [35]), which list experimentally validated miRNA-mRNA interactions (detected by HITS-CLIP [36], luciferase reporter or in vitro assays), to estimate the quality of the first level and to add already known interactions. The scan for interactions within these databases demonstrated that the 3 to 34 QTL-specific miRNAs target between 747 and 6,325 transcripts, covering 15–20% of the putative targets as shown in the circos plots in Figure 2A and B.
On the third level of target prediction, we performed expression profiling of metabolically active tissues (gWAT, BAT, and skeletal muscle) from the parental B6 and NZO mice that were crossed for the QTL analysis in order to further specify putative target genes (Figure 1 V.). Our array-based transcriptomics data (GEO accession ID: GSE111142) revealed a total number of 4,566 differentially expressed genes in gWAT, 2,378 in BAT, and 1,711 in skeletal muscle (Figure 2C). These differentially expressed genes were compared with the list of putative miRNA target genes resulting in a marked reduction of the number of interactions per miRNA to a median of 117 in gWAT, 53 in BAT, and 39 in skeletal muscle. The entire target gene sets of all 170 QTL-specific miRNAs for different tissues (gWAT, BAT, and skeletal muscle) are available as part of the online tool: https://146.107.176.32/miR-QTL-Scan/.
3.3. Selection of miRNAs expressed in metabolically relevant tissues
In order to focus on miRNAs that are expressed in the above listed tissues, we took advantage of the study described by Güller et al. providing data of miRNAs expressed in gWAT, BAT, and skeletal muscle from B6 mice which were one of the breeding partners used for our linkage analysis [26] (Figure 1 III.). As the traits involving fat mass and weight of fat depots can be linked to elevated body weight, we focused on miRNAs that are expressed in gWAT and BAT. For gWAT, four miRNAs (miR-15b, miR-31, miR-744, and miR-30b) with 1,525 putative interactions [37], [38], [39], [40] and for BAT six miRNAs (miR-491, miR-455, miR-423-5p, miR-132-3p, miR-365-3p, and miR-30b) with 1,188 targets [41], [42], [43], [44], [45] appear to play a role for the pathogenesis of obesity (Table 1).
Table 1.
List of miRNAs specific for gWAT and BAT in QTL, selected putative target genes and genetic variants in regulative elements identified by histone marks. Chromosomes, target genes, and traits linked to obesity and T2D are shown. Targets with experimental evidence by HITS-CLIP (*), in vitro assay (**), or luciferase assay (***). gWAT: gonadal white adipose tissue; scWAT: subcutaneous white adipose tissue; BAT: brown adipose tissue; NAFLD: Non-alcoholic fatty liver disease; BW: body weight; NASH: non-alcoholic steatohepatitis; DEG: differently expressed gene.
| miRNA | Chr. | Putative targets | Traits | Related | Target genes | Genetic variants |
Genomic organisation | Host gene DEG | |
|---|---|---|---|---|---|---|---|---|---|
| Promoter | Enhancer | ||||||||
| gWAT-specific miRNAs | |||||||||
| miR-15b | 3 | 402 | gWAT weight | NAFLD [37] | Prkag2 | – | 2 | host gene (Smc4) | No |
| miR-31 | 4 | 416 | BW & gWAT weight | diabetes [38] | Foxo1*, Slc2a4***, Prkaa1* | – | 2 | host gene (LOC106557447) | No |
| miR-744 | 11 | 528 | lean mass | NASH [39] | Lep, Il13, Il17b | – | 4 | host gene (Map2k4) | No |
| miR-30b | 15 | 179 | scWAT weight | beige fat & browning [40] | Socs3*, Tnf*, Pik3cd*, Ucp1*** | – | 10 | miRNA cluster | – |
| BAT-specific miRNAs | |||||||||
| miR-491 | 4 | 132 | BW & lean mass | – | – | – | 1 | host gene (Focad) | No |
| miR-455 | 4 | 291 | BW & lean mass | enhanced adipogenesis [41] | Irs3, mTor | – | 11 | host gene (Col27a1) | Yes |
| miR-423-5p | 11 | 401 | BAT weight | hepatic gluconeogenesis [72] | Hk3, Il17rb, Il18r1 | – | – | host gene (Nsrp1) | No |
| miR-132-3p | 11 | 72 | BAT weight | down regulated in obesity [43] | Map3k12, Sos1 | 7 | 37 | miRNA cluster | – |
| miR-365-3p | 11 | 113 | BAT weight | brown adipocyte differentiation [44] | – | – | – | intergenic | – |
| miR-30b | 15 | 179 | scWAT weight | beige fat & browning [40] | Socs3*, Tnf*, Pik3cd*, Ucp1*** | – | 12 | miRNA cluster | – |
Further enrichment analysis of target genes from gWAT (Figure 1 VI.) was performed with KEGG [46] and GO-Terms [47] and revealed an involvement of miR-31 in insulin signaling miscellaneous metabolic pathways. Pathway analysis of all QTL-specific miRNAs with their corresponding targets in different tissues (gWAT, BAT, and skeletal muscle) is available in our online tool which is connected to the DAVID website [25].
3.4. Polymorphic regulatory elements of miRNAs located in QTL
In order to clarify if cis-regulatory elements of the identified miRNAs carry genetic variants, the public available histone modification marks from Encode [48], [49] were used. For eight of the ten candidates several genetic variants were detected in putative enhancer regions (Table 1). Interestingly, six miRNAs were intragenic and located within a host gene but no genetic variants overlapping with histone modifications were detected for these candidates. However, Col27a1 the host gene of miR-455 was lower abundant in BAT of NZO mice in comparison to B6 mice (p = 0.026).
3.5. Evaluation of miR-31 as a relevant regulator in adipose tissue
The aim of the second part of our study was to prove that the strategy of the computational framework (miR-QTL-Scan) introduced above leads to the determination of metabolic relevant miRNAs and is indeed sensitive and specific for the identification of significant miRNA–mRNA relationships. As shown in Table 1, some of the detected miRNAs showed an involvement in metabolic diseases. We selected miR-31 as a candidate for further analysis as (1) it is located in a very prominent obesity QTL (Nob6; LOD-score for body weight in week 6 4.0; LOD-score for body weight in week 14 5.3; LOD score for gWAT weight 3.9; Figure 3A), (2) little is known about miR-31 in respect to body weight and insulin sensitivity, and (3) the pathway enrichment analysis of the predicted targets indicated that several genes were relevant for insulin signaling. Interestingly, miR-31 was described to be elevated in serum of T2D patients [38]. However, it has not been investigated in respect to metabolic dysfunction before.
Figure 3.
Position of miR-31 in the obesity QTL Nob6 on chromosome 4, the predicted and validated targets and differentially expressed genes in gWAT of B6 and NZO mice. (A) Peak regions of the QTL for body weight (BW) in week 6/14 and weight of gWAT. Peak of LOD-score is added to the bottom axis. (B) Comparison of differently expressed genes in gWAT with predicted targets and experimentally validated targets of miR-31. (C) Pathway enrichment analysis for the 416 target genes of miR-31 in gWAT, filtered for a fold enrichment >3 and p < 0.1. (D) Heat map for a two-dimensional genome scan with a two-QTL model for the NZOxB6 cross. The maximum LOD score for the interaction model (locus to locus interaction) in the upper left triangle. The maximum LOD score for the full model (two QTLs plus additive effect) is indicated in the lower right triangle. Black squares show an interaction between miR-31 and predicted genes (Foxo1, Hk2, Irs1, Glut4, Acaca), LOD-score >6 (dark red). gWAT: gonadal white adipose tissue.
miRNAs are known to be important regulators of adipose tissue functions [50]. Experimental evidence emphasizes that the adipose tissue is a major depot for miRNAs that are released from adipocytes into the blood via exosomes [51]. Therefore, we focused on the expression data and predicted targets of gWAT. As shown in the Venn diagram of Figure 3B, 260 differentially expressed genes were predicted by at least three tools as miR-31 targets. Among these genes, 52 were experimentally validated according to the databases, 208 were only predicted (Figure 3B). However, 156 differentially expressed genes with experimental evidence were not on the list of predicted genes but were included in further analysis.
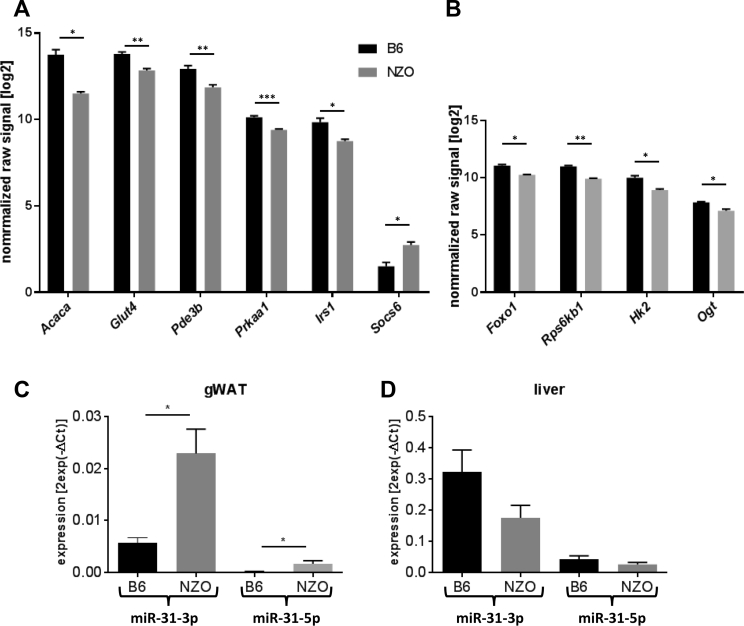
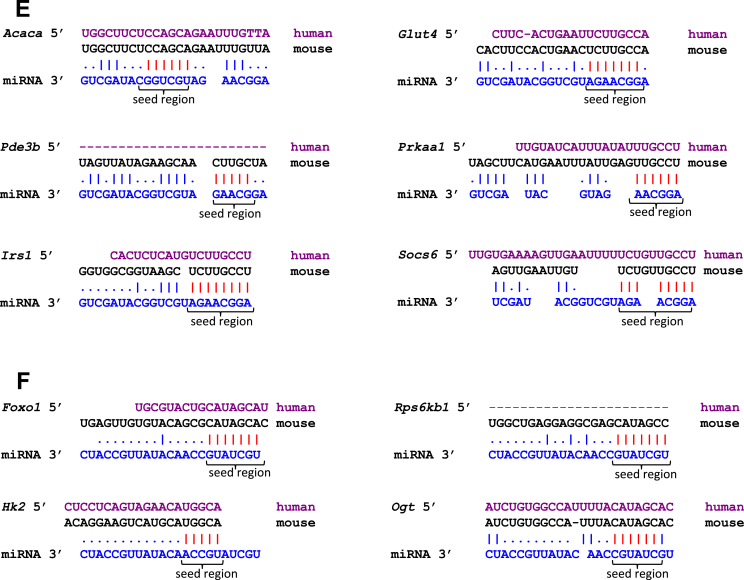
Taking into consideration the predicted and experimentally validated genes, miR-31 targets 416 genes in adipose tissue (Figure 3B) (Pearson's chi-square test p = 7.6e-14), which were used for pathway enrichment analysis [46], [47]. Ten genes showed a highly significant enrichment in the insulin signaling pathway (Acaca, Prkaa1, Rps6kb1, Glut4, Irs1, Pde3b, Hk2, Foxo1, Ogt, and Socs6) (p = 0.003; Figure 3C and Table S1) and were therefore evaluated further. Results of the transcriptome analysis revealed that nine of these genes exhibit a lower expression in gWAT of NZO in comparison to B6 mice (Figure 4A and B). Only Socs6 showed the opposite effect with an elevated expression in NZO gWAT (Figure 4A). This together with the results of qRT-PCR analysis demonstrating that miR-31-3p and miR-31-5p were significantly higher expressed in gWAT of NZO than in B6 mice supports the hypothesis that these genes are targets of miR-31 (Figure 4C). The action of miR-31 seemed to be specific for the adipose tissue as miR-31-3p and miR-31-5p were not differentially expressed in the liver of the two mouse strains (Figure 4D). Figure 4E and F shows the putative seed regions of the indicated target genes. Acaca, Prkaa1, Pde3b, Glut4, Irs1 and Hk2, Foxo1, Rps6kb1, and Ogt appear to be direct targets of miR-31-5p and miR-31-3p, respectively. The alignment also demonstrates that the seed regions of the targets except of Pde3b and Rps6kb1 are conserved between mouse and human.
Figure 4.
Expression of miR-31 target genes in gWAT and tissue-specific expression pattern of miR-31 in B6 and NZO mice. Differential expression of (A) miR-31-5p and (B) miR-31-3p target genes involved in insulin signaling pathway in gWAT detected by array analysis. (n = 4/group). (C) Expression of miR-31-5p and miR-31-3p in gWAT and (D) in liver of 6 weeks old B6 (n = 5) and NZO (n = 6) mice. (*p < 0.05, **p < 0.001, ***p < 0.0001). Data are presented as ± SEM. gWAT: gonadal white adipose tissue. (E) Partial sequences of the putative targets (human in magenta, mouse in black, miRNA in blue) of miR-31-5p and (F) of miR-31-3p. The seed regions of the targets are indicated as red dashes.
Using the two-dimensional genome scan with a two-QTL model, we investigated all interactions possible between miR-31 and the corresponding predicted genes listed in insulin signaling pathway. The upper left triangle of the two-dimensional genome scan depicts the weak interactions between loci with LOD-score <2. However, the lower right part of the heat map illustrates additive LOD-scores >6 between miR-31 and Foxo1, Glut4, Irs1, and Acaca (Figure 3D and Fig. S2). Taken together, our data suggest that downregulation of these genes is caused by elevated level of miR-31 in diabetes-susceptible strains.
3.6. miR-31 expression in adipose tissue of obese human subjects
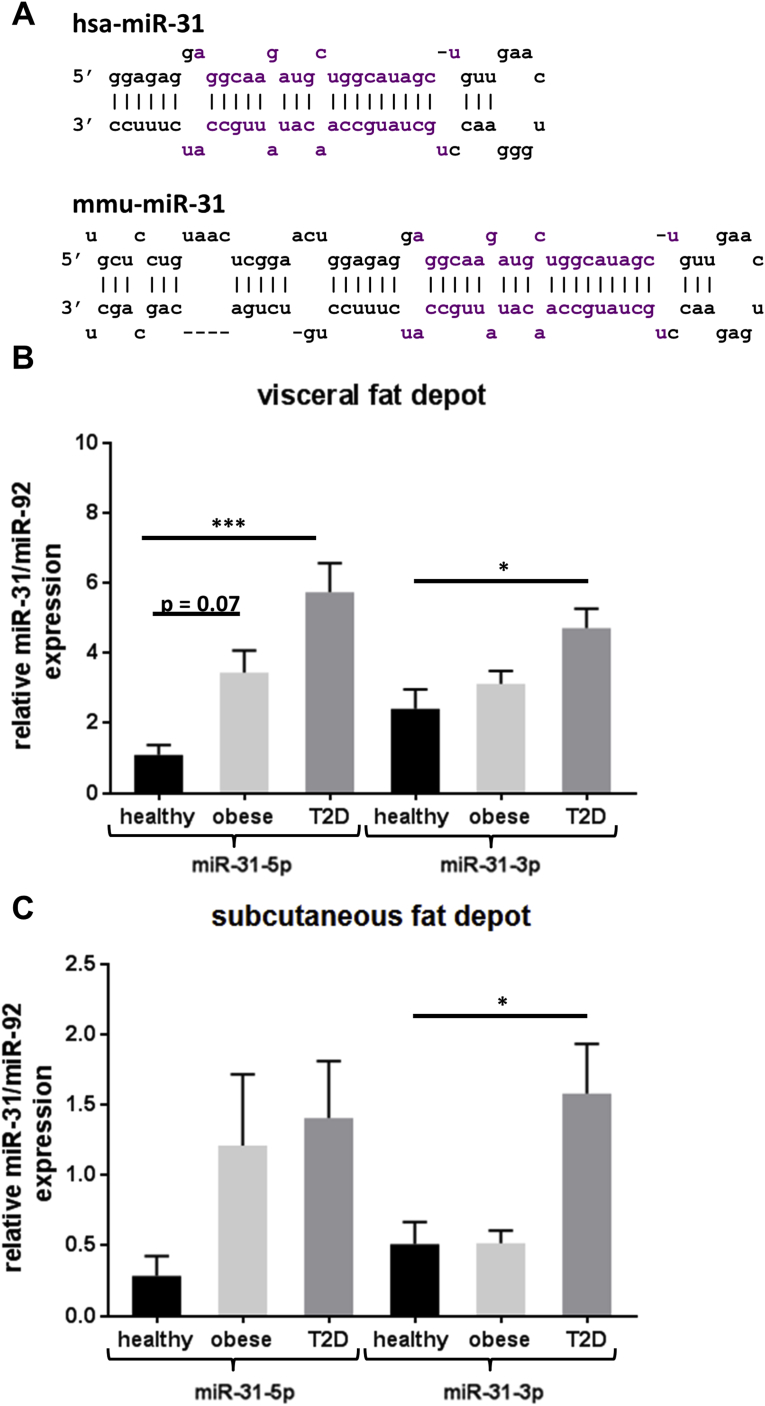
To test whether miR-31 is also affected in human obesity, we first investigated the sequence similarity between mmu-miR-31 and hsa-miR-31, which are located on chromosomes 4 and 9, respectively (Fig. S3, Figure 5A), using MiRBase 21 [27]. The mature sequences are identical suggesting that they equally target at least these genes in mice and human which are highly conserved. As shown in Figure 5B, expression of miR-31-5p and miR-31-3p was significantly higher in visceral adipose tissues of obese (p = 0.069) and T2D patients (p = 2.10-4) than in healthy subjects. In subcutaneous white adipose tissue (scWAT), only a higher expression of miR-31-5p was found in T2D patients (Figure 5C) (p = 0.017).
Figure 5.
Expression of miR-31-5p and miR-31-3p in human adipose tissue. (A) miR-31 sequence similarity between humans and mice. (B) miR-31-5p and miR-31-3p expression levels in visceral and (C) subcutaneous adipose tissue of healthy (n = 15–17), obese (n = 34–37), and diabetic (T2D, n = 33–35) subjects analyzed by qRT-PCR. Obese subjects had a BMI >30. (*p < 0.05, ***p < 0.0001). Data are presented as ± SEM.
3.7. Identification of miR-31 target genes in human SGBS cells
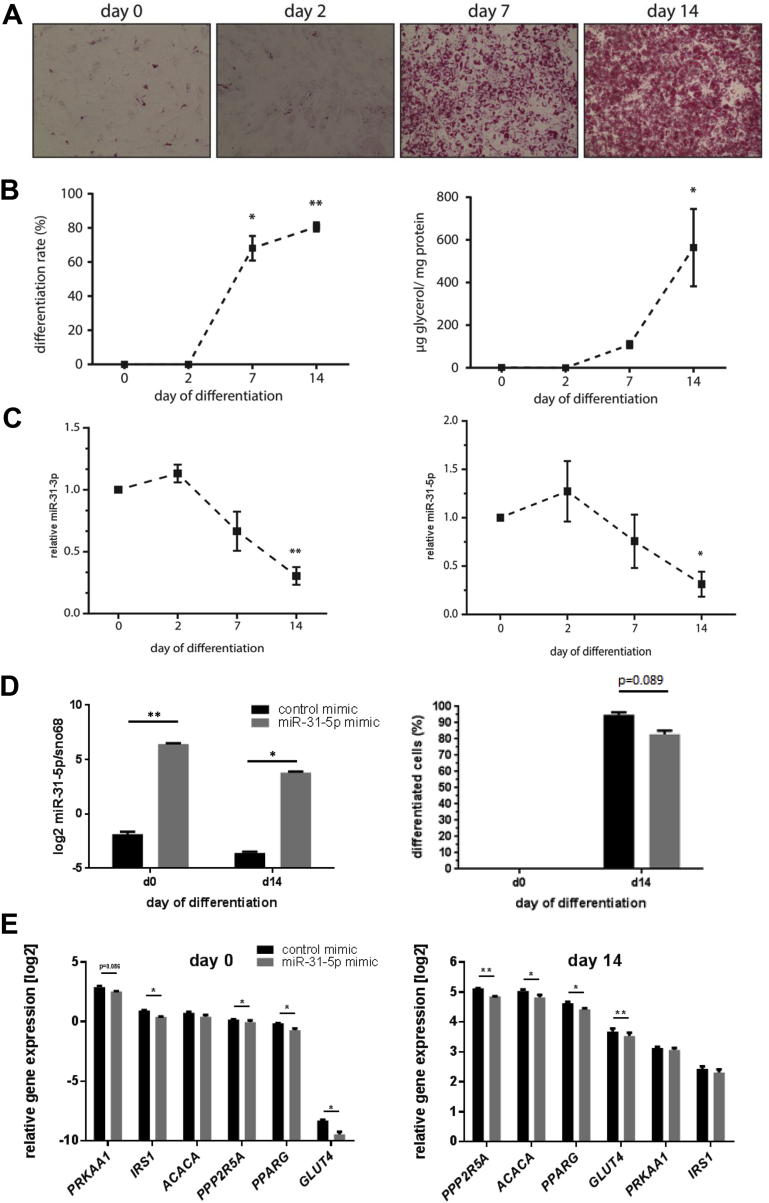
To gain further insights into the role of miR-31 in adipocytes, the human SGBS cell line was used as a model system. SGBS cells differentiate into mature adipocytes upon incubation in an adipogenic cocktail as seen by incorporation of lipids stained by Oil Red O and an upregulation of the adipogenic differentiation rate as well as the triglyceride content (Figure 6A and B). miR-31-5p and miR-31-3p expression was detected in the pre-adipocyte state (day 0), slightly increased two days after the induction of differentiation and decreased by approximately 70% on day 14 of differentiation (Figure 6C). As miR-31-5p showed a higher expression in human visceral fat depots (Figure 5A), we tested whether its manipulation affects the expression of predicted target genes and subsequently the differentiation of adipocytes. SGBS cells were transfected with a miR-31-5p mimic two days before applying the differentiation cocktail. The miR-31-5p mimic transfection significantly elevated the miR-31-5p levels in SGBS cells both in preadipocytes (day 0) as well as in differentiated cells (day 14). miR-31 mimic-transfected cells showed a tendency towards a reduction of the number of differentiated cells in comparison to control cells (Figure 6D). Elevated miR-31-5p levels significantly suppressed the expression of PPP2r5a, PPARy, IRS1, and GLUT4 at both time points (Figure 6E). PRKAA1 showed a tendency towards reduced expression on day 0 of differentiation in response to miR-31-5p mimic transfection. ACACA expression was only reduced in fully differentiated cells (Figure 6E). These data support our hypothesis that miR-31-5p targets genes involved in adipogenesis and insulin signaling.
Figure 6.
Impact of miR-31 mimic during differentiation in human adipocytes. (A) Oil red O staining of adipocytes in the culture dish of SGBS cells at the indicated days after induction of differentiation. (B) Percentage of differentiated adipocytes and glycerol levels during differentiation. (C) Expression levels of miR-31-5p and miR-31-3p in SGBS cells at indicated days after induction of differentiation (n = 3). (D) Expression of miR-31-5p at the indicated time points of differentiation after transfection with a miR-31-5p-specific mimic 2 days before treating SGBS cells with the differentiation cocktail. (E) mRNA expression of putative miR-31-5p targets at the indicated time points after differentiation. (*p < 0.05, **p < 0.01) Data are presented as ± SEM.
3.8. Identification of diabetes or obesity markers in PBMC
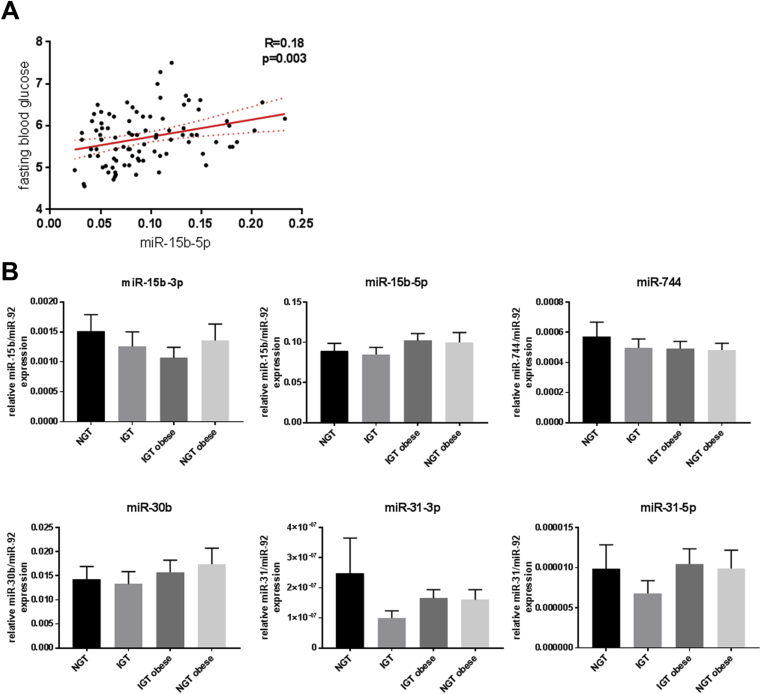
Since miRNAs can exhibit a similar pattern of expression in different tissues, the candidates miR-15b-5p/3p, miR-744, miR-30b, and miR-31-3p/5p were investigated in peripheral blood mononuclear cells (PBMC) of 89 participants, 25 healthy, 16 obese normoglycemic, and 48 diabetic subjects. A significant correlation of miR-15b-5p with fasted blood glucose levels was detected (Figure 7A, p = 0.003). However, no significant expression differences were detected between the groups (Figure 7B). All the other candidates did not show an altered expression or a correlation with specific parameters.
Figure 7.
Analysis of miRNAs in human PBMCs. (A) Correlation of miR-15b-5p expression with fasted blood glucose levels (n = 89). (B) Expression of miR-15b, miR-31, miR-744 and miR-30b in PBMCs of healthy (NGT; BMI < 30; n = 25), obese (NGT; BMI > 30; n = 16), diabetic (IGT; BMI < 30; n = 20) and diabetic and obese (IGT; BMI > 30; n = 28) patients. Data are presented as ± SEM.
4. Discussion
In the last decade, results from GWAS, QTL, transcriptomics, non-coding RNAs, and prediction of miRNA targets have been collected in order to clarify the genetic cause of complex polygenic diseases. miRNAs play a key role in the regulation of cellular functions such as proliferation, differentiation, and apoptosis, and they are involved in several biological processes like glucose homeostasis, inflammation, as well as in the pathophysiology of metabolic diseases [52]. Many cells and organs, including β-cells, liver, skeletal, and adipose tissue, have all been described to be affected by miRNAs [53]: miR-33a and miR-33b play crucial roles in cholesterol and lipid metabolism, miR-103 and miR-107 regulate hepatic insulin sensitivity. In β-cells, several miRNAs maintain the balance between differentiation and proliferation (miR-200 and miR-29 families) and insulin exocytosis in the differentiated state (miR-7, miR-375, and miR-335). Some miRNAs control white to brown adipocyte conversion or differentiation (miR-365, miR-133, miR-455). So far, most miRNAs were identified by experimental approaches, e.g. by miRNA profiling via microarrays, quantitative real time, and deep sequencing technologies [54], [55], [56]. For instance, Xie et al. reported 35 of the 576 detectable miRNAs to be differentially expressed in fat cells of lean and obese mice [57]. Microarray screening of human WAT tissues identified a number of miRNAs to be dysregulated in obese patients [58], [59].
Only a few studies have described computational tools for the identification of miRNAs and their impact on post-transcriptional gene regulation. Zhang et al. [60] screened for differentially expressed genes and miRNAs, and combined the results with a miRNA-transcription factor gene-regulatory network and identified 23 active regulatory pathways that were significantly linked to obesity-related inflammation.
miR-QTL-Scan, the computational framework described here applied different datasets including QTL, miRNA, histone marks, a combination of several prediction tools and expression profiles. To our knowledge, it is the first example of an online tool enabling users to screen for miRNAs located in murine QTL for obesity and diabetes, to trace their respective differentially expressed target genes in gWAT, BAT, and skeletal muscle, and to combine the results with pathway/GO-Term enrichment analyses providing an impression of a possible function of a selected miRNA. The procedure allows a first selection of miRNAs which might play a role for a specific disease such as obesity and T2D. That said, it is necessary to test the expression of candidate miRNAs, to manipulate their expression in an appropriate cell line, and to test its influence on putative target genes experimentally, as we did for miR-31. The limitation of miR-QTL-Scan is the large number of putative targets that result from the target prediction tools, even when at least three parallel tools are used. miR-QTL-Scan would benefit from data sets listing experimentally validated miRNA-RNA interactions. In theory, the approach is also useful for the translation of findings from mouse to human. However, it is important to align the miRNA sequences and to compare the seed regions of the targets as demonstrated in Figure 4E and F.
Similar to our approach, Shi et al. [61] created a complex heterogeneous network by integrating protein–protein interaction data, gene ontology data, miRNA–target relationships, disease phenotype data, and known miRNA-disease associations. Based on this network, a computational model was developed to identify miRNA associations to different types of cancer, myocardial infarction, and type 1 diabetes.
The successful application of our computational method was demonstrated in the second part of the study in which we focused on miRNAs in gWAT and PBMCs and investigated their impact on obesity. In gWAT, four candidate miRNAs (miR-15b, miR-744, miR-30b, and miR-31) and in BAT, six miRNAs (miR-423, miR-491, miR-132, miR-365, miR-455, and miR-30b) were identified that target metabolically relevant genes (Table 1). Further, for eight of ten miRNAs, putative variants in cis-regulatory elements were identified (Table 1). miR-15b-5p which was already described to be upregulated in WAT of NAFLD patients [37] was correlated with fasted blood glucose levels in human PBMCs (Figure 7A), highlighting its potential as a biomarker for diabetes. Additional focus was put on miR-31, which is located in a major obesity QTL on chromosome 4, the Nob6. miR-31 was selected for further analysis, since little was known about this miRNA. Furthermore, several of its putative targets were linked to insulin signaling such as Acaca (acetyl-CoA carboxylase alpha), Prkaa1 (protein kinase AMP-activated catalytic subunit alpha 1), Rps6kb1 (ribosomal protein S6 kinase B1), Glut4 (glucose transporter 4), Irs1 (insulin receptor substrate 1), Pde3b (phosphodiesterase 3B), Hk2 (hexokinase 2), Foxo1 (forkhead box O1), Socs6 (suppressor of cytokine signaling 6), and Ogt (O-linked N-acetylglucosamine (GlcNAc) transferase). Impaired insulin signaling is well known to negatively influence glucose and lipid metabolism [62]. In adipose tissue, insulin stimulates glucose uptake by inducing translocation of GLUT4 to the cell surface, it increases glycolysis rate by stimulating hexokinases (Hk2) and suppresses lipolysis (Acaca and Prkaa1) [63].
miR-31 exhibited a higher expression in gWAT of obese and diabetes-susceptible mice (NZO) as compared to lean and diabetes-resistant B6 mice (Figure 4C and D). As expected, its predicted target genes were found with lower abundance in gWAT of NZO than of gWAT of B6 mice (Figure 4A and B). The alignment of miR-31 on the mRNA sequences together with our in vitro data of the targets support its inhibitory potential (Figure 4E and F). However, it cannot be ruled out that the expression pattern of the predictive target genes is - at least in part - the consequence of another regulatory control, such as actions of transcription factors, histone modifications, or DNA methylation [64], [65], [66]. In order to demonstrate direct effects of miR-31 on targets like GLUT4 and ACACA, reporter assays would be required in which their 3′ UTR cloned in a luciferase vector would have to be co-transfected with a miR-31 expression plasmid and luciferase activity be measured.
miR-31 sequences in humans and mice are very well conserved (Figure 4C), and, as in mice, miR-31 levels were higher in visceral adipose depot of obese and diabetic patients compared to healthy subjects (Figure 5B). However, the question whether elevated miR-31 expression is the cause or the consequence of obesity or T2D needs to be answered and the mechanisms of its regulation also have to be clarified.
Several of the predicted miR-31 target genes have already been related to adipose tissue development, obesity, and diabetes [67], [68], [69]. Our in vitro study in human SGBS adipocytes showed direct effects of miR-31 manipulation on GLUT4, IRS1, ACACA, PRKAA1, and PPARγ expression. miR-31 seems to be implicated in two biological processes in adipose tissue; on the one hand, it affects genes of the insulin signalling (GLUT4 and IRS1) and on the other hand, genes which regulate adipocyte differentiation and lipogenesis (PPARγ [70], PRKAA1 [71], and ACACA [72]), which is in accordance to the location of miR-31 on QTL for gWAT weight and body weight (Figure 3A).
4.1. Conclusion
In summary, the introduced computational framework miR-QTL-Scan integrated miRNAs located in QTL, target-prediction tools and databases of experimentally validated targets, transcriptome profiles, and pathway enrichment analysis to identify miRNAs that can be related to obesity and T2D. Direct functional evidence for the achievement of this bioinformatics approach was provided by the detailed investigation of miR-31 and the correlation of miR-15b in PBMCs with fasted blood glucose levels. miR-31 exhibits a higher expression in adipose tissue of obese and diabetic mice and humans and the target genes are involved in adipogenesis and insulin signaling. Thus, a similar approach could also be applicable to discern the role of miRNA in other polygenic diseases.
Author contributions
P.G. developed and performed the computational analysis and edited the manuscript. M.J. provided computational expertise. M.O. and S.S. performed miR-31 experiments in mice and analyzed data, and M.O. edited the manuscript. A.K. and N.H. performed animal experiments. W.J. provided expertise and edited the manuscript. M.B. analyzed miR-31 expression in human adipose tissue samples. J.R. and P.F.-P. performed all experiments in SGBS cells. H.V. provided expertise, wrote, and edited the manuscript. L.S., A.F., H.H. and H.S measured and analyzed expression of miRNAs in blood cells. A.S. designed and directed the study, analyzed data, and wrote and edited the manuscript. All authors approved the final version of the manuscript.
Data availability
All data are available in our online tool at https://146.107.176.32/miR-QTL-Scan/. The data of transcriptome analyses are available at GEO, accession ID: GSE111142.
Acknowledgment
We thank Dirk Walther and Stefanie Hartmann for the contribution to the work by critical discussion and critical reading, and Ting Ting Cui for language editing. The study was supported by grants from the German Ministry of Education and Research and the Brandenburg state (82DZD00302, A.S.).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.03.005.
Contributor Information
Pascal Gottmann, Email: pascal.gottmann@dife.de.
Meriem Ouni, Email: meriem.ouni@dife.de.
Sophie Saussenthaler, Email: sophie.saussenthaler@dife.de.
Julian Roos, Email: julian.roos@uni-ulm.de.
Laura Stirm, Email: Laura.Stirm@med.uni-tuebingen.de.
Markus Jähnert, Email: markus.jaehnert@dife.de.
Anne Kamitz, Email: annekamitz@gmail.com.
Nicole Hallahan, Email: nicole.hallahan@dife.de.
Wenke Jonas, Email: wenke.jonas@dife.de.
Andreas Fritsche, Email: andreas.fritsche@med.uni-tuebingen.de.
Hans-Ulrich Häring, Email: Hans-Ulrich.Haering@med.uni-tuebingen.de.
Harald Staiger, Email: harald.staiger@med.uni-tuebingen.de.
Matthias Blüher, Email: Matthias.Blueher@medizin.uni-leipzig.de.
Pamela Fischer-Posovszky, Email: pamela.fischer@uniklinik-ulm.de.
Heike Vogel, Email: heikevogel@dife.de.
Annette Schürmann, Email: schuermann@dife.de.
Conflict of interest
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Golay A., Ybarra J. Link between obesity and type 2 diabetes. Best Practice & Research Clinical Endocrinology & Metabolism. 2005;19(4):649–663. doi: 10.1016/j.beem.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Davies J.L., Kawaguchi Y., Bennett S.T., Copeman J.B., Cordell H.J., Pritchard L.E. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371(6493):130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 3.Holmes E., Loo R.L., Stamler J., Bictash M., Yap I.K., Chan Q. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453(7193):396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClellan J., King M.C. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 5.Peters L.L., Robledo R.F., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L. The mouse as a model for human biology: a resource guide for complex trait analysis. Nature Reviews Genetics. 2007;8(1):58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- 6.Joost H.G., Schürmann A. The genetic basis of obesity-associated type 2 diabetes (diabesity) in polygenic mouse models. Mammalian Genome. 2014:401–412. doi: 10.1007/s00335-014-9514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. Mapping complex disease traits with global gene expression. Nature Reviews Genetics. 2009;10(3):184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 9.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. 2012:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-Hernando C., Ramírez C.M., Goedeke L., Suárez Y. MicroRNAs in metabolic disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(2):178–185. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 13.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P. miR-375 maintains normal pancreatic - and -cell mass. Proceedings of the National Academy of Sciences. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., MacDonald P.E. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 15.Vogel H., Scherneck S., Kanzleiter T., Benz V., Kluge R., Stadion M. Loss of function of Ifi202b by a microdeletion on chromosome 1 of C57BL/6J mice suppresses 11β-hydroxysteroid dehydrogenase type 1 expression and development of obesity. Human Molecular Genetics. 2012;21(17):3845–3857. doi: 10.1093/hmg/dds213. [DOI] [PubMed] [Google Scholar]
- 16.Kluth O., Matzke D., Kamitz A., Jähnert M., Vogel H., Scherneck S. Identification of four mouse diabetes candidate genes altering β-cell proliferation. PLoS Genetics. 2015;11(9) doi: 10.1371/journal.pgen.1005506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherneck S., Nestler M., Vogel H., Blüher M., Block M.D., Diaz M.B. Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity-associated diabetes contributed by mouse locus Nidd/SJL. PLoS Genetics. 2009;5(7) doi: 10.1371/journal.pgen.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander E., Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 19.Wabitsch M., Brenner R.E., Melzner I., Braun M., Möller P., Heinze E. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. International Journal of Obesity. 2001;25(1):8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 20.Fischer-Posovszky P., Newell F.S., Wabitsch M., Tornqvist H.E. Human SGBS cells - a unique tool for studies of human fat cell biology. Obesity Facts. 2008:184–189. doi: 10.1159/000145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos J., Enlund E., Funcke J.-B., Tews D., Holzmann K., Debatin K.-M. MiR-146a-mediated suppression of the inflammatory response in human adipocytes. Scientific Reports. 2016;6 doi: 10.1038/srep38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefan N., Machicao F., Staiger H., Machann J., Schick F., Tschritter O. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia. 2005;48(11):2282–2291. doi: 10.1007/s00125-005-1948-3. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Meltzer P., Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14(1):244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Güller I., McNaughton S., Crowley T., Gilsanz V., Kajimura S., Watt M. Comparative analysis of microRNA expression in mouse and human brown adipose tissue. BMC Genomics. 2015;16(1):820. doi: 10.1186/s12864-015-2045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozomara A., Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(D1) doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Research. 2013;41(Web Server issue) doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32(9):1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J., Li X., Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics. 2016;32(18):2768–2775. doi: 10.1093/bioinformatics/btw318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4(AUGUST2015) doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Research. 2015;43(D1):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Research. 2009;37(SUPPL. 1) doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou C.H., Chang N.W., Shrestha S., Hsu S. Da, Lin Y.L., Lee W.H. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Research. 2016;44(D1):D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson D.W., Bracken C.P., Goodall G.J. Experimental strategies for microRNA target identification. Nucleic Acids Research. 2011:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Cheng X., Lu Z., Wang J., Chen H., Fan W. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Research and Clinical Practice. 2013;99(3):327–334. doi: 10.1016/j.diabres.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Sebastiani G., Nigi L., Spagnuolo I., Morganti E., Fondelli C., Dotta F. MicroRNA profiling in sera of patients with type 2 diabetes mellitus reveals an upregulation of miR-31 expression in subjects with microvascular complications. Journal of Biomedical Science and Engineering. 2013;6(May):58–64. [Google Scholar]
- 39.Estep M., Armistead D., Hossain N., Elarainy H., Goodman Z., Baranova A. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Alimentary Pharmacology & Therapeutics. 2010;32(3):487–497. doi: 10.1111/j.1365-2036.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 40.Hu F., Wang M., Xiao T., Yin B., He L., Meng W. MiR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes. 2015;64(6):2056–2068. doi: 10.2337/db14-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Z., Liu J., Bian H., Cai J., Guo X. MIR-455 enhances adipogenic differentiation of 3T3-L1 cells through targeting uncoupling protein-1. Pharmamazie. 2016;71(11):625–628. doi: 10.1691/ph.2016.6734. [DOI] [PubMed] [Google Scholar]
- 42.Ortega F.J., Mercader J.M., Catalán V., Moreno-Navarrete J.M., Pueyo N., Sabater M. Targeting the circulating microRNA signature of obesity. Clinical Chemistry. 2013;59(5):781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 43.Ge Q., Brichard S., Yi X., Li Q. microRNAs as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. Journal of Immunology Research. 2014;2014:987285. doi: 10.1155/2014/987285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shamsi F., Zhang H., Tseng Y.H. MicroRNA regulation of brown adipogenesis and thermogenic energy expenditure. Frontiers in Endocrinology. 2017;8:205. doi: 10.3389/fendo.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W., Wang J., Chen Z., Chen J., Meng Y., Chen L. NFE2 induces miR-423-5p to promote gluconeogenesis and hyperglycemia by repressing the hepatic FAM3A-ATP-Akt pathway. Diabetes. 2017;66(7):1819–1832. doi: 10.2337/db16-1172. [DOI] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C. ENCODE data at the ENCODE portal. Nucleic Acids Research. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arner E., Mejhert N., Kulyté A., Balwierz P.J., Pachkov M., Cormont M. Adipose tissue MicroRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61(8):1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C. Corrigendum: adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;545(7653) doi: 10.1038/nature21365. 252–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto N., Tanaka T. Role of miRNAs in the pathogenesis and susceptibility of diabetes mellitus. Journal of Human Genetics. 2017;62(2):141–150. doi: 10.1038/jhg.2016.150. [DOI] [PubMed] [Google Scholar]
- 53.Vienberg S., Geiger J., Madsen S., Dalgaard L.T. MicroRNAs in metabolism. Acta Physiologica. 2017:346–361. doi: 10.1111/apha.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker M. MicroRNA profiling: separating signal from noise. Nature Methods. 2010;7(9):687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- 55.Dong H., Lei J., Ding L., Wen Y., Ju H., Zhang X. MicroRNA: function, detection, and bioanalysis. Chemical Reviews. 2013:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 56.Tian T., Wang J., Zhou X. A review: microRNA detection methods. Organic and Biomolecular Chemistry. 2015;13(8):2226–2238. doi: 10.1039/c4ob02104e. [DOI] [PubMed] [Google Scholar]
- 57.Xie H., Lim B., Lodish H.F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58(5):1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heneghan H.M., Miller N., McAnena O.J., O'Brien T., Kerin M.J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. Journal of Clinical Endocrinology and Metabolism. 2011;96(5) doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 59.Keller P., Gburcik V., Petrovic N., Gallagher I.J., Nedergaard J., Cannon B. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocrine Disorders. 2011;11(1):7. doi: 10.1186/1472-6823-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X.-M., Guo L., Chi M.-H., Sun H.-M., Chen X.-W. Identification of active miRNA and transcription factor regulatory pathways in human obesity-related inflammation. BMC Bioinformatics. 2015;16(1):76. doi: 10.1186/s12859-015-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H., Zhang G., Zhou M., Cheng L., Yang H., Wang J. Integration of multiple genomic and phenotype data to infer novel miRNA-disease associations. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0148521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saltiel A.R., Kahn C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 63.Dimitriadis G., Mitrou P., Lambadiari V., Maratou E., Raptis S.A. Insulin effects in muscle and adipose tissue. Diabetes Research and Clinical Practice. 2011;93(1):S52–S59. doi: 10.1016/S0168-8227(11)70014-6. [DOI] [PubMed] [Google Scholar]
- 64.Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 65.Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H. Targets and dynamics of promoter DNA methylation during early mouse development. Nature Genetics. 2010;42(12):1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 66.Bird A.P., Wolffe A.P. Methylation-induced repression— belts, braces, and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 67.Abel E.D., Peroni O., Kim J.K., Kim Y.-B., Boss O., Hadro E. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409(6821):729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 68.Munekata K., Sakamoto K. Forkhead transcription factor Foxo1 is essential for adipocyte differentiation. In Vitro Cellular and Developmental Biology - Animal. 2009;45(10):642–651. doi: 10.1007/s11626-009-9230-5. [DOI] [PubMed] [Google Scholar]
- 69.Dharuri H., t Hoen P.A.C., van Klinken J.B., Henneman P., Laros J.F.J., Lips M.A. Downregulation of the acetyl-CoA metabolic network in adipose tissue of obese diabetic individuals and recovery after weight loss. Diabetologia. 2014;57(11):2384–2392. doi: 10.1007/s00125-014-3347-0. [DOI] [PubMed] [Google Scholar]
- 70.Farmer S.R. Regulation of PPARgamma activity during adipogenesis. International Journal of Obesity. 2005;(29 Suppl 1):S13–S16. doi: 10.1038/sj.ijo.0802907. [DOI] [PubMed] [Google Scholar]
- 71.Daval M., Foufelle F., Ferré P. Functions of AMP-activated protein kinase in adipose tissue. The Journal of Physiology. 2006;574(1):55–62. doi: 10.1113/jphysiol.2006.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fullerton M.D., Galic S., Marcinko K., Sikkema S., Pulinilkunnil T., Chen Z.-P. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nature Medicine. 2013;19(12):1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in our online tool at https://146.107.176.32/miR-QTL-Scan/. The data of transcriptome analyses are available at GEO, accession ID: GSE111142.