Abstract
Circadian clock and senescence have been shown to tightly intertwined with each other in numerous eukaryotes, but the regulation of circadian oscillator on triggering leaf senescence, and vice versa, remains largely unknown in higher plants. Very recently, circadian system and leaf senescence were found to be highly interconnected in higher plants. Circadian clock was shown to regulate leaf senescence through a few cross signaling pathways including age-dependent, plant hormone mediated, and dark induced manners to trigger the onset of leaf senescence. By contrast, circadian clock itself also can be affected by the leaves senescing process. The eventually delineating cross networks between circadian clock and leaf senescence will lay the foundation for understanding the fitness of developmental dynamics of plants with aging.
Keywords: circadian clock, leaf senescence, CCA1, evening complex, MYC2
Introduction
By synchronizing the cellular events with external light-dark cycle resulted from daily rotation of earth, circadian clock regulates many aspects of plant growth and development, such as photosynthesis, pathogen defense, and flowering time, and provides a fitness benefit for higher plants (Dodd et al., 2005; Harmer, 2009; Greenham and McClung, 2015). The current consensus is that cell-autonomous core oscillator of circadian clock comprises three interlocking negative feedback loops. CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), suppress expression of PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9, while PRR9/7/5 can transcriptionally repress CCA1/LHY expression by recruiting TOPLESS family members and HISTONE DEACETLYLASE 6/19 (Wang et al., 2013), hence forming the morning loop of core oscillator (Pokhilko et al., 2012). TOC1 (TIMING OF CAB EXPRESSION 1) and CCA1/LHY reciprocally represses each other to form the central loop of core oscillator. In the evening loop, TOC1 represses the expression of GIGANTEA (GI) (Huang et al., 2012). Moreover, the evening complex (EC), consisting of EARLY FLOWERING 3 (ELF3), ELF4 and LUX ARRHYTHMO (LUX), suppresses the expression of TOC1, GI, and PRR9 (Nusinow et al., 2011; Herrero et al., 2012).
The key components of core oscillator mediate circadian outputs by both transcriptional and post-transcriptional mechanisms via timing numerous genes expression at appropriate phase. Recently, the accumulating evidence display that many components of circadian genetic circuit also serve as harbors to directly regulate the transcription of genes involved in clock-output pathways, such as abiotic stress response, photosynthesis and flowering time (Nusinow et al., 2011; Sawa and Kay, 2011; Nakamichi et al., 2012; Liu et al., 2013, 2016; Ezer et al., 2017). Moreover, many circadian core components also have been identified as key players to regulate hypocotyls growth and flowering time through protein–protein interaction, and affecting the abundance and activity of their interacting partners (Shin et al., 2012; Soy et al., 2016; Zhu et al., 2016; Hayama et al., 2017; Martin et al., 2018). Thus, circadian core oscillator serves as a master regulator in whole life span of plant (Sanchez and Kay, 2016). However, whether and how leaf senescence is modulated by circadian clock, or vice versa, remains to be discovered.
Leaf senescence not only means the inevitably death, as double edged sword, it also can trigger the reallocation of nutrients and energy to developing tissues or storage organs, thus properly timing leaf senescence onset is critical for the seed yield, fruit ripening and biomass production, especially for annual plants (Lim et al., 2007; Wu et al., 2012). The leaf senescence is under the control of an extremely coordinated cellular nework, which integrates the environmental information and internal signals such as plant hormone signaling, nutrition and energy status. Basically, onset of the leaf senescence is managed by developmental cues, and modulated by both the outer environmental conditions and inner signals independently and interdependently, such as dark (Sakuraba et al., 2014) and multiple hormone signaling such as JAs (Qi et al., 2015; for review see Kim et al., 2015), ethylene (Li et al., 2013; Qiu et al., 2015; also see review by Iqbal et al., 2017), ABA (Zhao et al., 2016), and strigolactone (Ueda and Kusaba, 2015). Accordingly, the signaling pathways of leaf senescence can be classified to age-dependent pathway, environmental conditions induced pathway, and internal hormones induced pathway, etc. In any environmental cues or inner signals trigged leaf senescence, this degenerative process was closely regulated by the coordinated expression profile of senescence-associated genes (SAGs). The collection of SAGs in Arabidopsis comprised of 192 genes involved in various regulatory networks underlying leaf senescence (Liu et al., 2011; Li et al., 2014). Very recently, a few core circadian components were found to be able to regulate the messenger RNA or protein abundance of SAGs at the transcriptional and post-transcriptional levels, respectively (Shin et al., 2012; Sakuraba et al., 2014; Song et al., 2018; Zhang et al., 2018). By contrast, circadian period were dramatically altered in the different ages of leaf, indicating the tightly reciprocal regulation between circadian clock and leaf senescence exists (Kim et al., 2016). Here, we reviewed the recent progresses on the interplay between circadian clock and leaf senescence, and proposed that the regulatory network of leaf senescence and circadian system is highly interconnected in higher plants. Unraveling this underlying interconnected mechanism will contribute to understand the complex regulatory networks for fitness and adaptive advantage of higher plants.
Circadian Clock Components Coordinates Age-Dependent Leaf Senescence
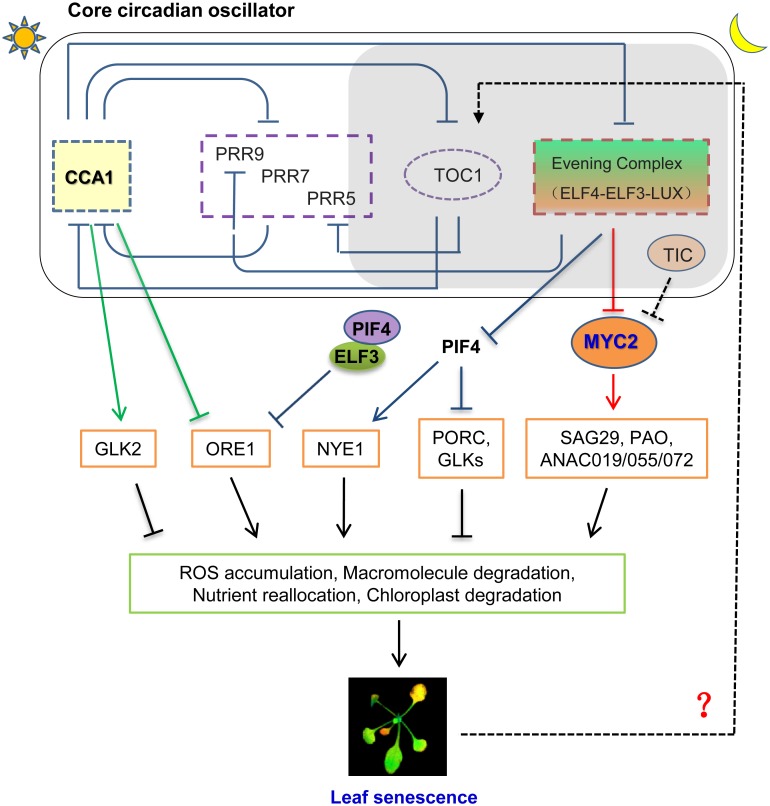
Age-dependent leaf senescence is regarded as an autonomous signaling pathway to trigger the initiation of leaf senescence via reprogramming transcriptome. ORESARA 1 (ORE1/ANAC092), a NAC-domain transcription factor, has been characterized as a positive regulator for age induced senescence (Kim et al., 2009; Rauf et al., 2013). Elevated expression level of ORE1 can trigger the onset of leaf senescence by inducing various SAGs expression. Importantly, ORE1 interacts with the G2-like transcription factors GLK1 and GLK2 and antagonizes their transcriptional activity, while GLKs are important for chloroplast development and maintenance (Rauf et al., 2013). At the transcriptional level, ORE1 expression is regulated by age and multiple plant hormones, such as ABA and ethylene (Li et al., 2013). Recently, the tight circadian control of ORE1 expression was unmasked to mediate circadian regulation on age dependent leaf senescence. By using high-resolution temporal profiling data, Song et al. (2018) showed that the expression level of ORE1 is continuously increased with aging under long day condition, while the corresponding abundance of CCA1 keeps reducing. In addition, they found that the expression level of GLK2 keep decreasing during leaf senescence process (Song et al., 2018), suggesting their highly coordination might coincidence with the processing of leaf senescence. They further found that CCA1, the core circadian transcription repressor, can directly bind to the promoter of ORE1 through its CBS element (CCA1-Binding site, AAMAATCT) and inhibits its transcription in the pre-senescence leaves (Song et al., 2018). Moreover, CCA1 is able to active GLK2 expression through binding the CBS element within the GLK2’s promoter (Song et al., 2018). Hence, they proposed that the age-declined CCA1 causes the gradual removal inhibition on ORE1 transcription and the lower activation of GLK2 to temporally promotes the onset of leaf senescence (Figure 1; Song et al., 2018).
FIGURE 1.
A proposed model for interplaying between circadian clock and leaf senescence in Arabidopsis. CCA1, one of core components of plant circadian oscillator, regulates age dependent leaf senescence by inhibiting ORE1 and activating GLK2 at the transcript level. Evening Complex (EC) modulates Jasmonate-induced leaf senescence through transcriptionally repressing MYC2, an essential component of JA signaling pathway to trigger leaf senescence. TIC, which regulates circadian rhythm with unknown mechanism, can modulate protein abundance of MYC2 with an unclear post-translational mechanism. ELF3 also regulates dark induced leaf senescence (DIS) through ORE1-dependent pathway by transcriptionally repressing PIF4. Whether other circadian clock components, such as Pseudo Response Regulators, are engaged into the regulation of leaf senescence awaits to be further investigated.
Song et al. (2018) also showed the mutation of ELF3 caused precocious leaf senescence, which might due to the aberrant expression of ORE1, because ELF3 forms transcription repressive complex together with ELF4 and LUX, to transcriptionally inhibit PIF4 and PIF5 expression. In turn, PIF4 can directly upregulate the transcription of ORE1 (Figure 1; Sakuraba et al., 2017). However, the messenger RNA abundance of PIF4 and PIF5 increases with aging (Song et al., 2018), in contrast to that of CCA1. Previously, they have identified PIF3, PIF4, and PIF5 as mediators of leaf senescence as their overexpression promotes leaf senescence, while their mutation confers longer leaf longevity (Song et al., 2014). Thus, higher PIF4 transcript level in elf3 mutant can in turn cause the higher level of ORE1 and bolster the onset of leaf senescence. Thus, it is reasonably to hypothesize that the higher level of PIFs in elf3 mutant boosts up the leaf senescence through increasing ORE1 expression, which need to be further verified experimentally. Taken together, the above lines of evidences strongly indicated the highly coordinated network underlying the circadian clock regulation on natural senescence, which remains to be fully investigated in the future.
Circadian System Directs Plant Hormones Dependent Leaf Senescence
Jasmonoyl-isoleucine (JA-Ile, hereafter was abbreviated to JAs) has long been known as a key plant hormone to induce leaf senescence (He et al., 2002; Qi et al., 2015). In the absence of JA, JA ZIM-domain (JAZ) repressors interact with and inhibit JA downstream transcription factors, such as bHLH subgroup IIIe factors MYC2, MYC3, and MYC4, by recruiting TOPLESS co-transcriptional repressor family members, adapted by NINJA (Pauwels et al., 2010). On perception of JAs, its receptor, CORONATINE INSENSITIVE1 (COI1), which is a F-box domain containing protein, can interact with and degrade JAZ proteins, thus releasing transcriptional activators including MYC2, MYC3, and MYC4 to mediate JA response by activating downstream gene expression (Pauwels et al., 2010). Upon JA signaling activated, the IIIe bHLH factors MYC2, MYC3, and MYC4 redundantly activate JA-induced leaf senescence. On the other hand, the IIId bHLH factors including bHLH03, bHLH13, bHLH14, and bHLH17 acting redundantly to repress JA-Induced leaf senescence by antagonizing with MYCs, through competing with their binding SAG29 promoter (Qi et al., 2015), suggesting the elaborative downstream regulation mechanism to mediate JA-induced leaf senescence. Nevertheless, MYC2, MYC3, and MYC4 play predominantly and positively regulatory roles in JA-induced leaf senescence.
A recent study from our laboratory details how Arabidopsis circadian system regulates JAs-induced senescence. We found that Evening Complex (EC), composed by ELF4-ELF3-LUX, locates at hub position to mediate circadian output regulation on JA-induced leaf senescence (Zhang et al., 2018). EC can directly bind to MYC2 promoter and gate its JA-induced expression profile. Genetically, mutation of MYCs abolished the hypersensitivity of JA-induced leaf senescence in EC mutants (Figure 1; Zhang et al., 2018). This finding not only delineated a key underlying mechanism for circadian gated JA signaling in triggering leaf senescence, but also closed the knowledge gap of a long sought-after transcriptional regulation between the circadian core oscillator and JA signaling. Interestingly, JA accumulation in plants is also under the circadian control with peaking at the middle of subjective day. This phase pattern anticipates the peak of the insect feeding behavior (Goodspeed et al., 2012). But the JA content is lower in EC mutant, reflects the feedback regulation of elevated JA signaling, and further emphasizes the JA signal, rather than JA accumulation, accounts for precocious leaf senescence in EC mutant.
Circadian clock also regulates JA signaling and response at post-transcriptional level. TIME FOR COFFEE (TIC) has been identified as a nuclear regulator for proper circadian function from middle to late subjective night (Hall et al., 2003; Ding et al., 2007). TIC also acts as a negative regulator in JA signaling by repressing MYC2 protein accumulation at the post-translational level (Shin et al., 2012). Hence, the mutation of TIC caused higher MYC2 protein abundance and might cause early senescence phenotype, which awaits to be experimentally verified. Moreover, plant circadian clock driven time-of-day susceptibility to necrotrophic fungal pathogen, Botrytis cinerea, is mediated by JA signaling (Ingle et al., 2015), implicating that multiple converging points mediated circadian regualtion on JA responses. It is unclear yet whether this related to leaf senescence or not.
Salicylic acid (SA) is another well known plant hormone to positively regulate leaf senescence, and closely interplays with circadian clock. Plants with defective SA signaling, such as npr1 (non-expressor of pathogenesis-related gene 1), display a delayed yellowing and reduced necrosis phenotype (Morris et al., 2000). Consistently, a few senescence enhanced genes, including SENESCENCE ASSOCIATED GENE 12 (SAG12), display lower transcript level in npr1 mutant, whereas SA treatment is capable of inducing a few senescence enhanced genes expression (Morris et al., 2000). Very recently, it was found that SA signal causes the reinforcement of circadian clock without circadian period change, instead of boosting circadian amplitudes of a few circadian core components via NPR1 (Zhou et al., 2015). Intriguingly, SA mediated plant immune response is influenced by circadian clock through the regulatory control of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) on expression of defense genes (Wang et al., 2011). However, whether SA mediated leaf senescence is governed by circadian clock or not still remains elusive, which will be of great interest to be fully investigated in the future.
Circadian Clock Components Tune Environmental Stresses Dependent Leaf Senescence
Unfavorable environmental stresses can initiate the onset of leaf senescence. Among the stresses, light deprivation trigged leaf senescence was considered as highly relevant with circadian system, especially given the highly interconnection between light signaling and circadian clock. The Pfr form of Phytochrome B can inhibit dark induced leaf senescence (DIS), probably through destabilizing PIF4 and PIF5, as the Pr form of Phytochrome B could be switched off by far-red light. PHYTOCHROME INTERACTION FACTORS (PIFs), the negative regulator of light signaling regulator, regulate a variety of red light signaling mediated developmental responses. By physically interacting with Phy A and/or Phy B, PIFs will be degraded. Consistently, mutants of PIF4 and PIF5, especially quadruple mutant of PIFs, displayed a significantly delayed DIS phenotype (Song et al., 2014; Sakuraba et al., 2017). ELF3 has known to be one of upstream regulators for PIF4 and PIF5 expression, in consistent with its mutant senesced much faster under dark induced condition. ELF3 functions as an adaptor to bridge ELF4 and LUX to form the ternary transcriptional complex to inhibit PIF4 and PIF5 expression in the early subjective night (Nusinow et al., 2011). Surprisingly, neither ELF4 nor LUX mutants showed the faster DIS phenotype as mutant of ELF3, indicating the altered senescence phenotype in elf3 mutant is not due to the transcription inhibition of PIF4 and PIF5, instead it might be caused by post-transcriptional regulation of PIFs (Figure 1). Strikingly, none of any other circadian mutants tested by Sakuraba et al. (2017) displayed the altered DIS phenotype, including CCA1, LHY, GI. Intriguingly, ELF3 does not rely on the function of PhyB to inhibit DIS, but through transcriptionally repressing PIFs. Given both ELF4 and LUX did not involve DIS processing, whether ELF3 transcriptional repression activity for PIFs recruits tissue specific partners in light deprivation leaves remains an open question. Nonetheless, it is reasonable to hypothesize that the orchestration of PIFs abundance by light signaling at the post-translation level and circadian clock at the transcriptional level to direct PIFs mediated DIS. Mechanistically, increased both ethylene biosynthesis and signaling may also mediate the downstream events of PIFs to trigger DIS (Song et al., 2014; Sakuraba et al., 2017). In addition, ABA signaling was also involved into downstream of PIFs to mediate DIS, as ABSCISIC ACID INSENSITIVE 5 (ABI5) is direct target genes of PIF4 and PIF5 (Sakuraba et al., 2017; Zhang et al., 2018). Notably, PIFs and ABI5 regulate the expression of ORE1, the key senescence promoter, hence forming a coherent feed forward regulation mechanism (for review see Liebsch and Keech, 2016).
Circadian Period Is Altered With Leaf Ages
Not only circadian clock regulates leaf senescence through a complex signaling network integrating the age-dependent, plant hormone mediated, and dark induced pathways to trigger the onset of leaf senescence, but also circadian clock itself was feedback regulated by the leaves senescing process. SENESCENCE 4(SEN4) is an age dependent marker gene whose expression is eventually decreasing in old leaves. While the younger leaves display the lower transcript level of SEN4 in single plant, they also showed longer circadian period, indicating the close reverse association between leaf senescence and circadian period exists (Kim et al., 2016). Similarly, the measuring circadian period of the detached third and fourth leaves from plants at different ages with dramatically distinct SEN4 expression level further confirmed that the younger leaves have a relatively longer period, while the old leaves with a shortened period (Kim et al., 2016). Intriguingly, the gradual decrease of circadian period correlates to the day length as the more pronounced circadian period change was found in long-day grown plants. Among the tested circadian mutant, toc1 mutants did not display shorter circadian period with aging, implicating TOC1 is an essential player to link leaf aging with circadian period change (Figure 1; Kim et al., 2016). However, the underlying mechanism how TOC1 mediates the link is unclear yet. Moreover, whether abiotic stresses or hormones induced senescent leaves will also alter circadian period needs to be further explored.
Perspectives and Remaining Questions
In conclusion, the intricate complex cellular network, but not linear signaling pathways, may govern the regulation of circadian clock on the initiation and processing of leaf senescence. As an example, besides promoting the DIS, PIFs also promote age-dependent leaf senescence which may act through activating ethylene and JA signaling pathway (Sakuraba et al., 2014; Song et al., 2014). This also could be true for explaining EC regulation on leaf senescence. As JA content does not increase with aging (Mao et al., 2017) and JA response declining with age, the early senescence phenotype in EC mutant under nature condition cannot simply caused by elevated expression level of MYC2. Actually, ABI5, WRKY22 and a few ethylene response regulators were also appreciably up-regulated in lux-6 mutant (Zhang et al., 2018).
On the other hand, since ABA-responsive genes are also circadian regulated, either as direct targets of the central clock or as a response of the rhythmic ABA signaling network (Seung et al., 2012; Liu et al., 2013; also for review see Sanchez and Kay, 2016). Whether ABA signaling is under the circadian control to timing leaf senescence awaits to be further investigated.
Moreover, Reactive oxygen species (ROS) homeostasis is essential for triggering leaf senescence, while the cellular redox state and circadian clock daily influence each other, partially via CCA1 (Lai et al., 2012). It is thus conceivable that circadian direct regulation on ROS homeostasis might also contribute the onset of leaf senescence bypass the known signaling pathways. Nevertheless, circadian clock acts as a master shepherd for timing and phasing the onset of leaf senescence, and the underlying regulatory network remains to be further discovered.
Another challenge will be to understand the cause and effect relationship between senescence and circadian clock. It remains unclear whether the age dependent change of circadian rhythm will induce downstream developmental program which further trigger the leaf senescence due to mis-synchronization or aged leaves has lower energy status and higher ROS accumulation which will cause the change of circadian period. It also cannot exclude the possibility that the leaves senescing process and circadian rhythm work interactively with aging. Recently, It was found that disruption peripheral circadian clock in mammals can reduce life span and accelerate aging (for review see Paschos and FitzGerald, 2017). Thus, circadian clock, the central pacemaker, tightly interconnected with regulatory network of leaf senescence by directing plant hormone response and ROS homeostasis, etc., to ensure the plant proper fitness and yield. The complex underlying regulatory network awaits to be further fully elucidated.
Author Contributions
YW, YZ, and LW wrote the manuscript and contributed to the discussion and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those authors whose excellent relevant work could not be cited due to space limitations.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Nos. 31670290 and 31570292), Youth Innovation Promotion Association CAS to YZ (No. 2017110), and ‘Youth 1000 Plan’ to LW.
References
- Ding Z., Millar A. J., Davis A. M., Davis S. J. (2007). Time for coffee encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536. 10.1105/tpc.106.047241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A. N., Salathia N., Hall A., Kevei E., Toth R., Nagy F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. 10.1126/science.1115581 [DOI] [PubMed] [Google Scholar]
- Ezer D., Jung J. H., Lan H., Biswas S., Gregoire L., Box M. S., et al. (2017). The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3:17087. 10.1038/nplants.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D., Chehab E. W., Min-Venditti A., Braam J., Covington M. F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U.S.A. 109 4674–4677. 10.1073/pnas.1116368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K., McClung C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16 598–610. 10.1038/nrg3976 [DOI] [PubMed] [Google Scholar]
- Hall A., Bastow R. M., Davis S. J., Hanano S., Mcwatters H. G., Hibberd V., et al. (2003). The time for coffee gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. 10.1105/tpc.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60 357–377. 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- Hayama R., Sarid-Krebs L., Richter R., Fernandez V., Jang S., Coupland G. (2017). Pseudo response regulators stabilize constans protein to promote flowering in response to day length. EMBO J. 36 904–918. 10.15252/embj.201693907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D. F., Gan S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128 876–884. 10.1104/pp.010843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., Kolmos E., Bujdoso N., Yuan Y., Wang M., Berns M. C., et al. (2012). Early flowering4 recruitment of early flowering3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24 428–443. 10.1105/tpc.111.093807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Perez-Garcia P., Pokhilko A., Millar A. J., Antoshechkin I., Riechmann J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336 75–79. 10.1126/science.1219075 [DOI] [PubMed] [Google Scholar]
- Ingle R. A., Stoker C., Stone W., Adams N., Smith R., Grant M., et al. (2015). Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 84 937–948. 10.1111/tpj.13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N., Khan N. A., Ferrante A., Trivellini A., Francini A., Khan M. I. R. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8:475. 10.3389/fpls.2017.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim Y., Yeom M., Lim J., Nam H. G. (2016). Age-associated circadian period changes in Arabidopsis leaves. J. Exp. Bot. 67 2665–2673. 10.1093/jxb/erw097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chang C., Tucker M. L. (2015). To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 6:20. 10.3389/fpls.2015.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Lai A. G., Doherty C. J., Mueller-Roeber B., Kay S. A., Schippers J. H., Dijkwel P. P. (2012). Circadian clock-associated 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109 17129–17134. 10.1073/pnas.1209148109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Peng J., Guo H., Luo J. (2014). LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res. 42 D1200–D1205. 10.1093/nar/gkt1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch D., Keech O. (2016). Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol. 212 563–570. 10.1111/nph.14217 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Liu T., Carlsson J., Takeuchi T., Newton L., Farre E. M. (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76 101–114. 10.1111/tpj.12276 [DOI] [PubMed] [Google Scholar]
- Liu T. L., Newton L., Liu M. J., Shiu S. H., Farre E. M. (2016). A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170 528–539. 10.1104/pp.15.01562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li Z., Jiang Z., Zhao Y., Peng J., Jin J., et al. (2011). LSD: a leaf senescence database. Nucleic Acids Res. 39 D1103–D1107. 10.1093/nar/gkq1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. B., Liu Y. Q., Chen D. Y., Chen F. Y., Fang X., Hong G. J., et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 8:13925. 10.1038/ncomms13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Rovira A., Veciana N., Soy J., Toledo-Ortiz G., Gommers C. M. M., et al. (2018). Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28 311.e315–318.e315. 10.1016/j.cub.2017.12.021 [DOI] [PubMed] [Google Scholar]
- Morris K., Mackerness S. A., Page T., John C. F., Murphy A. M., Carr J. P., et al. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23 677–685. 10.1046/j.1365-313x.2000.00836.x [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., et al. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109 17123–17128. 10.1073/pnas.1205156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D. A., Helfer A., Hamilton E. E., King J. J., Imaizumi T., Schultz T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475 398–402. 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos G. K., FitzGerald G. A. (2017). Circadian clocks and metabolism: implications for microbiome and aging. Trends Genet. 33 760–769. 10.1016/j.tig.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Barbero G. F., Geerinck J., Tilleman S., Grunewald W., Pérez A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464 788–791. 10.1038/nature08854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Fernandez A. P., Edwards K. D., Southern M. M., Halliday K. J., Millar A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8:574. 10.1038/msb.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Wang J., Huang H., Liu B., Gao H., Liu Y., et al. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27 1634–1649. 10.1105/tpc.15.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K., Li Z., Yang Z., Chen J., Wu S., Zhu X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11:e1005399. 10.1371/journal.pgen.1005399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M., Arif M., Dortay H., Matallana-Ramirez L. P., Waters M. T., Gil Nam H., et al. (2013). ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 14 382–388. 10.1038/embor.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Bulbul S., Piao W., Choi G., Paek N. C. (2017). Arabidopsis early flowering3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92 1106–1120. 10.1111/tpj.13747 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M. Y., Kim J., Paek N. C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- Sanchez S. E., Kay S. A. (2016). The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 8:a027748. 10.1101/cshperspect.a027748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Kay S. A. (2011). GIGANTEA directly activates flowering locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108 11698–11703. 10.1073/pnas.1106771108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Risopatron J. P., Jones B. J., Marc J. (2012). Circadian clock-dependent gating in ABA signalling networks. Protoplasma 249 445–457. 10.1007/s00709-011-0304-3 [DOI] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J. E., Davis S. J. (2012). Time for coffee represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24 2470–2482. 10.1105/tpc.111.095430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Jiang Y. P., Kuai B. K., Li L. (2018). circadian clock-associated 1 inhibits leaf senescence in Arabidopsis. Front. Plant Sci. 8:280. 10.3389/fpls.2018.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yang C., Gao S., Zhang W., Li L., Kuai B., et al. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7 1776–1787. 10.1093/mp/ssu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., Gonzalez-Schain N., Martin G., Diaz C., Sentandreu M., et al. (2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113 4870–4875. 10.1073/pnas.1603745113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Kusaba M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169 138–147. 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110 761–766. 10.1073/pnas.1215010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Barnaby J. Y., Tada Y., Li H., Tor M., Caldelari D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470 110–114. 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Y., Kuai B. K., Jia J. Z., Jing H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54 936–952. 10.1111/jipb.12005 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wei H., Li N., Tian W., Chong K., et al. (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11 326–337. 10.1016/j.molp.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chan Z., Gao J., Xing L., Cao M., Yu C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113 1949–1954. 10.1073/pnas.1522840113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wang W., Karapetyan S., Mwimba M., Marques J., Buchler N. E., et al. (2015). Redox rhythm reinforces the circadian clock to gate immune response. Nature 523 472–476. 10.1038/nature14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y., Oh E., Wang T., Wang Z. Y. (2016). TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7:13692. 10.1038/ncomms13692 [DOI] [PMC free article] [PubMed] [Google Scholar]