Summary
The flowers of most dicotyledons have petals that, together with the sepals, initially protect the reproductive organs. Later during development petals are required to open the flower and to attract pollinators. This diverse set of functions demands tight temporal and spatial regulation of petal development. We studied the functioning of the Arabidopsis thaliana TCP5‐like transcription factors (TFs) in petals. Overexpression of TCP5 in petal epidermal cells results in smaller petals, whereas tcp5 tcp13 tcp17 triple knockout lines have wider petals with an increased surface area. Comprehensive expression studies revealed effects of TCP5‐like TFs on the expression of genes related to the cell cycle, growth regulation and organ growth. Additionally, the ethylene biosynthesis genes 1‐amino‐cyclopropane‐1‐carboxylate (ACC) synthase 2 (ACS2) and ACC oxidase 2 (ACO2) and several ETHYLENE RESPONSE FACTORS (ERFs) are found to be differentially expressed in TCP5 mutant and overexpression lines. Chromatin immunoprecipitation–quantitative PCR showed direct binding of TCP5 to the ACS2 locus in vivo. Ethylene is known to influence cell elongation, and the petal phenotype of the tcp5 tcp13 tcp17 mutant could be complemented by treatment of the plants with an ethylene pathway inhibitor. Taken together, this reveals a novel role for TCP5‐like TFs in the regulation of ethylene‐mediated petal development and growth.
Keywords: Arabidopsis thaliana, petal, TCP5, ethylene, cell elongation, transcriptional regulation
Significance Statement
Comprehensive expression studies revealed effects of the Arabidopsis TCP5‐like transcription factors on the expression of cell cycle‐, growth regulation‐, and organ growth‐related genes. TCP5 is shown to directly inhibit ethylene biosynthesis by binding the ACS2 locus, which might explain the observed phenotypes in cell elongation during petal development. This adds TCP5 to the growing list of TCP genes involved in hormone biosynthesis and signalling.
Introduction
Flowers have been extensively studied throughout history as they are the most eye‐catching and, through fruit and seed production, economically important parts of the plant. Substantial knowledge has been acquired on identity specification and development of the different floral organs. The well‐known ABC model of flower development (Coen and Meyerowitz, 1991) explains how different genes and gene combinations specify floral organ identity. Apart from the Arabidopsis A‐class gene APETALA2 (AP2), all these genes encode members of the MADS box family of transcription factors (TFs), and their specific and unique interactions determine the identities of the four types of floral organs: carpels, stamens, petals and sepals (Coen and Meyerowitz, 1991; Immink et al., 2010).
Until recently, little was known about the control of floral organ growth and the determination of their final size and shape. Nevertheless, over recent years this topic has attracted more attention and insight has been gained into the cellular characteristics and underlying genetic factors controlling these traits. In the Arabidopsis flower, the final shape and size of sepals is largely determined by endoreduplication and the formation of giant cells (Roeder et al., 2012). Petals, on the other hand, have a morphology that requires differential regulation of cell proliferation and expansion in the basal and distal parts. The final shape and size of petals is mainly determined by cell elongation in the basal part, whereas the rate and direction of cell division determine the shape and size of the distal region (the blade) of the petal, which contains small and round conical cells (Hase et al., 2005; Irish, 2008).
Several key regulatory genes that ensure control of petal growth and development in Arabidopsis have been identified. JAGGED (JAG) for instance, is suggested to suppress premature cell‐cycle arrest in the distal part of an Arabidopsis petal (Dinneny et al., 2004; Schiessl et al., 2014). RABBIT EARS (RBE) is expressed in petal primordia, where it ensures cell proliferation in petal precursor cells (Takeda et al., 2004). AINTEGUMENTA (ANT) maintains cell proliferation in developing petals (Krizek et al., 2000; Mizukami and Fischer, 2000) and BIG PETAL (BPE) affects cell size by interfering with post‐mitotic cell expansion (Szecsi et al., 2006). A number of downstream genes have been identified, including members of the TEOSYNTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) TF family, which are thought to act downstream of JAG (Schiessl et al., 2014) and RBE (Huang and Irish, 2015).
The TCP TF family (Martín‐Trillo and Cubas, 2010) in Arabidopsis has 24 members, which can be divided into two classes based on a difference in the DNA‐binding TCP domain (Cubas et al., 1999). The class II TCPs are generally thought to act as repressors of cell division and inducers of cell differentiation (Efroni et al., 2008), and can be further subdivided into CINCINNATA (CIN)‐type and CYC/TB1‐type TCPs (Cubas et al., 1999). The roles of several of these class II TCPs in floral organ development have been studied. In Antirrhinum, for example, CYC is responsible for the asymmetric development of petals (Luo et al., 1995) whereas CIN promotes growth and cell division in these organs (Crawford et al., 2004).
The CIN‐type TCPs are represented in Arabidopsis by the Jagged and Wavy (JAW) TCPs (TCP2, ‐3, ‐4, ‐10 and ‐24) and the TCP5‐like genes (TCP5, ‐13 and ‐17). All five JAW TCPs are targeted by the same microRNA, miR319, which, upon overexpression in the jaw‐D mutant, simultaneously downregulates the expression of TCP2, ‐3, ‐4, ‐10 and ‐24 (Palatnik et al., 2003). This downregulation gives rise to a delay in the arrest of cell proliferation in the margin and distal end of organs, such as leaves and petals, resulting in overproduction of cells in these regions (Nath et al., 2003; Palatnik et al., 2003).
It has been suggested that the other CIN‐genes, TCP5, TCP13 and TCP17, although not targeted by miR319, are responsible for similar processes in a redundant manner (Efroni et al., 2008). However, mutants in the TCP5‐like genes show some phenotypic differences when compared with the JAW‐TCP mutants and have larger leaves, for example, but lack the jaw‐D characteristic crinkled phenotype. The constitutive overexpression of TCP5 results in a smaller petal area (Huang and Irish, 2015), whereas downregulating all three TCP5‐like genes by ectopically expressing an artificial micro‐RNA (also known as miR:3TCP) and a triple tcp5 tcp13 tcp17 knockout results in larger petals and leaves (Efroni et al., 2008; Huang and Irish, 2015). The single tcp13 and tcp17 mutants show no phenotypic alterations during petal development, whereas the single tcp5 mutant grows a slightly wider petal claw (Huang and Irish, 2015). This suggests that, of these three genes, TCP5 is the major player in petal development. Further analyses revealed that the reduced petal size in TCP5 overexpressing lines versus the increased size in miR:3TCP is attributed to the duration of growth and cell differentiation (Huang and Irish, 2015).
In this study, we aim to shed light on the molecular mode of action underlying the functioning of TCP5 in flowers, with a special focus on its function in petals. We show that TCP5 is expressed in petals of stage 9 flowers and onwards and hypothesize that it is involved in controlling cell expansion, which is initiated at this stage of petal development (Irish, 2008). Furthermore, we analysed gene expression by RNA‐sequencing analysis in whole inflorescences and petals of tcp5 tcp13 tcp17 knockout mutants and in constitutive and inducible TCP5 overexpression lines. This revealed a possible role for TCP5 in ethylene biosynthesis and signalling. Ethylene is known to play a role in the growth and elongation of (petal) cells (Chen et al., 2013; Ma et al., 2008; reviewed by Pierik et al., 2006). The involvement of TCP5 in ethylene biosynthesis was confirmed by an altered rate of ethylene accumulation in the analysed mutants, and complementation experiments with an ethylene pathway inhibitor were able to recover the tcp5 tcp13 tcp17 phenotype to wild type. Together with showing direct binding of TCP5 at the ACS2 promoter, these findings provide strong evidence for a central role of TCP5 in ethylene‐mediated control of petal growth.
Results
TCP5 is expressed during cell elongation stages of petal development, regulating the final shape and size of Arabidopsis petals
To visualize the spatial expression pattern of TCP5 in petals, a transgenic gTCP5‐GFP line was generated. Expression of TCP5 seems to be uniformly distributed in the petal and occurs from stage 9 onwards (Figure S1 in the online Supporting Information), which is slightly earlier than previously reported (Huang and Irish, 2015). However, expression in these early stages is very low in comparison with later developmental stages. Analysis of TCP5 expression during later developmental stages was done by quantitative (q)RT‐PCR and showed constant TCP5 expression until stage 15/16, when petal senescence occurs (Figure S1D; stages according to Smyth et al., 1990). Furthermore, the qRT‐PCR analyses revealed that the expression of TCP13 increased during later petal development whereas hardly any TCP17 expression was detected in any of the analysed stages.
To further understand the function of TCP5, we decided to study the effect of TCP5 overexpression in the epidermal layer of petals, because growth appears to be controlled and regulated to a large part from this cell layer (Anastasiou et al., 2007; Savaldi‐Goldstein et al., 2007; Urbanus et al., 2010). For this purpose, the promoter of the Arabidopsis L1‐specific gene MERISTEMLAYER1 (AtML1) was used; this drives expression in the epidermis of all above‐ground organs (Lu et al., 1996). In this experiment, TCP5 was tagged with GFP, enabling the protein inside tissues to be visualized and tracked (Figure S2A). Confocal analysis showed that the expression of TCP5‐GFP in ATML1 pro :TCP5‐GFP plants is limited to the epidermis, as expected, and that no migration to underlying layers occurs (most likely due to the nuclear entrapment of the TCP5‐GFP fusion protein; Figure S2B). Quantitative RT‐PCR analysis showed upregulation of TCP5 expression of approximately 3.5 times relative to Col‐0 (Figure S2C). Phenotypical analysis of ATML1 pro :TCP5‐GFP plants during the vegetative stage of development showed the development of long elongated leaves, from which the blade was curled downward at the periphery and had a smaller surface area (Figure S2D, E). This phenotype is completely opposite to the leaf phenotype of the triple tcp5 tcp13 tcp17 knockout mutant (Efroni et al., 2008; Huang and Irish, 2015). In addition, we observed significantly smaller petals of ATML1 pro :TCP5‐GFP flowers compared with wild‐type petals (Figure S3), which is opposite to the petal phenotype of the triple knockout line. Notably, the petal phenotype observed in the ATML1 pro :TCP5‐GFP plants is similar to the 35S pro :TCP5 phenotype previously observed by Huang and Irish (2015), revealing that specific ectopic expression of TCP5 in the epidermal layer is sufficient to trigger this developmental response.
Next, we phenotyped a single tcp5 mutant line and a tcp5 tcp13 tcp17 triple mutant. Detailed phenotyping showed that petals of the tcp5 tcp13 tcp17 mutant grow significantly longer and wider (Figure S3), which is in line with previously published data (Huang and Irish, 2015). The single knockout of tcp5 showed no differences from the wild type for these specific macroscopic characteristics (Figure S3).
TCP5 and TCP5‐likes alter conical cell morphology
To unravel the cellular causes of the observed petal phenotypes we took a closer look using scanning electron microscopy (SEM). We focused initially on the adaxial side of the distal part of the petal, where conical cells are located. In all three analysed lines (tcp5, tcp5 tcp13 tcp17 and ATML1 pro :TCP5‐GFP) the cells in the petal blade are bigger and more irregularly shaped than the round conical cells in wild‐type petals (Figure 1a, d). The shape of the conical cells was quantified by calculating the ratio of cell length to cell width, resulting in a measure of cell roundness. Conical cells at the distal end of the petal are expected to be (close to) perfectly round (Szécsi et al., 2014), which could be confirmed for Col‐0 (Figure 1e). In contrast, the ATML1 pro :TCP5‐GFP overexpressor plants and the T‐DNA knockout lines showed a decrease in conical cell roundness.
Figure 1.
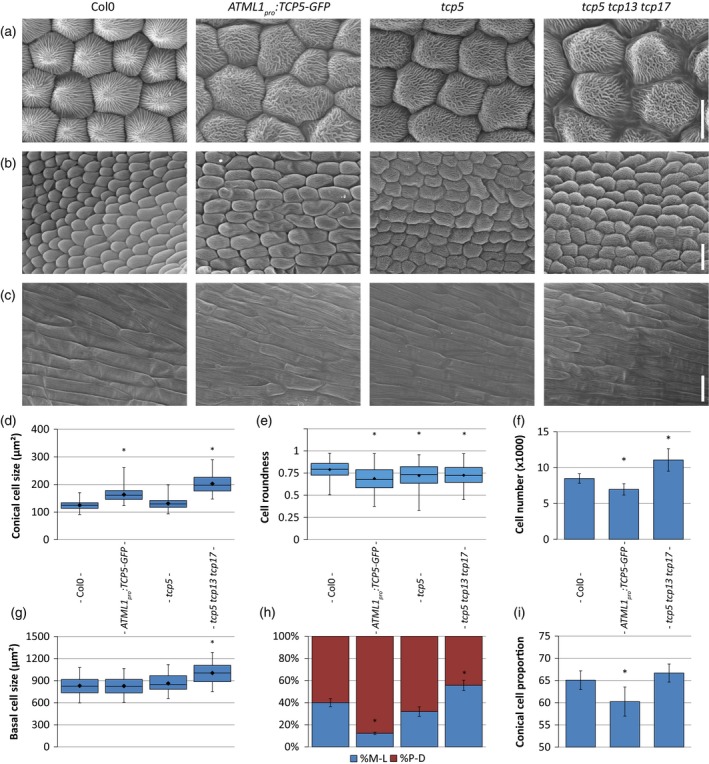
Overview of cellular phenotypes in petals of Col‐0, ATML1 pro :TCP5‐GFP, tcp5 and tcp5 tcp13 tcp17 mutant plants.
Scanning electron micrographs of the distal (a), central (b) and proximal (c) parts of the petal. Boxplots show the cell sizes of conical cells at the distal part of the petal in (d). (e) The roundness of the conical cells and (f) the total number of conical cells. (g) The size of basal cells. (h) The direction of cell elongation, medial–lateral (blue) or proximodistal (red). (i) The proportional area of conical cells in the petal. Scale bars: (a) 10 μm; (b), (c), 20 μm. Box plots (d, e and g) show mean (dot on horizontal line), median (middle horizontal line), second to third quartiles (box), and minimum and maximum ranges (vertical lines). The bars (f, h and i) indicate means and SDs. In both cases an asterisk indicates that the mean is significantly different from wild type (P < 0.05, Student's t‐test). At least 50 cells from 12 petals were analysed. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Subsequently, we combined the cell shape and size measurements to infer the directionality of cell elongation in petal conical cells. We noted that in the wild type the vector (direction) of elongation of the conical cells is quite random, which is expected for the more or less round petal blade cells (Figures 1 h and S4B, C). However, in ATML1 pro :TCP5‐GFP the direction appeared to be more proximodistally oriented, which explains the narrower petals. In the case of the tcp5 tcp13 tcp17 T‐DNA knockout line this directionality seemed to be more medial‐lateral (ML). This observation is in line with the observed wider petals in this mutant background (Figure S3A, C). The observation that the petal area is smaller in the ATML1 pro :TCP5‐GFP mutant compared with the wild type, although the cells are larger, can be explained by the reduction in total cell number in this mutant (Figure 1f).
Conical cells at the distal end of a petal possess a dome‐shaped structure with cuticular ridges that run from the edges of the cell (where the cell touches its neighbouring cells) to the top of the conical cell (Panikashvili et al., 2011) (Figure 1a). In the case of the single tcp5 knockout, these ridges are oriented similarly to wild‐type cells towards the tip of the cone, despite the differences in cell size and shape; however, in the case of ATML1 pro :TCP5‐GFP and the triple mutant, the ridges seem to be running more randomly and parallel to each other.
Finally, we investigated potential effects of TCP5 alterations on cell elongation at the base of the adaxial side of the petal and found that these cells are significantly larger in the tcp5 tcp13 tcp17 mutant (Figure 1c, g). Nonetheless, no significant differences were observed in either the single tcp5 mutant or the ATML1 pro :TCP5‐GFP overexpression line.
Molecular analysis of tcp5, tcp5 tcp13 tcp17 and ATML1 pro :TCP5‐GFP
To obtain insight into the potential molecular causes of the observed phenotypes, an RNA‐seq analysis was performed on dissected petals of stage 12 flowers of Col‐0 wild type, tcp5, tcp5 tcp13 tcp17 and ATML1 pro :TCP5‐GFP lines. We found a total of 2682 genes differentially expressed in ATML1 pro :TCP5‐GFP petals compared with the wild type, 1581 in tcp5 and 1519 in the tcp5 tcp13 tcp17 triple knockout. Of these, 345 differentially expressed genes (DEGs) were found in all three mutant backgrounds (Figure S5C, Table S1).
Subsequently, a Gene Ontology (GO) enrichment analysis was done to identify the biological and molecular processes affected by TCP5. To produce a first overview, the GO Slim method was chosen, which gives broad insight into ontology. A summary of GO terms found to be overrepresented in multiple samples is shown in Figure S6 (full list in Table S2). Previously, CIN‐TCPs have been described as being involved in cell growth and differentiation (Efroni et al., 2008), and the petal phenotypes of the TCP5 overexpressor and tcp5 tcp13 tcp17 mutants are in agreement with a function of TCP5‐like genes in these cellular processes (present work; Huang and Irish, 2015). Enrichment was found for GO terms such as cell wall (GO:0005618), anatomical structure morphogenesis (GO:0009653), membrane (GO:0016020) and the regulation of cell size (GO:0008361), cell differentiation (GO:0030154), cell growth (GO:0016049) and growth (GO:0040007). Nevertheless, steady‐state differential expression in stable mutant backgrounds does not provide information about the direct targets of the TCP5 TF. We therefore generated an inducible transgenic 35S pro :TCP5‐GR Arabidopsis line, which allowed us to use the glucocorticoid receptor to activate TCP5 at specific moments during development by the application of dexamethasone (DEX) (Aoyama and Chua, 1997). Continuous DEX treatment resulted in phenotypes in line with those of the stable ATML1 pro :TCP5‐GFP line (Figure S7). To shed light on the molecular mode of action of TCP5 in flower development, inflorescences were harvested after 2 and 8 h of DEX treatment. This two‐step analysis allows us to distinguish between the direct and indirect effects of TCP5 induction. We found 1057 genes differentially regulated after 2 h of treatment, which after 8 h had increased to 1350 genes. In these lists of significantly differential regulated genes upon induction of TCP5 we found more downregulated than upregulated genes (Figure S5B, Table S1).
For a detailed analysis of the differentially expressed genes, we performed a full GO term analysis for genes differentially expressed at 2 h after induction (T0–2) and genes differentially expressed between 2 and 8 h after induction (T2–8). Analysis of both time points revealed a profile strikingly rich in terms related to plant defence and hormonal responses (Figure 2a, b, Table S3).
Figure 2.
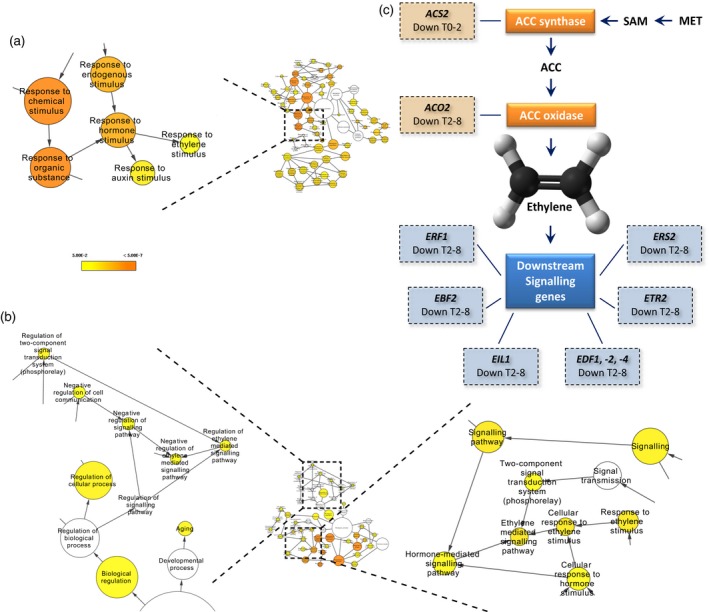
Over‐representation of ethylene signalling genes among the differentially expressed genes.
(a), (b) Hormone related Gene Ontology (GO) terms for (a) differentially expressed genes after 2 h induction (T0–2) and (b) differentially expressed genes after 8 h induction (T2–8). The colour bar in (a) and (b) indicates significance levels for the GO categories (FDR < 0.05), The circle size represents the number of genes present for a particular GO term. In the case of, for example, response to ethylene biosynthesis, the size corresponds to 12 and 17 genes, respectively.
(c) Differentially expressed genes in the ethylene biosynthesis and signalling pathway in either the first time point (T0–2) or the second (T2–8); lines indicate that, for example, ACS2 is a 1‐amino‐cyclopropane‐1‐carboxylate (ACC) synthase and link the different gene classes to individual genes in that class showing differential expression. SAM, S′‐adenosyl‐l‐methionine; MET, methionine.
Two hormones known to be growth regulators are found in our GO term overrepresentation analysis. Response to auxin stimulus (GO:0009733) is found in all samples, and a striking finding in both the 35S pro :TCP5‐GR induction as well as the stable ATML1 pro :TCP5‐GFP was the presence of the GO term response to ethylene stimulus (GO:0009723) (Figure 2, Table S3). Ethylene is known to influence cell expansion in petals (Pei et al., 2013), and therefore we focused our further experiments on this hormone. Mutual targets of both ethylene and auxin are the ARGOS and ARGOS‐like (ARL) genes, which all appear to be upregulated immediately after TCP5 induction (Figure 3). Additionally, in the list of differentially expressed genes we found the ethylene biosynthesis genes 1‐AMINOCYCLOPROPANE‐1‐CARBOXYLIC ACID SYNTHASE 2 (ACS2), which catalyses the rate‐limiting step in ethylene biosynthesis (van der Graaff et al., 2006), and ACC‐oxidase 2 (ACO2), which converts ACC into ethylene. Both ACS2 and ACO2 are upregulated in the tcp5 and tcp5 tcp13 tcp17 mutants and downregulated in the transgenic lines in which TCP5 is overexpressed. Similarly, many ETHYLENE RESPONSE FACTORS (ERFs), which are downregulated in our overexpressing lines, are upregulated in the knockout lines (Figure 3).
Figure 3.

Differentially expressed genes.
Heatmap of differentially expressed genes categorised by function and/or process involved. The colour scale indicates the log2(fold‐change), based on fragments per kilobase of transcript per million mapped reads (FPKM) values, for upregulated genes (green) through genes that show no difference (white) towards downregulated genes in red.
TCP5 inhibits ethylene biosynthesis
The differential gene expression analysis led to the hypothesis that TCP5 primarily inhibits ethylene biosynthesis and secondly ethylene response. We indeed observed a significant increase in accumulated ethylene in the inflorescences of the tcp5 and tcp5 tcp13 tcp17 knockout mutants versus a significant decrease in the ATML1 pro :TCP5‐GFP overexpressor (Figure 4a).
Figure 4.
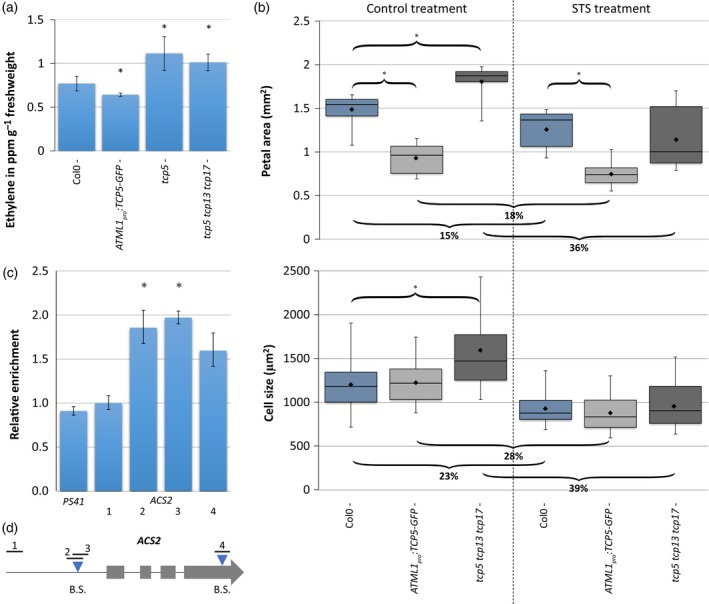
TCP5 inhibits ethylene biosynthesis.
(a), (b) Quantification of ethylene production of inflorescences (per hour per gram fresh weight) (a) and petal area and petal cell size analysis in control (left) and silver thiosulphate (STS) (right) treatment (b). Curly brackets with asterisks above indicate a significantly different area versus Col‐0 wild type. Curly brackets below show the percentage of decline in petal area and cell size in STS‐treated samples compared with the control treatment. Treatment with STS had a significant effect for all plants tested. Bars in (a) show the mean of at least five biological replicates. The boxplot in (b) shows mean (dot on horizontal line), median (middle horizontal line), second to third quartiles (box), minimum and maximum ranges (vertical lines). Twelve petals were analysed for petal area, and at least 50 cells from these petals were analysed for size. In both cases an asterisk indicates a significantly different mean from the wild type (P < 0.05, Student's t‐test).
(c) Binding of TCP5‐GFP to the ACS2 locus confirmed by chromatin immunoprecipitation (ChIP)‐qPCR. Binding of TCP5 is tested for regions 1–4, of which regions 2, 3 and 4 cover a putative binding site, indicated in (D). PS41 is the negative control. The data were normalized against ACT2. Means ± SEM of three biological replicates are shown. The asterisks above the columns indicate a significant difference between TCP5:GFP and input (P < 0.05, t‐test).
(d) Schematic diagram of the ACS2 locus. Grey boxes indicate the exons, blue triangles indicate the positions of putative TCP‐binding motifs (B.S.) and black lines show the amplified fragments in ChIP‐qPCR. [Colour figure can be viewed at http://wileyonlinelibrary.com]
If the disruption in ethylene biosynthesis/signalling is (at least partly) accountable for the observed phenotypes, exogenous alteration of the ethylene pathway should be able to restore mutant phenotypes. For that purpose, we conducted an experiment in which we added silver thiosulphate (STS), an inhibitor of the ethylene pathway (Beyer, 1979). After application of STS, which should block the enhanced ethylene response in the tcp5 tcp13 tcp17 mutant, petal and cell sizes returned to wild‐type dimensions (Figure 4b).
Ethylene has a well‐studied regulatory role in Arabidopsis leaf senescence (Weaver et al., 1998; Koyama, 2014; Kim et al., 2015), but a lot less is known about its role in petal senescence in Arabidopsis (Wagstaff et al., 2009; Rogers, 2013). The RNA‐seq data revealed a number of differentially expressed NAC TFs (Figure 3) known to act downstream of the ethylene signalling pathway and to be involved in senescence‐related processes: NAC019, NAP, SHG, NAC3, ORS1 and ORE1 (Kim et al., 2014). Their direction of differential regulation perfectly fits the increase or decrease in ethylene biosynthesis and signalling genes in our mutants, prompting us to take a closer look at petal senescence in our lines.
However, Arabidopsis petals have been questioned as suitable organs for studying senescence since a clear progress of senescence is lacking (Jones 2009). Pollination triggers senescence in these organs, after which they abscise without substantial wilting in a very short time, making differences hard to detect. Indeed, we could not observe any differences related to senescence in petals of the various mutant lines comparison with the Col‐0 wild type.
Finally, we tested the binding of TCP5 to putative TCP‐binding motifs in the promoter and genic regions of ACS2 (Figure 4d). We compared the immunoprecipitated DNA of inflorescences of gTCP5‐GFP with input DNA by qPCR and found a significant enrichment for the putative TCP‐binding site in the promoter (Figure 4c), showing direct binding of ACS2 by TCP5 in vivo.
Discussion
In this study, the roles of TCP5‐like genes in Arabidopsis petal development were studied with a special focus on the underlying molecular mechanisms. During organ development, cell differentiation is preceded by a period of rapid cell division. This holds true for most developing organs and tissues, but has been most extensively studied in leaves (Andriankaja et al., 2012). Petal cell differentiation seems to be a more gradual process along the proximodistal (base to tip) axis (Sauret‐Güeto et al., 2013), in contrast to the prompt transition from proliferation to cell differentiation in leaves (Andriankaja et al., 2012). This makes petals interesting organs in which to study growth and development. Petals grow from petal primordia that emerge from stage 5 flower buds (Smyth et al., 1990). Initially, the petal primordia remain small, but the cells start to divide faster from stage 7 and 8 of flower development onwards and cell division rate reaches a plateau around stages 9–11, after which it rapidly declines. During petal development, stage 9 marks the onset of cell expansion (Irish, 2008), which is exactly the moment when TCP5 expression becomes apparent.
A role for TCP5 in later petal developmental stages
Based on our results from the detailed phenotyping of petal development and previously published data (Huang and Irish, 2015), TCP5 seems to be involved in petal development from the onset of cell elongation (stage 9) and into the maturation phase until the later stages of flower development (stage 14–15). These observations led to the hypothesis that TCP5 is mainly involved in cell elongation, which was confirmed by whole transcriptome analysis using RNA‐seq and subsequent Gene Ontology analysis, in which we identified overrepresentation of the GO terms regulation of cell size (GO:0008361) and cell growth (GO:0016049). Hence, in line with the observed morphological changes in the mutants, the RNA‐seq results also point to cell elongation‐related processes, suggesting a major role for TCP5 in the regulation of petal growth. Furthermore, these results show which growth‐related genes are responsible for the observed altered phenotypes.
Previous research has shown that TCP5 is repressed by RBE during the early stages of petal development, limiting the function of TCP5 towards later stages of petal development (Huang and Irish, 2015). Interestingly, TCP4, a JAW TCP, is also repressed by RBE during the early stages of petal development (Li et al., 2016) and there is considerable overlap in the petal phenotypes of the different TCP5‐like and JAW TCP gene mutants. For example, a mutation in the CIN gene of Antirrhinum, which is orthologous to the CIN TCPs in Arabidopsis (TCP2, ‐3, ‐4, ‐5, ‐10, ‐13, ‐17 and ‐24; Uberti Manassero et al., 2013), results in flattening of the conical cells as well as an increase in cell size in certain petal regions (Crawford et al., 2004), which is in perfect agreement with our data on the tcp5 tcp13 tcp17 knockout. Furthermore, in the loss‐of‐function mutant miR319a 129, where the JAW TCPs are overexpressed, petals are significantly smaller and narrower (Nag et al., 2009), which is a phenocopy of the TCP5 overexpression phenotype.
It might be expected that the conical cell phenotype of jaw‐D plants resembles that of tcp5 tcp13 tcp17, which would further strengthen the hypothesis that the JAW‐like TCPs and the TCP5‐like TCPs share regulatory functions during petal development (Koyama et al., 2007; Efroni et al., 2008). Additionally, the similarity in phenotypes might be explained by the fact that TCP5‐like proteins preferentially interact with JAW‐like TCP proteins to form heterodimers (Danisman et al., 2013).
Another phenotype shared by the Antirrhinum cin mutant and our tcp5 mutant and overexpression lines is the clear difference in cuticle ridges on conical cells compared with that on conical cells of wild‐type petals. The precise mechanism that controls the patterning of the petal cuticle is still unknown but is thought to be linked to cell shape, because mutants defective in cutin biosynthesis show impaired cell expansion (Noda et al., 1994; Cominelli et al., 2008; Glover et al., 2016). This seems to be confirmed in our experiments, because we also observed effects on both cuticle ridge formation and conical cell sizes. Since the effect of altered TCP5 expression is primarily on genes involved in cell elongation, this implies that the cuticle patterning defect is a consequence, not the cause, of the altered petal cell size in the different tcp5 mutants.
Does the L1 layer have an important role in petal development related to TCP5 action?
The petal phenotypes observed in the ATML1 pro :TCP5‐GFP lines are similar to the 35S pro :TCP5 phenotypes published previously (Huang and Irish, 2015). Although TCP5 is expressed normally throughout the different cell layers of the petal, the fact that it can act non‐cell autonomously from the epidermal layer is in agreement with the hypothesis that control of organ growth is mediated to a large extent by the epidermal L1 cell layer (Savaldi‐Goldstein et al., 2007). Of note is the differential expression of several genes involved in epidermal specification in the ATML1 pro :TCP5‐GFP overexpression line, as well as in the tcp5 and tcp5 tcp13 tcp17 mutants, including genes such as CAPRICE‐LIKE MYB3 (CPL3) (Grebe, 2012), DEFECTIVE KERNEL1 (DEK1) and its downstream‐acting HD‐ZIP IV‐encoding genes HDG11 and HDG12 (Galletti et al., 2015) (Figure 3). Although the overexpression of TCP5 in the epidermal L1 layer is ectopic and petals are derived solely from L1 and L2 layers of the floral meristem (Jenik and Irish, 2000), the fact that this differential expression is also seen in the loss‐of‐function mutants suggests a specific growth regulatory function for TCP5 in, and coordinated from, the epidermis.
TCP5 controls petal growth and development via ethylene biosynthesis and signalling
Analysis of the genes differentially expressed after TCP5 induction showed enrichment for processes related to stress, defence responses and hormone responses. This might not be surprising, since TCPs have been described to act directly on hormonal pathways that regulate both defence responses and plant growth (reviewed by Nicolas and Cubas, 2016; Li, 2015; Danisman, 2016). Furthermore, it is well known that there is a trade‐off between growth and defence (Todesco et al., 2010). We therefore hypothesise that TCP5 primarily targets and regulates certain hormonal pathways which ultimately lead to the growth phenotypes observed here.
Further analysis of the RNA‐seq results pointed our attention towards ethylene signalling (GO:0009723), as it was overrepresented in the GO term analysis upon DEX induction of the 35S pro :TCP5‐GR plants. All samples showed a deregulation of ethylene biosynthesis genes as well as numerous downstream signalling elements. Analysis of ethylene production in the headspace of inflorescences confirmed the differential gene expression. We found a reduction of ethylene levels in the ATML1 pro :TCP5‐GFP overexpressor and an increase of ethylene in the headspace of both the tcp5 and tcp5 tcp13 tcp17 knockout mutants, suggesting a role for TCP5 as an inhibitor of ethylene biosynthesis. Alternatively, these alterations can be caused by effects on the expression of ethylene biosynthesis and signalling genes, because the ethylene signalling pathway is known to have feedback regulatory loops (Rai et al., 2015; Prescott et al., 2016) and hence effects downstream of ethylene can ultimately result in differences in ethylene production.
Next to ethylene, the response to auxin (GO:0009733) was also overrepresented in our differential gene lists. Both auxin (Varaud et al., 2011) and ethylene (Ma et al., 2008; Chen et al., 2013; Pei et al., 2013) have been shown to function in cell proliferation and elongation during petal development. Analysis of the two time points upon TCP5 activation revealed differential expression in ethylene biosynthesis in the first time point (T0–2), whereas only the second time point (T2–8) shows differential expression in downstream ethylene signalling genes (Figure 2c). This suggests that TCP5 has a direct effect on the biosynthesis of ethylene, which is further confirmed by our finding that TCP5 binds the promoter of ACS2 in vivo (Figure 4c).
Previous research has shown that reduced ethylene signalling leads to an increase in petal size due to an increase in conical cell size (e.g. in the loss‐of‐function mutant ein2) (Pei et al., 2013). This effect on petal development was confirmed by us by an analysis of petal size in ein2 and two other mutant lines defective in the ethylene signalling and transcription cascade, ein3 and etr1 (Figure S8). However, there is evidence to suggest that ethylene can both induce cell elongation (reviewed by Van de Poel et al., 2015; Feng et al., 2017) and repress cell growth, depending on the exact concentration and cellular context (Pierik et al., 2006; Dugardeyn and Van Der Straeten, 2008). As a consequence, a low ethylene concentration can have the same effect on cell elongation as a high concentration (Abts et al., 2014; Lv et al., 2018).
The latter results seem to corroborate the data from our mutants that all show an increase in cell size, regardless of an increase or decrease in ethylene biosynthesis. We suggest that there is a tight balance between the ethylene concentration and its effect on cell size, as opposed to a linear relationship, linked to an ‘on’ or ‘off’ status for cell elongation. A deviation from ‘normal’ physiological ethylene concentrations then results in an enhanced cell elongation phenotype, as previously seen in root cells (Abts et al., 2014; Lv et al., 2018). This is further strengthened by the fact that we revert the phenotype of the tcp5 tcp13 tcp17 line (which produces more ethylene but grows bigger petals with bigger cells) to a wild‐type phenotype by blocking the ethylene signalling pathway with STS, demonstrating that a large part of the mutant phenotype is caused by altered ethylene signalling. Moreover, blocking the ethylene pathway by STS also reduced petal growth in our Col‐0 control plants, albeit to a much lesser extent than that of the tcp5 tcp13 tcp17 mutant.
The perturbation in the ethylene balance might also lead to the observed differential direction of cell elongation, which is known to be influenced by the effect that ethylene has on microtubule orientation (Le et al., 2004; Plett et al., 2009). In addition, genes involved in directional cell growth, such as MICROTUBULE ASSOCIATED PROTEIN 18 (MAP18) (Wang et al., 2007), were found to be deregulated. This could in part also explain the defect in cuticle patterning, as the composition of cuticular ridges has been linked to differences in cell morphology (Shi et al., 2011) and microtubule orientation was recently shown to control conical cell shape (Ren et al., 2017).
Downstream of ethylene biosynthesis are numerous genes involved in cell elongation processes, which might account in part for the phenotypes observed in our mutants. For example, auxin is known to induce the production of ethylene (Tsuchisaka and Theologis, 2004; Pierik et al., 2006). Interestingly, the ARGOS gene family is involved in a negative feedback loop in ethylene signalling, downstream of ethylene biosynthesis (Rai et al., 2015). Upregulated by both ethylene and auxin, it inhibits a proper downstream ethylene signalling response. In our induction experiment, ARGOS and ARGOS LIKE 1, ‐2, and ‐3 (ARL1, ‐2 and ‐3) were upregulated after the first time point. At the second time point, however, these genes were downregulated. This might point towards initial upregulation by TCP5 after which the ethylene biosynthesis is downregulated, possibly both through the activity of ARGOS and the ARLs and directly by TCP5. Consequently, downregulation of ethylene biosynthesis would then downregulate ARGOS and the ARLs.
In conclusion, we have demonstrated that TCP5 is an important regulator of growth and development of the Arabidopsis petal. However, in contrast to directly regulating growth regulatory genes we show here that one of the functions of TCP5 is to act as a regulator of ethylene biosynthesis, after which the downstream targets of ethylene signalling are responsible for various of the observed developmental phenotypes. Several TCP TFs have been described to regulate hormone synthesis, transport and signal transduction for a number of key plant hormones (reviewed by Nicolas and Cubas, 2016). However, no TCP protein has yet been linked to ethylene, and we provide proof in this study for a tight association between TCP5 and ethylene biosynthesis and signalling.
Experimental Procedures
Plant materials and growth conditions
The triple T‐DNA insertion mutant tcp5 tcp13 tcp17 contains the mutant alleles tcp5‐1 (SM_3_29639), tcp13 (SM_3_23151) and tcp17 (SALK_147288), all three of which have insertions in the coding regions. The tcp5‐1 single mutant was kindly provided by Dr Koyama and the triple tcp5 tcp13 tcp17 was a gift from Professor Eshed. All three lines defective in the ethylene signalling and transcription cascade (etr1‐1, ein2‐5 and ein3‐2) were obtained through the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info/). Plants were grown under long‐day conditions (16 h/8 h light/dark cycle) at 21°C on Rockwool and received 1 g l−1 Hyponex plant food solution twice a week.
Constructs and transformation
The coding sequence (without a STOP codon) of TCP5 was amplified by PCR and recombined into the modified pK7FWG2 destination vector, containing the AtML1 (AT4G21750) promoter in place of CaM35S (Urbanus et al., 2010), resulting in ATML1 pro :TCP5‐GFP. Simultaneously, the coding sequence was recombined in pARC146 (Danisman et al., 2012), resulting in the destination vector 35S pro :TCP5‐GR. Next, a 3‐kb promoter region was cloned together with the TCP5 coding sequence and recombined into pMDC204 (Curtis and Grossniklaus, 2003) resulting in the destination vector pTCP5:TCP5‐GFP. All primer sequences can be found in Table S4. All three constructs where transformed into Agrobacterium tumefaciens strain C58C1‐PMP90. Arabidopsis transformation was conducted by the floral dip method (Clough and Bent, 1998). The T1 seeds were selected on germination medium containing 30 μg ml−1 kanamycin for 2 weeks, after which rooting green T 1 seedlings were transferred to Rockwool and grown until seed set. The following T 2 generation was checked for expression of the transgene by reverse‐transcription PCR. Col‐0 was used as the wild type and reference in all experiments.
Petal and cell measurements and analyses
Petals were collected from fully grown flowers at stage 14–15 (Smyth et al., 1990) and subjected to further analysis – either by epidermal imprinting for cellular phenotype analysis or by overall shape and size phenotyping. We used three plants, of which 12 petals were analysed from four or five flowers; 50 cells from each petal were subject to analysis.
Petals destined for SEM were fixed in paraformaldehyde (4% in phosphate buffer) and dehydrated in absolute ethanol, critical point dried (Balzers CPD 030, https://www.oerlikon.com/balzers/com/en/), mounted onto metallic stubs and gold‐sputtered (with 40‐nm colloidal gold, Balzers SCD 004). Observation and documentation were performed in a LEO 435 ( http://www.leo-usa.com/) VP scanning electron microscope. Digital images were obtained with LEOUIF software. Samples destined for optical microscopy were mounted in a drop of glycerol on a microscopy side, covered with a cover slip, observed and documented under a Zeiss Axioskope optical microscope ( https://www.zeiss.com/) equipped with a digital camera.
The microscopic drawings of the abaxial petal epidermis were scanned for digitisation. Conical cells from the most distal third of the petal were digitised, but the epidermal cells at the petal margin were not considered. At least 19 petals were imaged for each genotype. Data on cell area and cell roundness were collected from digital images, essentially following the protocol described by Andriankaja et al. (2012). Colour gradients according to cell area as well as vectors of maximum and minimum cell diameters were assessed from each cell image using the ImageJ macro language ( http://rsbweb.nih.gov/ij/). At least 10 000 cells from each genotype were analysed. A two‐sample t‐test was used to distinguish between mutant and wild type.
Epidermal imprinting
An addition‐reaction silicone elastomer polyvinyl siloxane (dental resin kit) was used to obtain imprints of petals epidermis. Equal amounts of the two types of paste from the kit were mixed together to make the working dental resin. The resin was placed onto a glass slide with the help of a toothpick, forming a layer 1–2 mm thick. Using tweezers, petals were placed on the resin layer with the adaxial surface facing the resin for at least 5 min or until the resin solidified. After gently removing the petals from the resin the impression was left to fully set for 5 min. Then clear nail polish was applied to the surface of the resin impression and left to completely dry for approximately 20 min. The nail polish layer was carefully peeled off from the resin and placed onto a new glass slide with the imprinted surface face up. The imprints were observed and imaged using a differential interference contrast microscope for further image analysis by ImageJ.
Tissue sampling and RNA isolation for qRT‐PCR and RNA‐sequencing
Petals of stage 12 flowers (Smyth et al., 1990) were harvested from 50 flowers per biological replicate. For induction of 35S pro :TCP5‐GR, inflorescences were treated with a DEX induction solution (2 μm dexamethasone, 0.01% (v/v) ethanol, and 0.01% Silwet L‐77) or with an identical mock solution that lacked DEX. Whole inflorescences were harvested 0, 2 and 8 h after induction and RNA was isolated using the InviTrap® Spin Plant RNA Mini Kit (Stratec Molecular, https://www.molecular.stratec.com/) according to the manufacturer's protocol. TURBO™ DNase (ThermoFisher Scientific, https://www.thermofisher.com/) was used to clean the RNA samples from DNA.
iScript reverse transcriptase (Bio‐Rad, https://www.bio-rad.com) was used for cDNA synthesis. The complementary DNA made this way was used for qRT‐PCR using the SYBR Green mix from Bio‐Rad ( http://www.bio-rad.com/). The reference genes used for all expression analyses were a SAND family gene, At2G28390, and the TIP41‐like gene At4G34270, both ‘superior reference genes’ (Czechowski et al., 2005).
Library preparation for whole‐genome RNA sequencing was done using the Illumina Truseq Library Preparation Kit. Library quality was evaluated using a Bioanalyzer and an RNA Nano 6000 kit (Agilent, http://www.agilent.com/). RNA concentrations were determined using the Xpose ‘DSCVRY’ (Trinean, https://www.trinean.com). The libraries were then sequenced with the Hi‐Seq 2500 system (Illumina, https://www.illumina.com/).
Gene expression and gene set enrichment analysis
Libraries of three biological replicates were sequenced and analysed using the Bowtie–Tophat–Cuffdiff (BTC) pipeline (Trapnell et al., 2012). Differential gene expression was based on FPKM (fragments per kilobase of transcript per million mapped reads) values and determined for all samples using Col‐0 as the control. The cut‐off was set at a false discovery rate (FDR) < 0.05 in all analyses performed. The RNA‐seq data are made available via NCBI and can be accessed through GEO accession number GSE103762.
The BINGO 3.03 plug‐in (Maere et al., 2005), implemented in CYTO‐SCAPE 2.81 (Shannon et al., 2003), was used to determine and visualize the GO enrichment according to both GO Slim (Table S2) and GO enrichment (Table S3) categorization. A hypergeometric distribution statistical testing method was applied to determine the enriched genes and the Benjamini–Hochberg correction was performed in order to limit the number of false positives (FDR < 0.05).
Ethylene treatment and quantification
Flowering plants with an inflorescence of approximately 4 cm were treated every other day for 1 week with a 50 μm STS solution by floral dipping. Petals were harvested from stage 14–15 flowers and subjected to further analysis, either by epidermal imprinting for cellular phenotype analysis or by overall shape and size phenotyping. We used three plants, 12 petal of which were analysed from four or five flowers.
Ethylene production (EP) was measured by putting the top 1.5 cm of an Arabidopsis inflorescence in a 5‐ml headspace vial (one inflorescence per vial). To prevent wilting of the inflorescence, 1 ml of MS10 (Murashige and Skoog, 1962) was poured into the vial into which the inflorescence was placed. Before being sealed, the headspace vials were kept open for 1 h to allow the wound‐induced ethylene burst to subside. After closure the vials were kept for 7 h at 21°C to accumulate sufficient endogenous ethylene. Then, to determine the ethylene content, 0.5 ml of headspace gas was injected into a Thermo Focus Gas Chromatograph (Thermo Electron, https://www.thermofisher.com/) fitted with a Valco sample valve and analysed using a Restek RT QPLOT column, (0.53 mm ID × 15 m; Interscience B.V., http://www.interscience.nl/) at a column temperature of 50°C and flame ionisation detection. Quantitative data were obtained using a certified calibration gas, namely 1.01 p.p.m. ethylene in synthetic air (Linde Gas Benelux, http://www.linde-gas.com/).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (van Mourik et al., 2015) on gTCP5‐GFP inflorescences, using μMACS Anti‐GFP (Miltenyi, http://www.miltenyibiotec.com/). Primers used for qPCR can be found in Table S4. Regions of ACTIN 2 (ACT2, At3g18780), a peptidase S41 family protein (At4g17740) and ACS2 (At1g01480) without a TCP5‐binding site were used as negative controls. ACT2 was used as a normaliser for DNA quantity and PS41 as a background control for immunoprecipitated DNA. The same results were obtained when PS41 was used as a normaliser and ACT2 as a background control. Three biological replicates were used and a t‐test (P < 0.05) was done to calculate significant enrichment in the ChIP‐qPCR. Four primer combinations were designed in the promoter and genic regions of ACS2 (Figure 4d). Primer combination 1 was used as a negative control since no consensus TCP‐binding site was present. Primer combinations 2 and 3 cover a single putative TCP‐binding site in the promoter of ACS2, and primer combination 4 covers a putative TCP‐binding region in the fourth exon of ACS2. We used the PlantPAN 2.0 website for promoter analysis to search for putative binding sites (Chow et al., 2016).
Supporting information
Figure S1. Expression pattern of TCP5 in Arabidopsis petals during early and later stages of flower development.
Figure S2. Construction, expression and leaf phenotype of ATML1 pro :TCP5‐GFP.
Figure S3. Floral phenotypes in TCP5 overexpression and mutant lines.
Figure S4. Graphic representation accompanying the cell morphology phenotyping.
Figure S5. Summary of RNA sequencing results.
Figure S6. Gene Ontology term analysis.
Figure S7. Phenotypes of the 35S pro :TCP5‐GR mutant after dexamethasone treatment.
Figure S8. Phenotypical alterations in petals of mutants deficient in ethylene reception and signal transduction.
Table S1. Lists of differentially expressed genes after RNA‐seq analysis of all samples used in this study.
Table S2. List of GO Slim annotation results.
Table S3. List of Gene Ontology terms (full analysis).
Table S4. Primers used in this study.
Acknowledgements
We thank Dr Alice Pajoro for her help with establishing the ChIP protocol on gTCP5:GFP inflorescences, Dr Koyama for providing the tcp5‐1 mutant and Professor Y. Eshed for sending us the triple tcp5 tcp13 tcp17 T‐DNA insertion line. We thank A. C. van de Peppel for help with the ethylene measurements. Our work is supported by grants from the Dutch Scientific Organization (NWO); (NWO‐JSTP grant 833.13.008), CAPES/NUFFIC (no. 010/07) and CAPES/NUFFIC (no. 033/2012). The authors declare no conflict of interest.
References
- Abts, W. , Van de Poel, B. , Vandenbussche, B. and De Proft, M.P. (2014) Ethylene is differentially regulated during sugar beet germination and affects early root growth in a dose‐dependent manner. Planta, 240, 679–686. [DOI] [PubMed] [Google Scholar]
- Anastasiou, E. , Kenz, S. , Gerstung, M. , MacLean, D. , Timmer, J. , Fleck, C. and Lenhard, M. (2007) Control of plant organ size by KLUH/CYP78A5‐dependent intercellular signaling. Dev. Cell, 13, 843–856. [DOI] [PubMed] [Google Scholar]
- Andriankaja, M. , Dhondt, S. , De Bodt, S. et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not‐so‐gradual process. Dev. Cell, 22, 64–78. [DOI] [PubMed] [Google Scholar]
- Aoyama, T. and Chua, N.H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Beyer, E.M. (1979) Effect of silver ion, carbon‐dioxide, and oxygen on ethylene action and metabolism. Plant Physiol. 63, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Yin, X. , Wang, L. , Tian, J. , Yang, R. , Liu, D. , Yu, Z. , Ma, N. and Gao, J. (2013) Involvement of rose aquaporin RhPIP1;1 in ethylene‐regulated petal expansion through interaction with RhPIP2;1. Plant Mol. Biol. 83, 219–233. [DOI] [PubMed] [Google Scholar]
- Chow, C.N. , Zheng, H.Q. , Wu, N.Y. et al. (2016) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coen, E.S. and Meyerowitz, E.M. (1991) The war of the whorls: genetic interactions controlling flower development. Nature, 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Cominelli, E. , Sala, T. , Calvi, D. , Gusmaroli, G. and Tonelli, C. (2008) Over‐expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53, 53–64. [DOI] [PubMed] [Google Scholar]
- Crawford, B.C.W. , Nath, U. , Carpenter, R. and Coen, E.S. (2004) Cincinnata controls both cell differentiation and growth in petal lobes and leaves of antirrhinum. Plant Physiol. 135, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P. , Lauter, N. , Doebley, J. and Coen, E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- Curtis, M. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.‐R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. (2016) TCP transcription factors at the interface between environmental challenges and the plant's growth responses. Front. Plant Sci. 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van der Wal, F. , Dhondt, S. et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van Dijk, A.D. , Bimbo, A. , van der Wal, A. , Van Der, F. , Hennig, L. , Folter, S.De , Angenent, G.C. and Immink, R.G.H. (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny, J.R. , Yadegari, R. , Fischer, R.L. , Yanofsky, M.F. and Weigel, D. (2004) The role of JAGGED in shaping lateral organs. Development, 131, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dugardeyn, J. and Van Der Straeten, D. (2008) Ethylene: fine‐tuning plant growth and development by stimulation and inhibition of elongation. Plant Sci. 175, 59–70. [Google Scholar]
- Efroni, I. , Blum, E. , Goldshmidt, A. and Eshed, Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell, 20, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , Xu, P. , Li, B. et al. (2017) Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl Acad. Sci. 114, 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti, R. , Johnson, K.L. , Scofield, S. , San‐Bento, R. , Watt, A.M. , Murray, J.A.H. and Ingram, G.C. (2015) DEFECTIVE KERNEL 1 promotes and maintains plant epidermal differentiation. Development, 142, 1978–1983. [DOI] [PubMed] [Google Scholar]
- Glover, B.J. , Airoldi, C.A. and Moyroud, E. (2016) Epidermis: outer Cell Layer of the Plant. In: eLS. Chichester: John Wiley & Sons Ltd. [Google Scholar]
- van der Graaff, E. , Schwacke, R. , Schneider, A. , Desimone, M. and Kunze, R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe, M. (2012) The patterning of epidermal hairs in Arabidopsis‐updated. Curr. Opin. Plant Biol. 15, 31–37. [DOI] [PubMed] [Google Scholar]
- Hase, Y. , Fujioka, S. , Yoshida, S. , Sun, G. , Umeda, M. and Tanaka, A. (2005) Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J. Exp. Bot. 56, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Huang, T. and Irish, V.F. (2015) Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 25, 1765–1770. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H. , Kaufmann, K. and Angenent, G.C. (2010) The “ABC” of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21, 87–93. [DOI] [PubMed] [Google Scholar]
- Irish, V.F. (2008) The Arabidopsis petal: a model for plant organogenesis. Trends Plant Sci. 13, 430–436. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D. and Irish, V.F. (2000) Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development, 127, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Jones, M.L. , Stead, A.D. , Clark, D.G. (2009) Petunia flower senescence. In Petunia, Evolutionary, Developmental and Physiological Genetics (Gerats T., and Strommer J., eds) 2nd edn. New York, NY: Springer‐Verlag, pp. 1–445. [Google Scholar]
- Kim, H.J. , Hong, S.H. , Kim, Y.W. et al. (2014) Gene regulatory cascade of senescence‐associated NAC transcription factors activated by ETHYLENE‐INSENSITIVE2‐mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Chang, C. and Tucker, M.L. (2015) To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T. (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 5, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T. , Furutani, M. , Tasaka, M. and Ohme‐Takagi, M. (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary‐specific genes in Arabidopsis. Plant Cell, 19, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A. , Prost, V. and Macias, A. (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell, 12, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, J. , Vandenbussche, F. , van der Straeten, D. and Verbelen, J.P. (2004) Position and cell type‐dependent microtubule reorientation characterizes the early response of the Arabidopsis root epidermis to ethylene. Physiol. Plant. 121, 513–519. [Google Scholar]
- Li, S. (2015) The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal. Behav. 10, e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Wang, Y. , Zhang, Y. , Wang, W. , Irish, V.F. and Huang, T. (2016) RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 67, 6473–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. , Porat, R. , Nadeau, J.A. and O'Neill, S.D. (1996) Identification of a meristem L1 layer‐specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell, 8, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D. , Carpenter, R. , Vincent, C. , Copsey, L. and Coen, E. (1995) Origin of floral asymmetry in Antirrhinum. Nature, 383, 794–799. [DOI] [PubMed] [Google Scholar]
- Lv, B. , Tian, H. , Zhang, F. , Liu, J. , Lu, S. , Bai, M. , Li, C. and Ding, Z. (2018) Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 14, e1007144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N. , Xue, J. , Li, Y. , Liu, X. , Dai, F. , Jia, W. , Luo, Y. and Gao, J. (2008) Rh‐PIP2;1, a rose aquaporin gene, is involved in ethylene‐regulated petal expansion. Plant Physiol. 148, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S. , Heymans, K. and Kuiper, M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics, 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Martín‐Trillo, M. and Cubas, P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y. and Fischer, R.L. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, F. and Skoog, T. (1962) A revised medium for rapid growth and bioassys with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nag, A. , King, S. and Jack, T. (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl Acad. Sci. USA 106, 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, U. , Crawford, B.C.W. , Carpenter, R. and Coen, E. (2003) Genetic control of surface curvature. Science, 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Nicolas, M. and Cubas, P. (2016) TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. [DOI] [PubMed] [Google Scholar]
- Noda, K. , Glover, B.J. , Linstead, P. and Martin, C. (1994) Flower colour intensity depends on specialized cell shape controlled by a Myb‐related transcription factor. Nature, 369, 661–664. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F. , Allen, E. , Wu, X.L. , Schommer, C. , Schwab, R. , Carrington, J.C. and Weigel, D. (2003) Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Panikashvili, D. , Shi, J.X. , Schreiber, L. and Aharoni, A. (2011) The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol. 190, 113–124. [DOI] [PubMed] [Google Scholar]
- Pei, H. , Ma, N. , Tian, J. et al. (2013) An NAC transcription factor controls ethylene‐regulated cell expansion in flower petals. Plant Physiol. 163, 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik, R. , Tholen, D. , Poorter, H. , Visser, E.J.W. and Voesenek, L.A.C.J. (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11, 176–183. [DOI] [PubMed] [Google Scholar]
- Plett, J.M. , Mathur, J. and Regan, S. (2009) Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana . J. Exp. Bot. 60, 3923–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, A.M. , McCollough, F.W. , Eldreth, B.L. , Binder, B.M. and Abel, S.M. (2016) Analysis of network topologies underlying ethylene growth response kinetics. Front. Plant Sci. 7, 1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, M.I. , Wang, X. , Thibault, D.M. , Kim, H.J. , Bombyk, M.M. , Binder, B.M. , Shakeel, S.N. and Schaller, G.E. (2015) The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H. , Dang, X. , Cai, X. , Yu, P. , Li, Y. , Zhang, S. , Liu, M. , Chen, B. and Lin, D. (2017) Spatio‐temporal orientation of microtubules controls conical cell shape in Arabidopsis thaliana petals. PLoS Genet. 13, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, A.H.K. , Cunha, A. , Ohno, C.K. and Meyerowitz, E.M. (2012) Cell cycle regulates cell type in the Arabidopsis sepal. Development, 139, 4416–4427. [DOI] [PubMed] [Google Scholar]
- Rogers, H.J. (2013) From models to ornamentals: how is flower senescence regulated? Plant Mol. Biol. 82, 563–574. [DOI] [PubMed] [Google Scholar]
- Sauret‐Güeto, S. , Schiessl, K. , Bangham, A. , Sablowski, R. and Coen, E. (2013) JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol. 11, e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi‐Goldstein, S. , Peto, C. and Chory, J. (2007) The epidermis both drives and restricts plant shoot growth. Nature, 446, 199–202. [DOI] [PubMed] [Google Scholar]
- Schiessl, K. , Muiño, J.M. and Sablowski, R. (2014) Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip‐related cell cycle inhibitors. Proc. Natl Acad. Sci. USA 111, 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N.S. , Wang, J.T. , Ramage, D. , Amin, N. , Schwikowski, B. and Ideker, T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J.X. , Malitsky, S. , de Oliveira, S. , Branigan, C. , Franke, R.B. , Schreiber, L. and Aharoni, A. (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 7, e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R. , Bowman, J.L. and Meyerowitz, E.M. (1990) Early flower development in Arabidopsis. Plant Cell, 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecsi, J. , Joly, C. , Bordji, K. , Varaud, E. , Cock, J.M. , Dumas, C. and Bendahmane, M. (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 25, 3912–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi, J. , Wipperman, B. and Bendahmane, M. (2014) Genetic and phenotypic analyses of petal development in Arabidopsis In Methods in Molecular Biology. (Riechmann J. and Wellmer F., eds). New York, NY: Humana Press, pp. 191–202. [DOI] [PubMed] [Google Scholar]
- Takeda, S. , Matsumoto, N. and Okada, K. (2004) RABBIT EARS, encoding a SUPERMAN‐like zinc finger protein, regulates petal development in Arabidopsis thaliana . Development, 131, 425–434. [DOI] [PubMed] [Google Scholar]
- Todesco, M. , Balasubramanian, S. , Hu, T.T. et al. (2010) Natural allelic variation underlying a major fitness trade‐off in Arabidopsis thaliana . Nature, 465, 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. et al. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka, A. and Theologis, A. (2004) Unique and overlapping expression patterns among the Arabidopsis 1‐amino‐cyclopropane‐1‐carboxylate synthase gene family members. Plant Physiol. 136, 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti Manassero, N.G. , Viola, I.L. , Welchen, E. and Gonzalez, D.H. (2013) TCP transcription factors: architectures of plant form. Biomol. Concepts, 4, 111–127. [DOI] [PubMed] [Google Scholar]
- Urbanus, S.L. , Martinelli, A.P. , Dinh, Q.D. , Aizza, L.C.B. , Dornelas, M.C. , Angenent, G.C. and Immink, R.G.H. (2010) Intercellular transport of epidermis‐expressed MADS domain transcription factors and their effect on plant morphology and floral transition. Plant J. 63, 60–72. [DOI] [PubMed] [Google Scholar]
- Van de Poel, B. , Smet, D. and van der Straeten, D. (2015) Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mourik, H. , Muiño, J.M. , Pajoro, A. , Angenent, G.C. and Kaufmann, K. (2015) Characterization of in vivo DNA‐binding events of plant transcription factors by ChIP‐seq: experimental protocol and computational analysis In Plant Functional Genomics. Methods in Molecular Biology (Alonso J. and Stepanova A., eds), vol. 1284. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Varaud, E. , Brioudes, F. , Szécsi, J. , Leroux, J. , Brown, S. , Perrot‐Rechenmann, C. and Bendahmane, M. (2011) AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell, 23, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, C. , Yang, T.J.W. , Stead, A.D. , Buchanan‐Wollaston, V. and Roberts, J.A. (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J. 57, 690–705. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhu, L. , Liu, B. , Wang, C. , Jin, L. , Zhao, Q. and Yuan, M. (2007) Arabidopsis MICROTUBULE‐ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell Online, 19, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, L.M. , Gan, S. , Quirino, B. and Amasino, R.M. (1998) A comparison of the expression patterns of several senescence‐associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression pattern of TCP5 in Arabidopsis petals during early and later stages of flower development.
Figure S2. Construction, expression and leaf phenotype of ATML1 pro :TCP5‐GFP.
Figure S3. Floral phenotypes in TCP5 overexpression and mutant lines.
Figure S4. Graphic representation accompanying the cell morphology phenotyping.
Figure S5. Summary of RNA sequencing results.
Figure S6. Gene Ontology term analysis.
Figure S7. Phenotypes of the 35S pro :TCP5‐GR mutant after dexamethasone treatment.
Figure S8. Phenotypical alterations in petals of mutants deficient in ethylene reception and signal transduction.
Table S1. Lists of differentially expressed genes after RNA‐seq analysis of all samples used in this study.
Table S2. List of GO Slim annotation results.
Table S3. List of Gene Ontology terms (full analysis).
Table S4. Primers used in this study.


