Abstract
Rapalogs, inhibitors of mTORC1 (mammalian target of rapamycin complex 1), increase life span and delay age-related phenotypes in many species. However, the molecular mechanisms have not been fully elucidated. We determined gene expression changes comparing 6- and 24-month-old rats in the kidney, liver, and skeletal muscle, and asked which of these changes were counter-regulated by a clinically-translatable (short-term and low-concentration) treatment, with a rapalog (RAD001). Surprisingly, RAD001 had a more pronounced effect on the kidney under this regimen in comparison to the liver or skeletal muscle. Histologic evaluation of kidneys revealed that the severity of chronic progressive nephropathy lesions was lower in kidneys from 24-month-old rats treated with RAD001 compared with vehicle. In addition to other gene expression changes, c-Myc, which has been shown to regulate aging, was induced by aging in the kidney and counter-regulated by RAD001. RAD001 caused a decrease in c-Myc protein, which could be rescued by a proteasome inhibitor. These findings point to settings for use of mTORC1 inhibitors to treat age-related disorders, and highlight c-Myc regulation as one of the potential mechanisms by which mTORC1 inhibition is perturbing age-related phenotypes.
Keywords: Aging, Rapamycin, RAD001, Kidney, Myc
Aging is the strongest risk-factor for many serious diseases and comorbidities, including cancer, heart disease, kidney disease, dementia, Alzheimer’s disease, frailty, and sarcopenia (1–5). Increasing evidence suggests that aging occurs in a regulated manner, and that perturbation of discrete cell signaling pathways can extend life span and delay age-related diseases and comorbidities (6–12). Inhibition of the mammalian target of rapamycin (mTORC1) pathway is perhaps the best-validated pharmacological intervention to forestall age-related phenotypic changes; inhibition of mTORC1 using rapamycin can extend murine life span, including in settings where the treatment is initiated relatively late in life (9,10,12). As to mechanism, treatment with rapamycin has been shown to reverse signs of senescence in stem cells (7,13,14) and decrease inflammation (8,15).
Quite a few other mechanisms have been put forth as potentially regulating age-related conditions, and life span (16). Interestingly, it was recently demonstrated that c-Myc inhibition could increase life span (17); haplo-insufficient c-Myc +/- mice were shown to live longer, and they experienced a delay in age-related phenotypes, coincident with a decrease in activation of nutrient sensing pathways, which included Akt and mTOR pathways (17).
It was unknown whether mTORC1 inhibition affects aging or its consequences in human beings. However, a recent clinical trial in elderly subjects demonstrated that a rapalog (RAD001) ameliorated immuno-senescence, as had been shown in mice (7), and thereby improved the response of elderly humans to influenza vaccination (18). The intervention in this case made use of relatively low or intermittent doses of the pharmacologic agent, and lasted only 6 weeks (18). The study in humans established a treatment basis for a further understanding of mechanism by which rapalogs function when given at similarly low levels and shorter durations, a treatment paradigm which in this study we aimed to reverse-translate to animals.
Understanding the age-related changes that occur in multiple tissues upon drug treatment is a useful way to determine the site of action of particular interventions, and where more intervention might be called for. We were curious to know whether examination of kidney, liver, and skeletal muscle could lead to new insights into the possible mechanism by which mTORC1 inhibition counter-regulates age-related changes, which might be causal for age-related pathology.
Methods
Animal Maintenance, RAD001 Treatment, and Tissue Collection
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the Novartis Institutes for Biomedical Research, Cambridge, MA, USA. Sprague Dawley (SD) male rats were purchased and aged at Envigo (Indianapolis, IN) under specific pathogen-free (SPF) conditions. Once imported, rats were maintained at the SPF facilities with controlled temperature and light (22°C, 12-hour light/12-hour dark cycle: lights on at 0600hours/lights off at 1800 hours) and with ad libitum access to food and water. The mortality rate of these rat cohorts at 21 months is ~22% and at 24 months is ~30%. Male SD rats were imported at 3 and 21 months of age, housed singly and maintained on a 2014 Teklad Global 14% Protein diet (Envigo) ad libitum. Rats were acclimated for at least 4 weeks before experiments commenced.
RAD001 (Novartis) was formulated as a custom made micro-emulsion preconcentrate at 2% (w/w) and diluted to a final concentration prior to dosing per os. Rats were treated with RAD001 diluted in water, supplemented with 0.25% methylcellulose. Microemulsion preconcentrate (adequate to a dose) diluted in water with 0.25% methylcellulose served as a vehicle control.
For RAD001 treatment, 22.5-month-old rats received per os either a vehicle daily (N = 12) or RAD001 once a week (N = 12) (seven doses of RAD001 in total) and vehicle daily for the rest of the week for 6 weeks. During the experiment, five aged rats died and one rat was removed from the experiment due to a malignancy. Four and a half month old rats treated with vehicle daily for 6 weeks served as young adult controls. Rats were given vehicle daily, because this study was a part of the larger study, that included groups of rats treated with other compounds daily. Blood glucose levels were measured in rats fasted from 0600 hours to 1200 hours using Embrance glucometer. Fifty-two hours following the last dose of RAD001 or 4 hours after the last vehicle dose, rats were anesthetized with 3.5% isoflurane and killed by exsanguination and thoracotomy. Kidneys, livers, gastrocnemius muscles, and spleens were collected and frozen in liquid nitrogen. One kidney from each animal was cryo-preserved in OCT for histology.
RNA Extraction, cDNA Synthesis, and Real-Time Quantitative polymerase chain reaction (RT-qPCR)
Snap frozen kidneys, livers, and gastrocnemius muscles were ground in liquid nitrogen by mortar and pestle and total RNA was extracted from ~30 mg of tissue powder using miRNeasy Micro Kit (Qiagen, 217084). The RNA concentration was quantified using NanoDrop Spectrophotometer (NanoDrop Technologies, USA) and the integrity validated by the OD260/OD280 absorption ratio (>1.8) and by RIN score (>8) using Agilent 2100 Bioanalyzer, RNA 6000 Nano LabChip kit and Agilent 2100 Expert Software (Agilent Technologies, Inc., Santa Clara, CA). Samples with RIN scores >8 were designated for gene arrays (RNAseq). cDNA was synthesized from total RNA using TaqMan reverse transcription reagents (Applied Biosystems, Forster City, CA). RT-qPCR was performed using an Applied Biosystems 7900 fast real-time PCR system. All the Taqman probes were optimized by Applied Biosystems and are summarized in Supplementary Table 1. The PCR reactions used FastStart Universal Probe Master Mix (Roche Diagnostics, IN) and incubated in a 384-well optical plate at 50°C for 2 minutes, 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60° for 1 minute. Data generated by the RT-qPCR were analyzed in GraphPad using ANOVA followed by Dunnett’s multiple comparison tests, where means from all groups were compared to the old vehicle treated group. p Value cutoff of .05 was used to determine significant changes. Data are presented as means with standard deviations. Animal numbers per group are indicated in the figure legends.
Transcriptomic Analyses With RNAseq
For RNAseq, 76 bp paired-end reads were mapped to rat genome rn5 using STAR (19) with default parameters. A genome-wide transcription signal map was derived by counting reads coverage in 10 bp step after normalizing to 1 million total reads. Stranded information was used to separate counts from sense and antisense strands. For gene annotation, we first clustered the Refseq transcripts based on the sharing of splicing sites to get a master set of gene loci representing all alternative isoforms. Signal from sense-strand were averaged across the exonic regions for each gene locus to measure the transcription level. This measure is equivalent to the RPKM “reads per kilobase per million” in other RNAseq analyses. Expression profiles were quantile-normalized to enable cross-sample comparison.
Age-regulated genes were identified by comparing the vehicle treated old samples with young samples, and RAD001-regulated genes were identified by comparing RAD001-treated with vehicle treated old samples, using a t test. p Values were adjusted for multiple tests and a false discovery rate (FDR) cutoff of .05 was used to identify significant changes. Pathway enrichment was tested with a Hypergeometric test, and FDR-adjusted p value cutoff of .001 was used to identify significant enrichment based on pathway annotation from MsigDB (20).
Evaluation of Kidney Tissues by Light Microscopy and Semiquantitative Scoring
Semiquantitative scoring of hematoxylin and eosin (H&E)-stained kidney cryo-sections was performed according to standard methods for assessment of chronic progressive nephropathy (CPN) in aged laboratory rats (21,22). In this schema, a 0 signifies no or nil lesions, 1 a minimal grade, 2 mild, 3 low-moderate, 4 mid-moderate, 5 high-moderate, 6 low-severe, 7 high-severe, and 8 end-stage. Grades from minimal to high-moderate represent a progressive increase in the number of CPN lesions as focal changes; low-severe the point where foci begin to coalesce into areas of cortical tubule change; high-severe where a majority of the cortical parenchyma is affected by CPN change; and end-stage where no, or almost no, normal parenchyma remains. Scoring of all tissue specimens by light microscopy was performed without knowledge of treatment group (ie, blinded/masked evaluation). Data generated by semiquantitative scoring was evaluated using a nonparametric Mann–Whitney test for comparison of histopathology scores from 6- and 24-month-old rats treated with vehicle and for comparison of histopathology scores from 24-month-old rats treated with vehicle and 24-month-old rats treated with RAD001.
Experiments With HEK293 Cells
HEK293 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Life Technologies, USA; 11995) supplemented with 10% fetal bovine serum (Hyclone, Utah; SH30070.03) and penicillin-streptomycin, 100 U/mL (Life Technologies; 15140-122), at 37°C in 5% CO2 until they were 60% confluent. Cells were treated with RAD001 (Novartis), Emetine (Sigma Aldrich, USA; E2375) and MG132 (Sigma Aldrich; M7449).
Protein Extraction and Immunoblotting
For protein extraction, spleens were lysed in RIPA buffer (Thermo, IL; 89901) and HEK293 cells were lysed in M-PER buffer (Thermo; 78501), supplemented with complete EDTA free protease inhibitor and PhosSTOP phosphatase inhibitor tablets (Roche, Manheim, Germany), and centrifuged at 13,000g for 20 minutes at 4°C. The resultant supernatant was used for immunoblotting. Protein was quantified with BCA protein assay (Thermo Scientific, MA). Samples were resolved on 4%–20% Criterion TGX Precast Midi Protein gels (Bio-Rad, CA) and transferred onto nitrocellulose membranes (Bio-Rad) using a Trans Turbo Blot system (Bio-Rad). Immunoblotting was performed with antibodies to Myc (#5605, #9402), p-S6K1(Thr389) (#9205) and t-S6K1 (#9202) all from Cell Signaling Technologies (all 1:1,000 in TBS-T with 5% BSA). The “p” and “t” prefixes signify “phosphorylated” and “total” forms respectively. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) detected with an anti-GAPDH antibody (#5174, Cell Signaling Technologies, MA) and HPRT1 detected with an anti-HPRT1 antibody (15059-1-AP Proteintech, USA) were used as protein loading controls. HRP-conjugated secondary antibodies against rabbit (#7074) were from Cell Signaling Technologies. The chemiluminescence signal was generated using SuperSignal West Femto Enhanced Chemiluminescent Substrate (#34095, Thermo Scientific) or Western Lightning® Plus-ECL Enhanced Chemiluminescence Substrate (NEL103001EA, Perkin Elmer, MA) and was captured using the ChemiDoc MP Imaging System (Bio-Rad). Resultant digital images were converted into a TIFF format and quantified using ImageJ software.
Results
Transcriptional Profile of Kidney, Liver, and Gastrocnemius Muscles in Old Rats Treated With RAD001
We sought to understand how gene expression is affected by age and whether use of a rapalog (RAD001), at doses and treatment duration similar to those used in human subjects (18), would perturb age-related gene expression profiles in various tissues. The Sprague-Dawley (SD) rat lives approximately 2.5–3 years under laboratory conditions (23). The mortality rate of our male SD rat cohorts at 24 months (2 years) is ~30% and we have previously shown that at this age rats exhibit pronounced age-related loss of muscle mass and function (sarcopenia) associated with transcriptional changes of many genes in this organ (24). Thus in the present experiment we chose 24 months to be the endpoint of the study with 6-month-old rats used as young controls. RAD001 treatment was introduced at 22.5 months. Rats were treated for 6 weeks, with a dosing schedule and RAD001 concentration aimed at being the rat equivalent of the dose regimen previously used in humans (18). An intermittent (1 mg/kg once a week) RAD001 dose used in the present study for treating old rats is equivalent to a weekly (5 mg/week) human dose (18). This RAD001 dose was well-tolerated by old rats; no change in the body weight or the baseline glucose level was observed (Supplementary Figure 1A, B). We performed transcriptome profiling of kidney, liver and gastrocnemius muscles by RNA-seq and interrogated the expression of protein coding genes (Figure 1).
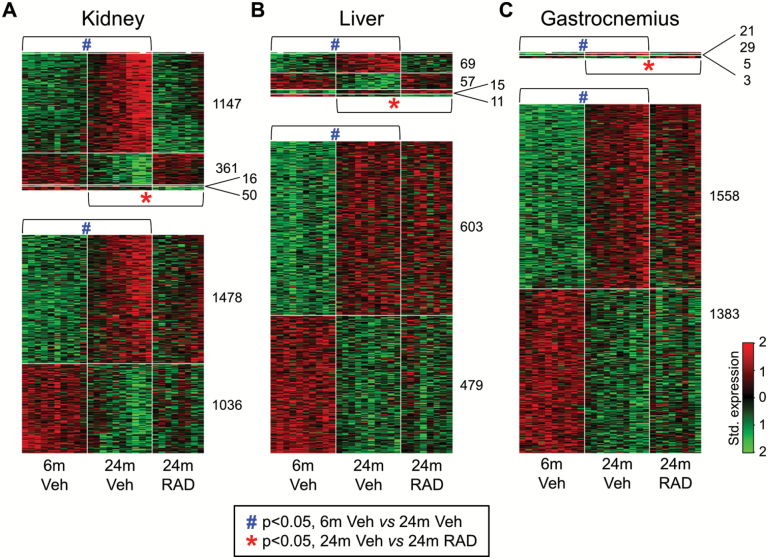
Figure 1.
Genes up- and down-regulated by aging from 6 months to 24 months in rat kidney, liver, and gastrocnemius muscle, and the impact of RAD001 on the expression of these age-perturbed genes at 24 months. Rats aged 4.5 month and 22.5 months were treated with vehicle and rats aged 22.5 months were treated with RAD001 for 6 weeks, with a read-out at 6 and 24 months. Transcriptional profiling in the kidney, liver and gastrocnemius muscle was carried out by RNA-seq. Heat maps (A, B, C) depict genes that are differentially expressed between 6- and 24 month-old tissues (p < .05 adjusted for false discovery rate [FDR]) and the impact of RAD001 on the expression profile of these age-regulated genes. The blue hash (#) indicates genes that are significantly different between 6- and 24-month-old vehicle treated rats (p < .05 adjusted for FDR) and the red asterisk (*) indicates genes that are significantly different between 24-month-old vehicle treated and 24-month-old RAD001 treated rats (p < .05 adjusted for FDR). Standardized expression levels, from low to high, are colored from green to red: thus gene up-regulation is shown by a transition from green to red, and down regulation by a transition from red to green. In each heat map image, each column represents a single rat and each row represents a single gene. N = 8–10 rats per group.
We first identified genes that change with aging in a particular direction (either upward or downward) in three organs (kidney, liver, and gastrocnemius) by comparing transcriptomes between 6- and 24-month-old rats treated with vehicle. Aging differentially regulated expression of 4,088 genes in the kidney, 1,234 genes in the liver and 2,999 genes in the gastrocnemius muscle (t test, FDR < 0.05, Figure 1A–C). We next sought to establish which age-regulated genes are counter-regulated, back toward a “youthful” direction, by mTORC1 inhibition, using a rapalog. Comparison of transcriptomics between 24-month-old rats treated with vehicle and RAD001 revealed that RAD001 treatment counter-regulated expression of 37% of the age-regulated genes in the kidney (1,508 out of total 4,088) and 10% of age-regulated genes in the liver (126 out of 1,234; Figure 1A and B). In the gastrocnemius muscle, only 50 out of 2,999 age-regulated genes were counter-regulated by a rapalog treatment, using this dosing regimen (Figure 1C).
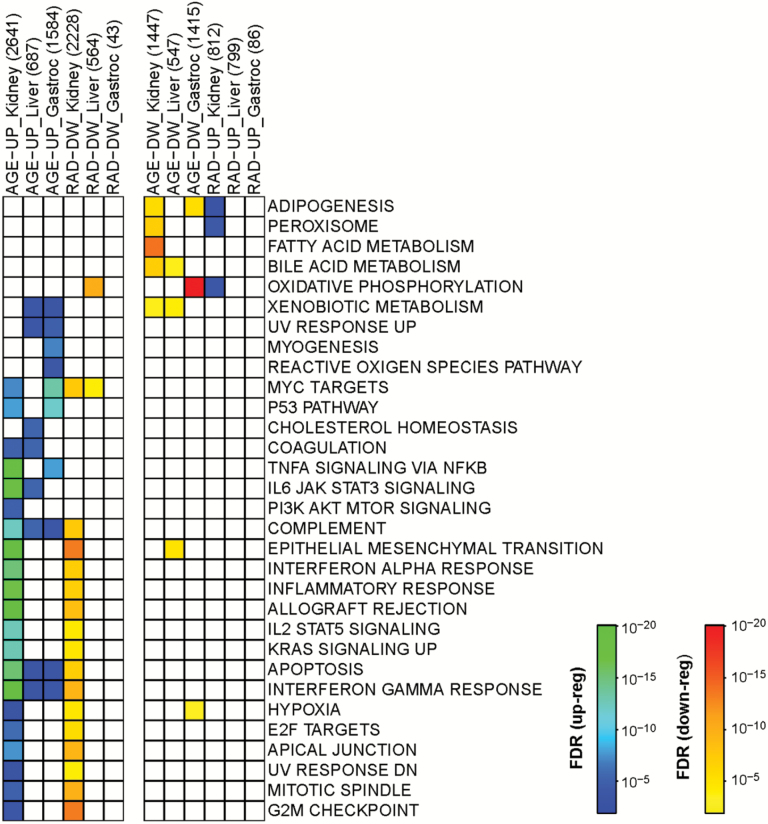
Next we wanted to determine gene pathways that were regulated by age and counter-regulated by RAD001. Gene pathway analyses are displayed in Figure 2; we report those pathways which are either up or down regulated with age, and those which are counter-regulated by RAD001—thus, for a pathway up-regulated by age we show which of these are down-regulated by RAD001; for those pathways down-regulated by age, we show which are up-regulated by RAD001 (Figure 2). Gene pathways up-regulated by age in all three tissues were complement genes, apoptosis and interferon gamma response pathway genes, albeit this upregulation was highly significant (hypergeometric test, p < 10−12–10−20) in the old kidney and less significant in the old liver and skeletal muscle (Figure 2). In the kidney, inflammation-related pathways were particularly noteable of the pathways perturbed by age (TNFα signaling, IL6/Jak/Stat signaling, Interferon alpha and gamma responsive genes, IL2/Stat5 signaling, etc; Figure 2). Of the three tissues studied, RAD001 had a particularly notable effect on the age-regulated gene pathways in the kidney (Figure 2; compare column 1 to column 4; compare column 7 to column 10). Strikingly, RAD001 treatment counter-regulated the majority of the age-regulated pathways in the kidney. Of particular interest was counter-regulation of the following pathways: inflammatory response, interferon alpha and interferon gamma response, complement and allograft rejection, epithelial to mesenchymal transition and apoptosis—as these pathways were strongly up-regulated by aging (Figure 2). Surprisingly, RAD001 did not impact age-related gene pathways in the liver or the muscle as much using this treatment paradigm (Figure 2). It should be noted that there are trends towards counter-regulation of age-perturbed gene pathways by RAD001 in skeletal muscle and liver, but these did not reach statistical significance once the data were corrected for FDR (false-discovery rate).
Figure 2.
Functional annotation of gene pathways differentially regulated by age (6-months vs 24-months vehicle-treated groups) and by RAD001 (24-month vehicle vs 24-month RAD001-treated groups) in kidney, liver, and gastrocnemius (gastroc) muscle of rats. Boxes with a color transition from green to blue represent up-regulated pathways (AGE-UP or RAD-UP) with p < 10–20 (green) – p < 10–3 (blue). Boxes with a color transition from red to yellow represent down-regulated pathways (AGE-DW or RAD-DW) with p < 10–20 (red) – p < 10–3 (yellow). Numbers of genes that are regulated in either direction by age or RAD001 are indicated in brackets: eg, AGE-UP_Kidney (2641).
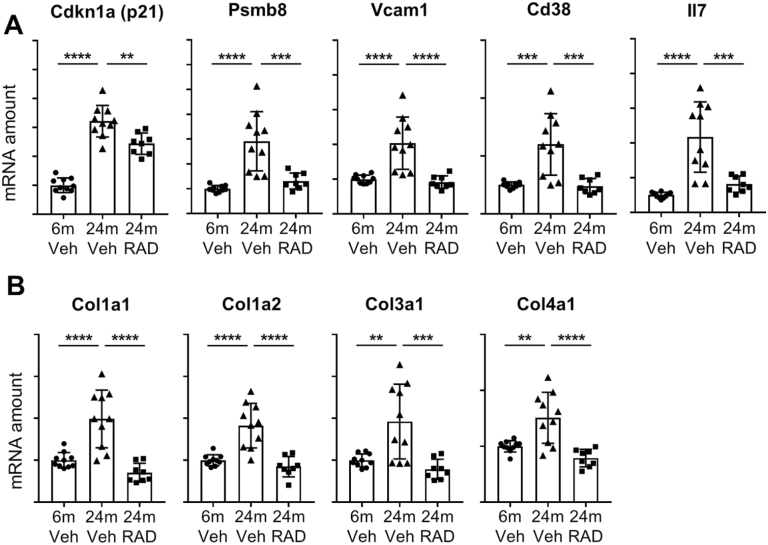
Since the response to RAD001 treatment was the most striking in the aged rat kidney, we validated kidney-perturbed genes; the pathways included those that were tagged as being part of the interferon gamma response (Figure 3A and Supplementary Figure 2) and genes involved in epithelial to mesenchymal transition (Figure 3B and Supplementary Figure 3) by RT-qPCR. Validation of these pathways confirmed that RAD001 counter-regulated gene markers of inflammation and fibrosis (eg, collagen isoforms; Figure 3A and B).
Figure 3.
Validation of interferon gamma (A) and (B) epithelial to mesenchymal transition pathways in the rat kidney. mRNA amounts of the selected genes were quantified by real-time quantitative polymerase chain reaction in the kidneys from 6-month-old rats treated with vehicle and 24-month-old rats treated with vehicle or RAD001. mRNA amounts of the genes of interest were normalized to a geometric mean of reference genes Cox7a2, Vps26a, and TATA-box-binding protein. Data are mean ± SD. **p ≤ .01; ***p ≤ .001; ****p ≤ .0001. Y-axes represent arbitrary units for normalized mRNA amounts. N = 8–10 rats per group.
RAD001 Treatment Reduces the Severity of Chronic Progressive Nephropathy Lesions in Aged Rats
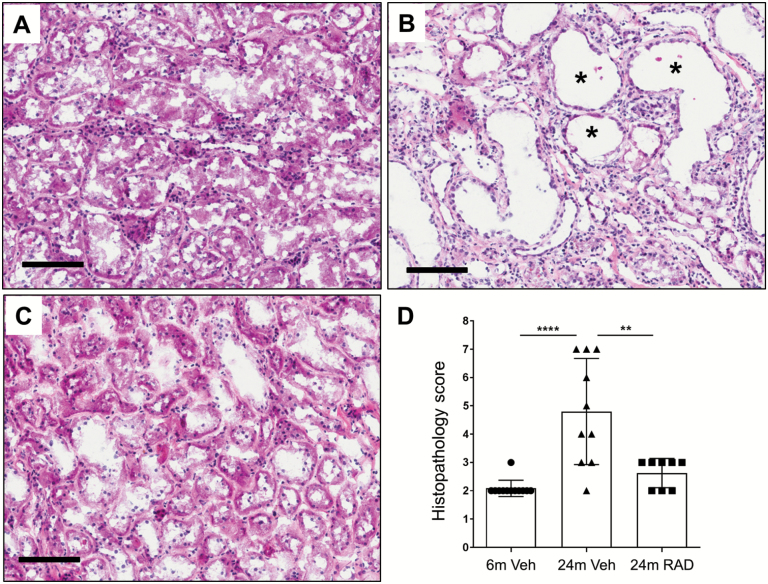
Since RAD001 counter-regulated the largest number of age-related genes and gene pathways in aged rat kidneys, we assessed the effect of this treatment on kidney morphology. Semiquantitative histopathology scoring of hematoxylin and eosin (H&E)-stained longitudinal sections of frozen kidneys for chronic progressive nephropathy lesions (25) was performed by light microscopy according to standard methods (21,22). As expected, the histopathology scores were greater in 24-month-old rats treated with vehicle compared with 6-month-old rats treated with vehicle (Figure 4A, B and D). Compared with 24-month-old rats treated with vehicle, animals treated with RAD001 had a significantly lower mean histopathology score, indicating prevention of age-related pathology occurred (Figure 4B, C and D).
Figure 4.
Representative photomicrographs illustrating longitudinal sections of hematoxylin and eosin (H&E)-stained kidneys (cortex region) from a 6-month-old rat treated with vehicle (A), a 24-month-old rat treated with vehicle (B) and a 24-month-old rat treated with RAD001 (C). Asterisks (*) in panel B indicate dilated renal tubules in the kidney from a 24-month-old rat treated with vehicle. Scale bars, 100 μm. (D) Graphical summary of semiquantitative histopathology scores. Data are mean ± SD. ****p ≤ .0001, **p≤ = .01. N = 8–10 rats per group.
Age Increases and RAD001 Suppresses the Myc Pathway in Old Rats
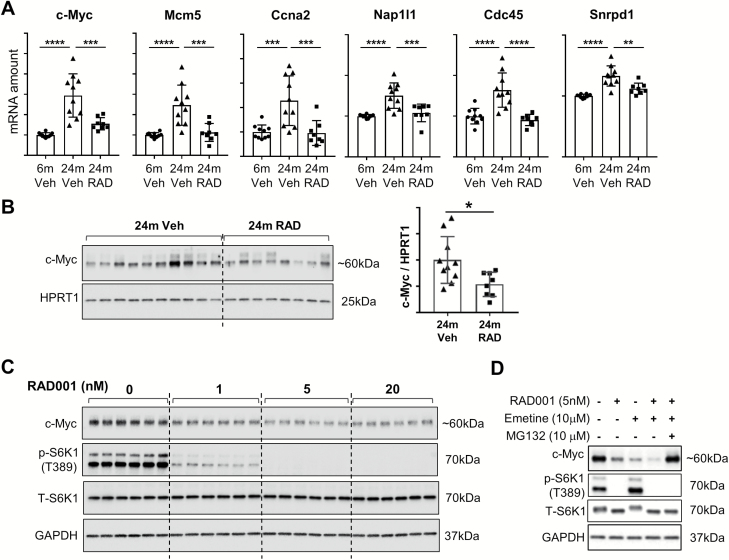
Myc target genes were found to be upregulated by age and suppressed by RAD001 in the old rat kidney (Figure 2), which we found interesting because mice that are hypomorphic for c-Myc have been shown to have extended lifespans (17). Validation of this pathway with RT-qPCR confirmed that expression of c-Myc itself, along with c-Myc target genes, was upregulated during aging in the rat kidney and counter-regulated by a rapalog (Figure 5A). To begin exploring the mechanism by which mTORC1 regulates Myc, we determined whether rapalog treatment perturbed c-Myc protein amounts. Two anti-c-Myc antibodies from Cell Signaling Technologies, which were first validated for specificity to c-Myc, were used to detect c-Myc protein in the rat kidney, but c-Myc protein was below detectable levels in this organ. Since spleen is one tissue that has a high c-Myc mRNA expression level in rats (www.biogps.org), we next performed immunoblotting on spleens from old rats treated with a vehicle in comparison to old rats treated with RAD001 (Figure 5B). Treatment with RAD001 resulted in lower c-Myc levels in 24-month-old animals (Figure 5B; compare 24 months Veh to 24 months RAD).
Figure 5.
(A) Validation of Myc target genes in the rat kidney. mRNA amounts of selected genes were quantified by real-time quantitative polymerase chain reaction in the kidneys from 6-month-old rats treated with vehicle and 24-month-old rats treated with vehicle or RAD001. mRNA amounts of genes of interest were normalized to a geometric mean of reference genes Cox7a2, Vps26a, and TATA-box-binding protein. Data are mean ± SD. **p ≤ .01; ***p ≤ .001; ****p ≤ .0001. Y-axes represent arbitrary units for normalized mRNA amounts. N = 8–10 rats per group. (B) c-Myc protein detected by immunoblotting in the spleen from 24-month-old rats treated with either vehicle alone or with RAD001. For the spleen protein samples, HPRT1 was used as a loading control. Densitometric quantification of c-Myc protein bands relative to HPRT1 (used as a loading control) is shown on the right hand side (B). *p ≤ .05, data are mean ± SD. N = 8–10 rats per group. (C) HEK293 cells were treated with vehicle (DMSO) as a negative control, or with increasing concentrations of RAD001 (1 nM, 5 nM, and 20 nM) for 24 hours and c-Myc protein was detected by immunoblotting (panel 1). (D) c-Myc protein amounts detected in HEK293 cells 6 hours following treatment with RAD001 (5 nM), Emetine (10 μM), and MG132 (10 μM) (panel 1 in D). In cases where either of these compounds was omitted, cells were treated with blank carriers: DMSO for RAD001 and MG132 and water for Emetine.
Immunoblotting for p-S6K1(T389) and t-S6K1 was performed to demonstrate inhibition of the mTORC1 pathway by RAD001 (panels 2 and 3 in C and D). GAPDH is shown as a protein loading control (panel 4 in C and D).
To validate an in vitro setting for exploring the mechanism, we then tested whether mTORC1 inhibition with RAD001 perturbs c-Myc protein levels in HEK293 (Human Embryonic Kidney) cells. HEK 293 cells have readily detectable c-Myc levels, by immunoblotting. HEK293 cells were treated with 1 nM, 5 nM, and 20 nM of RAD001 and c-Myc protein amounts were measured 24 hours following treatment. All doses of RAD001 reduced c-Myc protein amounts, and there was an apparent dose–response (Figure 5C).
To determine whether RAD001 is perturbing c-Myc translation or degradation, HEK293 cells were treated with: RAD001 alone; Emetine (a protein synthesis inhibitor) alone; the combination of RAD001 and Emetine, and the combination plus a proteasome inhibitor (MG132) (Figure 5D). RAD001 alone and Emetine alone both reduced c-Myc protein levels. Interestingly, c-Myc protein levels were further reduced with the combination of RAD001 and Emetine, demonstrating that RAD001 can decrease c-Myc further, even when translation is blocked; this suggested a mechanism distinct from solely inhibiting protein translation, the usual mechanism for mTORC1 inhibition. To examine protein degradation, a proteasome inhibitor was also tested—MG132. The loss of c-Myc protein induced by the rapalog was rescued by the addition of MG132 (Figure 5D). These results indicate that at least one of the mechanisms by which RAD001 reduces c-Myc protein is by promoting its degradation by the proteasome, since a proteasome inhibitor can entirely block the effects of RAD001.
Discussion
Aging dramatically increases the likelihood for the most serious diseases that affect humans, including cancer, heart disease, dementia, sarcopenia, and chronic kidney disease. It seems plausible that the gene-changes associated with aging could contribute to the onset of these conditions, either by creating a permissive environment for further pathology, or by directly inducing these conditions. Rapalogs have been shown to extend life span (10,12) and forestall many of these age-related morbid phenotypes (7,8,11,26,27), including in humans (18). Therefore, we were curious to determine age-related genes, and the gene pathways, which were counter-regulated by a rapalog, RAD001. We used clinically relevant low doses of the rapalog RAD001, and a short (6-week) time course of treatment, since these had been shown to be effective in a prior human study, where 6 weeks of treatment was sufficient to counter-regulate age-related declines in responses to vaccination (18). It is important to determine whether lower doses and shorter time-frames are sufficient to induce meaningful change—since lifetime treatment at high doses of this mechanism is not feasible.
The results of the gene expression component of the study were surprising: of the three tissues studied, there was a much greater effect of RAD001 on the kidney in comparison to liver and skeletal muscle with this low-dose and short-term treatment paradigm, pointing toward the therapy as fairly kidney-selective. From a safety point of view, perhaps the relative lack of a strong effect on skeletal muscle at these doses is fortuitous, given the concern of perturbing gene expression downstream of inhibiting protein synthesis pathways, which contribute to muscle maintenance, in settings of sarcopenia (the age-related loss of skeletal muscle mass and function) (5). This relative kidney-selective effect on gene expression occurred despite the fact that the drug was able to get into liver and skeletal muscle and perturb downstream signaling at 1 mg/kg (the dose given), as evidenced by inhibition of S6K1 and S6 phosphorylation in skeletal muscle and liver (data not shown).
One gene-set that was upregulated in rat kidney when comparing 6- to 24-month-old animals was c-Myc target genes (Figure 2). This finding was confirmed by examining specific genes in the pathway by RT-qPCR (Figure 5A). In the kidney, c-Myc mRNA itself was upregulated with age, and counter-regulated by 6-week treatment with the rapalog, as were genes that fall under the Myc-regulated pathway. This was of particular interest for a couple of reasons: first, it was recently shown that Myc haplo-insufficient mice had prolonged lifespans, and demonstrated a downregulation of several pathways, including mTORC1 signaling (17). A distinct study very recently demonstrated that transient upregulation of the Yamanaka factors, the four genes which are required to epigentically reprogram cells to their dedifferentiated form, inducing the formation of induced-pluripotent cells, could significantly increase life span (28). One of those genes is c-Myc (28). The combination of these studies and our current work point to a key role Myc may be playing in regulating the epigenetic changes associated with aging, and the potential therapeutic value of inhibiting those Myc-induced changes to treat age-related diseases, also highlighting mTORC1 inhibition as a strategy to intervene when Myc or Myc-induced genes are elevated. Of course, it is also of interest to note that part of the mechanism by which mTORC1 inhibition perturbs aging might be its ability to inhibit Myc, and this is a mechanism not previously implicated for rapalogs.
We went on to determine the mechanism by which RAD001 perturbs c-Myc protein levels, both in kidney cells in vitro (HEK293 cells) and in the rat spleen in vivo. A dose response of RAD001 treatment decreased c-Myc protein levels significantly. This might in part be due to decreases in c-Myc translation, or it might also be due o signaling changes, leading to a relative increase in c-Myc protein turnover. We investigated the mechanism further, thinking that since mTOR controls protein translation, inhibition of mTORC1 might decrease the levels of c-Myc by this mechanism; however, even in the presence of a protein translation inhibitor, Emetine, the rapalog further decreased c-Myc protein levels. Furthermore, a proteasome inhibitor—MG132—entirely rescued c-Myc levels from rapalog-induced turnover; this data seemingly justifies adding mTORC1 inhibition to the growing list of mechanisms which regulate c-Myc protein stability (29).
There are many studies that demonstrate inflammatory signaling increases with age (8,30,31), and this was seen in the kidney in the current study. Multiple pathways that can be considered “inflammatory” were counter-regulated by transient and low-dose RAD001 treatment, including the inflammatory response gene pathway, interferon alpha and gamma targets, and the epithelial to mesenchymal transition genes, which might be early signs of fibrosis. In addition to changes observed in gene expression profiling, treatment of 24-month-old rats with RAD001 for six weeks reduced the severity of chronic progressive nephropathy lesions observed post-treatment. This rescue of age-induced pathology in the kidney is probably happening by preventing or delaying the onset of pathological changes in the kidney, since it is unlikely that kidney regeneration can easily occur, given the multiple cellular lineages involved.
Age-related kidney disease is a significant problem in the elderly adults (32), affecting 46.8% of the people aged 70 years and older (32). The risk is increased in individuals with diabetes and/or hypertension—the incidence of which is also increased as a result of an increase in the prevalence of obesity. Thus, there is a significant need for intervention in age-related kidney failure. It has been noted that an inflammatory state exists in chronic kidney disease (33) as a function of aging, a finding that was recapitulated in the present study. Thus, the ability of the rapalog to counter-regulate inflammatory processes might be translatable to the human. The prior experience in the clinic, showing improvement of immune function (18), also suggests that other tissues are sensitive to rapalogs.
Further study is needed to examine other tissues in a similar manner, including immune cells, and to determine if distinct pharmacological therapy can be added to an mTORC1 inhibition treatment regimen to increase the coverage of age-related gene changes that might be counter-regulated by intervention, but this study demonstrates the potential tissue-specific mechanisms by which transient mTORC1 inhibition may be counter-regulating age-related pathology.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was supported by Novartis.
Conflict of Interest
The authors were employees of Novartis at the time this work was conducted; some are stockholders in the company.
Acknowledgments
The authors thank Danuta Lubicka (NIBR) for formulating RAD001, Sebastien Ronseaux (NIBR) for pharmacokinetic modeling of RAD001 doses, Yunyu Zhang (NIBR) for the assistance in bioinformatics analyses and NIBR study support associates for the assistance in animal experimentation. The authors thank Paola Capodieci, Kristie Wetzel and Karen Niss (NIBR) for tissue histological processing and also thank Sam Cadena (NIBR) for support. The authors thank the entire Age-Related Disorders group, and the NIBR community for their enthusiastic support.
References
- 1. Armanios M, de Cabo R, Mannick J, Partridge L, van Deursen J, Villeda S. Translational strategies in aging and age-related disease. Nat Med. 2015;21:1395–1399. doi:10.1038/nm.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi:10.1038/nature11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kirkland JL, Tchkonia T. Clinical strategies and animal models for developing senolytic agents. Exp Gerontol. 2015;68:19–25. doi:10.1016/j.exger.2014.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22:R741–R752. doi:10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 5. Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49:59–68. doi:10.3109/10409238.2013.857291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bitto A, Ito TK, Pineda VV et al. . Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. eLife. 2016;5:e16351. doi:10.7554/eLife.16351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi:10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flynn JM, O’Leary MN, Zambataro CA et al. . Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12:851–862. doi:10.1111/acel.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller RA, Harrison DE, Astle CM et al. . Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi:10.1093/gerona/glq178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller RA, Harrison DE, Astle CM et al. . Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi:10.1111/acel.12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Bokov A, Gelfond J et al. . Rapamycin extends life and health in C57BL/6 mice. J Gerontol A Biol Sci Med Sci. 2014;69:119–130. doi:10.1093/gerona/glt056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison DE, Strong R, Sharp ZD et al. . Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi:10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. García-Prat L, Martínez-Vicente M, Perdiguero E et al. . Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi:10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- 14. Gu Z, Tan W, Ji J et al. . Rapamycin reverses the senescent phenotype and improves immunoregulation of mesenchymal stem cells from MRL/lpr mice and systemic lupus erythematosus patients through inhibition of the mTOR signaling pathway. Aging (Albany NY). 2016;8:1102–1114. doi:10.18632/aging.100925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herranz N, Gallage S, Mellone M et al. . mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17:1205–1217. doi:10.1038/ncb3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi:10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- 17. Hofmann JW, Zhao X, De Cecco M et al. . Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–488. doi:10.1016/j.cell.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mannick JB, Del Giudice G, Lattanzi M et al. . mTOR inhibition improves immune function in the elderly. Sci Transl Med. 2014;6:268ra179. doi:10.1126/scitranslmed.3009892 [DOI] [PubMed] [Google Scholar]
- 19. Dobin A, Davis CA, Schlesinger F et al. . STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi:10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi:10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hard GC, Khan KN. A contemporary overview of chronic progressive nephropathy in the laboratory rat, and its significance for human risk assessment. Toxicol Pathol. 2004;32:171–180. doi:10.1080/01926230 490422574 [DOI] [PubMed] [Google Scholar]
- 22. Hard GC, Seely JC. Recommendations for the interpretation of renal tubule proliferative lesions occurring in rat kidneys with advanced chronic progressive nephropathy (CPN). Toxicol Pathol. 2005;33:641–649. doi:10.1080/01926230500299716 [DOI] [PubMed] [Google Scholar]
- 23. Sengupta P. The laboratory rat: Relating its age with human’s. Int J Prev Med. 2013;4:624–630. PMID:23930179. [PMC free article] [PubMed] [Google Scholar]
- 24. Ibebunjo C, Chick JM, Kendall T et al. . Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol Cell Biol. 2013;33:194–212. doi:10.1128/MCB.01036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barthold SW. Chronic progressive nephropathy in aging rats. Toxicologic Pathology. 1979;7:1–6. [Google Scholar]
- 26. Halloran J, Hussong SA, Burbank R et al. . Chronic inhibition of mammalian target of rapamycin by rapamycin modulates cognitive and non-cognitive components of behavior throughout lifespan in mice. Neuroscience. 2012;223:102–113. doi:10.1016/j.neuroscience. 2012.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilkinson JE, Burmeister L, Brooks SV et al. . Rapamycin slows aging in mice. Aging Cell. 2012;11:675–682. doi:10.1111/j.1474-9726.2012.00832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ocampo A, Reddy P, Martinez-Redondo P et al. . In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016;167:1719–1733.e12. doi:10.1016/j.cell.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farrell AS, Sears RC. MYC degradation. Cold Spring Harbor Perspectives in Medicine. 2014;4:a014365. doi:10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. doi:10.1093/gerona/glu057 [DOI] [PubMed] [Google Scholar]
- 31. O’Brown ZK, Van Nostrand EL, Higgins JP, Kim SK. The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLoS Genet. 2015;11:e1005734. doi:10.1371/journal.pgen.1005734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stevens LA, Viswanathan G, Weiner DE. Chronic kidney disease and end-stage renal disease in the elderly population: Current prevalence, future projections, and clinical significance. Adv Chronic Kidney Dis. 2010;17:293–301. doi:10.1053/j.ackd.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silverstein DM. Inflammation in chronic kidney disease: Role in the progression of renal and cardiovascular disease. Pediatr Nephrol. 2009;24:1445–1452. doi:10.1007/s00467-008-1046-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.