Abstract
Protein coding sequences represent only 2% of the human genome. Recent advances have demonstrated that a significant portion of the genome is actively transcribed as non-coding RNA molecules. These non-coding RNAs are emerging as key players in the regulation of biological processes, and act as "fine-tuners" of gene expression. Neurological disorders are caused by a wide range of genetic mutations, epigenetic and environmental factors, and the exact pathophysiology of many of these conditions is still unknown. It is currently recognized that dysregulations in the expression of non-coding RNAs are present in many neurological disorders and may be relevant in the mechanisms leading to disease. In addition, circulating non-coding RNAs are emerging as potential biomarkers with great potential impact in clinical practice. In this review, we discuss mainly the role of microRNAs and long non-coding RNAs in several neurological disorders, such as epilepsy, Huntington disease, fragile X-associated ataxia, spinocerebellar ataxias, amyotrophic lateral sclerosis (ALS), and pain. In addition, we give information about the conditions where microRNAs have demonstrated to be potential biomarkers such as in epilepsy, pain, and ALS.
Keywords: microRNA, Gene regulation, Molecular biomarkers
Introduction
Recent developments have indicated that numerous non-coding sequences present in the human genome are actively transcribed as non-coding RNA (ncRNA) molecules (1). These ncRNAs may be grouped into different classes and classified according to size and function. They have emerged as key players in the regulation of many biological processes and the fine-tune control of gene expression (2).
It is not surprising that the complexity of neurological disorders is determined by different molecular mechanisms, including genetic mutations and epigenetic factors. In this context, changes in ncRNA gene expression regulation have emerged as a putative mechanism in a variety of neurological disorders such as epilepsy, neurodegenerative disorders, and autoimmune conditions (3,4). Specific processes by which ncRNAs may influence disease vary widely and include quantitative changes in coding and ncRNA expression, induction of abnormal RNA species, and others (2,5). Furthermore, circulating ncRNAs may act as disease biomarkers, contributing to early disease diagnosis and treatment follow-up (6).
In this review, we discuss the classification, biogenesis, and mechanisms of action of ncRNAs. We also review key studies that show associations between microRNA (miRNA) and long non-coding RNA (lncRNA) dysregulation and different early and adult onset neurological disorders, as well as the use of circulating miRNAs as biomarkers and potential therapeutic strategies based on manipulating ncRNAs. The role of ncRNAs in aging-related neurological disorders, such as Alzheimer's or Parkinson's disease, are thoroughly reviewed elsewhere and are not the focus of the present review (7 –9).
Structure, function, and classification of non-coding RNAs
ncRNAs are defined as RNA molecules transcribed from genomic DNA that are not translated into proteins (10). The earliest recognized members of this category of RNA molecules were transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) (10). More recently, an increasing number of other ncRNAs have been detected and characterized, leading to the discovery that at least two thirds of the mammalian genome is actively transcribed (1).
ncRNAs are, in a broader sense, classified as long or small RNAs. lncRNAs are molecules ranging from ∼200 nucleotides (nt) to more than 20 kilobases. The major components of this category are rRNAs, tRNAs, X-chromosome inactivation RNAs (XIST RNAs) and regulatory lncRNAs (2). However, lncRNAs are an ever-increasing category, with more components than the four mentioned above (2). Small ncRNAs have lengths ranging from 20 to 200 nt, including small regulatory miRNAs, small nucleolar RNAs (snoRNAs), and piwi interacting RNAs (piRNAs) (11,12).
The molecular machinery responsible for miRNA biogenesis and interaction with mRNAs (Figure 1) is better elucidated than that underlying the activity of other ncRNAs. miRNA genes are transcribed by RNA polymerase II or III. This process generates a molecule, the pri-miR, that folds itself into a hairpin conformation and is 5′ capped and 3′ polyadenylated (13,14). The pri-miR molecule is recognized by the DROSHA RNAse III enzyme and cleaved, forming a 60- to 100-nt hairpin molecule, the pre-miR, that is exported from the nucleus to the cytoplasm (14,15). In the cytoplasm, the pre-miR is cleaved by the DICER enzyme, yielding a double-stranded ∼22nt RNA molecule (16). One of the strands of the formed 22-nt miRNA molecule is loaded into an RNA-induced silencing complex (RISC) protein to serve as the template for target mRNA recognition (17).
Figure 1. Main processes involved in the biogenesis and mechanism of action of microRNAs. DROSHA: Drosha ribonuclease III; DICER: Dicer 1; Ago1-4: Argonaute 1-4.
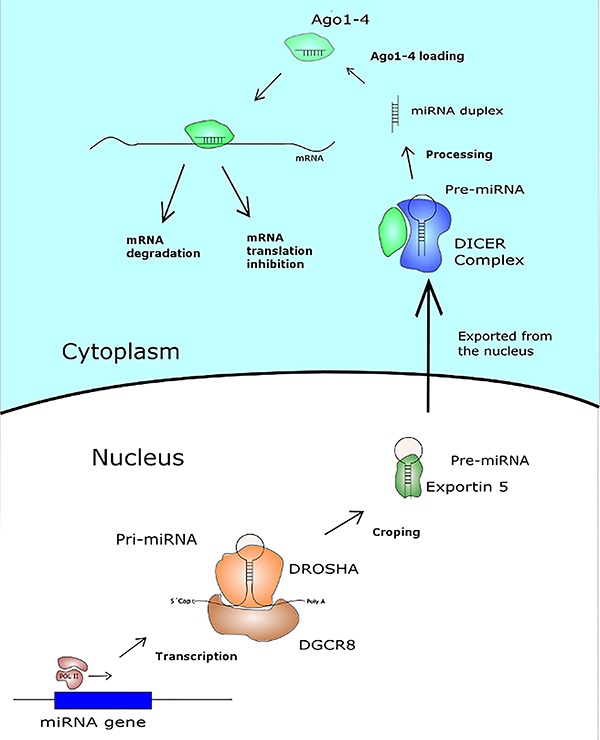
Mature miRNA molecules loaded into RISCs have two mechanisms of action. Perfect or near-perfect base pairing of the entire miRNA molecule to a complementary region within an mRNA leads to mRNA degradation by RISC (18). Perfect base pairing of almost all 22 nt is an uncommon scenario in animals. The more common scenario involves imperfect pairing, or pairing of a 5–8 nt ‘seed’ region of the miRNA, which leads to reduced translation or destabilization of the target mRNA (19). A single miRNA molecule may regulate multiple genes that contain a sequence complementary to the miRNA seed, and a given mRNA may be regulated by different miRNAs (20). Notably, the administration of exogenous nucleic acid sequences can mimic miRNA action (mimic-miRs), and employ the endogenous cellular machinery for miRNA-mediated gene silencing (21). Another possibility is the administration of stabilized exogenous nucleic acid sequences that are complementary to endogenous miRNAs, such as antagomirs, resulting in the inhibition of target cellular miRNAs (22).
miRNAs are also present and enriched in the plasma and serum. Furthermore, these RNAs are especially resistant to degradation (23). Blood circulating miRNAs are contained in microvesicles known as exosomes or are associated with Argonaute 2 complexes and, as a consequence, are protected from degradation (6,24). Because circulating miRNAs may originate from many different tissues throughout the body and may reflect normal function, changes in the circulating levels of these miRNAs may constitute a useful and easily accessible biomarker of many different pathological conditions. Moreover, it is feasible to quantify the levels of such circulating miRNAs by RT-PCR or even high throughput techniques such as micro-arrays or RNA-sequencing. The dysregulation of miRNA expression is well established in some tumors, and circulating miRNAs are indeed emerging as promising biomarkers in this field (23,25). The search for circulating miRNAs as biomarkers is also being applied to neurological disorders.
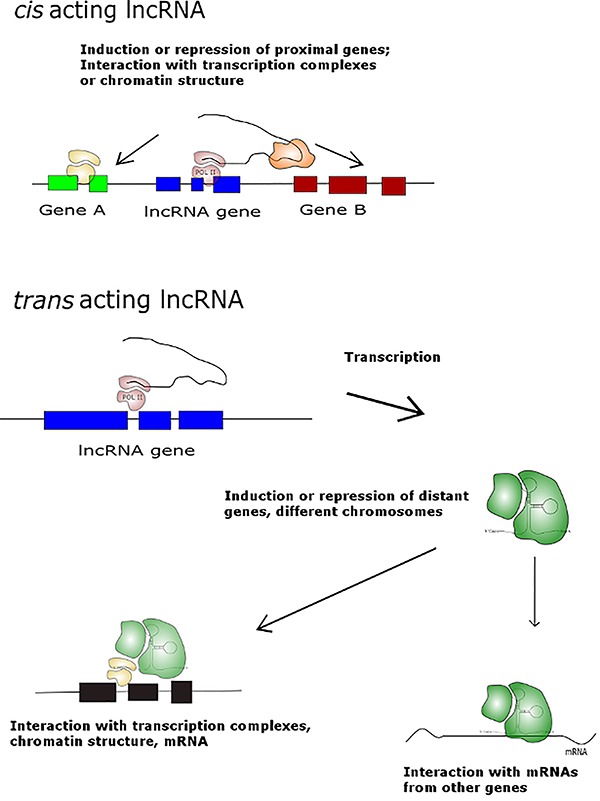
lncRNAs boast distinct and diverse molecular machinery involved in the regulation of gene expression (Figure 2). Most of these ncRNAs are RNA polymerase II products that lack open reading frames but are generally 5′ capped and 3′ polyadenylated (26,27). lncRNAs are numerous, with estimates in the range of thousands of lncRNA coding genes (28). Briefly, lncRNAs may act in cis, silencing or enhancing the expression of proximal genes on the same chromosome. For example, the lncRNA HOTTIP gene is present in the HOXA gene cluster, and its expression enhances the expression of other component genes in the same cluster (20). lncRNAs may also act in trans, silencing or enhancing the expression of genes on different chromosomes. One example of an lncRNA acting in trans is Six3OS. This lncRNA was shown to activate the targets of the retinal development involving the Six3 transcription factor (29). Another mechanism of action for lncRNAs is the regulation of other ncRNAs. lncRNA can act as a ‘sponge' or decoy target. The lncRNA lincRNA-RoR mechanism of action illustrates this mechanism: this lncRNA has a binding site for miR-145, and the presence of lincRNA-RoR inhibits miR-145 action by interacting directly with lncRNA miRNA (30). The mechanisms of lncRNA-mediated regulation of protein-coding gene transcription are explored in more detail in the current literature (26,27).
Figure 2. Mechanisms by which long non-coding RNAs (lncRNAs) can regulate gene expression.
Role of non-coding RNAs in disease
Table 1 presents a list of ncRNAs associated with mechanisms underlying selected neurological disorders.
Table 1. List of ncRNAs associated with different mechanisms underlying selected neurological disorders.
| Disorder | Gene Affected | Proposed mechanisms associated with Noncoding RNAs | References |
|---|---|---|---|
| FXTAS | FMR1; FMR4 | Sequestration of RNA binding protein; antisense transcript | Tassone et al. 2004 58 |
| DM1 | DMPK | Sequestration of RNA binding protein; antisense transcript | Rau et al. 2011 66 |
| SCA1 | ATXN1 | Altered miRNA pathway | Galka-Marciniak et al. 2012 56 |
| SCA3 | ATXN3 | An auxiliary toxic long CAG repeat RNA; altered miRNA pathway | Galka-Marciniak et al. 2012 56 |
| SCA7 | ATXN7 | Antisense transcript repress sense ataxin-7 | Tan et al. 2014 63 |
| SCA8 | ATXN8OS; ATXN8 | Sequestration of RNA binding protein; antisense transcript | Daughters et al. 2009 61; Moseley et al. 2006 62 |
| HDL2 | JPH3 | Antisense transcript; polyQ toxicity | Wojciechowska and Krzyzosiak, 2011 5 |
| MTLE | P2X7 | Down-regulation by miR-22 | Jimenez-Mateos et al. 2015 3 |
| HD | HTT | An auxiliary toxic long CAG repeat RNA; altered miRNA pathway | Wojciechowska and Krzyzosiak, 2011 5 |
| MTLE | Genes involved with inflammation | Up-regulation of miR-146a expression | Aronica et al. 2010 37 |
| ALS | SOD1 and others | An artificial microRNA may extend survival and delays paralysis; Up regulation of miR-206. | Stoica et al. 2016 79; Takahashi et al. 2015 81 |
| Cortical dysplasia | Lis1 | Dysregulation of miR-139-5p | Huang et al. 2014 90 |
| Pain | Inflammation, neural processing | Dysregulation of miR-1, -16, and -206 | Kusuda et al. 2011 86 |
Epilepsy
Epilepsy is a neurological condition with a high prevalence in the population (1.5–2%). A common feature of different epileptic conditions is the occurrence of seizures (31,32). The mechanism responsible for epileptogenesis (the process by which normal nervous tissue becomes epileptic) is complex and multifactorial (33). Evidence in the literature, as reviewed below, indicates that ncRNAs may have critical roles in the molecular mechanisms associated with epilepsy (34).
Hippocampal tissue from patients with mesial temporal lobe epilepsy (MTLE) who underwent temporal lobe resection for the control of seizures has been shown to have a reduction in the overall expression of miRNAs when compared with normal hippocampus from autopsy controls (35). Moreover, MTLE is associated with inflammation, and changes in the expression of miRNAs involved in the regulation of inflammation have been demonstrated in samples from MTLE patients (36,37). For example, miR-146-a, a miRNA involved in inflammation, is upregulated in resected hippocampus from MTLE patients (37).
In animal models of epilepsy, the dysregulation of miRNAs has been explored more extensively. miRNA expression studies were performed, using high-throughput platforms, in the animal model induced by lithium-pilocarpine, systemic kainic acid, and by intra-amygdalar kainic acid injection (38 –40). Based on such studies, an extensive list of candidate miRNAs was found, but relatively few miRNAs were consistent among different studies. One example of replicable findings is mir-34a, which was found to be differentially expressed in two independent studies (38,41). mir-134 is another promising miRNA that may be involved in the molecular mechanisms of epilepsy. mir-134 was found to be differentially expressed in an epilepsy animal model, and the reduction in its expression by antagomir administration was shown to reduce cell death and seizure severity (42). In addition, downregulation of mir-132 in an animal model reduced seizure-induced neuronal death (40).
More recently, Jimenez-Mateos et al. (3) demonstrated that miR-22 downregulates the purinergic P2X7 receptor, a key component of the inflammatory response, in a mouse model of focal onset status-epilepticus. Furthermore, an increase in miR-22 activity by the administration of a Mir-22 mimic molecule reduced spontaneous seizures in these mice (3).
The role of lncRNAs has also been explored in the context of experimental animal models of epilepsy. Lee et al. (43) explored the expression of lncRNAs in two animal epilepsy models, pilocarpine- and kainic acid-induced seizures (43). These authors found hundreds of lnRNAs that were differentially expressed when comparing nervous tissue from controls with that of treated mice. Of these differentially expressed lncRNAs, 54 (for pilocarpine) and 14 (for kainic acid) were close to protein-coding genes and appear to induce significant changes in gene expression, thus indicating a possible cis effect of these lncRNAs (43).
The first evidence for the potential use of miRNAs as biomarkers in epilepsy also came from studies in experimental animal models. Liu et al. (44) demonstrated the differential regulation of several miRNAs isolated from the blood of rats that received the chemoconvulsant kainic acid. More recently, Roncon et al. (45) found 27 miRNAs to be differentially expressed in the plasma of rats treated with pilocarpine. In humans, Wang et al. (46), using RNA-sequencing and subsequent RT-PCR validation, found four upregulated and two downregulated blood circulating miRNAs when comparing epilepsy patients to healthy controls. Among the differentially expressed miRNAs, miR-106b-5p had the highest sensitivity and specificity (46). Furthermore, in a subsequent study, there were five circulating miRNAs identified as potential biomarkers of drug-resistant epilepsy, and miR-301a-3p had the highest sensitivity and specificity (47). We have identified that miR-134 is a circulating biomarker for patients with mesial temporal lobe epilepsy regardless of their response to treatment, which may help in the diagnosis of this type of epilepsy (48).
In focal cortical dysplasia, a cortical malformation frequently associated with refractory seizures, miR-4521 has been shown to be upregulated in the plasma of patients compared to control subjects (49).
Neurodegenerative and neuromuscular disorders
Neurodegenerative disorders are associated with a wide range of genetic mutations and epigenetic and environmental factors. Among genetic mutations, trinucleotide repeat expansion is increasingly recognized as the cause of a large subset of these conditions. Trinucleotide repeat expansions account for more than 30 neurological and neuromuscular diseases that are categorized into coding and non-coding repeat expansion disorders, depending on the genetic location of their causative mutations (50 –52). Disorders such as Huntington's disease (HD), spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7, 8, and 17, dentatorubral-pallidoluysian atrophy, and spinal and bulbar muscular atrophy are typically associated with a protein gain-of-function mechanism (53). In contrast, diseases such as myotonic dystrophy type 1 (DM1) (54,55), fragile X-associated tremor ataxia syndrome (FXTAS), myotonic dystrophy type 2 (DM2), SCA31, SCA10, SCA8, and, more recently, amyotrophic lateral sclerosis and frontotemporal sclerosis have been associated with an RNA gain-of-function mechanism in which the trinucleotide expansion leads to the formation of nuclear RNA foci that sequester specific RNA-binding proteins (5,56,57).
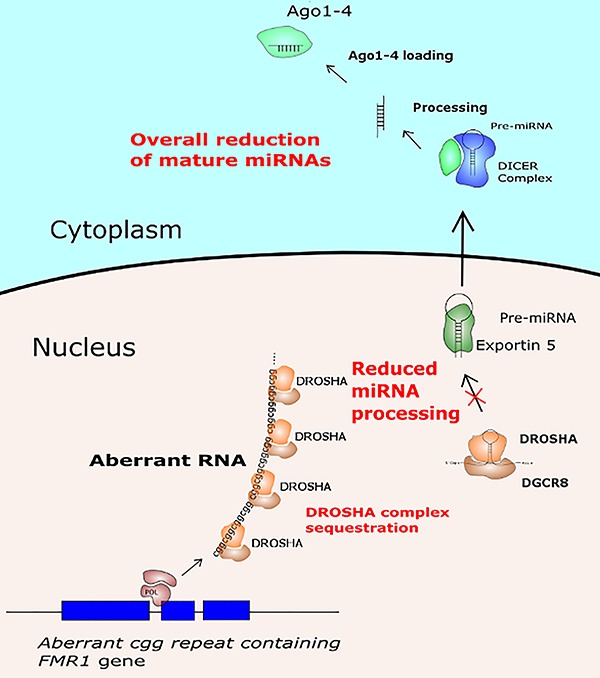
Studies of FXTAS have established that the sequestration of RNA-binding proteins due to the expression of pathogenic RNA with expanded repeats is involved in disease pathogenesis (58) (Figure 3). A recent study identified that the double-stranded RNA-binding protein DGCR8 binds to expanded CGG repeats, resulting in the partial sequestration of DGCR8 and its partner, DROSHA, within CGG RNA aggregates. Consequently, the processing of miRNAs is reduced, resulting in decreased levels of mature miRNAs in neuronal cells expressing expanded CGG repeats such as in brain tissue from patients with FXTAS (59).
Figure 3. Mechanism involved in microRNA machinery sequestration by aberrant RNA species produced in a triplet repeat disease, fragile-X associated tremor ataxia syndrome (FXTAS). DICER: Dicer 1; DROSHA: Drosha ribonuclease III; Ago1-4: Argonaute 1-4.
SCA8 is a dominantly inherited, slowly progressive neurodegenerative disorder caused by a CTG CAG repeat expansion (60). In pathological samples from SCA8 patients, bidirectional (sense and antisense) expression of the SCA8 CTG·CAG expansion produces toxic non-coding CUG expansion in RNAs from the Ataxin 8 opposite strand (ATXN8OS) and a nearly pure polyglutamine expansion protein encoded by ATXN8 (61,62). In SCA7, the tissue-specific alterations caused by CAG repeat expression in the ATXN7 gene seems to be related to cross-talk between the lncRNA lnc-SCA7, the ATXN7 mRNA, and mir-124. Mutant ATXN7 disrupts this crosstalk and is itself upregulated, since it is not repressed by ncRNAs (63).
Recent studies have suggested that alterations in small regulatory ncRNAs, such as miRNAs, could contribute to the pathogenesis of several neurodevelopmental disorders. Some studies have found a relationship between miRNAs and DM1 (64). Alterations in the miRNA expression patterns have been observed in muscle-specific miRNAs (myomiRs). Given the small distance between the seed binding sites of miR-206 and 148a in the DMPK 3′ UTR, Koscianska et al. (65) analyzed the binding mechanism of both miRNAs. They discovered cooperative binding; the joint binding of miRs 206 and 148a increased the negative regulation of DMPK mRNA. These findings provide mechanistic insights into the miRNA-mediated regulation of the DMPK transcript. In this regard, the dysregulation of DM1-associated miRNAs has also been linked to alterations in their predictive target expression, showing that miRNA dysregulation in DM1 is functionally relevant and may contribute to disease pathology (66,67). Furthermore, RNA toxicity has been confirmed in transgenic mice harboring long triplet repeats in the dmpk gene. Seznec et al. (68) showed that mice develop multi-system abnormalities mimicking the human DM phenotype, with predominant involvement of muscles and the central nervous system (CNS). Pathway and function analysis highlighted the involvement of the miRNA-dysregulated mRNAs in multiple aspects of DM2 pathophysiology as well (4,69).
Huntington's disease is characterized by widespread mRNA dysregulation, especially in the striatum and cortical regions and alterations in miRNA-mediated post-transcriptional regulation could be an important mechanism contributing to mRNA dysregulation in HD (70). In addition, there is evidence that abnormal neurodevelopment might also have a critical role in HD (71). These emerged from studies using mouse embryonic stem cells and patient-derived induced pluripotent stem cells (The HD iPSC Consortium, 2012) showing that chromatin modifications and DNA methylation status support the hypothesis that wild-type and mutant Huntingtin might affect key chromatin regulators such as DNA and histone methyltransferases, and demethylases (72 –74). In fact, a growing body of evidence suggests that alterations of epigenetic modifications constitute a basic molecular mechanism caused by the HD mutation and are responsible for early features of the pathological process (75). Furthermore, a recent genome-wide screen of miRNAs in post mortem brains highlighted miRNAs that were differentially expressed in HD patients, especially miRNAs in the HOX family, which have been associated with early brain development (76). Indeed, there are several classes of lncRNAs that are potentially involved in developmental processes and that were found to be dysregulated in brain tissue from patients with HD such as TUG1, NEAT1, MEG3, and DGCR5 (77).
Amyotrophic lateral sclerosis (ALS) is a widespread motor neuron disorder causing injury and death of lower and upper motor neurons. Familial ALS (∼10% of all ALS cases) is inherited as a dominant trait, and 20% of these cases have mutations in the gene encoding Cu/Zn cytosolic superoxide dismutase 1 (SOD1) (78). A recent study demonstrated that an AAV9-delivered SOD1-specific artificial miRNA is an effective and translatable therapeutic approach to ALS (79). Another promising miRNA with a possible therapeutic use in ALS is mir-155. It was demonstrated that this inflammation-associated miRNA is upregulated in the mutant SOD1 mouse model and that reduction in the expression of mir-155 significantly extended the life span of this mouse (80).
In addition, expression levels of certain miRNAs, such as miR-4649-5p and hsa-miR-4299, were significantly correlated with disease progression and might be useful as prognostic biomarkers (81). Another potential biomarker was mir-206, found to be upregulated in the plasma of SOD1-G93A mice, an experimental ALS model, and in patients with confirmed ALS (82). In addition, there is evidence of dysregulation of miRNAs extracted from leukocytes from sporadic ALS patients (83). More recently, we have demonstrated that among 11 miRNAs identified as differently expressed in muscle of patients with ALS, only two, miR-214 and miR-424, correlated with clinical deterioration over time in these patients (84).
Pain
Conditions leading to chronic pain are related to multiple etiologic factors, ranging from maladaptive neuronal plasticity to diverse inflammatory pathways (85). Due to the complexity of chronic pain, some studies have explored the possible role of ncRNAs in different experimental pain models. Kusuda et al. (86) observed a change in the expression of three miRNAs, miRs 1, 16, and 206, in different pain conditions such as peripheral inflammation, nerve ligation, or axotomy. Other studies have employed low-density TaqMan arrays to profile the expression pattern of miRNAs after spinal nerve ligation in rats and found 63 altered miRNAs (87).
A possible role for lncRNAs has been explored in experimental models of neuropathic pain. A microarray analysis demonstrated hundreds of differentially expressed lncRNAs and mRNAs in the spinal cords of mice subjected to spinal nerve ligation. As demonstrated in other experiments, 35 differentially regulated lncRNAs were in genomic regions proximal to differentially regulated genes from the same dataset (88).
Non-coding RNAs as target treatments for neurologic disorders
The use of ncRNAs as therapeutic tools in human disorders is still in its early stages. To date, there is only one therapeutic use of human miRNA for the treatment of hepatitis C (HCV) that has passed phase IIa clinical trials (89). The clinical trial data showed the efficacy of the employed anti-miRNA in reducing viral load and showed good treatment tolerability, thus indicating the feasibility of similar strategies for other clinical uses such as in the case of neurological conditions.
Animal experiments already indicate some promising targets for the use of ncRNAs as therapeutic tools in disorders affecting the CNS. In epilepsy, the use of miR antagonists for miR-134 or mimic-miRs for miR-22 was capable of reducing neuronal death and seizure severity in animal models (3,42). These and other examples of pre-clinical uses of miRNAs for the treatment of neurological conditions need further study; however, due to the good tolerability already shown in the existing human clinical trial for HCV, there is optimism about the possible utility of ncRNAs in the treatment of neurological conditions in the future. However, several challenges remain for the efficient delivery of ncRNA molecules into the CNS, thus most of the pre-clinical studies still use invasive techniques for administering these molecules (4,44)
Conclusions
In conclusion, ncRNAs are emerging as key players in the field of neurological disorders. ncRNAs are involved in many conditions, either as part of the molecular mechanisms underlying disease or as biomarkers that may be used for improved diagnosis or assessment of disease progression. ncRNAs are also promising targets for new therapeutic strategies to be employed in the treatment of neurological conditions.
Acknowledgments
A.S.V. is supported by a grant from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP; #2016/22447-5), Brazil. D.B.D. is supported by a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil. I.L-C is supported by grants from FAPESP (#2011/50680 and #2013/07559-3) and from Conselho Nacional de Pesquisa (CNPq), Brazil.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez-Mateos EM, Arribas-Blazquez M, Sanz-Rodriguez A, Concannon C, Olivos-Ore LA, Reschke CR, et al. microRNA targeting of the P2X7 purinoceptor opposes a contralateral epileptogenic focus in the hippocampus. Sci Rep. 2015;5:17486. doi: 10.1038/srep17486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greco S, Perfetti A, Fasanaro P, Cardani R, Capogrossi MC, Meola G, et al. Deregulated microRNAs in myotonic dystrophy type 2. PloS One. 2012;7:e39732. doi: 10.1371/journal.pone.0039732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Femminella GD, Ferrara N, Rengo G. The emerging role of microRNAs in Alzheimer's disease. Front Physiol. 2015;6:40. doi: 10.3389/fphys.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z. Long non-coding RNAs in Alzheimer's disease. Curr Top Med Chemy. 2016;16:511–519. doi: 10.2174/1568026615666150813142956. [DOI] [PubMed] [Google Scholar]
- 9.Majidinia M, Mihanfar A, Rahbarghazi R, Nourazarian A, Bagca B, Avci CB. The roles of non-coding RNAs in Parkinson's disease. Mol biol rep. 2016;43:1193–1204. doi: 10.1007/s11033-016-4054-3. [DOI] [PubMed] [Google Scholar]
- 10.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006:R17–R29. doi: 10.1093/hmg/ddl046. 15 Spec No 1. [DOI] [PubMed] [Google Scholar]
- 12.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 13.McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 18.Gu S, Kay MA. How do miRNAs mediate translational repression? Silence. 1:11. doi: 10.1186/1758-907X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannell IG, Kong YW, Bushel M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 20.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol Biol. 2011;676:211–223. doi: 10.1007/978-1-60761-863-8. [DOI] [PubMed] [Google Scholar]
- 22.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Nat Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Engel J., Jr Mesial temporal lobe epilepsy: what have we learned? The Neuroscientist. 2001;7:340–352. doi: 10.1177/107385840100700410. [DOI] [PubMed] [Google Scholar]
- 32.Annegers JF, Rocca WA, Hauser WA. Causes of epilepsy: contributions of the Rochester epidemiology project. Mayo Clin Proc. 1996;71:570–575. doi: 10.4065/71.6.570. [DOI] [PubMed] [Google Scholar]
- 33.Pitkänen A, Lukasiuk K. Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 2011;10:173–186. doi: 10.1016/S1474-4422(10)70310-0. [DOI] [PubMed] [Google Scholar]
- 34.Dogini DB, Avansini SH, Vieira AS, Lopes-Cendes I. MicroRNA regulation and dysregulation in epilepsy. Front Cell Neurosci. 2013;7:172. doi: 10.3389/fncel.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, Sano T, et al. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PloS One. 2012;7:e35921. doi: 10.1371/journal.pone.0035921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vezzani A, Friedman A, Dingledine RJ. The role of inflammation in epileptogenesis. Neuropharmacology. 2013;69:16–24. doi: 10.1016/j.neuropharm.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, et al. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Euro J Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 38.Hu K, Xie YY, Zhang C, Ouyang DS, Long HY, Sun DN, et al. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012;13:115. doi: 10.1186/1471-2202-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.McKiernan RC, Jimenez-Mateos EM, Sano T, Bray I, Stallings RL, Simon RP, et al. Expression profiling the microRNA response to epileptic preconditioning identifies miR-184 as a modulator of seizure-induced neuronal death. Exp Neurol. 2012;237:346–354. doi: 10.1016/j.expneurol.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, et al. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012;3:e287. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee DY, Moon J, Lee ST, Jung KH, Park DK, Yoo JS, et al. Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem Biophys Res Commun. 2015;462:433–440. doi: 10.1016/j.bbrc.2015.04.149. [DOI] [PubMed] [Google Scholar]
- 44.Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roncon P, Soukupova M, Binaschi A, Falcicchia C, Zucchini S, Ferracin M, et al. MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy--comparison with human epileptic samples. Sci Rep. 2015;5:14143. doi: 10.1038/srep14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Yu JT, Tan L, Tian Y, Ma J, Tan CC, et al. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci Rep. 2015;5:9522. doi: 10.1038/srep09522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Tan L, Tan L, Tian Y, Ma J, Tan CC, et al. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci Rep. 2015;5:10201. doi: 10.1038/srep10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avansini SH, de Sousa Lima BP, Secolin R, Santos ML, Coan AC, Vieira AS, et al. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS One. 2017;12:e0173060. doi: 10.1371/journal.pone.0173060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Sun Y, Tan Z, Che N, Ji A, Luo X, et al. Serum MicroRNA-4521 is a Potential Biomarker for Focal Cortical Dysplasia with Refractory Epilepsy. Neurochem Res. 2016;41:905–912. doi: 10.1007/s11064-015-1773-0. [DOI] [PubMed] [Google Scholar]
- 50.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 51.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 52.Ranum LP, Day JW. Dominantly inherited, non-coding microsatellite expansion disorders. Curr Opin Genet Dev. 2002;12:266–271. doi: 10.1016/S0959-437X(02)00297-6. [DOI] [PubMed] [Google Scholar]
- 53.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 54.Wang LC, Chen KY, Pan H, Wu CC, Chen PH, Liao YT, et al. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell Mol Life Sci. 2011;68:1255–1267. doi: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011;39:8938–8951. doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galka-Marciniak P, Urbanek MO, Krzyzosiak WJ. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol Chem. 2012;393:1299–1315. doi: 10.1515/hsz-2012-0218. [DOI] [PubMed] [Google Scholar]
- 57.Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 59.Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikeda Y, Shizuka-Ikeda M, Watanabe M, Schmitt M, Okamoto K, Shoji M. Asymptomatic CTG expansion at the SCA8 locus is associated with cerebellar atrophy on MRI. J Neurol Sci. 2000;182:76–79. doi: 10.1016/S0022-510X(00)00446-9. [DOI] [PubMed] [Google Scholar]
- 61.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 63.Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, Curtis HJ, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21:955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner C, Hilton-Jones D. The myotonic dystrophies: diagnosis and management. J Neurol Neurosurg Psychiatry. 2010;81:358–367. doi: 10.1136/jnnp.2008.158261. [DOI] [PubMed] [Google Scholar]
- 65.Koscianska E, Witkos TM, Kozlowska E, Wojciechowska M, Krzyzosiak WJ. Cooperation meets competition in microRNA-mediated DMPK transcript regulation. Nucleic Acids Res. 2015;43:9500–9518. doi: 10.1093/nar/gkv849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer M. C, Jost B, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nature Struct Molec Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 67.Perbellini R, Greco S, Sarra-Ferraris G, Cardani R, Capogrossi MC, Meola G, et al. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul Disorder. 2011;21:81–88. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 69.Deng JH, Deng P, Lin SL, Ying SY. Gene silencing in vitro and in vivo using intronic microRNAs. Meth Molecular Biol. 12015;1218:321–340. doi: 10.1007/978-1-4939-1538-5. [DOI] [PubMed] [Google Scholar]
- 70.Hoss AG, Lagomarsino VN, Frank S, Hadzi TC, Myers RH, Latourelle JC. Study of plasma-derived miRNAs mimic differences in Huntington's disease brain. Mov Disorder. 2015;30:1961–1964. doi: 10.1002/mds.26457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humbert S. Is Huntington disease a developmental disorder? EMBO Rep. 2010;11:899. doi: 10.1038/embor.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci USA. 2013;110:2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biagioli M, Ferrari F, Mendenhall EM, Zhang Y, Erdin S, Vijayvargia R, et al. Htt CAG repeat expansion confers pleiotropic gains of mutant huntingtin function in chromatin regulation. Hum Mol Genet. 2015;24:2442–2457. doi: 10.1093/hmg/ddv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kerschbamer E, Biagioli M. Huntington's disease as neurodevelopmental disorder: altered chromatin regulation, coding, and non-coding RNA transcription. Front Neurosci. 2015;9:509. doi: 10.3389/fnins.2015.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoss AG, Kartha VK, Dong X, Latourelle JC, Dumitriu A, Hadzi TC, et al. MicroRNAs located in the Hox gene clusters are implicated in huntington's disease pathogenesis. PLoS Genet. 2014;10:e1004188. doi: 10.1371/journal.pgen.1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 78.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 79.Stoica L, Todeasa SH, Toro Cabrera G, Salameh JS, ElMallah MK, Mueller C, et al. Adno associated virus delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Butovsky O, Jedrychowski MP, Cialic R, Krasemann S, Murugaiyan G, Fanek, et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol. 2015;77:75–99. doi: 10.1002/ana.24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi I, Hama Y, Matsushima M, Hirotani M, Kano T, Hohzen H, et al. Identification of plasma microRNAs as a biomarker of sporadic amyotrophic lateral sclerosis. Mol Brain. 2015;8:67. doi: 10.1186/s13041-015-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toivonen JM, Manzano R, Olivan S, Zaragoza P, Garcia-Redondo A, Osta R. MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PloS One. 2014;9:e89065. doi: 10.1371/journal.pone.0089065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Felice B, Guida M, Guida M, Coppola C, De Mieri G, Cotrufo R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508:35–40. doi: 10.1016/j.gene.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 84.de Andrade HM, de Albuquerque M, Avansini SH, de S Rocha C, Dogini DB, Nucci A, et al. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J Neurol Sci. 2016;368:19–24. doi: 10.1016/j.jns.2016.06.046. [DOI] [PubMed] [Google Scholar]
- 85.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, et al. Differential expression of microRNAs in mouse pain models. Mol Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.von Schack D, Agostino MJ, Murray BS, Li Y, Reddy PS, Chen J, et al. Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain. PloS One. 2011;6:e17670. doi: 10.1371/journal.pone.0017670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang BC, Sun WX, He LN, Cao DL, Zhang ZJ, Gao YJ. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain. 2015;11:43. doi: 10.1186/s12990-015-0047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–9. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 90.Huang Y, Jiang J, Zheng G, Chen J, Lu H, Guo H, et al. miR-139-5p modulates cortical neuronal migration by targeting Lis1 in a rat model of focal cortical dysplasia. Int J Mol Med. 2014;33:1407–1414. doi: 10.3892/ijmm.2014.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]


