A YTH domain protein acts as a reader of m6A mRNA modifications, thereby playing crucial roles in trichome branching, likely by acting on mRNAs in the cytosol.
Abstract
Methylations at position N6 of internal adenosines (m6As) are the most abundant and widespread mRNA modifications. These modifications play crucial roles in reproduction, growth, and development by controlling gene expression patterns at the posttranscriptional level. Their function is decoded by readers that share the YTH domain, which forms a hydrophobic pocket that directly accommodates the m6A residues. While the physiological and molecular functions of YTH readers have been extensively studied in animals, little is known about plant readers, even though m6As are crucial for plant survival and development. Viridiplantae contains high numbers of YTH domain proteins. Here, we performed comprehensive evolutionary analysis of YTH domain proteins and demonstrated that they are highly likely to be actual readers with redundant as well as specific functions. We also show that the ECT2 protein from Arabidopsis thaliana binds to m6A-containing RNAs in vivo and that this property relies on the m6A binding pocket carried by its YTH domain. ECT2 is cytoplasmic and relocates to stress granules upon heat exposure, suggesting that it controls mRNA fate in the cytosol. Finally, we demonstrate that ECT2 acts to decode the m6A signal in the trichome and is required for their normal branching through controlling their ploidy levels.
INTRODUCTION
Gene expression regulation is a multilayered process that takes place at the transcriptional and posttranscriptional levels and is crucial for organism development, growth, and survival. Recently, chemical modification of mRNAs has emerged as an additional and important layer in the control of gene expression. The repertoire of these transcriptomic modifications represents what is now called the cell RNA epigenome (He, 2010) or epitranscriptome (Saletore et al., 2012). Methylation at position N6 of internal adenosines (m6As) is the most abundant and widespread of these modifications. This modification is conserved and found in mRNAs of most eukaryotes, such as animals (Dominissini et al., 2013), yeast (Schwartz et al., 2013), and vascular plants (Li et al., 2014b; Luo et al., 2014; Wan et al., 2015), and represents some 1.5% of the total number of adenosines on mRNAs. In plants, as in other eukaryotes, m6As are not evenly distributed on an mRNA molecule. They are found at the RRACH (R=G/A, H: U>A>C) consensus site (GAC in 70% of the cases) and almost exclusively on exons, with a very strong enrichment in terminal exons and 3′ untranslated regions (Meyer et al., 2012; Dominissini et al., 2013; Schwartz et al., 2013, 2014; Ke et al., 2015). m6As are cotranscriptionally deposited by a so-called writer complex and can be reverted to unmodified adenosines by so-called erasers. The core of the heteromultimeric writer complex contains METTL3, the active methylase, METTL14, a degenerated methylase (Śledź and Jinek, 2016; Wang et al., 2016), and WTAP, a stabilizing cofactor required for m6A deposition (Schwartz et al., 2014) and localization of the complex to nuclear speckles (Ping et al., 2014). In animals, this complex was also found to contain the KIAA1429 (fruit fly [Drosophila melanogaster] Virilizer) (Lence et al., 2016) protein, which associates with METTL3 (Schwartz et al., 2014). The core components of the writer complex are also present in Arabidopsis thaliana, with the MTA (METTL13) and MTB (METTL14) methylases, the FIP37 (WTAP) and VIRILIZER (VIR; KIAA1429) cofactors, plus the E3 ubiquitin ligase HAKAI (Zhong et al., 2008; Bodi et al., 2012; Růžička et al., 2017), which is also likely to be part of the mammalian writer complex (Horiuchi et al., 2013). The biological consequences of m6A methylation are multiple, but a common feature is that it is required in fundamental processes in all organisms studied to date (Roignant and Soller, 2017). In Arabidopsis, so far the sole plant where m6A roles have been reported, downregulation of components of the writer complex leads to reduced relative levels of m6A in mRNAs and shared pleitropic phenotypes. Deletion of MTA, MTB, FIP37, or VIR stops embryogenesis at the globular stage (Vespa et al., 2004; Zhong et al., 2008; Bodi et al., 2012; Shen et al., 2016; Růžička et al., 2017). Hypomorphic mta, mtb, fip37, or vir mutants show growth delay, aberrant shoot apical meristem proliferation, reduced root growth, and aberrant gravitropic responses, the severity of which is directly proportional to the diminution of the m6A:A ratio (Shen et al., 2016; Růžička et al., 2017). Moreover, downregulation of MTA or overexpression of FIP37 leads to aberrant trichome formation, with an increase in the number of trichomes with four to six branches (Vespa et al., 2004; Bodi et al., 2012). Assessment of the DNA content of FIP37 overexpressor lines supports that this phenotype might be the consequence of excessive rounds of endoreduplication (Vespa et al., 2004).
Studies conducted in animals clearly established that, under constitutive growth conditions, m6A marks trigger mRNA turnover (Wang et al., 2014; Du et al., 2016; Ke et al., 2017; Shi et al., 2017) and stimulate translation (Wang et al., 2015; Hsu et al., 2017; Li et al., 2017; Shi et al., 2017). These modifications also affect, although to a limited extent, alternative splicing control (Lence et al., 2016; Ke et al., 2017) and alternative poly(A) site choice (Ke et al., 2015). How m6A modifications of mRNAs control plant development at the cellular level remains so far largely unknown. One study in Arabidopsis demonstrated that m6A in plants negatively controls the stability of at least two key regulators of stem cell differentiation (Wushel and STM), spatially and temporally confining their expression to control shoot apical meristem proliferation (Shen et al., 2016).
At the molecular level, the m6A mark directly influences the recruitment of RNA Binding Proteins (RBPs) to the transcript, acting in particular as anchors for so-called m6A readers (Patil et al., 2018). The most widespread and well characterized of these reader proteins are those sharing the YTH (YT512-B homology) domain (Zhang et al., 2010). YTH domains form two evolutionary clades, named DC and DF, which adopt a similar α-helix-β-sheet fold, forming a hydrophobic pocket containing two to three aromatic residues essential for m6A mononucleoside binding (Li et al., 2014a; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014; Zhu et al., 2014). Three of the DF-type (YTHDF1 to YTHDF3) and two of the DC-type (YTHDC1 and YTHDC2) proteins are present in mammals (Wang and He, 2014). The YTHDF1 to YTHDF3 proteins act in a cooperative and integrated manner in the cytoplasm of mammalian cell lines to convey the constitutive roles of m6A on mRNA translation and stability control (Shi et al., 2017). In mouse (Mus musculus) testis, YTHDC2 promotes the translation and triggers the decay of a few hundred transcripts (Hsu et al., 2017). In human (Homo sapiens) and mouse cell lines, YTHDC1 mediates the role of m6A in transcriptional repression through binding to the XIST long noncoding RNA at its m6A sites (Patil et al., 2016). In fruit fly, Ythdc1 mediates the role of m6A in a few alternative splicing events, in particular that of the sex-lethal transcript (Haussmann et al., 2016; Lence et al., 2016). Consistent with their m6A reader functions, the physiological roles of these YTH proteins overlap with those of the writer complex (Ivanova et al., 2017).
How m6A controls mRNA fate and expression in plants at the molecular level is currently unclear. Nevertheless, the presence of YTH domain proteins suggests that, here again, it can act as an anchor to RBPs. Eleven of the 13 members of the Arabidopsis YTH protein family were initially recognized as proteins that share a highly conserved C terminus and were named ECT1 to ECT11 for Evolutionarily Conserved C Terminus (Ok et al., 2005). ECT1 and ECT2 were found to physically interact (via their YTH domain) to the CIPK1 serine/threonine kinase in yeast-two hybrid assays and (for ECT1) in vitro assays (Ok et al., 2005). ECT1 was found (in onion [Allium cepa] epidermal cells) to display a nucleocytoplasmic distribution, and its YTH domain was found to be necessary for its nuclear localization. Moreover, its nuclear targeting is triggered in the presence of exogenous calcium, supporting the notion that ECT1 function might be regulated by stress. However, the m6A reading activity of YTH domain proteins and the physiological and molecular importance of such a property are currently unclear.
Here, we performed comprehensive evolutionary analysis of YTH domains from Viridiplantae and functional characterization of a putative plant m6A reader, the ECT2 protein. We show that ECT2 is mainly cytosolic, relocalizes to stress granules (SGs) upon heat stress, and forms a complex in vivo with m6A RNA only when carrying an unaltered aromatic m6A binding cage. Plants depleted of ECT2 show a trichome-branching defect, likely related to an excessive number of endoreduplication cycles, identical to that of hypomethylated plants (Bodi et al., 2012) or FIP37 overexpressor plants (Vespa et al., 2004). This phenotype cannot be complemented with an ECT2 allele with a defective m6A binding pocket. Therefore, we identified a physiological role for a plant YTH protein and demonstrated that trichome formation requires that the m6A mark is decoded by at least 1 of the 13 Arabidopsis YTH proteins.
RESULTS
Evolutionary Analysis of YTH Domain Proteins from Viridiplantae
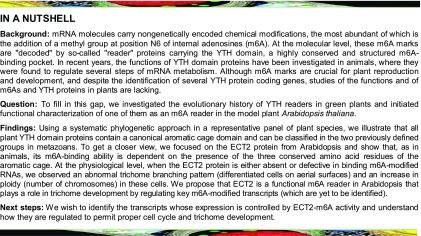
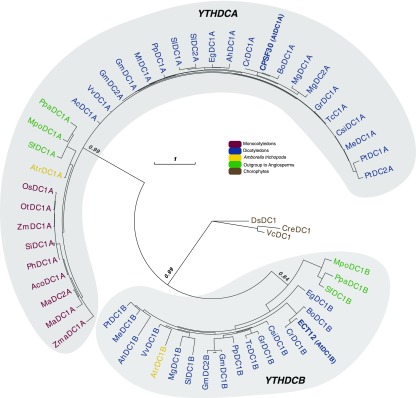
Using a phylogenetic approach, we first compared representative Viridiplantae YTH domains with several yeast and metazoan YTHs, including those from human, fruit fly, the yeast Saccharomyces cerevisiae, and the fission yeast Schizosaccharomyces pombe. To represent the Viridiplantae lineage, we initially collected YTH-containing proteins from a chlorophyte species (Chlamydomonas reinhardtii), from species that are outgroups for the angiosperms (the liverwort Marchantia polymorpha and the moss Physcomitrella patens), at the root of the angiosperm lineage (Amborella trichopoda), and from Arabidopsis. We confirm by this analysis that Viridiplantae YTH domains have a common ancestry with yeast and metazoan domains and can be classified in the two previously defined YTH-DC and YTH-DF groups (Roundtree and He, 2016) (Figure 1; Supplemental Figure 1). Based on this result, and to adopt a systematic nomenclature, we decided to name each YTH-containing protein by the initials of its species of origin, followed by either DF or DC, by a number, and possibly by a letter (A, B, or C) referring to a subgroup when appropriate (see below). When another name already existed in the literature, it was kept and its systematic nomenclature name was added in parentheses (see Supplemental Data Set 1 for all YTH sequences and for the list of all name codes used in this study).
Figure 1.
Phylogenetic Relationships among Viridiplantae, Metazoan, and Yeast Core (β1 to α3) YTH Domains.
The phylogenetic tree was built using representative YTH motifs from Viridiplantae compared with several yeast and metazoan YTHs, including those from human, fruit fly, the yeast S. cerevisiae, and the fission yeast S. pombe (see Supplemental Data Set 1 for YTH sequences and for a list of name codes and Supplemental Figure 1 for the alignment used to build the tree). The two clades, YTHDF and YTHDC, are indicated. The color code is as follows: brown for yeast species, green for plant species, blue for chlorophyte species, and red for metazoan species. Statistical supports of key nodes that are important for our argumentation, calculated with the approximate likelihood-ratio test (aLRT), are indicated. The scale bar indicates a length of 0.1 substitution per site.
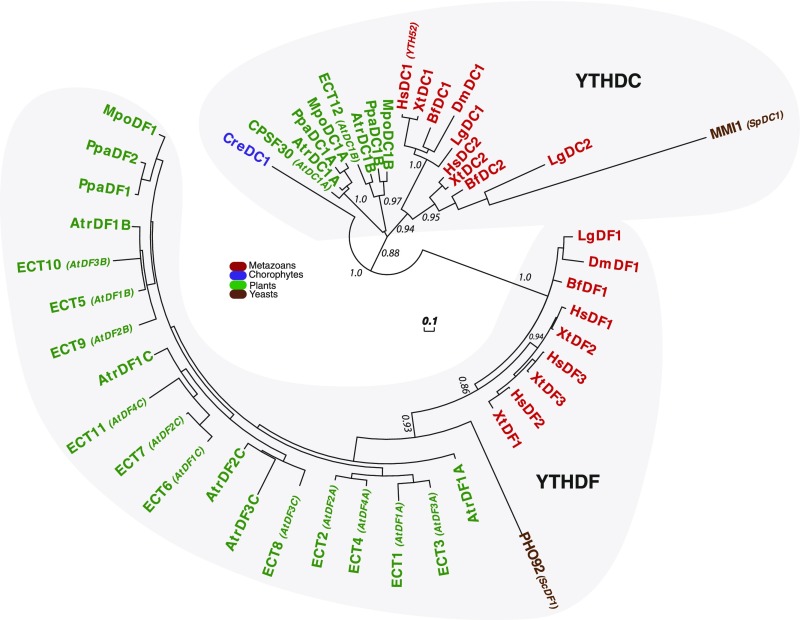
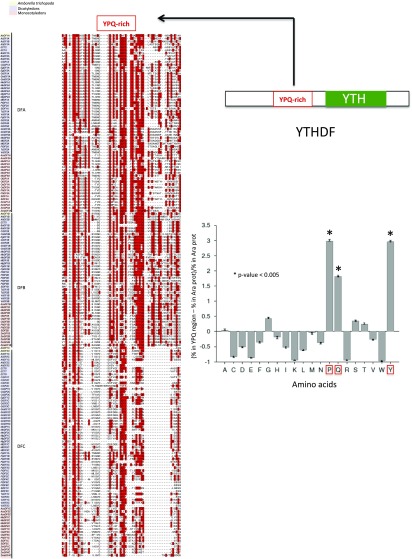
To have a closer look at the evolutionary history of Viridiplantae YTH domains, we next identified and collected 297 YTH-containing proteins from 32 representative species and found that 57 of these domains belong to the DC group and 240 to the DF group. Our phylogenetic analysis of the DF group showed that plant DF proteins can be further separated into three subgroups (DFA, DFB, and DFC), with the exception of species that are outgroups for the angiosperms, where a single DF version is present, and chlorophyte species, which lack DF proteins (Figure 2; Supplemental Figure 2). The conservation of at least one protein version from each DF subgroup in all angiosperm species strongly suggests a scenario in which a gene duplication event occurred in the common ancestor and was followed by the neofunctionalization of the duplicated copies. This also supports the notion that DF-containing proteins from the different subgroups are likely not completely redundant. Consistent with a putative neofunctionalization of the DF-type domains, we observe that an aspartic acid (position 401 in human proteins) from helix α1 is systematically replaced by an asparagine (Asp for Asn) in the clade of the DFB domains (see position labeled with an asterisk in Supplemental Figure 2). This substitution was shown through in vitro approaches conducted with the YTH domain from the human YTHDF1 protein to increase the affinity for an m6A target 15-fold (Xu et al., 2015). Therefore, plant DFB proteins are likely to have a higher affinity for m6A than proteins belonging to the two other subgroups. Apart from the YTH motif, plant DF proteins do not contain other highly conserved amino acid regions, except for a small domain (around 50 amino acids), N terminal to most YTHDF motifs, that presents a bias in amino acid composition in favor of tyrosine, proline, and glutamine (YPQ-rich; Figure 3). This bias in composition is similar to the one found in the three human DF proteins that possess a PQ-rich region, N-terminal to their YTH domain, that is likely to promote HsYTHDF2 aggregation in processing bodies as well as protein-protein interactions (Wang et al., 2014).
Figure 2.
Phylogenetic Relationships among Plant YTHDF Proteins.
The phylogenetic tree was built using the YTH domain of 240 DF proteins from 29 species representing the diversity of the Viridiplantae lineage (see Supplemental Data Set 1 for YTH sequences and for a list of name codes and Supplemental Figure 2 for the alignment used to build the tree). The three clades, DFA, DFB, DFC, are indicated. The color code is as follows: green for outgroup species to angiosperms, orange for A. trichopoda, blue for dicotyledon species, and dark red for monocotyledon species. Statistical supports of key nodes that are important for our argumentation, calculated with the aLRT, are indicated. The scale bar indicates a length of 0.1 substitutions per site.
Figure 3.
Identification of a YPQ-Rich Region in Plant YTHDF Proteins.
Alignment of the YPQ-rich region present in most plant YTHDF proteins. The three amino acids are highlighted in red. The histogram shows the bias in composition of the YPQ-rich region compared with all Arabidopsis proteins. The y axis represents the proportion of each amino acid in the YPQ-rich region compared with a database of all Arabidopsis proteins. The YPQ amino acids are statistically (P < 0.005) overrepresented in this region. Statistical significance associated with a specific enrichment or depletion is estimated using the two-sample t test including a Bonferroni correction according to the Composition Profiler website (http://www.cprofiler.org).
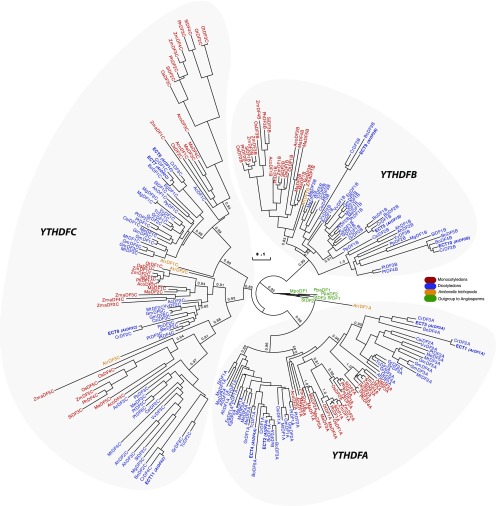
The phylogenetic tree of the Viridiplantae DC group also revealed two subgroups, DCA and DCB (Figure 4; Supplemental Figure 3). The DCA subgroup includes Arabidopsis CPSF30 (AtDC1A), which was shown to be a member of the plant polyadenylation complex (Addepalli and Hunt, 2007). As for CPSF30, all plant DCA proteins possess, in addition to the YTH domain, three highly conserved N-terminal zinc fingers (Supplemental Figure 4). Surprisingly, while DCA and DCB proteins are present in all species that are outgroups to the angiosperms and in all dicotyledon species, no DCB protein was found in the monocotyledon lineage, suggesting that this version of the YTH motif was lost in the common ancestor of monocotyledons.
Figure 4.
Phylogenetic Relationships among Plant YTHDC Proteins.
The phylogenetic tree was built using 57 YTHDC proteins from 32 species representing the diversity of the Viridiplantae lineage (see Supplemental Data Set 1 for YTH sequences and for a list of name codes and Supplemental Figure 3 for the alignment used to build the tree). The two clades, DCA and DCB, are indicated. The color code is as follows: brown for chlorophyte species, green for outgroup species to angiosperms, yellow for A. trichopoda, blue for dicotyledon species, and dark red for monocotyledon species. Statistical supports of key nodes that are important for our argumentation, calculated with the aLRT, are indicated. The scale bar indicates a length of one substitution per site.
To obtain an initial understanding of YTH domain protein function in plants, we performed an initial functional analysis of the ECT2 (AtDF2A) protein (AT3G13460), whose transcript, according to public databases, appears to be the most abundant and ubiquitously expressed of all YTH domain-containing protein transcripts.
Characterization of Two ect2 Loss-of-Function Mutants and ECT2-Specific Antibodies
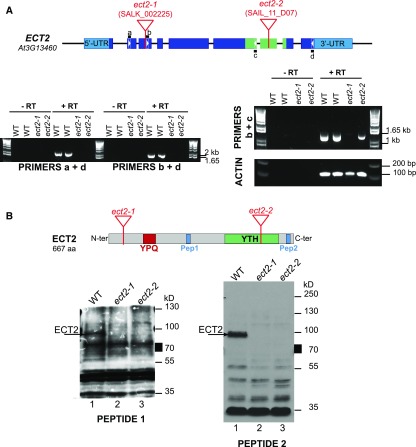
To study the physiological and molecular functions of ECT2, we collected two T-DNA insertion lines from Arabidopsis stock centers and determined whether they are full loss-of-function (lof) mutants. Through sequencing, we mapped the positions of the T-DNAs and found that they are inserted in exon 3 (at position +524 from the ATG) in line SALK_002225 and in exon 7 (at position +2402 from the ATG) in line SAIL_11_D07. We named these lines ect2-1 and ect2-2, respectively (Figure 5A). Using an RT-PCR approach with three sets of primers, we analyzed the steady-state levels of ECT2 mRNA in the wild-type and mutant backgrounds. No PCR signal was amplified from cDNAs prepared from either mutant line using primers respectively located at the 5′ and 3′ ends of the coding region and allowing the full-length ECT2 mRNA to be amplified (primers a and d, Figure 5A). The ect2-1 mutant does not seem to produce any stable mRNA, as no PCR signal was detected even when using a primer pair located downstream of the T-DNA insertion site, suggesting that it is a null allele. Conversely, the ect2-2 allele appeared to produce a truncated mRNA corresponding to the coding region located upstream of the T-DNA insertion site, suggesting that a truncated form of ECT2 might be expressed in this mutant. To explore this possibility and to confirm that ect2-1 does not produce any full-length or truncated protein, we raised antibodies against two synthetic peptides respectively located in the central part upstream of the YTH domain (Pep 1) and at the very C terminus of the protein downstream of the YTH domain (Pep 2) (Figure 5B). We found that both antibodies enabled the detection of a protein with an apparent size of 100 kD in wild-type extracts (Figure 5B). In extracts prepared from the ect2-1 and ect2-2 mutant lines, this signal was no longer detected regardless of the antibody used, supporting that it corresponds to the endogenous ECT2 protein. In addition, no signal specific to one or both mutant lines was detected, suggesting that neither mutant allele produces a full-length or truncated ECT2 protein. Considering the positions of the peptides (both downstream of the T-DNA location in ect2-1), one could argue that a small protein corresponding to the first 50 amino acids of ECT2 could remain undetected. Even if this was the case, this small protein could not support ECT2 function. Hence, we conclude that ect2-1 and ect2-2 are both lof mutants.
Figure 5.
Characterization of Two ECT2 Insertion Mutants.
(A) RT-PCR analyses of steady-state levels of ECT2 mRNA in wild-type (WT), ect2-1 (SALK_002225), and ect2-2 (SAIL_11_D07) seedlings. The top panel shows a schematic representation of the ECT2 genomic locus; boxes represent exons, with light blue corresponding to untranslated regions (UTR) and dark blue to coding regions. In addition, green boxes label the region coding for the ECT2 YTH domain. The positions of primers used for the PCR assays are reported as well as the T-DNA insertion sites in ect2-1 and ect2-2.
(B) Protein gel blot analyses of steady-state levels of ECT2 protein in wild-type, ect2-1, and ect2-2 plants. Custom-made antibodies raised against a centrally (pep1, pos. 281 to 295) and a C-terminally (pep2, pos. 642 to 656) located peptide, respectively, were used.
ECT2 Expression Is Likely Regulated at the Posttranscriptional Level
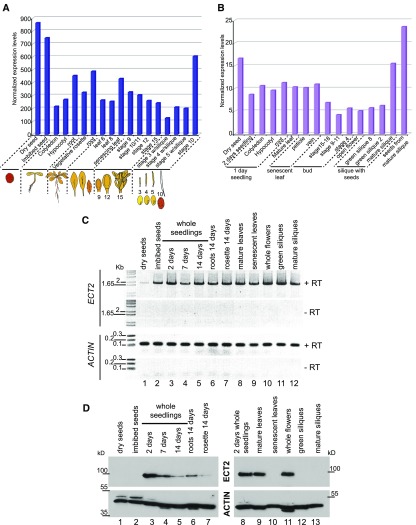
Analysis of ECT2 mRNA expression profiles in publicly available databases (ATH1 Affymetrix data at the eFP browser [http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi] [Winter et al., 2007] and RNA-seq data at TraVA db [http://travadb.org/]) (Figures 6A and 6B) suggested that this gene is ubiquitously expressed across plant development, with its highest level of expression in embryos at the green cotyledon stage and in dry and imbibed seeds. To confirm this observation, we monitored ECT2 full-length transcript and protein levels across plant development from dry seed to mature siliques (Figures 6C and 6D). The RT-PCR assays, consistent with the data in public databases, supported the notion that ECT2 transcript is ubiquitously expressed (Figure 6C), while protein gel blot analysis showed that this is not the case for the ECT2 protein (Figure 6D). Indeed, ECT2 was detected in extracts from whole seedlings, roots, rosettes, mature leaves, and whole flowers (Figure 6D, lanes 3 to 7, 9, and 11) but was undetectable in dry and imbibed seeds, senescent leaves, and green and mature siliques (Figure 6D, lanes 1, 2, 10, 12, and 13), a set of tissues where ECT2 transcript is present (Figure 6C). Since we utilized the same biological samples to analyze ECT2 mRNA and protein accumulation, we can exclude the possibility that these discrepancies are the result of distinct growth conditions and propose that ECT2 expression is tightly regulated at the posttranscriptional level.
Figure 6.
ECT2 Has a Tissue-Specific Expression Profile.
(A) and (B) ECT2 mRNA expression levels across plant development. Data were selected from publicly available databases at the eFP browser (microarray) (A) and TraVA db (RNA-seq) (B).
(C) RT-PCR monitoring of ECT2 mRNA levels across development; primers a and d (Figure 5A) were used for reverse transcription. ACTIN mRNA was used as a control. RNAs were prepared from the same tissues as those used for protein gel blot analysis in (D).
(D) Protein gel blot analyses of ECT2 steady-state levels across development and in various tissues. Sample tissue from 2-d-old whole seedlings was used as a reference and loaded on both gels 1 (left) and 2 (right) (lanes 3 and 8). ACTIN levels were used as a loading control.
ECT2 Protein Is Required for Normal Trichome Branching and Ploidy Levels
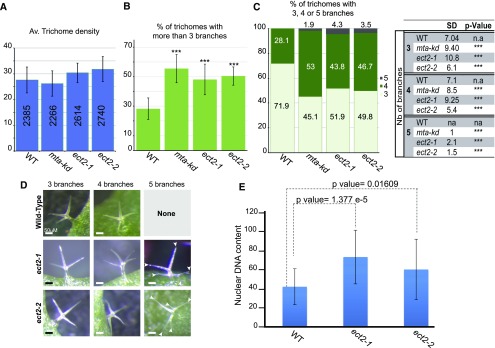
We next looked for growth or developmental phenotypes linked to ect2 lof and did not observe any obvious morphological defects at the macroscale level (Supplemental Figures 5A to 5F). Next, we looked for more subtle phenotypes. As a potential m6A reader, ECT2 should perform at least some physiological roles of m6A; hence, its lof mutants should display at least some phenotypes of hypomethylated mutants (Růžička et al., 2017), such as that of the ABI3Pro:MTA (mta-kd) hypomorph (Bodi et al., 2012). Considering that we found high levels of ECT2 in flowers, we looked at the floral architecture of ect2 lof mutants because mta-kd stamens often show partial conversion to petals (Bodi et al., 2012). Nonetheless, a close examination of ect2 mutant flowers did not reveal any significant defect (Supplemental Figure 5G). A second subtle defect of the mta-kd mutant is an increase in the percentage of abnormal trichomes compared with the wild type. Bodi et al. (2012) found that, in the mta-kd background, 60% of the observed trichomes have four or more branches versus 24% in wild-type plants. Such a phenotype was also observed in plants overexpressing FIP37, a core component and cofactor of the methylation complex (Vespa et al., 2004; Zhong et al., 2008; Shen et al., 2016; Růžička et al., 2017). Vespa et al. (2004) found that overexpressor lines display on average twice as many (42 to 45%) trichomes with four branches as the wild type and that 5 to 10% of trichomes have five or six branches, a population undetected in normal plants. Hence, we monitored trichome morphology in 3-week-old mta-kd (as a reference), ect2-1, ect2-2, and wild-type plants (Figures 7A to 7C). Wild-type plants displayed an average of 71.9 and 28.1% trichomes with three and four branches, respectively, whereas no trichomes with five or six branches (as previously reported) were detected (Vespa et al., 2004; Bodi et al., 2012). We reproduced the mta-kd phenotype, observing an increase in the frequency of trichomes with more than three branches (55.1%), and found that, as was the case for the FIP37 overexpressor line, five-branched trichomes appeared at a frequency of 1.9% on average (Figure 7C). Both ect2 lof mutants displayed a clear trichome developmental defect. First, they showed a significant increase in the frequency of trichomes with more than three branches (47.3% for ect2-1 and 50.4% for ect2-2). Second, looking at the distribution of trichomes according to the number of branches, we observed that the loss of ECT2 appeared to trigger the appearance of five-branched trichomes at a frequency of 4.3 and 3.5% in ect2-1 and ect2-2, respectively. These frequency variations are not related to discrepancies in trichome density between genotypes, as each line displayed an average of 25 to 30 trichomes per leaf (Figure 7A). Notably, besides the appearance of supernumerary branches, trichomes from the mutant lines did not appear to display additional morphological defects (Figure 7D).
Figure 7.
ECT2 Is Required for Normal Trichome Branching and Ploidy Levels.
(A) to (C) Histogram representation of the average trichome density per leaf (A), the percentage of trichomes with more than three branches (B), and the percentage of trichomes with three (light green), four (green), and five branches (dark green) (C) in wild-type (WT), mta knockdown (mta-kd), and ect2-1 and ect2-2 lof lines. sd values were calculated from 11 biological replicates (one biological replicate corresponds to the analysis of four plants of each genotype sown and grown together and issued from seed stocks harvested at the same time and stored together). P values were obtained using Dunnett’s test to determine whether mutant values are statistically distinct from wild-type values (***P < 10E-4). The table in (C) reports the sd and P values related to the values represented in the histogram shown in (C). na, not applicable.
(D) Representative photographs of each type of trichome observed in each genotype. Bars = 50 μm.
(E) Histogram representation of the average DNA content of trichomes with three branches in the wild type and trichomes with four branches in ect2 mutants. DNA content was calculated based on the fluorescence intensity of guard cells that are known to have a 2C content. sd was calculated from three biological replicates. P values were calculated with Student’s t test to determine whether mutant values are statistically different from wild-type ones.
Trichomes are differentiated protodermal cells that have stopped mitosis and undergo replication cycles without cell division (a process known as endoreduplication). Mature leaf trichomes are branched single cells, most with three branches, with a DNA content of 32C (resulting from four endoreduplication cycles) (Hülskamp, 2004), with the number of branches directly correlated to ploidy levels (Vespa et al., 2004). To determine whether the supernumerary branches of the ect2 lof mutant result from extra rounds of endoreduplication, we measured the nuclear DNA content of four-branched trichomes of ect2-1 and ect2-2 compared with the ploidy level of normal three-branched trichomes from wild-type plants (Figure 7E). Individual trichomes were isolated and their DNA content evaluated through epifluorescence imaging. We used the average fluorescence intensity of the nucleus of guard cells, which are known to have a 2C ploidy level, to normalize the fluorescent signals of wild-type and mutant trichomes and deduce their ploidy levels. The average DNA content was higher in both mutants compared with the wild type, with an average ploidy of 40C for the wild type and 60 to 70C for the ect2 mutants. This result indicates that ect2 trichomes underwent extra rounds of endoreduplication. To determine whether this increase was restricted to trichome cells, we monitored DNA content in cells from various organs, including roots, cotyledons, and cauline leaves, by flow cytometry and observed no increase in the ect2 lof mutant nuclei relative to the wild type. Therefore, we conclude that ECT2 is required for normal trichome branching and for the control of the number of endoreduplication cycles in developing trichomes.
ECT2 Associates with m6A-Containing RNAs in Vivo
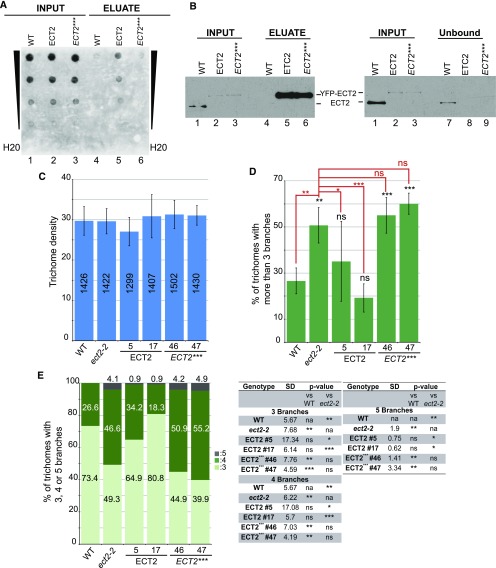
Considering our evolutionary analyses of the plant YTH domain proteins (Figures 1, 2, and 4), we reasoned that Arabidopsis YTH proteins might possess m6A reading activity. To test this possibility for ECT2 protein, we prepared a point mutant allele (ECT2***) where the tryptophans (Trp-464, Trp-521, and Trp-526 on ECT2) (Supplemental Figure 2) homologous to those that form the m6A binding aromatic cage in mammalian and yeast proteins were substituted with alanines (Theler et al., 2014). We next constructed transgenic lines expressing YFP-tagged versions of the wild-type and triple point mutant proteins in the ect2-2 ko background under the control of the ECT2 upstream genomic sequences and selected two independent lines per construct. Each of these four lines expressed the YFP fusions at levels higher than that of the endogenous protein, demonstrating that the mutations do not affect its stability (Supplemental Figure 6). To determine whether ECT2 can interact with m6A-containing RNAs in vivo, we performed RNA immunoprecipitation (RIP) assays (Figures 8A and 8B). RNA-protein complexes were immunopurified from YFP-ECT2 and YFP-ECT2*** crude extracts, and the presence of m6A-containing RNAs in the eluate fractions was monitored by dot blots using anti-m6A antibodies. This experiment was repeated over four independent biological replicates, which gave identical results (Figures 8A and 8B). The m6A-containing RNAs were immunoprecipitated together with the YFP-ECT2 fusion, as demonstrated by the presence of an m6A signal in lane 5 of Figure 8A and no signal in lane 4 corresponding to the negative control, even though similar levels of m6A-containing RNAs were detected in both inputs (Figure 8A, lanes 1 and 2). When experiments were conducted with YFP-ECT2*** crude extracts, the protein, which accumulated to levels similar to that of the YFP-ECT2 fusion in the inputs (compare lanes 2 and 3, Figure 8B), was immunoprecipitated as efficiently as the unmutated version (lanes 5 and 6, Figure 8B). Yet, m6A-containing RNAs were no longer significantly enriched in the eluate fraction (lane 6, Figure 8A).
Figure 8.
m6A Binding by ECT2 Is Required for Normal Trichome Formation.
(A) and (B) RIP assays.
(A) Dot blot analysis of the input (lanes 1 to 3) and eluate (lanes 4 to 6) fractions obtained from wild-type (WT), ect2-2 (YFP-ECT2; ECT2), and ect2-2 (YFP-ECT2***; ECT2***) 15-d-old seedlings. Serial dilutions of the input and eluate fractions were spotted onto the membrane as follows: 1:300, 1:1500, 1:7500, and 1:37.5e-3 for the input and 1:6, 1:30, 1:150, and 1:750 for the eluate. The blot was probed with an anti-m6A antibody.
(B) Protein gel blot monitoring of the levels of proteins that were respectively present in each input, unbound, and eluate fraction. Totals of 1:100 of the input and unbound fractions and 1:6 of the eluate fractions were loaded for each genotype. The blots were probed with the anti-ECT2 antibody (pep2). Images presented correspond to one replicate over four independent replicates.
(C) to (E) Monitoring of trichome-branching defects. Histograms show the density of trichomes for each genotype (C). The total number of trichomes monitored is reported for each genotype. The percentages of trichomes with more than three branches (D) and three, four, or five branches (E) are shown. ECT2 represents the ect2-2 lof mutant expressing the YFP-ECT2 transgene. Two independent transgenic lines were tested (nb 5 and 17). ECT2*** represents transgenic ect2-1 lines expressing the triple ECT2 point mutant (as in [A] and [B]) fused to the YFP tag. Two independent lines (nb 46 and 47) were analyzed. The experiment was conducted over six independent replicates. Four pairs of leaves were analyzed in each replicate. Error bars represent the sd calculated over the six replicates. P values reported over the histograms in (D) and in the table in (E) were obtained with Dunnett’s test to determine if the values are significantly distinct from the wild type (labeled in black) or from ect2-2 (labeled in red and with red bars). *P < 0.05, **P < 0.005, and ***P < 0.0001; ns, not significant; na; not applicable.
We conclude from these RIP assays that ECT2 forms a complex in vivo with m6A-containing RNAs and that its formation is dependent upon the presence of an unmutated aromatic cage on its YTH domain. Given the very high conservation of ECT2 at the primary sequence level over a large evolutionary time, we propose that its YTH domain is an m6A reading domain.
ECT2 m6A Reading Activity Is Required for Normal Trichome Branching
To explore the possibility that the trichome-branching defect in ect2 results from the loss of ECT2 m6A reading activity, we monitored the trichome morphology of ect2-2 lines expressing either a wild-type or a triple (Trp-to-Ala) mutant allele of ECT2 (Figures 8C to 8E) at levels similar to that of the endogenous protein (Supplemental Figure 6). Lines expressing a wild-type ECT2 allele displayed a trichome morphology similar to that of wild-type plants, with an average level of four-branched trichomes of 35 and 19% in the transgenic lines and 26% in the wild type. Moreover, the population of five-branched trichomes was significantly reduced in lines expressing a wild-type ECT2 allele compared with the ect2-2 mutant (0.9 versus 4.1%) (Figure 8E). These results indicate that the trichome defects of ect2-1 and ect2-2 are a direct consequence of the full inactivation of ECT2 and that the YFP and GFP translational fusions of ECT2 are functional. When the mutated version was the sole allele of ECT2 expressed, the trichome morphology of ect2-2 was not restored to normal. Trichomes of the point mutant lines displayed more than three branches at a frequency identical to that of the ect2-2 mutant and significantly above that of wild-type plants (55 and 60% for lines 46 and 47, respectively). In addition, five-branched trichomes were also detected at frequencies similar to that of the lof mutants (4.2 and 4.9%) (Figure 8E). These results show that an ECT2 protein with a defective YTH domain was not able to restore normal trichome branching.
We postulate from the results of these and RIP experiments that the trichome-branching defect of ect2 null mutants is linked to the loss of ECT2 m6A reading activity.
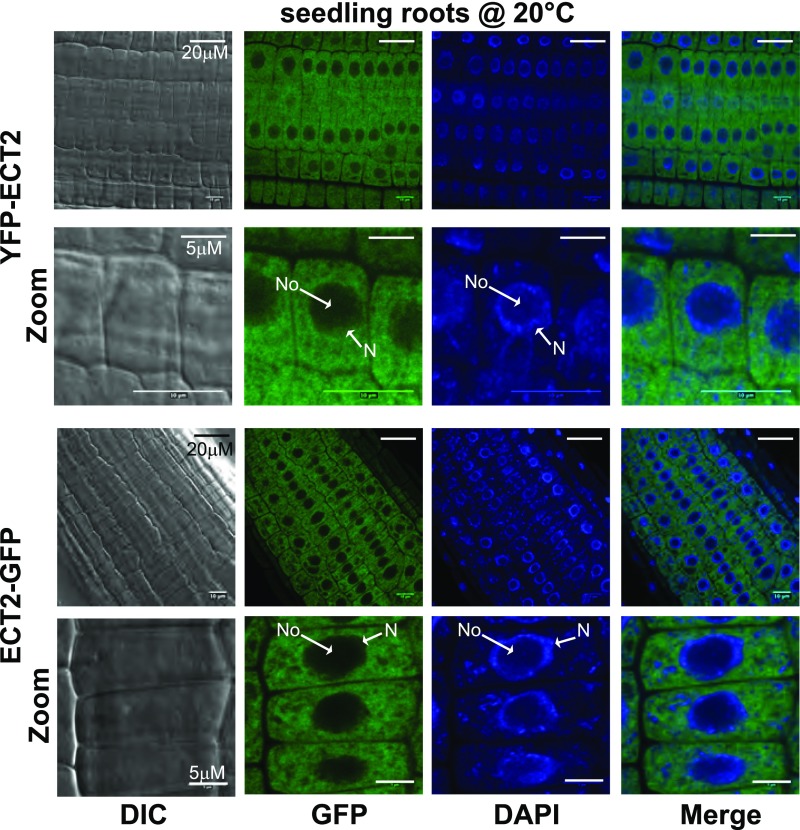
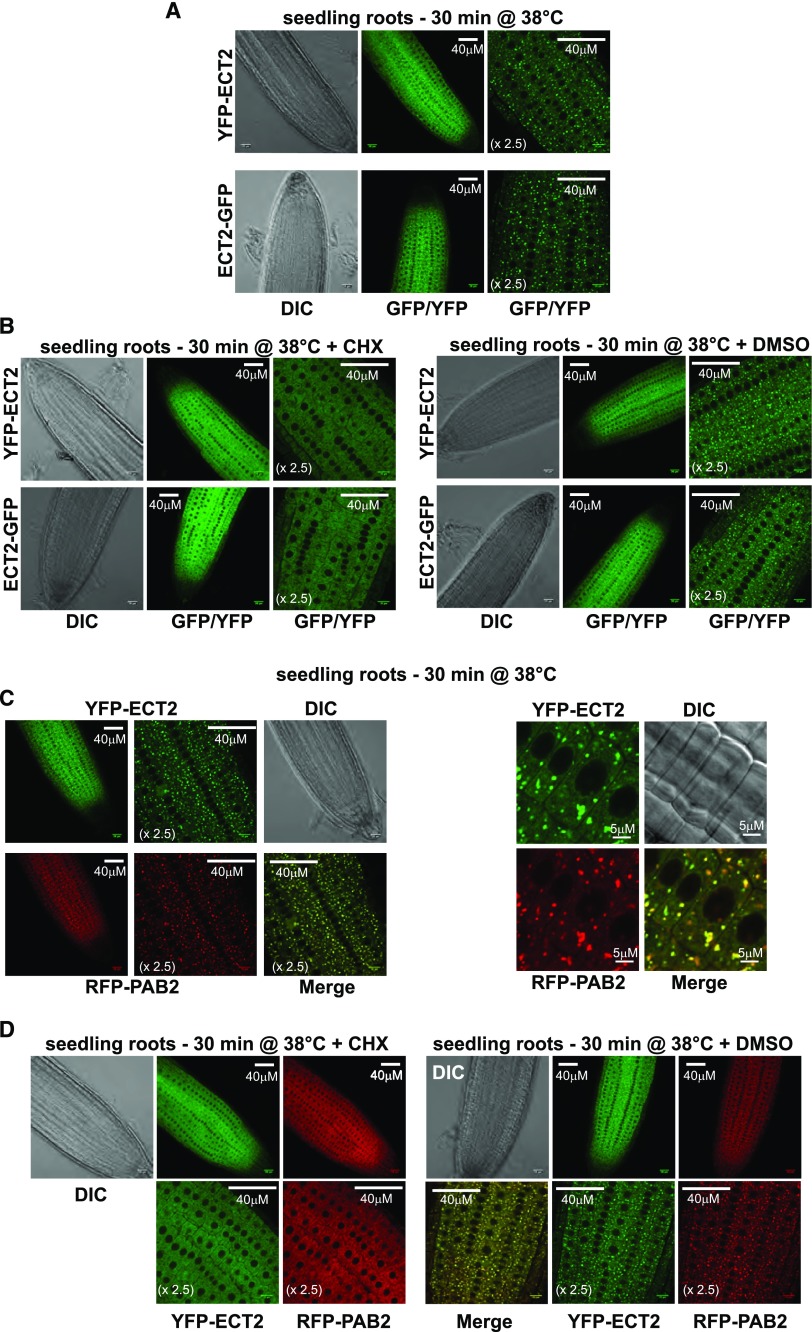
ECT2 Is Mainly Cytoplasmic and Relocates to SGs upon Heat Treatment
Next, to obtain hints about the molecular roles of ECT2, we characterized its subcellular distribution in the root tips of 7-d-old seedlings grown under normal conditions (Figure 9). To this end, we used transgenic lines expressing fluorescently labeled ECT2 and found that it complemented ect2-2 trichome-branching defects (Figures 8C to 8E). The N-terminal YFP and C-terminal GFP fusions displayed identical subcellular distributions (Figure 9). Under normal growth conditions, they mostly accumulated in the cytoplasm with a diffuse pattern. Counterstaining with 4′,6-diamidino-2-phenylindole to visualize the limits of the nucleus suggested that some of the YFP/GFP signal might also be present in the nucleus. mRNA binding proteins located in the cytoplasm and involved in translation or stability control often aggregate together with mRNPs into membraneless supramolecular structures. There are two major types of these bodies in the cytoplasm: processing bodies (P-bodies) and SGs (Anderson and Kedersha, 2008; Erickson and Lykke-Andersen, 2011). In addition to various RBPs, P-bodies contain players in the 5′-mRNA turnover process, while SGs contain the 48S preinitiation complex and various proteins involved in translation, such as the Poly(A) Binding Protein. Unlike SGs, P-bodies in mammalian and plant cells (Kedersha et al., 2005; Weber et al., 2008) can be detected under normal growth conditions, while stress situations that block translation initiation increases their size and number and trigger SG formation (Weber et al., 2008; Sfakianos et al., 2016). We previously found that exposure to 38°C for 30 min triggers the formation of such aggregates in Arabidopsis seedlings (Merret et al., 2013). Hence, we exposed transgenic plantlets to such heat stress, monitored the fluorescent signal in roots, and observed that the tagged ECT2 proteins formed punctate foci in the cytosol (Figure 10A). Since the formation of SGs and P-bodies is inhibited when translation elongation is blocked, we repeated an identical experiment but in the presence of cycloheximide (a translation elongation inhibitor) (Figure 10B). While ECT2 still formed foci in response to heat under mock treatment (DMSO), it showed a diffuse pattern in the presence of cycloheximide, supporting the notion that ECT2 aggregates correspond to P-bodies or SGs. Since ECT2 does not appear to form foci under normal growth conditions, we postulated that it associates in SGs rather than P-bodies. We next ran colocalization experiments using a stable line coexpressing both YFP-ECT2 and RFP-PAB2, a canonical marker of SGs (Weber et al., 2008; Merret et al., 2017) (Figures 10C and 10D). As expected, both the YFP and RFP signals appeared as discrete foci in the cytosol in response to heat stress, and the vast majority of them overlapped (merged panels in Figure 10C). Here again, cycloheximide treatment prevented foci formation at 38°C (Figure 10D). All in all, these findings allow us to propose that, under normal growth conditions, ECT2 is mostly present in the cytosol and that, upon stress-triggered inhibition of translation initiation, it relocates to SGs. Consistent with this conclusion, Arribas-Hernández et al. (2018) show that ECT2 foci do not contain VCS protein, a cofactor of the decapping holoenzyme that is exclusively found in P-bodies (Xu et al., 2006).
Figure 9.
ECT2 Mainly Accumulates in the Cytosol under Normal Growth Conditions.
Monitoring of N-terminally and C-terminally tagged versions of ECT2 (YPF-ECT2 and ECT2-GFP, respectively) expressed in the ect2-2 mutant background. The YFP or GFP signals of roots of 7-d-old seedlings were observed under a confocal microscope and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The transgenic lines used are the same as those used to monitor ECT2’s ability to complement the trichome-branching defect of the ect2-2 lof mutant. DIC, differential interference contrast; N, nucleus; No, nucleolus.
Figure 10.
ECT2 Relocates to SGs under Heat Stress Conditions.
Root tips of 7-d-old seedlings were monitored after 30 min of exposure to 38°C (heat stress), 30 min of exposure to 38°C in the presence of 100 μM cycloheximide (CHX), or 30 min of exposure to 38°C in the presence of DMSO for mock treatment.
(A) and (B) Subcellular distribution of YFP-ECT2 and ECT2-GFP. The same transgenic lines as those used in Figure 8 were analyzed.
(C) and (D) Confocal monitoring of the colocalization of YFP-ECT2 and RFP-PAB2 using the ect2-2 (YFP-ECT2 and RFP-PAB2) line.
One representative image is shown for each transgenic line in each environmental condition. Each experiment was conducted over three independent replicates, and at least four roots were monitored for each condition and replicate. The sizes of the scale bars are indicated. Images labeled × 2.5 correspond to a 2.5× enlargement of images in the panels immediately to the left. DIC, differential interference contrast.
DISCUSSION
We performed a systematic evolutionary analysis of YTH domains from Viridiplantae and retraced their evolutionary history. We demonstrate that, like mammals, vascular plants and bryophytes systematically display both DF- and DC-type motifs, while chlorophytes only possess the DC type (Figures 1, 2, and 4). Moreover, we found that YTH motifs carry the amino acids required to bind RNA and accommodate m6A and that they are predicted to adopt the same structural fold as animal and yeast YTH domains (Supplemental Figures 2 and 3). In S. pombe, the Mmi1 protein carries a YTH domain that unambiguously belongs to the DC group (Figure 1, statistical support of 1) and could (based on this analysis) be considered to be an m6A binding motif. Yet, this domain lost this ability in the absence of such an epitranscriptomic mark in fission yeast (Wu et al., 2017). Mmi1 YTH adopts a structural fold highly similar to that of m6A binding motifs and displays a putative m6A aromatic cage (Touat-Todeschini et al., 2017; Wu et al., 2017). Nevertheless, while bona fide binding cages create a hydrophobic environment to accommodate m6A that consists of three amino acids with hydrophobic side chains [W/W/(Y, W, or L)], the S. pombe cage has a positively charged histidine in the third position that is likely to hinder m6A binding. All Viridiplantae YTH domains display a canonical aromatic cage sequence (Supplemental Figures 2 and 3), and hence, we propose that these YTHs carry bona fide m6A binding pockets. Our evolutionary analyses also demonstrated that YTH proteins containing the DC- and DF-type motifs from the green lineage are further classified into three and two subclades, respectively, suggesting that a neofunctionalization process has most likely taken place. Thus, it is likely that YTH domain proteins from plants are not fully redundant, as is the case for animal proteins. In agreement with this hypothesis, we found that domains from different clades are likely to have significantly different m6A binding affinities and that this feature was conserved across evolution.
We also performed an initial functional characterization of a putative plant reader: the Arabidopsis ECT2 protein. We demonstrated that, in vivo, the full-length protein is in a complex with m6A-containing RNAs. When the three tryptophan residues of the ECT2 m6A binding pocket were substituted with alanines, m6A RNAs were no longer coimmunoprecipitated by ECT2. Considering that these point mutations do not alter the level of ECT2*** protein and that structural studies demonstrated that these tryptophans are directly involved in the recognition of the m6A residues (Theler et al., 2014), we propose that ECT2 directly binds m6A in vivo and that this binding relies on its YTH domain.
We uncovered a physiological role for an m6A reader by demonstrating that ect2 lof mutants display abnormal trichome development. Trichomes formed in the absence of ECT2 displayed the same morphological defect as that observed when m6A levels are reduced or when FIP37, a core component of the writer complex, is overexpressed. Moreover, we clearly showed that the m6A binding ability of ECT2 is necessary for normal trichome development, as expression of an allele with an inactive m6A binding pocket did not restore normal trichome morphology. Hence, we propose that ECT2 functions in the trichome development process to allow m6A to perform molecular roles in the control of gene expression. Considering that we cannot evaluate the impact of a full loss of m6A, we cannot exclude the possibility that other readers also allow m6A to function in trichome development, but it appears that their functions are not fully redundant, at least in this developmental process.
One puzzling observation is that the overexpression of FIP37 phenocopies ect2 and mta-kd trichome-branching defects (Vespa et al., 2004). FIP37 is a conserved core component of the writer complex required for m6A deposition, as shown by the strong decrease in the m6A:A ratio in fip37 hypomorphic lines (Růžička et al., 2017). Whether overexpression of FIP37 increases this ratio has not been tested, but if this were the case, one could propose that, not only is m6A necessary for trichome branching, but its levels must also be finely balanced. WTAP, the mammalian homolog of FIP37, is required for the proper localization of the catalytic subunits of the writer complex (Ping et al., 2014) and, hence, is thought to act as a regulatory component. As such, appropriate stoichiometric levels of WTAP/FIP37 might be necessary for proper functioning of the writer, leading us to propose an alternative scenario where FIP37 overexpression would have a dominant negative effect and provoke a decrease in m6A levels.
Trichomes are elongated cells that consist of a stalk with three to four branches and a DNA content of 32 to 64C. The onset of their formation involves the exit of pluripotent epidermal cells from the mitotic program and their transit into the endocycle. These cells undergo the first DNA replication event coupled to an arrest of their mitotic cycle at the G2-to-M transition. They then enter an endocycle during which they undergo three to four endoreduplication events before DNA replication ceases (Breuer et al., 2014). The number of branches of a trichome is directly correlated to its ploidy, and mutants with overbranched trichomes often have an elevated DNA content related to an excessive number of endocycles (Schellmann and Hülskamp, 2005). We demonstrate here that overbranched trichomes of the ect2 lof mutants display a significant increase in their DNA content and ploidy levels. We conclude from these observations that the overbranched phenotype of ect2 lof mutants is likely the direct consequence of extra rounds of endoreduplication. Trichomes of plants overexpressing FIP37 display an elevated DNA content, with 44 and 9% of mutant trichomes with DNA contents of 128 and 256C, respectively, compared with wild-type trichomes, which peak at 32C (Vespa et al., 2004). This supports the notion that misregulation of m6A levels or of their decoding triggers excessive rounds of endoreduplication, resulting in the formation of overbranched trichomes.
The endoreduplication process that occurs in protodermal cells in the course of trichome formation is controlled at the entry step, during its progression, and at the exit (Breuer et al., 2014). These steps are regulated through the precise control of cyclin (CYC) and cyclin-dependent kinase (CDK) levels (Breuer et al., 2014; Edgar et al., 2014). Several studies have demonstrated the role of m6A (Horiuchi et al., 2006, 2013) and of its readers (Zhao et al., 2017) in the control of cell cycle progression through posttranscriptional control of cyclin mRNA expression in animals and animal cell lines. The observation that, in plants, m6A and at least one of its readers are required to control the ploidy level and, hence, the number of endocycles necessary for trichome formation supports the notion that the role of the m6A reader system in the control of cell cycle progression was conserved across evolution. It is tempting to propose that ECT2, through its m6A reading activity, could be required for the posttranscriptional regulation of CYC and/or CDK expression levels in protodermal cells and/or in trichomes.
The molecular roles of ECT2, which type of m6A-containing RNAs it binds, and how it influences m6A-containing RNA fate and/or function are still largely unexplored. Amino acid sequence analysis of regions outside of its YTH domain did not allow us to determine if ECT2 is the ortholog of one of the three mammalian YTHDF proteins, nor to propose a molecular mode of action of ECT2 on its targets. Indeed, apart from the YTH, a motif search conducted against a Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) did not allow us to identify any remarkable protein domains in ECT2, as well as in any other plant DF-type protein, with the exception of a YPQ-rich region reminiscent of that found in human YTHDF proteins and in aggregation-prone factors such as TIA-1, TIAR, and prion proteins (Wang et al., 2014) (Figure 3). The presence of such an aggregation-prone domain in the N-terminal domain of ECT2 is consistent with the observation that this protein is a component of mRNP aggregates. Indeed, we clearly showed that, while ECT2 is mainly localized to the soluble fraction of the cytosol under normal growth conditions, it relocates to SGs in response to heat stress. Considering that, so far, long noncoding RNAs or other noncoding RNAs modified with m6A have not been shown to be present in SGs, we hypothesize that ECT2 binds to mRNAs. Nevertheless, at this stage, we cannot exclude the possibility that, like YTHDC1, ECT2 might play a dual role and also bind m6A-containing noncoding RNAs.
METHODS
Sequence Selection, Multiple Sequence Alignments, and Phylogenetic Reconstruction
BLAST searches (blastp and tblastn) were performed starting from known Arabidopsis thaliana YTH domains on 32 species representing the diversity of the Viridiplantae lineage at the JGI Phytozome (V11) genomic resource (https://phytozome.jgi.doe.gov/pz/portal.html). Each time a new YTH was found in a given species, it was itself used as a query in a new BLAST search. YTH sequences were aligned using the multiple sequence comparison by log expectation (MUSCLE v3.7) software (Edgar, 2004) using standard parameters. Trees were reconstructed using the fast maximum likelihood tree estimation program PHYML (Guindon and Gascuel, 2003) with the LG amino acid replacement matrix (Le and Gascuel, 2008). Statistical support for the major clusters was obtained using the aLRT (Anisimova and Gascuel, 2006). Bias in amino acid composition was detected using the Composition Profiler website (http://www.cprofiler.org/).
Arabidopsis Lines and Growth Conditions
Columbia-0 was used as the wild-type ecotype. The ect2-1 (SALK_002225) and ect2-2 (SAIL_11_D07) lines were obtained from the Nottingham Arabidopsis Stock Centre. Stable transgenic lines were obtained using the floral dip technique (Clough and Bent, 1998; Zhang et al., 2006). Plants were either grown in soil or cultivated in vitro on plates containing 4.41 g/L synthetic Murashige and Skoog (MS) (Duchefa) medium, pH 5.7, and 8 g/L agar. Transgenic seed selection was performed on MS medium containing 25 μg/L gentamycin. Seeds were surface sterilized, sown on plates, incubated for 48 h at 6°C in the dark, and placed in a growth cabinet at 20°C with a 16-h-day/8-h-dark cycle and 130 μE m−2 s−1 light (LEDs with white 4500K spectrum, purchased from Vegeled). For growth in soil, plants were placed in a growth chamber with a 16-h-day/8-h-dark cycle, 100 μE m−2 s−1 light (fluorescent bulbs with white 6500K spectrum, purchased from Sylvania), 60 to 75% humidity, and 20°C.
Cloning
To obtain the p825 plasmid, the genomic region upstream of the ECT2 (AT3G13460) coding region (spanning from position −2000 to −1 from the ATG) was PCR amplified with primers 1156 and 1157, digested with EcoRV and KpnI, and cloned into restrictions sites Ecl136II and KpnI of a derivative of pPZP221 upstream of a Gateway receptor cassette and bearing the gentamycin resistance gene aacC1 (Hajdukiewicz et al., 1994). The ECT2 coding region was PCR amplified from cDNAs reverse transcribed from total RNA with an oligo(dT) primer. To obtain plasmid p819, the ECT2 coding sequence was PCR amplified with primers 1152 and 1153 and inserted at SpeI and EcoRV sites downstream of the YFP tag into a derivative of pBluescript between the AttL1 and AttL2 Gateway donor sites. To obtain the p827 plasmid, the ECT2 coding sequence was PCR amplified with primers 1152 and 1154 and inserted at sites SpeI and EcoRV upstream of the GFP tag into a pBluescript derivative between the AttL1 and AttL2 Gateway donor sites. The final binary vectors respectively expressing YFP-ECT2 and ECT2-GFP under the control of ECT2 genomic sequences were obtained through Gateway recombination. The triple point mutant coding sequence labeled ect2*** codes for a mutated version of ECT2 bearing three tryptophan-to-alanine substitutions (TGG to GCT) at positions 464, 521, and 526. It was custom made and ordered from Biomatik (https://www.biomatik.com/). This sequence was fused downstream of the YFP tag and inserted through Gateway recombination into plasmid p825 downstream of the ECT2 genomic sequences.
Protein Gel Blot Analyses
Total proteins were extracted with Laemmli denaturing buffer, separated by SDS-PAGE (6.5% acrylamide), and electrotransferred onto a PVDF membrane. Custom-made antibodies generated from rabbit with two ECT2-specific peptides were ordered from Eurogentec (double-X immunization program): pep1-TVSSRNQNYRSNSH, pos. 281 to 295, and pep2-TSSDVKVAENGSVAK, pos. 642 to 656. The anti-pep1 antibody was used at 1:100 dilution and anti-pep2 at 1:2000 dilution. The anti-ACTIN antibodies were used at 1:6 × 105 dilution. Primary antibodies were incubated on membranes overnight at 4°C in TBS-1% Tween-5% dry milk buffer, and secondary antibodies (anti-rabbit horseradish peroxidase at 1:7500 for ECT2 detection and anti-mouse horseradish peroxidase at 1:5000 for ACTIN detection) were incubated in the same buffer for 45 min at room temperature.
RT-PCR Assays
Total RNAs were extracted from various plant tissues using the plant RNeasy mini kit from Qiagen according to the manufacturer’s instructions. cDNAs were prepared from 0.5 μg of DNase-free total RNA using an oligo(dT)18 primer and a SuperScript IV reverse transcriptase kit (Invitrogen) following the manufacturer’s instructions. PCR was conducted using 1:20 dilutions of cDNA with GoTaq (Promega) polymerase and the appropriate primers (Supplemental Data Set 2) according to the manufacturer’s instructions. PCR was conducted in a thermocycler with an initial step at 95°C for 3 min followed by 25 cycles at 95°C for 30 s, at Tm for 30 s, and at 72°C for 1 kb/min.
RIP and m6A Dot Blot Assays
Whole 15-d-old seedlings were flash frozen and crushed in liquid nitrogen, and 2.1 g of frozen powder was homogenized in 15.75 mL of lysis buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% IGEPAL, and 1% plant protease inhibitor cocktail from Sigma). The crude extract was clarified by centrifugation for 10 min at 3000g at 4°C. The immunoprecipitation was performed using 11.25 mL of crude extract incubated for 1.5 h at 4°C on a rotating wheel with 120 μL of GFP-trap magnetic beads (Chromotek). To monitor protein immunoprecipitation efficiency, a parallel immunoprecipitation experiment was conducted using 700 μL of crude extract incubated with 7.5 μL of GFP-trap. The beads were washed six times with 9 mL of lysis buffer at 4°C. RNAs were eluted from the beads with 400 μL of guanidium extraction buffer and precipitated overnight at −20°C with 800 μL of 100% ethanol. RNAs were precipitated by centrifugation, resuspended in 350 μL of RTL buffer (Qiagen RNEasy Micro kit), purified according to the manufacturer’s instructions, and eluted in 15 μL of RNase-free water. The RNA solution was concentrated into a 6 μL volume of RNase-free water using the RNA clean and concentrator kit R1015 from Zymo Research. To monitor RNAs from the input fraction, RNAs were extracted from 200 μL of crude extract following the same procedure. Notably, RNAs from both the eluate and input fractions were treated with DNase as part of the Qiagen RNEasy purification procedure. To elute the proteins, the beads were resuspended in 15 μL of Laemmli buffer and incubated for 5 min at 95°C, and the supernatant was collected from the beads using a magnetic device. The dot blot analysis was performed as follows: 1 μL of RNA solutions corresponding to 1:6, 1:30, 1:150, and 1:750 of the RNA eluate and 1:300, 1:1500, 1:7500, and 1:37.5 e-3 of the input were spotted onto and UV cross-linked to a Hybond N+ membrane (GE Healthcare) according to the manufacturer’s instructions. The membrane was saturated for 2 h at room temperature in TBS-1% Tween-5% fat-free dried milk before being incubated with an anti-m6A antibody (Synaptic Systems) diluted at 1:1000 overnight at 4°C.
The experiment was repeated four times independently. For each replicate, whole RIP and protein immunoprecipitation experiments were conducted from freshly prepared and harvested seedlings.
Trichome Counting and Ploidy Measurements
To monitor trichome-branching defects, trichomes from the second pair of leaves of soil-grown (see above for culture conditions) 3-week-old plants were observed under a binocular loupe. The number of trichomes with three, four, and five branches was counted for each plant and respectively expressed as a percentage over the total number of trichomes. One biological replicate corresponds to the analysis of four plants of each genotype sown and grown together and issued from seed stocks harvested at the same time and stored together. For each genotype, trichomes were monitored and counted on the second pair of leaves (leaves number 3 and 4) on the rosettes of four plants. The value of one replicate corresponds to the average value:total number of trichomes counted in one category divided by 4 for each genotype. The statistical analysis was performed with ANOVA followed by Dunnett’s posthoc test using XLSTAT software (Addinsoft) (Supplemental Figure 7). To analyze trichome DNA content, individual trichomes were isolated from fully expanded rosette leaves as described (Zhang and Oppenheimer, 2004). Briefly, fully expanded rosette leaves were incubated overnight at 4°C in PBS EGTA solution (50 mM, pH 9) supplemented with 0.05% (v/v) Triton X-100. Trichomes were removed from leaves using a paintbrush. After staining with Hoechst 3342 solution (10 µg/mL), trichomes and leaf epidermis fragments were imaged using an epifluorescence microscope (Zeiss, Axioimager Z.2). Average fluorescence intensity was measured for 20 nuclei for each genotype and normalized to the average fluorescence intensity of guard cells, which are known to have a 2C DNA content. One biological replicate corresponds to one set of all genotypes grown independently. Average values were obtained from two biological replicates.
Confocal Microscopy and Heat Treatment
Root tips from 7-d-old seedlings cultivated on solid MS medium in vitro (as described above) were used to monitor the subcellular distribution of fluorescently labeled ECT2. To perform cycloheximide and/or heat stress treatment, seedlings were transferred to a six-well plate containing 3 mL of liquid MS medium with 100 μM cycloheximide or 0.1% DMSO (for mock treatment) and incubated for 30 min at 20 or 38°C (heat stress). Plantlets were vacuum infiltrated for 5 min in 1 mL of fixation solution (2% paraformaldehyde, 1× MTSB [50 mM PIPES, pH 7, 5 mM EGTA, and 5 mM MgSO4], and 0.2% Triton X-100) and further incubated at room temperature for 10 min. The seedlings were washed twice for 5 min in 2 mL of 1× MTSB and stored at 4°C prior to microscopy analyses. The subcellular distribution of tagged ECT2 was assessed with an LSM700 (Zeiss) confocal microscope with the following excitation/emission wavelengths: GFP, 488 nm/490 to 540 nm; YFP, 514 nm/520 to 550 nm; and RFP, 561 nm/580 to 640 nm.
Accession Numbers
Sequence data from this article can be found in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Alignment of the Yeasts, Metazoans, Chlorophytes, and Plants Core (β1 to α3) YTH Motif Used to Build the Phylogenetic Tree Shown in Figure 1.
Supplemental Figure 2. Alignment of the Viridiplantae YTHDF Used to Build the Phylogenetic Tree Shown in Figure 2.
Supplemental Figure 3. Alignment of the Viridiplantae YTHDC Used to Build the Phylogenetic Tree Shown in Figure 4.
Supplemental Figure 4. Alignment of a Conserved Portion of the N-Terminal Region of Plant YTHDCA Proteins with Three Zinc Fingers.
Supplemental Figure 5. ect2 Loss of Function Does Not Trigger Macroscale Developmental Changes or Growth Delay.
Supplemental Figure 6. Transgenic ECT2 Protein Levels.
Supplemental Figure 7. ANOVA Tables.
Supplemental Data Set 1. Amino Acid Sequences of YTH Domains and Name Codes and Accession Numbers of YTH Domain Proteins from Viridiplantae.
Supplemental Data Set 2. List of Primers Used for Cloning, Genotyping, and RT-PCRs.
Acknowledgments
We thank Rupert Fray (University of Nottingham) for sharing the ABI3Pro:MTA70 hypomorphic line and Soizik Berlivet (LGDP, Perpignan) for critical reading of the manuscript. This work was supported by the Agropolis Foundation (international mobility program, Contract 1502-003), the CNRS, the University of Perpignan Via Domitian, the Institut Universitaire de France, the Bio-Environnement platform through utilization of the confocal microscope, and l’Agence Nationale pour la Recherche (ANR HEAT-EpiRNA, ANR-17-CE20-0007-01, and ANR HEAT-Adapt, ANR-14-CE10-0015). We acknowledge the IPS2 Imaging Facility for epifluorescence microscopy analysis. J.S. was the recipient of a PhD grant from the University of Perpignan Via Domitia, Doctoral School ED305.
AUTHOR CONTRIBUTIONS
C.B.-A. and R.M. designed and supervised the work. J.-M.D. ran the evolutionary analyses. V.J. genotyped the ect2 T-DNA insertion lines, prepared the transgenic lines, and did the crosses between the YFP-ECT2 and RFP-PAB2 lines. J.S. set up the ECT2 antibodies, verified that the ect2-1 and -2 lines were KO, did the cloning, ran the trichome phenotypings, and conducted the confocal analyses with the help of J.-J.F. C.R. and M.B. measured the DNA content of trichomes. R.M. conducted the RIP-m6A assays. C.B.-A. wrote the manuscript, with the help of J.-M.D.
Footnotes
Articles can be viewed without a subscription.
References
- Addepalli B., Hunt A.G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35: 4453–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. [DOI] [PubMed] [Google Scholar]
- Arribas-Hernández L., Bressendorff S., Hansen M.H., Poulsen C., Erdmann S., Brodersen P. (2018). Control of developmental timing and morphogenesis in Arabidopsis by an m6A-YTH module. Plant Cell 30: 952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z., Zhong S., Mehra S., Song J., Graham N., Li H., May S., Fray R.G. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Braidwood L., Sugimoto K. (2014). Endocycling in the path of plant development. Curr. Opin. Plant Biol. 17: 78–85. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8: 176–189. [DOI] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7: 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., Gutierrez C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Erickson S.L., Lykke-Andersen J. (2011). Cytoplasmic mRNP granules at a glance. J. Cell Sci. 124: 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M. (2016). m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304. [DOI] [PubMed] [Google Scholar]
- He C. (2010). Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 6: 863–865. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Umetani M., Minami T., Okayama H., Takada S., Yamamoto M., Aburatani H., Reid P.C., Housman D.E., Hamakubo T., Kodama T. (2006). Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. USA 103: 17278–17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. (2013). Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288: 33292–33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.J., et al. (2017). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5: 471–480. [DOI] [PubMed] [Google Scholar]
- Ivanova I., Much C., Di Giacomo M., Azzi C., Morgan M., Moreira P.N., Monahan J., Carrieri C., Enright A.J., O’Carroll D. (2017). The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67: 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29: 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Pandya-Jones A., Saito Y., Fak J.J., Vågbø C.B., Geula S., Hanna J.H., Black D.L., Darnell J.E. Jr., Darnell R.B. (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31: 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Lence T., Akhtar J., Bayer M., Schmid K., Spindler L., Ho C.H., Kreim N., Andrade-Navarro M.A., Poeck B., Helm M., Roignant J.Y. (2016). m6A modulates neuronal functions and sex determination in Drosophila. Nature 540: 242–247. [DOI] [PubMed] [Google Scholar]
- Li A., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27: 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhao D., Wu J., Shi Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24: 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang X., Li C., Hu S., Yu J., Song S. (2014). Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 11: 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Tong L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA 111: 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.Z., MacQueen A., Zheng G., Duan H., Dore L.C., Lu Z., Liu J., Chen K., Jia G., Bergelson J., He C. (2014). Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R., Descombin J., Juan Y.T., Favory J.J., Carpentier M.C., Chaparro C., Charng Y.Y., Deragon J.M., Bousquet-Antonelli C. (2013). XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 5: 1279–1293. [DOI] [PubMed] [Google Scholar]
- Merret R., Carpentier M.C., Favory J.J., Picart C., Descombin J., Bousquet-Antonelli C., Tillard P., Lejay L., Deragon J.M., Charng Y.Y. (2017). Heat shock protein HSP101 affects the release of ribosomal protein mRNAs for recovery after heat shock. Plant Physiol. 174: 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ok S.H., Jeong H.J., Bae J.M., Shin J.S., Luan S., Kim K.N. (2005). Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol. 139: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D.P., Pickering B.F., Jaffrey S.R. (2018). Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X.L., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J.Y., Soller M. (2017). m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 33: 380–390. [DOI] [PubMed] [Google Scholar]
- Roundtree I.A., He C. (2016). RNA epigenetics: chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 30: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y., Meyer K., Korlach J., Vilfan I.D., Jaffrey S., Mason C.E. (2012). The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Hülskamp M. (2005). Epidermal differentiation: trichomes in Arabidopsis as a model system. Int. J. Dev. Biol. 49: 579–584. [DOI] [PubMed] [Google Scholar]
- Schwartz S., et al. (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos A.P., Whitmarsh A.J., Ashe M.P. (2016). Ribonucleoprotein bodies are phased in. Biochem. Soc. Trans. 44: 1411–1416. [DOI] [PubMed] [Google Scholar]
- Shen L., Liang Z., Gu X., Chen Y., Teo Z.W., Hou X., Cai W.M., Dedon P.C., Liu L., Yu H. (2016). N(6)-Methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 38: 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27: 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P., Jinek M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5: e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42: 13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touat-Todeschini L., et al. (2017). Selective termination of lncRNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 36: 2626–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L., Vachon G., Berger F., Perazza D., Faure J.D., Herzog M. (2004). The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 134: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Tang K., Zhang D., Xie S., Zhu X., Wang Z., Lang Z. (2015). Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 16: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2014). N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., He C. (2014). Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 11: 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Doxtader K.A., Nam Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. (2015). N(6)-Methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C., Nover L., Fauth M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56: 517–530. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Xu J., Su S., Liu H., Gan J., Ma J. (2017). Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1. Biochem. Biophys. Res. Commun. 491: 310–316. [DOI] [PubMed] [Google Scholar]
- Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10: 927–929. [DOI] [PubMed] [Google Scholar]
- Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. (2015). Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290: 24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J.Y., Niu Q.W., Chua N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Oppenheimer D.G. (2004). A simple and efficient method for isolating trichomes for downstream analyses. Plant Cell Physiol. 45: 221–224. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Theler D., Kaminska K.H., Hiller M., de la Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnicki J.M., Allain F.H., Stamm S. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285: 14701–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C. (2017). m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542: 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y. (2014). Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24: 1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]