Metabolic allocation of diterpenoids in rice leads to the discovery of a novel physiological function for diterpenoids in plant disease resistance.
Abstract
Among their responses to microbial infection, plants deploy an arsenal of natural antibiotic products. Historically these have been identified on the basis of their antibiotic activity in vitro, which leaves open the question of their relevance to defense in planta. The vast majority of such natural products from the important crop plant rice (Oryza sativa) are diterpenoids whose biosynthesis proceeds via either ent- or syn-copalyl diphosphate (CPP) intermediates, which were isolated on the basis of their antibiotic activity against the fungal blast pathogen Magnaporthe oryzae. However, rice plants in which the gene for the syn-CPP synthase Os-CPS4 is knocked out do not exhibit increased susceptibility to M. oryzae. Here, we show that knocking out or knocking down Os-CPS4 actually decreases susceptibility to the bacterial leaf blight pathogen Xanthomonas oryzae. By contrast, genetic manipulation of the gene for the ent-CPP synthase Os-CPS2 alters susceptibility to both M. oryzae and X. oryzae. Despite the secretion of diterpenoids dependent on Os-CPS2 or Os-CPS4 from roots, neither knockout exhibited significant changes in the composition of their rhizosphere bacterial communities. Nevertheless, rice plants allocate substantial metabolic resources toward syn- as well as ent-CPP derived diterpenoids upon infection/induction. Further investigation revealed that Os-CPS4 plays a role in fungal non-host disease resistance. Thus, examination of metabolic allocation provides important clues into physiological function.
INTRODUCTION
Plants deploy an arsenal of antimicrobial natural products during pathogen infection (Bednarek and Osbourn, 2009). This is considered to be an effective means of defense. However, historically the underlying studies most often rely on antimicrobial activity demonstrated with isolated compounds. Such antibiotics whose production is strongly induced by infection are termed phytoalexins, while those constitutively present are termed phytoanticipans (VanEtten et al., 1994). The relevance of such natural products to plant defense is most rigorously supported by genetic evidence using gene knockout lines in which the compound(s) of interest can no longer be produced, but the remainder of the plant defense response has been retained. Such evidence has been provided in at least a few cases (Ahuja et al., 2012). For example, camalexin and certain sesquiterpenes in Arabidopsis thaliana, as well as triterpenoid saponins in oat (Avena sativa), all clearly play a role in plant defense against infection by certain pathogens (Papadopoulou et al., 1999; Thomma et al., 1999; Huang et al., 2012). Nevertheless, in other cases, such genetic analysis does not support the relevance of at least some phytoalexins to the plant defense response. Of particular relevance here, despite being the first identified rice (Oryza sativa) phytoalexin against the fungal blast agent Magnaporthe oryzae (Cartwright et al., 1981), the momilactones do not seem to play an important role in rice plant defense against this fungal pathogen (Xu et al., 2012).
Almost all of the identified antibiotic natural products from the important crop plant rice are diterpenoids (Schmelz et al., 2014), more specifically, labdane-related diterpenoids, whose biosynthesis can be traced back to that of the gibberellin phytohormones (Zi et al., 2014). Most of these compounds, specifically momilactones A and B, oryzalexins A-F, oryzalexin S, and phytocassanes A-F, were originally isolated and identified on the basis of their activity against M. oryzae (Peters, 2006). Infection by M. oryzae further strongly induces accumulation of these labdane-related diterpenoids (Hasegawa et al., 2010), which then seem to be phytoalexins. The remaining labdane-related diterpenoid antibiotics, including oryzalide A and B, oryzalic acid A and B, and oryzadione I to III (Toyomasu, 2008), all of which are termed oryzalide-related here, were isolated and identified on the basis of their activity against the bacterial leaf blight pathogen Xanthomonas oryzae pv oryzae (Xoo). The production of these labdane-related diterpenoids is only weakly induced by Xoo infection (Watanabe et al., 1996), and they then seem to be phytoanticipans.
As with the vast majority of phytoalexins, these rice labdane-related diterpenoids have been assumed to be relevant to plant defense on the basis of their antibiotic activity in vitro. However, it should be noted that the momilactones were actually first isolated on the basis of their plant growth suppressing activity (Kato et al., 1973) and are constitutively secreted from the roots into the soil, where they have long been suggested to act as allelochemicals, most specifically momilactone B (Kato-Noguchi and Peters, 2013). This offers an alternative rationale for production of at least the momilactones, although it has been shown that phytocassanes are secreted from the roots as well (Toyomasu et al., 2008). Nevertheless, momilactones, as well as phytocassanes, accumulate in leaves upon infection by M. oryzae (Hasegawa et al., 2010). In addition, both the momilactones and phytocassanes have recently been shown to accumulate specifically at the site of Xoo infection in rice leaves (Klein et al., 2015).
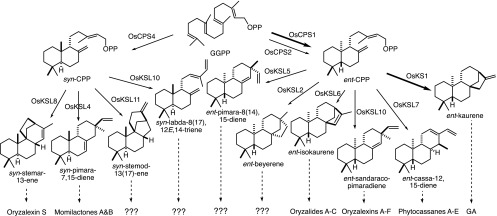
The defining characteristic of labdane-related diterpenoids is their biosynthetic origin from bicyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) catalyzed by class II diterpene cyclases (Peters, 2010). This most often results in the production of the eponymous labdadienyl/copalyl diphosphate (CPP), with the relevant enzymes then termed CPP synthases (CPSs; Figure 1). All vascular plants seem to produce the labdane-related diterpenoid gibberellins as phytohormones, which depends on the production of ent-CPP (Hedden and Thomas, 2012). Rice produces two different stereoisomers of CPP, both the requisite ent-CPP, and the diastereomer syn-CPP. The production of these is catalyzed by three distinct CPSs in rice. Two of these produce ent-CPP, both Os-CPS1 and Os-CPS2, while the third, Os-CPS4, produces syn-CPP (Otomo et al., 2004; Prisic et al., 2004; Xu et al., 2004). Of the two ent-CPP producing CPSs, only Os-CPS1 seems to be involved in gibberellin metabolism (Sakamoto et al., 2004). On the other hand, transcript levels of Os-CPS2 and Os-CPS4 are inducible, and both have been suggested to play a role in the production of more specialized labdane-related diterpenoids that serve as phytoalexins and/or phytoanticipans (Peters, 2006; Toyomasu, 2008). The diterpenes whose hydrocarbon structures define the various families of derived diterpenoids are produced from ent- or syn-CPP by class I diterpene synthases, which are related to the ent-kaurene synthase required for gibberellin phytohormone biosynthesis and have been termed kaurene synthase-like (KSLs; Figure 1).
Figure 1.
Rice Labdane-Related Diterpenoid Biosynthetic Network.
Thicker arrow indicates enzymatic reactions specifically involved in gibberellin phytohormone metabolism. Dashed arrows indicate multiple enzymatic reactions leading to families of diterpenoid natural products derived from the shown diterpene olefins where known (??? indicates presumed unknown derivatives).
Given the potential redundancy stemming from the presence of two ent-CPP producing CPSs, previous work has focused on analysis of the syn-CPP derived labdane-related diterpenoids in rice, whose biosynthesis depends only on Os-CPS4. However, of the identified antibiotic labdane-related diterpenoids from rice, only momilactones A and B and oryzalexin S are derived from syn-CPP, while the rest are derived from ent-CPP (Figure 1). Previous work with a T-DNA insertion mutant of Os-CPS4 indicated that these knockout Os-cps4ko plants are not more susceptible to infection with M. oryzae than the parental/wild-type line. Additional analyses, including the use of a mutant rice line in which the Os-KSL4 that acts downstream of Os-CPS4 in momilactone biosynthesis (Figure 1) is similarly knocked out by T-DNA insertion, indicate that these diterpenoids act as allelochemicals (Xu et al., 2012). However, another study, using rice plants in which Os-CPS4 expression is knocked down, although not completely abolished, suggested that OsCPS4-dependent labdane-related diterpenoids may play a role in defense against M. oryzae (Toyomasu et al., 2014). This discrepancy may reflect the different genetic backgrounds of the rice lines and/or fungal strains used in each of these studies. For example, it has been noted that susceptible cultivars produce less momilactones and phytocassanes than resistant cultivars, at least at early time points (Hasegawa et al., 2010; Klein et al., 2015), and Os-cps4ko is derived from a susceptible cultivar (ssp japonica cv Zhonghua 11). Accordingly, such reduced and/or delayed production (i.e., even in the parental/wild-type line) may underlie the observed lack of effect with Os-cps4 plants.
Here, genetic evidence is provided indicating that Os-CPS2-dependent diterpenoids (i.e., those derived from ent-CPP) are relevant to rice plant defense against both the fungal pathogen M. oryzae and bacterial pathogen Xoo. In addition, although Os-CPS4 dependent diterpenoids (i.e., those derived from syn-CPP) do not seem to be relevant to rice plant defense against either M. oryzae or Xoo, the equivalent metabolic allocation toward such natural products prompted further analysis, which indicated that Os-CPS4 is relevant to non-host disease resistance against Magnaporthe poae, which is adapted to wheat (Triticum aestivum) rather than rice. Accordingly, investigation of metabolic allocation has provided insight into the roles of diterpenoids in the microbial defense response of the important crop plant rice, and this approach may be applicable to inference of the physiological function of natural products more broadly.
RESULTS
Characterization of Os-cps2 Rice
The production of ent-CPP for more specialized diterpenoid metabolism in rice has been hypothesized to largely depend on Os-CPS2. Here, a reverse genetics approach was undertaken to investigate the biological function of ent-CPP derived diterpenoids (other than gibberellins). Only a single rice plant line with an insertion in an exon of Os-CPS2 was identified from the Salk Research Institute Rice Functional Genomic Express database (Krishnan et al., 2009). This was obtained and verified to be a homozygous Os-cps2ko line, as characterized by PCR analysis of the transposon inserted into the sixth exon of the gene. As expected, Os-CPS2 transcripts could no longer be detected in Os-cps2ko plants (Supplemental Figure 1A), even following induction with methyl jasmonate (MeJA), which has been shown to induce accumulation of transcripts for both Os-CPS2 and Os-CPS4 (Prisic et al., 2004; Xu et al., 2004). To supplement this single knockout line, Os-CPS2 was also targeted by RNAi to generate knockdown Os-cps2i lines. To compare this more directly to the previously reported Os-cps4ko line, these were constructed in that genetic background (i.e., ssp japonica cv Zhonghua 11). Three Os-cps2i lines were selected for further analysis based on their reduced expression of Os-CPS2 (relative to their parental/wild-type cultivar) following MeJA induction (Supplemental Figure 1B). Consistent with the previously suggested predominant role of Os-CPS1 in gibberellin metabolism (Sakamoto et al., 2004), none of these Os-cps2 lines were found to exhibit any significant reduction in growth (Supplemental Figure 2A), although there is a slight delay (∼1 week) in flowering time.
Os-cps2 Rice Exhibits Increased Susceptibility to M. oryzae and X. oryzae pv oryzae
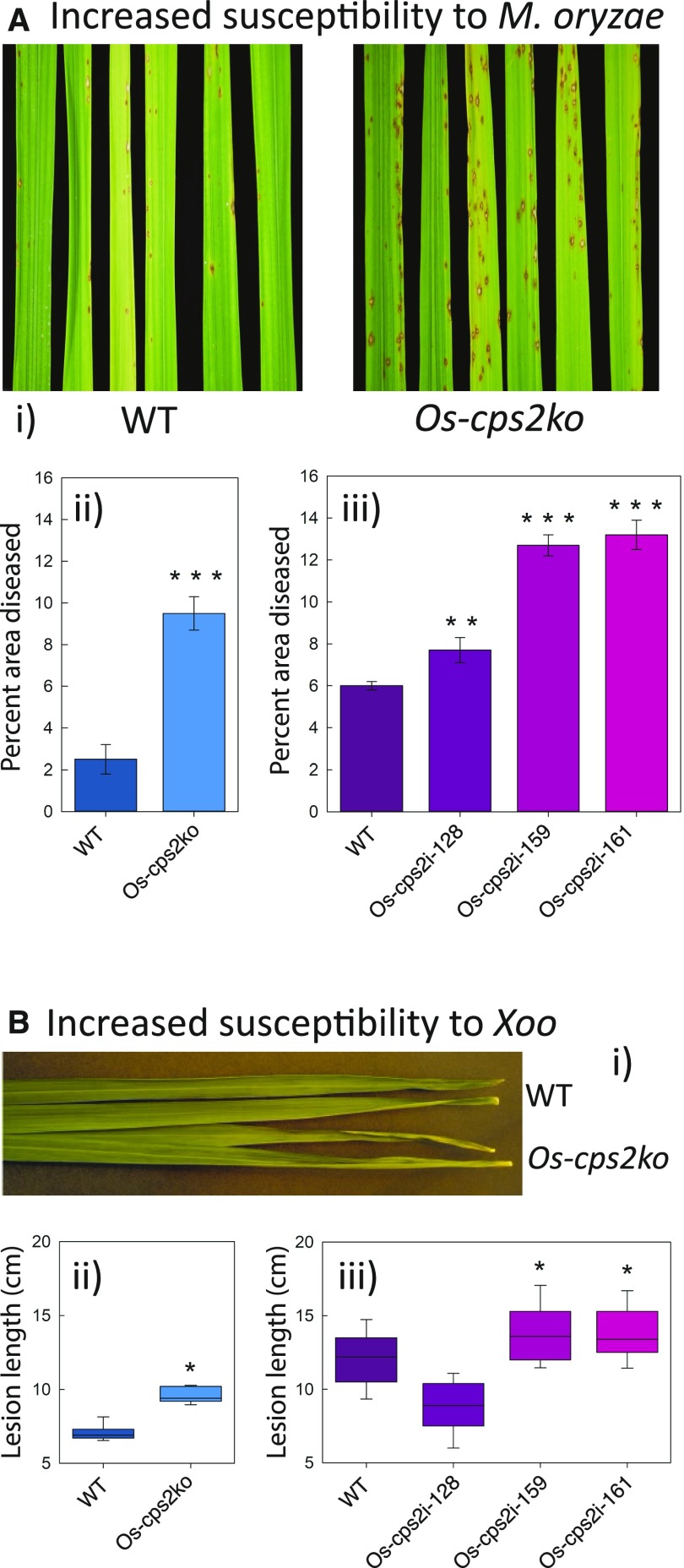
Given that some ent-CPP-dependent diterpenoids have been suggested to serve as phytoalexins against M. oryzae (Peters, 2006), while others have been suggested to be phytoanticipans against Xoo (Toyomasu, 2008), the susceptibility of Os-cps2 rice relative to the parental/wild-type cultivar was investigated with both microbial pathogens. Infecting leaves with a rice isolate of M. oryzae (Guy 11) causes disease symptoms on Os-cps2 lines and the parental/wild-type cultivar. However, there was a significant difference in the percentage of infected/necrotic area of leaves from Os-cps2 plants relative to that for the parental/wild-type cultivar (Figure 2A). Thus, ent-CPP-derived diterpenoids seem to serve an effective role in rice plant defense against this fungal pathogen. In addition, when infected with Xoo via a leaf clipping method Os-cps2 rice exhibited significantly longer lesion lengths than the parental/wild-type cultivar (Figure 2B). Accordingly, ent-CPP-derived diterpenoids seem to serve an effective role in rice plant defense against this bacterial pathogen as well. These results are consistent with the reported antifungal activity of the ent-cassadiene derived phytocassanes and antibacterial activity of the ent-(iso)kaurene-derived oryzalide-related diterpenoids.
Figure 2.
Susceptibility of Os-cps2 Knockout (Os-cps2ko) and RNAi Knockdown (Os-cps2i) Rice Plants Relative to Their Parental/Wild-Type Lines.
(A) Os-cps2 exhibits increased susceptibility to M. oryzae fungal blast disease (rice isolate Guy11). (i) Representative leaves after M. oryzae infection for Os-cps2ko versus parental/wild-type plants (as indicated). (ii) Percentage of diseased (discolored) area for Os-cps2ko compared with its parental/wild-type plants. (iii) Percentage of diseased (discolored) area for Os-cps2i lines compared with their parental/wild-type plants. **P < 0.01 and ***P < 0.001, calculated using the Mann-Whitney (Wilcoxon) W-test, for Os-cps2 plants relative to the relevant parental/wild-type line. No statistically significant differences were found between Os-cps2ko and Os-cps2i lines.
(B) Os-cps2 plants exhibit increased susceptibility to Xoo bacterial blight disease. (i) Representative leaves after Xoo infection of Os-cps2ko versus parental/wild-type plants (as indicated). (ii) Lesion lengths for Os-cps2ko compared with its parental/wild-type plants. (iii) Lesion lengths for Os-cps2i lines compared with their parental/wild-type plants. *P < 0.05 and **P < 0.01, calculated using post-ANOVA pairwise analysis of significance with the Tukey’s honestly significant difference test, for Os-cps2 plants relative to the relevant parental/wild-type line.
Os-cps4 Rice Exhibits Increased Resistance to X. oryzae pv oryzae
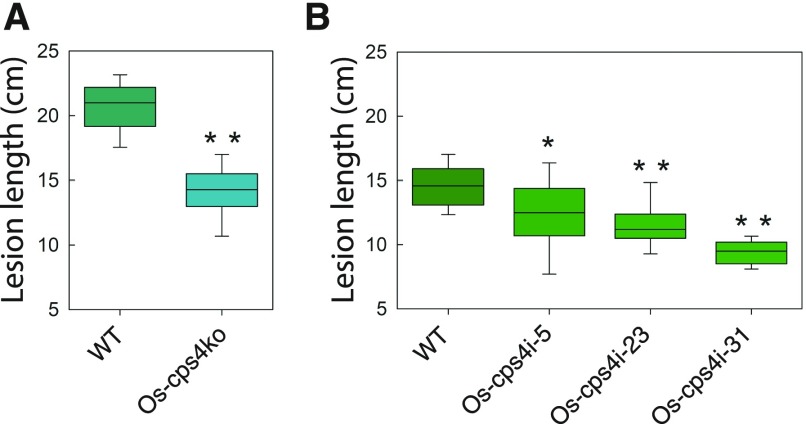
Previous results indicated that a T-DNA insertion (knockout) mutant of Os-CPS4, Os-cps4ko, is no more susceptible than its parental/wild-type line (ssp japonica cv Zhonghua 11) to infection with M. oryzae, as well as another rice fungal pathogen, Fusarium fujikuroi (Xu et al., 2012). To investigate the role of syn-CPP-derived diterpenoids in resistance to Xoo, analogous infections were performed with Os-cps4ko and its parental/wild-type line. Intriguingly, Os-cps4 plants exhibited significantly shorter lesion length relative to those from its parental/wild-type line, indicating that this mutant has increased resistance to the bacterial leaf blight disease (Figure 3A). To further support this surprising effect, Os-CPS4 was also targeted by RNAi to generate knockdown Os-cps4i lines. To compare this directly to Os-cps2ko rice, these were constructed in the same genetic background (i.e., ssp japonica cv Nipponbare). Again, three Os-cps4i lines were selected for further analysis based on their reduced expression of Os-CPS4 (relative to the parental/wild-type cultivar), even following MeJA induction (Supplemental Figure 1C). Similar to Os-cps4ko, all three Os-cps4i lines also exhibited increased resistance to Xoo (Figure 3B).
Figure 3.
Characterization of Os-cps4 Knockout and RNAi Knockdown Rice Plants Relative to Their Parental/Wild-Type Lines.
(A) Os-cps4ko exhibits reduced susceptibility to Xoo bacterial blight disease. Histogram depicting lesion lengths for Os-cps2ko compared with its parental/wild-type line.
(B) Os-cps4i lines exhibit reduced susceptibility to Xoo bacterial blight disease. Box plot depicting lesion lengths for Os-cps4i lines compared with their parental/wild-type. *P < 0.05 and **P < 0.01, calculated using post-ANOVA pairwise analysis of significance with the Tukey’s honestly significant difference test, for Os-cps4 plants relative to the relevant parental/wild-type line.
Os-CPS2-Overexpressing Rice Exhibits Increased Resistance to X. oryzae pv oryzae
The observation that Os-cps4 rice exhibits increased resistance to Xoo suggests that blocking metabolic flux to syn-CPP-derived diterpenoids leads to reallocation of GGPP toward ent-CPP-derived diterpenoids, which are effective antibiotics against Xoo. To further explore this possibility, Os-CPS2 was overexpressed in ssp japonica cv Kitaake. A line whose enhanced expression of Os-CPS2 was verified by RT-PCR was then selected for further characterization (Supplemental Figure 1D). This Os-CPS2OE line exhibited normal growth and development (Supplemental Figure 2B), indicating that its gibberellin phytohormone levels were not affected. When infected with Xoo, this Os-CPS2OE rice line exhibited significantly shorter lesion lengths relative to the parental/wild-type line (P < 0.01 in an unpaired t test), indicating increased resistance to bacterial leaf blight (Figure 4). This result further supports a role for ent-CPP-derived diterpenoids as antibacterial agents in rice plant defense, at least against Xoo, consistent with the known anti-Xoo activity of the oryzalide-related diterpenoids, and also suggests that the increased resistance of Os-cps4 rice to Xoo infection may be due to increased allocation toward these phytoanticipans.
Figure 4.
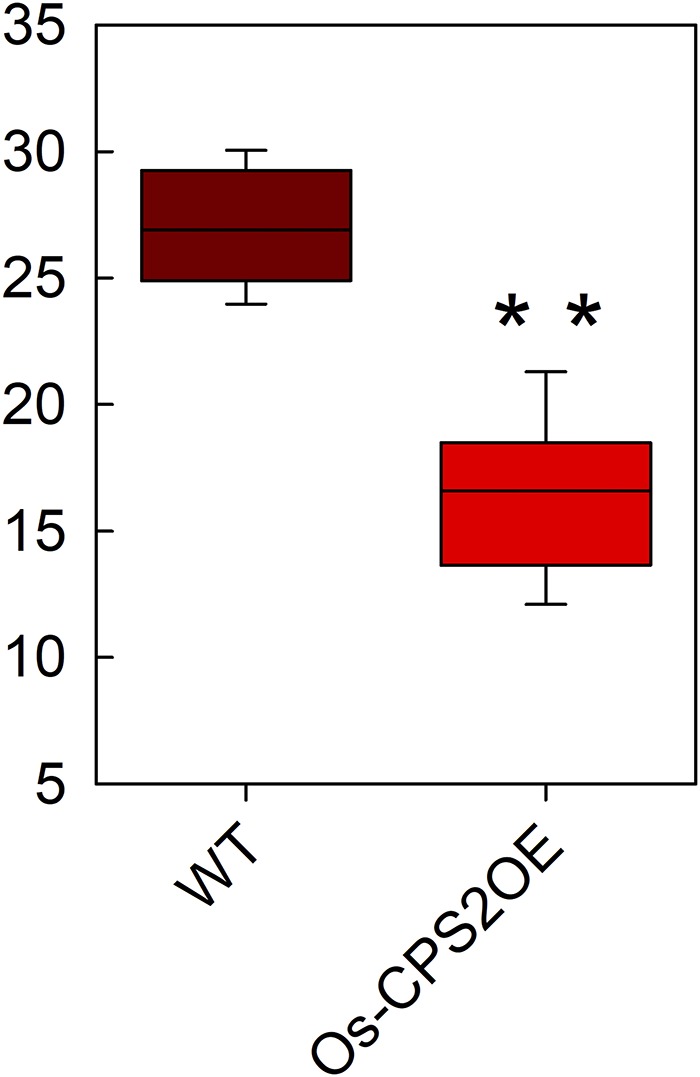
Decreased Susceptibility of the Os-CPS2OE Line to Xoo Blight Disease.
Box plot contrasting lesion lengths of Os-CPS2OE versus its parental/wild-type line infected with Xoo (**P < 0.01, calculated using post-ANOVA pairwise analysis of significance with the Tukey’s honestly significant difference test).
Os-CPSs Do Not Influence the Composition of Root-Associated Bacterial Communities
Given the secretion of both momilactones (Os-CPS4 dependent) and phytocassanes (Toyomasu et al., 2008), which are presumably at least partially Os-CPS2 dependent, along with the effects that knocking these out had on susceptibility to Xoo, it seemed likely that this also might have an effect on the composition of the bacterial community associated with roots. This was tested with the two knockout lines and the relevant parental/wild-type cultivars. To provide a relevant inoculum, this was investigated by growing seedlings in rice paddy soil, much as previously described (Shrestha et al., 2010). Environmental DNA was isolated from bulk soil, the rhizosphere, and roots (for analysis of ectophytes and endophytes), from which a variable region of the bacterial 16S ribosomal was then amplified and subjected to sequencing. Much as recently reported (Edwards et al., 2015), rice seems to select for certain bacteria in its rhizosphere and roots, with clear differentiation between the species found therein relative to both bulk soil and each other (Supplemental Figure 3). However, no clear difference was observed between either Os-cps2ko or Os-cps4ko and/or their parental/wild-type lines. While initial analysis suggested that a number of species might be over- or underrepresented in Os-CPS knockouts relative to their parental/wild-type cultivars, when properly corrected for multiple comparisons by use of Benjamini-Hochberg false discovery rate analysis, none met the usual threshold for statistical significance (i.e., P < 0.05). Moreover, even those that came closest to meeting this cutoff were from comparison of Os-cps2ko relative to its parental/wild-type cultivar. Most notably, there may be ∼6-fold increase in a bacterial genus from the TM7-1 class in the Os-cps2ko rhizosphere (albeit these make up <0.05% of the community), as suggested by Benjamini-Hochberg false discovery rate corrected P values of <0.1 at the phylum (TM7) and class level, with corrected P values < 0.2 down to the genus level and no change in relative frequency (i.e., this remains constant from the class level on down; Supplemental Table 1).
Os-CPS4 Plays a Role in Fungal Non-Host Disease Resistance
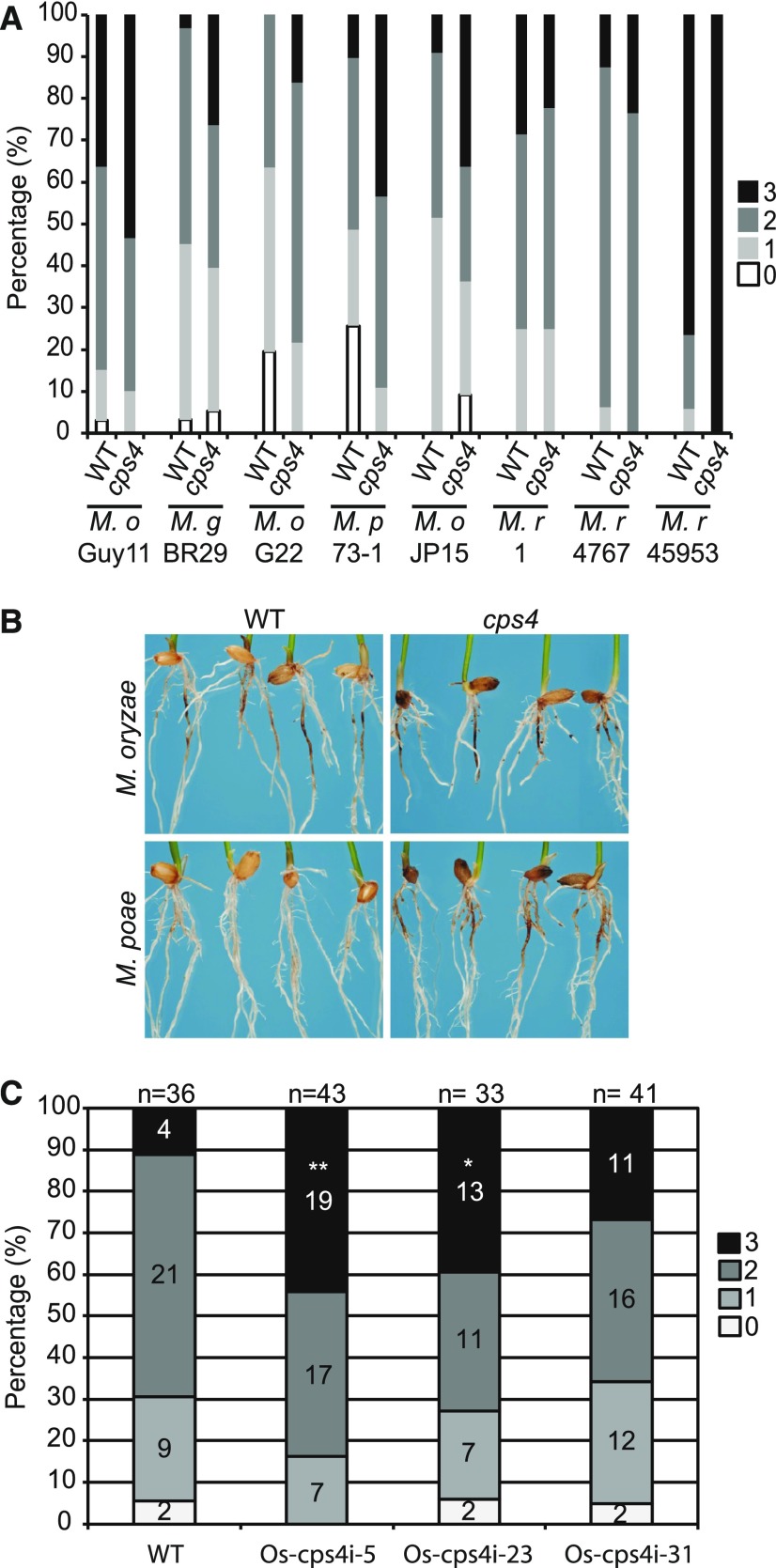
Strikingly, the results presented above indicate that syn-CPP-derived diterpenoids do not have a role in defense against either M. oryzae or Xoo nor in shaping the bacterial community of either the rhizosphere or roots. Indeed, the most consistent role indicated for these natural products is as allelochemicals (Xu et al., 2012; Toyomasu et al., 2014), which raises the question of why Os-CPS4 transcripts accumulate in response to either elicitation with the fungal cell wall component chitin (Okada et al., 2007), or the defense signaling molecule MeJA (Xu et al., 2004), along with further elaborated syn-CPP-derived diterpenoids (Schmelz et al., 2014). Given that allelopathic activity has been convincingly assigned to the more specifically Os-KSL4 dependent momilactones (Xu et al., 2012), it also is unclear why rice produces additional syn-CPP derived diterpenoids (e.g., oryzalexin S; Figure 1). It has been shown that rice is resistant to a number of other fungal pathogens from the Magnaporthe genus other than rice isolates of M. oryzae (Faivre-Rampant et al., 2008). Hypothesizing that Os-CPS4 might be a factor in such non-host disease resistance, a range of M. oryzae pathovars (isolated from alternative host plants), and other Magnaporthe species (M. grisea, M. rhizophila, and M. poae) were tested with Os-cps4ko and its parental/wild-type line. Given that M. rhizophila and M. poae infect only underground tissues, and that M. oryzae can infect rice through the roots as well (Sesma and Osbourn, 2004), these infections were directed at the roots. As previously reported (Xu et al., 2012), no significant differences were observed with the Guy 11 strain (rice isolate) of M. oryzae (Figure 5A). However, with two other pathovars/strains of M. oryzae (G22, isolated from Eragrostis curvula, and JP15, isolated from Elusine coracana), as well as M. grisea (strain BR29, isolated from Digitaria sanguinalis), M. rhizophila (strain 1, isolated from Poa pratensis) and M. poae (strain 73-1, isolated from wheat), Os-cps4ko lines exhibited varying increases in susceptibility relative to the parental/wild-type cultivar (Figure 5A), perhaps most strongly with M. poae (Figure 5B). Similar significantly decreased resistance to M. poae was observed with the Os-cps4i lines as well (Figure 5C). Thus, these results indicate a role for syn-CPP-derived diterpenoids in the non-host disease resistance of rice, at least against fungi from the Magnaporthe genus, providing a role for Os-CPS4 and, hence, syn-CPP-derived diterpenoids, in plant defense against microbial pathogens.
Figure 5.
Effect of Os-cps4 on Non-Host Disease Resistance.
(A) Infection of Os-cps4ko and wild-type rice lines (17 < n < 46) with different Magnaporthe species and strains (M. o. indicates M. oryzae, M. p. indicates M. poae, and M. r. indicates M. rhizophila, with specific strains indicated below). Numbers indicate the degree of infection area (% indicates the proportion of the total fall into the given disease degree score bin).
(B) Representative effect of Os-cps4ko on M. poae non-host disease susceptibility.
(C) Os-cps4i lines (33 < n < 43) exhibit increased susceptibility to infection with M. poae relative to their parental/wild-type plants (% indicates the proportion to the total disease degree of each score). n = number of total seeds analyzed. *P < 0.05 and **P < 0.005, calculated using the χ2 test.
Phytochemical Analyses
To investigate which diterpenoids might be responsible for the observed effect on defense/resistance, a liquid chromatography-tandem mass spectrometry method targeted at analysis of these phytochemicals was developed. This enabled detection of not only the momilactones and phytocassanes that have been previously investigated via genetic approaches (Shimura et al., 2007; Wang et al., 2012; Xu et al., 2012; Toyomasu et al., 2014), but also oryzalexins A-E, based on the availability of authentic standards from a metabolic engineering approach used to elucidate their biosynthesis (Wu et al., 2013; Kitaoka et al., 2016). To simplify analysis, this was focused on root exudates, in which it was possible to detect all three of these families of labdane-related diterpenoids. However, particularly in order to detect oryzalexins, elicitation of these phytoalexins was required. After examination of several different methods of induction, including the previously described use of methyl jasmonate or UV irradiation (Peters, 2006), it was discovered that the application of 0.5 mM CuCl2 in the growth media of hydroponically grown seedlings provided the most reliable means of elicitation.
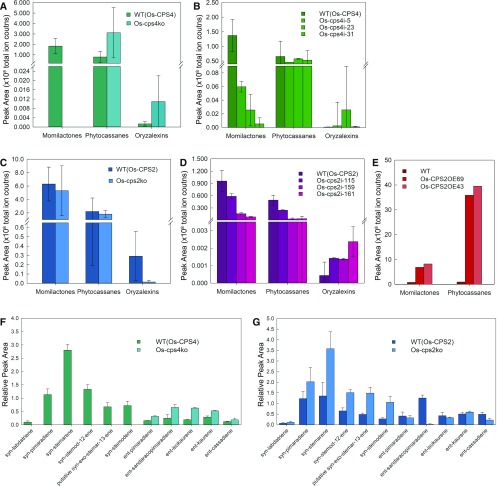
Much as previously reported (Xu et al., 2012), no momilactones were evident in root exudates from the Os-cps4ko line (Figure 6A), and momilactone levels were very significantly reduced in the Os-cps4i lines as well (Figure 6B). While the amounts of phytocassanes and oryzalexins seemed to be somewhat increased in these Os-cps4 lines, this effect was not statistically significant in all lines.
Figure 6.
Effect of Genetically Manipulating Os-CPS on Phytochemical Composition.
(A) Effect of Os-cps4ko on levels of diterpenoid natural products in induced root exudates.
(B) Effect of Os-cps4i on levels of diterpenoid natural products in induced root exudates.
(C) Effect of Os-cps2ko on levels of diterpenoid natural products in induced root exudates.
(D) Effect of Os-cps2i on levels of diterpenoid natural products in induced root exudates.
(E) Effect of Os-cps2OE on levels of diterpenoid natural products in induced root exudates.
(F) Effect of Os-cps4ko on levels of diterpene precursors in induced leaf tissue (values reflect indicated diterpene peak area relative to that for the internal standard).
(G) Effect of Os-cps2ko on levels of diterpene precursors in induced leaf tissue (values reflect indicated diterpene peak area relative to that for the internal standard).
In the case of the Os-cps2 lines, due to the presence of the biochemically analogous (i.e., ent-CPP producing) Os-CPS1, the amount of phytocassanes was reduced, but not eliminated, relative to the relevant parental/wild-type cultivar. However, somewhat surprisingly, this reduction was not significant for the Os-cps2ko line (Figure 6C), although it was for the Os-cps2i lines (Figure 6D). However, none of these lines exhibited consistently significant effects on oryzalexin levels nor, as might have been expected, the amount of momilactones. On the other hand, the Os-CPS2OE line was found to exude much greater amounts of phytocassanes relative to its parental/wild-type cultivar (∼40-fold increase), although some increase in momilactones were also observed (<10-fold; Figure 6E).
Despite extensive investigation, it was not possible to detect oryzalide-related diterpenoids. Thus, to investigate potential effects on the production of these, as well as the other unidentified labdane-related diterpenoids presumably made by rice (Figure 1), the array of upstream olefin precursors was analyzed by gas chromatography-mass spectrometry (GC-MS). As previously described (Morrone et al., 2011), these hydrocarbons were selectively targeted by fractionation over silica gel of extracts of MeJA-induced leaves. In addition to the various labdane-related diterpenes shown in Figure 1, this targeted metabolite analysis also found ent-casbene, a diterpene olefin that is directly formed from GGPP and is the expected intermediate in the biosynthesis of the recently identified rice diterpenoid phytoalexin (+)-10-oxodepressin (Inoue et al., 2013). To simplify comparison, only analysis of the knockout lines is presented here.
As expected, the Os-cps4ko line exhibits a complete loss of all syn-CPP-derived diterpenes, along with significant increases in the levels of most ent-CPP-derived diterpenes, with no change in the amount of ent-casbene (Figure 6F). By contrast, the Os-cps2ko line exhibits significant reductions only in the levels of ent-cassadiene and, especially ent-sandaracopimaradiene, which is no longer observed, but also exhibits significant increases in the levels of several syn-CPP-derived diterpenes, along with a decrease in the amount of ent-casbene (Figure 6G).
DISCUSSION
The sheer number of rice diterpenoid phytoalexins isolated on the basis of antibiotic activity against just M. oryzae raises questions regarding their relevance to plant defense. This was heightened by a report that the Os-CPS4-dependent diterpenoids do not appear to play a significant role against M. oryzae, instead acting as allelochemicals, a role that was more specifically assigned to the Os-KSL4-dependent momilactones (Xu et al., 2012). It was then unclear what role other syn-CPP-derived diterpenoids might play. Moreover, the role of the many ent-CPP-derived diterpenoids also had not yet been proven. Here, genetic manipulation of Os-CPS2 and Os-CPS4 was employed to demonstrate that, although Os-CPS4 further does not play a role in defense against the bacterial pathogen Xoo, Os-CPS2 is relevant to rice plant defense against both Xoo and the fungal pathogen M. oryzae. Despite secretion of both Os-CPS2- and Os-CPS4-dependent diterpenoids from roots, these do not appear to significantly affect the composition of the bacterial communities in the rice rhizosphere or roots.
Although the ent-CPP product of Os-CPS2 could serve as a precursor to the gibberellin phytohormones, manipulation of Os-CPS2 does not seem to greatly affect normal rice plant growth and development. The lack of such effect in Os-cps2 rice is consistent with the previously reported dominant role of Os-CPS1 in gibberellin metabolism (Sakamoto et al., 2004). In addition, the normal development of the Os-CPS2OE plants is consistent with previous results in Arabidopsis, where overexpression of the endogenous At-CPS led to increases in ent-kaurene, but no increase in gibberellins (Fleet et al., 2003). While similar increases in ent-kaurene were observed here (e.g., Figure 6F), this also may serve as a precursor of the antibacterial oryzalide-related diterpenoids (Supplemental Figure 4), providing an alternative metabolic fate that would be consistent with the observed increased resistance of Os-CPS2OE rice to Xoo (Figure 4).
The results reported here suggest that phytocassanes may play a particularly key role in rice defense against M. oryzae and Xoo, as alterations in the level of this family of labdane-related diterpenoids seem to correlate with the observed changes in susceptibility, although the alterations in phytocassane levels are not statistically significant in all lines. Indeed, decreasing Os-CPS2 expression only has a small effect on phytocassane and oryzalexin content. This is due to the ability of Os-CPS1 to also produce ent-CPP (Figure 1). Nevertheless, Os-CPS2 is important for plant defense, as evidenced by the increased susceptibility of the Os-cps2 lines to both Xoo and M. oryzae. This disparity may reflect the differential expression patterns of the two ent-CPP producing CPSs, as Os-CPS1 is selectively expressed in the vascular bundle, while Os-CPS2 is largely expressed in epidermal cells (Toyomasu et al., 2015). Thus, in the absence (or upon suppression) of Os-CPS2 expression, ent-CPP-derived phytoalexins may not be produced in the epidermal tissue, allowing establishment of pathogen infection. Presumably the observed production of these compounds occurs in the vascular bundle where they do not serve as an effective defense. However, a role for the oryzalide-related diterpenoids also cannot be ruled out.
The results reported here further indicate that genetic manipulation of the rice CPSs leads to reallocation of GGPP from conversion to one stereoisomer of CPP to the other (i.e., ent versus syn; Figure 1), at least at the level of the immediately subsequently formed olefins (compare Figures 6F and 6G). This suggests that there may be a distinct pool of GGPP that is tapped for more specialized diterpenoid metabolism in rice plant leaves, which seems to be separate from that utilized for gibberellin biosynthesis or in the production of photosynthetic pigments (this latter presumably reflects the predominant metabolic fate for GGPP overall). These distinct pools also may reflect the separation of these various metabolic processes into distinct tissues, as Os-CPS1 is selectively expressed in the vascular bundle where gibberellin metabolism occurs, while Os-CPS2, and presumably Os-CPS4, are largely expressed in epidermal cells (Toyomasu et al., 2015). These tissues are further separate from the mesophyll where photosynthesis is localized.
Notably, the observed significant metabolic allocation toward syn-CPP-derived diterpenoids (which are dependent on Os-CPS4), indicated that these natural products must play some role in plant defense. This prompted studies that revealed at least some of these compounds are important in non-host disease resistance against Magnaporthe species of fungi that are not adapted to rice. Indeed, M. oryzae has already been shown to metabolize momilactones (Hasegawa et al., 2010), which presumably is at least partially responsible for its ability to infect rice. Thus, rice labdane-related diterpenoid natural products seem to play a number of roles, including acting as antibiotics against adapted bacterial and fungal pathogens (i.e., Xoo and M. oryzae, respectively), as well as in fungal non-host disease resistance, along with acting as allelochemicals. Accordingly, the complexity of rice diterpenoid metabolism seems to be matched by the variety of physiological roles played by these natural products. It will now be of interest to determine if further dissection of the roles played by rice diterpenoids, and the clusters of biosynthetic genes that (at least partially) underlie the production of these natural products (Yamane, 2013; Zi et al., 2014), supports this hypothesis.
More broadly, the results reported here suggest that evolution is unlikely to allow allocation of significant metabolic effort without physiological function. Accordingly, investigation of metabolic partitioning, particularly in combination with various stresses (e.g., biotic and/or abiotic elicitation), then provides a means of inferring potential roles. In conjunction with biochemical analyses defining the relevant enzymatic genes, the inferred relevance to various physiological functions can be further investigated via reverse genetic approaches. Critically, much as shown here for the role of Os-CPS4 in fungal non-host disease resistance, the inferred relevance (here to plant defense against microbial pathogens) can be used to stimulate further productive investigations even in the absence of evidence for the originally proposed function (here a role in resistance against the rice fungal pathogen M. oryzae).
METHODS
Chemicals
Unless otherwise noted, all molecular biology reagents were purchased from Invitrogen and all other chemicals from Fischer Scientific. The sequences of the primers used in the PCR experiments described below can be found in Supplemental Table 2.
Plant Materials
Rice plants (Oryza sativa ssp japonica cv Nipponbare, Kitaake, or Zhonghua 11) were germinated and grown in standard growth chambers cycling between 12 h light at 28°C and 12 h dark at 24°C. For germination, surface-sterilized rice seeds were placed on 0.5× Murashige and Skoog plates (2.22 g Murashige and Skoog medium, 15 g sucrose, and 8 g agar per L) for 7 d. These germinated seedlings were then transferred to 4-inch square pots filled with soil for continued growth (unless otherwise indicated). The plants were fertilized weekly with a solution of Peters Excel All-Purpose Formulation fertilizer (15%N-5%P-15%K; two cubic centimeters in 7 liters of water).
The Os-cps4ko T-DNA insertion mutant RMD_03Z11UZ33, and selection of a homozygous line, has been previously described (Xu et al., 2012). This mutant is in the ssp japonica cv Zhonghua 11 background. The Os-cps2ko T-DNA insertion mutant RGT1043_5.1 (insert in the sixth exon of Os-CPS2; TIGR ID, LOC_ Os02g36210; Supplemental Figure 1A) was obtained from the UC-Davis Transposable Element Insertion Database. This mutant is in the ssp japonica cv Nipponbare background. Homozygous Os-cps2ko plants were selected by RT-PCR of Os-CPS2 (i.e., the absence of any cDNA) from the obtained seeds. Homozygous Os-cps2ko plants were verified in the next generation and propagated to provide seeds for the experiments described here.
RNAi Vector Construction and Rice Transformation
Binary vectors were developed for RNAi-mediated repression of Os-CPS2 and Os-CPS4 transcription using target regions of 374 and 435 bp in length, respectively, cloned in both sense and antisense orientation, separated by a 1.13-kb intron sequence derived from the Arabidopsis thaliana FAD2 gene (Okuley et al., 1994) and positioned downstream of the constitutive polyubiquitin-1 promoter from maize (Zea mays) with its cognate intron (Christensen et al., 1992). The specific target sequences selected for CPS2 (GenBank accession number AY602991) correspond to nucleotides 2386 to 2760 of the coding sequence plus an additional 153 bp of contiguous 3′ untranslated region sequence; the specific target sequences selected for CPS4 (GenBank accession number AY530101) correspond to nucleotides 2192 to 2627 of the coding sequence plus an additional 224 bp of contiguous 3′ untranslated region sequence. Os-CPS2 and Os-CPS4 target regions flanked by EcoRI (5′ end) and BamHI (3′ end) restriction sites were first generated by PCR amplification using rice (O. sativa cv Nipponbare) genomic DNA as template, to facilitate direct ligation with EcoRI- and BamHI-digested pUbi-IF2 (DNA Cloning Service), with RNAi-1 primer pairs (Supplemental Table 2). The resulting intermediate constructs were then digested with BsrGI and MluI, and Os-CPS2 and Os-CPS4 target regions flanked by BsrGI (5′ end) and MluI (3′ end) were also generated in a second round of PCR amplifications with RNAi-2 primer pairs (Supplemental Table 2). Following digestion with BsrGI and MluI, the PCR products were ligated with their corresponding intermediate vectors, resulting in the final intermediate vectors, pUbi-CPS2 and pUbi-CPS4, containing the complete hpRNA-generating transgene cassettes for Os-CPS2 and Os-CPS4 as confirmed by DNA sequencing. Finally, pUbi-CPS2 and pUbi-CPS4 were digested with SfiI, then the RNAi cassette-containing fragments were gel purified and ligated with SfiI-digested pLH7000 (Hausmann and Toepfer, 1999). The resulting binary vectors contain the hpRNA-generating cassettes arranged in a head-to-tail orientation with respect to the phosphinothricin N-acetyltransferase-selectable marker cassette and were designated pCPS2-RNAi and pCPS4-RNAi. For the generation of transgenic rice events, Agrobacterium tumefaciens strain EHA101 was transformed by electroporation with either pCPS2-RNAi or pCPS4-RNAi, and the resulting recombinant strains used to transform immature embryos of rice, as previously described (Toki, 1997). Lines exhibiting decreased expression of the targeted Os-CPS were selected by semi-quantitative RT-PCR, with seeds from T1 plants used in all subsequent analyses.
Overexpression Vector Construction and Rice Transformation
The open reading frame for Os-CPS2 was amplified from cDNA by PCR using CPS2-OE primers (Supplemental Table 2), which also introduce unique EcoRI and SpeI restriction sites at the 5′ and 3′ ends, respectively (Supplemental Table 2). The PCR fragment was cloned into the EcoRI and SpeI restriction sites of the binary vector pBY02, such that expression is under the control of a CaMV 35S promoter. The resulting pCPS2-OE construct was transformed into Agrobacterium EHA105 by electroporation. Immature embryo-derived callus cells of rice (cv Kitaake) were transformed by adapting a previously described protocol (Hiei et al., 1994). Briefly, transformed calli and regenerated plantlets were selected on media containing hygromycin B, with the resulting plants grown to seed, and Os-CPS2 overexpression confirmed by PCR. Selection was performed for three more generations to obtain nonsegregating homozygous Os-CPS2OE plants. One of the lines that exhibited strong overexpression of Os-CPS2 (Supplemental Figure 1D) was selected for further analysis.
Disease Resistance Assays
Magnaporthe infection assays were performed as previously described (Sesma and Osbourn, 2004). Briefly, for leaf infection assays, rice seedlings were grown until the stage of the third leaf emergence. All RNAi lines were pregerminated in the presence of BASTA to ensure retention of the recombinant insert. Three pots of 10 plants were used per line and per experiment. Each pot was sprayed with 2 mL of a suspension of 105 conidia mL−1. The plants were further incubated at the same growth conditions (85% relative humidity, 25°C, and a 16-h-light/8-h-dark photoperiod) for 6 d to score disease symptoms. Leaf lesions were scanned and analyzed with Assess 2.0 software (automatic parameters; American Phytopathological Society). For root infection assays, a 50-mL centrifuge tube was filled with 30 cm of moist vermiculite, followed by a mycelial plug of the same diameter as the centrifuge tube, a further layer of 5 cm of moist vermiculite, and five rice seeds covered with another 5 cm layer of moist vermiculite. Then, the tube was sealed with Parafilm to prevent loss of humidity. Fungal lesions on roots were scored after 15 d of incubation at 25°C and a 16-h-light/8-h-dark photoperiod. For the analysis of Os-cps4i lines, a total 153 seeds were analyzed (33< n < 43 per line). Disease symptoms on rice roots were scored at 15 d postinfection. Blast lesions were evaluated based on color intensity (for roots), lesion number (for leaves), and lesion extension (for both) of disease symptoms as previously reported (Tucker et al., 2010). A Mann-Whitney (Wilcoxon) W-test with a P value cutoff of 0.01 was used to compare medians that are statistically significant in Figure 2A. The χ2 test was applied to compare wild-type and different Os-cps4i lines using all the scores, and it highlighted significant differences with a P value < 0.05 (Figures 5A and 5C). When the same χ2 test was applied a second time using the score groups independently and the Yates’ correction (applied for 2 × 2 data with less than five values as was the case for the data shown in Figure 5C), only “score 3” of Os-cps4i_5 and Os-cps4i_23 was significantly different with a P value < 0.05, strongly supporting the significance found among these rice lines.
Xoo infections were carried using the leaf-clipping method (Kauffman et al., 1973), using the fully expanded leaves of 8-week-old rice plants and inoculating with Xoo strain PXO99A. Xoo was grown for 2 to 3 d on TSA (1% [w/v] Tryptone, 1% sucrose, 0.1% glutamic acid, and 1.5% agar, pH 6.6 to 6.8) plates at 28°C, scraped off, and resuspended in sterile deionized water. This suspension was then adjusted to an optical density of 0.5 at 600 nm before use for infection. After clipping, symptoms consisting of grayish chlorotic coloration moved down the inoculated leaf along the main vein from the cut tip. Disease progression was analyzed by measuring the length of these lesions (discoloration) 14 d after inoculation. Ten plants were used for infection and the disease assay was repeated thrice. One-way ANOVA statistical analyses were performed on the lesion measurements. The Tukey’s honestly significant difference test was used for post-ANOVA pairwise analysis of significance, set at 5% (P < 0.05).
Analysis of Rhizosphere/Plane Bacteriome
Sterile germinated seedlings were grown in rice paddy soil “microcosms” much as described (Shrestha et al., 2010). Soil taken from drained paddy fields at the Italian Rice Research Institute in Vercelli, Italy, in 2006 was a kind gift from Ralf Conrad (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany). This was first passed through a 1.2-mm sieve to remove organic matter, and 40 kg mixed with 15 liters of sterile water and 15 liters of fertilizer solution designed to mimic common rice field application (0.87 g KH2PO4, 1.85 g KCl, and 2.3 g urea per liter), with the resulting slurry (∼40 liters) divided between 46 pots (∼0.5 liters/16 cm square) and allowed to settle overnight. To generate a “flooding” depth of ∼5 cm, 200 mL of sterile water was added to each pot, and this depth was maintained for 3 d. To avoid submersion, the depth was then reduced to ∼2 cm for transplantation of 16-d-old seedlings, with three plants of the same genotype in each pot, then raised back to ∼5 cm 11 d later. Six pots were unplanted and served as the controls. Samples from these microcosms were taken 17 d after transplantation, with collection of the entire root system from each plant. The rhizosphere was considered to be the soil adhering to the roots, which was washed off by repeated plunges into ∼30 mL of sterile water, with centrifugation of the soil (5′ × 4000g at 4°C) to remove the water. The resulting “clean” roots were retained for analysis of ectophytic and endophytic bacteria. In addition, three 1-mL bulk soil samples were taken from the remaining soil in each pot following stirring, again followed by centrifugation to remove water. These samples were frozen at −20°C and then environmental DNA was isolated using PowerSoil DNA kits (Qiagen).
Samples were sequenced by an external provider (Mr. DNA) using the 16s rRNA primers Gray28F and Gray519R (Ishak et al., 2011), with barcodes included in the forward primer. Individual libraries were amplified via PCR, using the HotStarTaq Plus Maser Mix Kit (Qiagen). Cycle parameters were as follows: 94°C for 3 min, 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, with a final elongation step at 72°C for 5 min. After 28 amplification cycles, the PCR products were checked via a 2% agarose gel. Libraries were pooled together in equal proportions, purified, and sequenced in two runs on a MiSeq system, using 300-bp paired-end sequence reads following the manufacturer´s guidelines. Sequences were joined after q25 trimming of the ends, barcodes were removed, and sequences with ambiguous bases, as well as reads shorter than 200 bp, were removed.
High-quality sequences were demultiplexed in Qiime version 1.8.0 (Caporaso et al., 2010a), and reads from both runs were concatenated. Sequences were then clustered de novo into OTUs (operational taxonomic units) at 97% identity using UPARSE (Edgar, 2013). Chimeras were removed from the data set using UCHIME (Edgar et al., 2011) and GOLD as the reference database (Reddy et al., 2015). Clusters were assigned taxonomy in Qiime using the RDP classifier (Wang et al., 2007) and Greengenes database released in May, 2013 (DeSantis et al., 2006), with 0.8 confidence levels. Representative sequences were aligned with PyNast (Caporaso et al., 2010b), and a phylogenetic tree was built with FastTree (Price et al., 2009), both implemented in Qiime. An OTU table was constructed describing the abundance of bacterial OTUs within each sample. This table was filtered to eliminate nonbacterial OTUs (e.g., mitochondria and chloroplast derived sequences) and samples with <615 reads. Removal of these sequences only represented OTUs comprising <0.005% of the rarefied library within each genotype sample. Phyla level taxonomy was summarized in a separate OTU table for visualization.
Beta-diversity matrixes (e.g., abundance-weighted and unweighted UniFrac distances) were calculated with the OTU table after rarefying it to 28,000 sequences with Qiime. We statistically tested whether the bacterial communities differed between bulk soil, rhizosphere, and rhizoplane, as well as between the knockouts and their parental/wild-type lines, by independently analyzing the matrixes with one-way analyses of similarities and Adonis. To test whether any bacterial taxa was overrepresented in the knockouts compared with their parental lines, we performed linear discriminant analyses at all taxonomical levels, using the Galaxy (Afgan et al., 2016) implementation of LEfSe (Segata et al., 2011), with a minimum logarithmic LDA score of 2 and a maximum P value of 0.05.
Phytochemical Analyses
To determine the effect of CPS genetic manipulation on diterpenoid levels, germinated rice seedlings were placed in glass tubes, five seedlings in each tube, with ample amounts of rice growth media consisting of 1.43 mM NH4NO3, 1.0 mM CaCl2, 0.32 mM NaH2PO4·2H2O, 0.51 mM K2SO4, 1.64 mM MgSO4·7H2O, 9.47 μM MnCl2·4H2O, 0.075 μM (NH4)6Mo7O24·4H2O, 0.018 mM H3BO3, 0.15 μM ZnSO4·7H2O, 0.155 μM CuSO4·5H2O, 0.036 mM FeCl3.6H2O, and 0.077 mM C6H8O7. After another week, the seedlings were induced by the addition of CuCl2 to a final concentration of 0.5 mM, and 72 h later diterpenoids were extracted from the exudate by filtering the media from individual tubes through a 0.1-g Bond Elut C18 spin cartridge (Agilent) and eluting with 100% methanol. This was dried down under a stream of N2 and resuspended in 80% methanol for analysis by liquid chromatography-mass spectrometry.
Liquid chromatography-mass spectrometry analysis was performed with 10-μL samples (10 µL) over a Supelco (Sigma-Aldrich) Ascentis C18 column (10 cm × 2.1 mm; 3μm) using an Agilent 1200 HPLC coupled to a Bruker Solarix FT-ICR MS located in the Iowa State University W.M. Keck Metabolomics Research Laboratory. A binary gradient was used, consisting of acetonitrile with 0.1% (v/v) acetic acid (buffer A) and water with 0.1% (v/v) acetic acid (buffer B). The solvent gradient elution was programmed as follows: initial, 40% buffer A; 0 to 2 min isocratic elution by 40% buffer A; 2 to 17 min, a linear gradient from 40% buffer A to 100% buffer A; 17 to 20 min isocratic elution by 100% buffer A; 20 to 22 min a linear gradient from 100% buffer A to 40% buffer A. Molecules were ionized in positive mode with 0.5 V. Rice phytoalexins were isolated based on the calculated monoisotopic mass of each ion plus one [M+H]+ with an isolation selection window of ±2 m/z. The isolated masses were reionized and the resulting mass spectra recorded.
The parent ions isolated and quantified secondary ions for the rice phytoalexins with their corresponding retention times (RT), based on optimization with the available authentic standards, were as follows: momilactone A, m/z 315.2→271.2, RT 11.85 min; momilactone B, m/z 331.2→269.2, RT 9.93 min; oryzalexin A, m/z 303.3→285.2, RT 10.98 min; oryzalexin C, m/z 301.2→283.2, RT 12.41 min; oryzalexin D, m/z 287.2→269.2, RT 10.33 min; oryzalexin E, m/z 287.2→269.2, RT 12.21 min; oryzalexin S, m/z 305.2→287.2, RT 11.3 min; phytocassane A, m/z 317.2→299.2, RT 9.39 min; phytocassane B, m/z 335.2→317.2, RT 8.11 min; phytocassane C, m/z 319.2→301.2, RT 7.57 min; phytocassane D, m/z 317.2→299.2, RT 10.78 min; phytocassane E, m/z 317.2→299.2, RT 10.17 min; phytocassane F, m/z 333.3→315.2, RT 8.1 min, much as previously reported (Riemann et al., 2013; Horie et al., 2015; Miyamoto et al., 2016). For the diols oryzalexins D and E, the molecular ion was not observed, leading to the use of a dehydrated fragment for isolation, as previously reported for the similar diol oryzalexin S (Toyomasu et al., 2014; Horie et al., 2015).
Due to the coelution of some compounds at the same RT, every sample was run twice, each time isolating half of the phytoalexins. The first run isolated phytocassane C at 0 to 7.72 min; phytocassane B at 7.72 to 8.5 min; phytocassanes A, D, and E at 8.5 to 10.71 min; oryzalexin A at 10.71 to 11.47 min, momilactone A at 11.47 to 11.95 min, and oryzalexin E at 11.95 min to the end of the program. The second run isolated phytocassane C at 0 to 7.72 min, phytocassane F at 7.72 to 8.5 min, momilactone B at 8.5 to 10.1 min, oryzalexin D at 10.1 to 10.71 min, oryzalexin S at 10.71 to 11.47 min, momilactone A at 11.47 to 11.95 min, and oryzalexin C at 11.95 min to the end of the program.
The exuded amount of each diterpenoid was quantified from the corresponding total peak area divided by total number of seedlings from the relevant tube. Samples were run at a minimum in triplicate per line. Momilactone A was the largest and most regular peak in every sample (except those for Os-cps4ko). Accordingly, if a sample differed by more than a factor of 3 from the average in momilactone A content, it was excluded from subsequent analysis. The Student’s one-tailed two samples with heteroscedastic t test was used to determine statistically relevant significance on all data sets.
To determine the effect of CPS genetic manipulation on diterpene olefin levels, rice plants were cultivated to the 6th leaf stage. For induction, detached leaves (3 g) were sprayed with 10 mL 0.2% MeJA (in 0.1% Tween 20), wrapped with aluminum foil, and incubated in the dark at room temperature (24°C) for 72 h. The leaves were then frozen and ground to powder in liquid nitrogen, and the residue suspended in 200 mL hexanes, with the addition of 53 μg of the diterpene cembrene A (Sigma-Aldrich) as an internal standard (note that cembrene A is not found in rice) per gram of fresh leaf weight. This slurry was stirred for 12 h at room temperature. The hexane was collected by filtration through Whatman filter paper and the residue extracted twice more with 200 mL hexane. The pooled 600 mL hexane extract was then dried by rotary evaporation. The residue was resuspended in 12 mL hexane. The organic extract was fractionated by passage over an open column filled with 20 mL silica gel (J.T. Baker), which was then washed with a further 60 mL of hexane, with 6 × 12-mL fractions collected in glass tubes throughout. Each fraction was dried under nitrogen gas, and the residue resuspended in 100 μL of hexane for analysis by GC-MS.
GC-MS was performed over an HP-5ms column (Agilent; 0.25-μm film, 0.25-mm i.d., 30-m length) using a Varian 3900 GC with detection via a Saturn 2100 ion trap mass spectrometer in electron ionization (70 eV) mode. Samples (1 μL) were injected in splitless mode at 50°C, and after holding for 3 min at 50°C, the oven temperature was raised at a rate of 18°C/min to 194°C. Then, the oven temperature was raised at a rate of 0.1°C/min from 194°C to 195°C. After that, the temperature was raised at a rate 20°C/min to 300°C, where it was held for an additional 2 min. MS data from m/z 90 to 300 were collected starting 10 min after injection until the end of the run. Diterpenes were identified by comparison of their retention times and mass spectra with those of authentic standards. For each sample, all fractions containing diterpenes were pooled together (generally fractions 2–4) for relative quantification, with normalization accomplished by comparison of the total ion peak areas for the rice diterpenes to that of the internal standard.
Due to the partial overlap of certain diterpenes in the method described above, a longer temperature ramp was employed to separate these, particularly syn-labdatriene, as well as verify that the recently reported ent-beyerene product of Os-KSL2 (Tezuka et al., 2015) was not detected in our samples. This was accomplished by injecting 1-μL samples in splitless mode at 60°C and, after holding for 3 min at 50°C, the oven temperature was raised at a rate of 3°C/min to 246°C. After that, the temperature was raised at a rate of 20°C/min to 300°C, where it was held for an additional 2 min. MS data from m/z 90 to 300 were collected starting 5 min after injection until the end of the run. Again, diterpenes were normalized by comparison of their peak area to that of the internal standard.
Accession Numbers
Sequence data from the microbiome analysis reported in this article can be found in the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.339k00b). The rice genes targeted here can be found in GenBank under the following accession numbers: Os-CPS2, AY602991; Os-CPS4, AY530101.
Supplemental Data
Supplemental Figure 1. Characterization of genetic manipulations of rice CPSs by RT-PCR of plants after MeJA treatment.
Supplemental Figure 2. Growth data for genetic manipulation of Os-CPS2.
Supplemental Figure 3. Primary component analysis plot of bacterial composition.
Supplemental Figure 4. Representative of putative oryzalide-related diterpenoid biosynthetic pathway.
Supplemental Table 1. Data for TM7-1 bacteria in the rhizosphere of Os-cps2ko versus wild-type/parental rice.
Supplemental Table 2. Primers used in this study.
Acknowledgments
This work was supported by a grant from the USDA-NIFA (2014-67013-21720 to R.J.P. and B.Y.), by a fellowship from the China Postdoctoral Council (to J.Z.), by a fellowship from the Alexander von Humboldt Foundation (to R.J.P.), by the Max Planck Society (A.B. and J.G.), and by grants to A.S. and J.R.-R. from the Spanish Research Council (MINECO, BIO2014-53211-R and BIO2014-54233-JIN) and the Community of Madrid (S2013/ABI-2734).
AUTHOR CONTRIBUTIONS
X.L., J.Z., B.B., R.L., J.R.-R., A.B., B.L., X.M., D.L., and X.P. were involved in designing and carrying out experiments. S.R.B., J.G., Z.L., A.S., B.Y., and R.J.P. were involved in conceptual design and obtaining financial support. X.L., J.Z., and R.J.P. were primarily responsible for writing the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Afgan E., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I., Kissen R., Bones A.M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17: 73–90. [DOI] [PubMed] [Google Scholar]
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., Knight R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright D.W., Langcake P., Pryce R.J., Leworthy D.P., Ride J.P. (1981). Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry 20: 535–537. [Google Scholar]
- Christensen A.H., Sharrock R.A., Quail P.H. (1992). Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18: 675–689. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z.P., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., Eisen J.A., Sundaresan V. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112: E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre-Rampant O., Thomas J., Allègre M., Morel J.B., Tharreau D., Nottéghem J.L., Lebrun M.H., Schaffrath U., Piffanelli P. (2008). Characterization of the model system rice--Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 180: 899–910. [DOI] [PubMed] [Google Scholar]
- Fleet C.M., Yamaguchi S., Hanada A., Kawaide H., David C.J., Kamiya Y., Sun T.-P. (2003). Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 132: 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Mitsuhara I., Seo S., Imai T., Koga J., Okada K., Yamane H., Ohashi Y. (2010). Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant Microbe Interact. 23: 1000–1011. [DOI] [PubMed] [Google Scholar]
- Hausmann L., Toepfer R. (1999). Development of plasmid vectors. In Bioengineering of Custom-Tailored Rape Varieties, Brauer D., Roebbelen G., Toepfer R., eds (Goettingen, Germany: Gesellschaft fuer Pflanzenzuechtung; ), pp. 155–172. [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Horie K., Inoue Y., Sakai M., Yao Q., Tanimoto Y., Koga J., Toshima H., Hasegawa M. (2015). Identification of UV-induced diterpenes including a new diterpene phytoalexin, phytocassane F, from rice leaves by complementary GC/MS and LC/MS approaches. J. Agric. Food Chem. 63: 4050–4059. [DOI] [PubMed] [Google Scholar]
- Huang M., Sanchez-Moreiras A.M., Abel C., Sohrabi R., Lee S., Gershenzon J., Tholl D. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 193: 997–1008. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Sakai M., Yao Q., Tanimoto Y., Toshima H., Hasegawa M. (2013). Identification of a novel casbane-type diterpene phytoalexin, ent-10-oxodepressin, from rice leaves. Biosci. Biotechnol. Biochem. 77: 760–765. [DOI] [PubMed] [Google Scholar]
- Ishak H.D., Plowes R., Sen R., Kellner K., Meyer E., Estrada D.A., Dowd S.E., Mueller U.G. (2011). Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb. Ecol. 61: 821–831. [DOI] [PubMed] [Google Scholar]
- Kato T., Kabuto C., Sasaki N., Tsunagawa M., Aizawa H., Fujita K., Kato Y., Kitahara Y., Takahashi N. (1973). Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett. 14: 3861–3864. [Google Scholar]
- Kato-Noguchi H., Peters R.J. (2013). The role of momilactones in rice allelopathy. J. Chem. Ecol. 39: 175–185. [DOI] [PubMed] [Google Scholar]
- Kauffman H.E., Reddy A.P.K., Hsieh S.P.Y., Merca S.D. (1973). Improved technique for evaluating resistance of rice varieties to Xanthomonas-Oryzae. Plant Dis. Rep. 57: 537–541. [Google Scholar]
- Kitaoka N., Wu Y., Zi J., Peters R.J. (2016). Investigating inducible short-chain alcohol dehydrogenases/reductases clarifies rice oryzalexin biosynthesis. Plant J. 88: 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.T., Yagnik G.B., Hohenstein J.D., Ji Z., Zi J., Reichert M.D., MacIntosh G.C., Yang B., Peters R.J., Vela J., Lee Y.J. (2015). Investigation of the chemical interface in the soybean-aphid and rice-bacteria interactions using MALDI-mass spectrometry imaging. Anal. Chem. 87: 5294–5301. [DOI] [PubMed] [Google Scholar]
- Krishnan A., et al. (2009). Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 149: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., et al. (2016). Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 87: 293–304. [DOI] [PubMed] [Google Scholar]
- Morrone D., Hillwig M.L., Mead M.E., Lowry L., Fulton D.B., Peters R.J. (2011). Evident and latent plasticity across the rice diterpene synthase family with potential implications for the evolution of diterpenoid metabolism in the cereals. Biochem. J. 435: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A., Shimizu T., Okada K., Kuzuyama T., Koga J., Shibuya N., Nojiri H., Yamane H. (2007). Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 65: 177–187. [DOI] [PubMed] [Google Scholar]
- Okuley J., Lightner J., Feldmann K., Yadav N., Lark E., Browse J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo K., Kenmoku H., Oikawa H., König W.A., Toshima H., Mitsuhashi W., Yamane H., Sassa T., Toyomasu T. (2004). Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39: 886–893. [DOI] [PubMed] [Google Scholar]
- Papadopoulou K., Melton R.E., Leggett M., Daniels M.J., Osbourn A.E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 96: 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.J. (2006). Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317. [DOI] [PubMed] [Google Scholar]
- Peters R.J. (2010). Two rings in them all: the labdane-related diterpenoids. Nat. Prod. Rep. 27: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S., Xu M., Wilderman P.R., Peters R.J. (2004). Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136: 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T.B.K., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C. (2015). The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 43: D1099–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., et al. (2013). Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74: 226–238. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134: 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz E.A., Huffaker A., Sims J.W., Christensen S.A., Lu X., Okada K., Peters R.J. (2014). Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 79: 659–678. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesma A., Osbourn A.E. (2004). The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431: 582–586. [DOI] [PubMed] [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018. [DOI] [PubMed] [Google Scholar]
- Shrestha M., Shrestha P.M., Frenzel P., Conrad R. (2010). Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J. 4: 1545–1556. [DOI] [PubMed] [Google Scholar]
- Tezuka D., Ito A., Mitsuhashi W., Toyomasu T., Imai R. (2015). The rice ent-KAURENE SYNTHASE LIKE 2 encodes a functional ent-beyerene synthase. Biochem. Biophys. Res. Commun. 460: 766–771. [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Nelissen I., Eggermont K., Broekaert W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171. [DOI] [PubMed] [Google Scholar]
- Toki S. (1997). Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Report. 15: 16–21. [Google Scholar]
- Toyomasu T. (2008). Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci. Biotechnol. Biochem. 72: 1168–1175. [DOI] [PubMed] [Google Scholar]
- Toyomasu T., et al. (2014). Reverse-genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol. Plant. 150: 55–62. [DOI] [PubMed] [Google Scholar]
- Toyomasu T., et al. (2015). Transcripts of two ent-copalyl diphosphate synthase genes differentially localize in rice plants according to their distinct biological roles. J. Exp. Bot. 66: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T., Kagahara T., Okada K., Koga J., Hasegawa M., Mitsuhashi W., Sassa T., Yamane H. (2008). Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci. Biotechnol. Biochem. 72: 562–567. [DOI] [PubMed] [Google Scholar]
- Tucker S.L., Besi M.I., Galhano R., Franceschetti M., Goetz S., Lenhert S., Osbourn A., Sesma A. (2010). Common genetic pathways regulate organ-specific infection-related development in the rice blast fungus. Plant Cell 22: 953–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten H.D., Mansfield J.W., Bailey J.A., Farmer E.E. (1994). Two classes of plant antibiotics: Phytoalexins versus “Phytoanticipins”. Plant Cell 6: 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Hillwig M.L., Okada K., Yamazaki K., Wu Y., Swaminathan S., Yamane H., Peters R.J. (2012). Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287: 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Kono Y., Esumi Y., Teraoka T., Hosokawa D., Suzuki Y., Sakurai A., Watanabe M. (1996). Studies on a quantitative analysis of Oryzalides and Oryzalic acids in rice pants by GC-SIM. Biosci. Biotechnol. Biochem. 60: 1460–1463. [Google Scholar]
- Wu Y., Wang Q., Hillwig M.L., Peters R.J. (2013). Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 454: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hillwig M.L., Prisic S., Coates R.M., Peters R.J. (2004). Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39: 309–318. [DOI] [PubMed] [Google Scholar]
- Xu M., Galhano R., Wiemann P., Bueno E., Tiernan M., Wu W., Chung I.M., Gershenzon J., Tudzynski B., Sesma A., Peters R.J. (2012). Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 193: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H. (2013). Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 77: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Zi J., Mafu S., Peters R.J. (2014). To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65: 259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]