Proteomic profiling of SUMO conjugation identifies a large catalog of SIZ1-dependent targets important for stress protection, many of which are related to transcription and chromatin assembly.
Abstract
The posttranslational addition of small ubiquitin-like modifier (SUMO) is an essential protein modification in plants that provides protection against numerous environmental challenges. Ligation is accomplished by a small set of SUMO ligases, with the SAP-MIZ domain-containing SIZ1 and METHYL METHANESULFONATE-SENSITIVE21 (MMS21) ligases having critical roles in stress protection and DNA endoreduplication/repair, respectively. To help identify their corresponding targets in Arabidopsis thaliana, we used siz1 and mms21 mutants for proteomic analyses of SUMOylated proteins enriched via an engineered SUMO1 isoform suitable for mass spectrometric studies. Through multiple data sets from seedlings grown at normal temperatures or exposed to heat stress, we identified over 1000 SUMO targets, most of which are nuclear localized. Whereas no targets could be assigned to MMS21, suggesting that it modifies only a few low abundance proteins, numerous targets could be assigned to SIZ1, including major transcription factors, coactivators/repressors, and chromatin modifiers connected to abiotic and biotic stress defense, some of which associate into multisubunit regulatory complexes. SIZ1 itself is also a target, but studies with mutants protected from SUMOylation failed to uncover a regulatory role. The catalog of SIZ1 substrates indicates that SUMOylation by this ligase provides stress protection by modifying a large array of key nuclear regulators.
INTRODUCTION
The covalent attachment of small ubiquitin-like modifier (SUMO) to other proteins provides an essential mechanism for controlling the activity, localization, and turnover of many intracellular effectors in eukaryotes (Hay, 2013; Hendriks and Vertegaal, 2016). Besides regulating development and cellular homeostasis under normal growth conditions, SUMOylation plays a central role in defense against genotoxic stress and a variety of abiotic and biotic challenges. As examples, SUMOylation in plants has been connected genetically to thermotolerance, resistance to cold, salt, and drought stress, the phosphate starvation response, and innate immunity (Yoo et al., 2006; Miura et al., 2007a; Castro et al., 2012; Park and Yun, 2013). Some of these outcomes are linked to the stress hormones salicylic acid and abscisic acid (ABA) and their associated signaling pathways (Catala et al., 2007; Lee et al., 2007; van den Burg et al., 2010; Zheng et al., 2012).
Most notable is the rapid and reversible accumulation of SUMO conjugates during stress, which for heat stress is one of the fastest molecular responses observed, suggesting that specific SUMOylation events directly help mitigate damage (Kurepa et al., 2003; Saracco et al., 2007). Indeed, SUMOylation of the transcription factors PHOSPHATE STARVATION RESPONSE1 (PHR1), INDUCER OF CBF EXPRESSION1, heat shock factor A2 (HSFA2), ABA-INSENSITIVE5, MYB domain protein 30 (MYB30), and FLOWERING LOCUS D (FLD) are associated with tolerance to phosphate starvation, extreme cold and heat survival, ABA signaling, and flowering time, which is often accelerated by stress (Miura et al., 2005, 2007b, 2009; Jin et al., 2008; Cohen-Peer et al., 2010; Zheng et al., 2012). Stress-induced SUMOylation of the DELLA protein family in particular provides a mechanism for gibberellin-independent growth restraint under stress (Conti et al., 2014). Additionally, SUMOylation of the BCL-2-ASSOCIATED ATHANOGENE7 (BAG7) cochaperone has been linked recently to heat tolerance, where this modification promotes the unfolded protein response by helping translocate BAG7 to the nucleus (Li et al., 2017). Outside of stress, SUMOylation of phytochrome B, nitrate reductase 1 (NIA1) and NIA2, and DNA chromomethylase 3 (CMT3) has been connected to light signaling, enhanced nitrogen assimilation, and the epigenetic regulation of gene expression, respectively (Park et al., 2011; Kim et al., 2015; Sadanandom et al., 2015).
Most plant species express a small family of SUMO isoforms (SUMO1, SUMO2, SUMO3, and SUMO5 in Arabidopsis thaliana), with the highly related SUMO1/SUMO2 subfamily (83% similarity) being dominant (Augustine et al., 2016; Hammoudi et al., 2016). Their addition is driven by a three-step reaction cascade in which the SUMO moiety is first adenylated and then bound via a high-energy thioester linkage to the heterodimeric SUMO-activating enzyme (E1), composed of the SAE1 and SAE2 subunits (Colby et al., 2006; Saracco et al., 2007). The activated SUMO is then transferred to the SUMO-conjugating enzyme 1 (SCE1) (E2) via transesterification, and finally donated to substrate proteins, often with assistance from a SUMO-protein ligase (E3). The end result is SUMO covalently linked through an isopeptide bond between its C-terminal glycine and specific lysine(s) within the target. In many cases, additional SUMOs become attached, sometimes by using previously bound SUMOs to concatenate poly-SUMO chains (Miller et al., 2010; Hendriks and Vertegaal, 2016).
Once bound, the SUMO moieties alter the function(s) of their targets, which can include changes in intracellular location, activity, and/or interactions with other cellular factors, including proteins bearing SUMO-interacting motifs (SIMs) (Elrouby and Coupland, 2010; Hay, 2013; Aguilar-Martinez et al., 2015; Li et al., 2017). In some instances, the SUMO moiety provides sites for subsequent ubiquitylation that directs proteasomal turnover of the modified proteins (Miller et al., 2010, 2013; Hay, 2013). Additionally, SUMO conjugation can be reversed by a collection of deSUMOylating proteases that specifically cleave the intervening isopeptide bond to release both the protein and SUMO moieties intact (Conti et al., 2008; Hickey et al., 2012).
Akin to protein modification by ubiquitin, the SUMO ligases are thought to confer specificity to the system, where they help connect the activated SUMO-E2 intermediate to specific substrates and then promote SUMO transfer. Ultimately, a myriad of proteins become SUMOylated (Hendriks and Vertegaal, 2016). In Arabidopsis, previous proteomic studies identified over 350 SUMO targets modified by the SUMO1/SUMO2 isoforms, many of which appear to be dynamically regulated (Miller et al., 2010, 2013). Most are localized to the nucleus and have functions related to DNA modification, chromatin assembly and structure, transcription, and RNA processing, export, and turnover.
Whereas the ubiquitin system employs a large and diverse array of ligases with strong substrate specificities to direct conjugation (>1000 in Arabidopsis; Vierstra, 2009), the SUMO system appears to engage a much more limited collection (Geiss-Friedlander and Melchior, 2007). For example, only four Arabidopsis SUMO ligases have been described thus far; SAP AND MIZ1 DOMAIN-CONTAINING LIGASE1 (SIZ1) (Miura et al., 2005; Cheong et al., 2009), METHYL METHANESULFONATE-SENSITIVE21 (MMS21 or HIGH PLOIDY2) (Huang et al., 2009b; Ishida et al., 2009), and the PROTEIN INHIBITORS OF ACTIVATED STATs-LIKE1 (PIAL1) and PIAL2 (Tomanov et al., 2014). That such a small set of ligases theoretically influences such a large array of proteins implies that substrate specificity is also endowed by other features, such as target location, assembly into protein complexes, and/or possibly through direct recognition of the target by the E2 (Hay, 2013; Jentsch and Psakhye, 2013).
Important steps toward understanding the molecular ramifications of SUMOylation would be the development of target catalogs for each SUMO isoform and ligase, and definition of how SUMO addition might alter the activity, interactions, location, and/or half-life of these targets. Here, in a step toward this goal, we developed a proteomic strategy to help assign individual SUMO ligases to specific Arabidopsis proteins. This strategy combines mutations eliminating specific ligases with a purification background in which the SUMO1 and SUMO2 isoforms responsible for most SUMOylation are genetically replaced with a variant [6His-SUMO1(H89-R)] engineered to enable affinity purification (Miller et al., 2010). This SUMO1 variant fully complements sumo1 sumo2 null mutants, including rapid conjugation in response to temperature and other stresses, indicating that it retains full functionality (Miller et al., 2010, 2013). A stringent three-step purification protocol based on the 6His tag and anti-SUMO1 antibodies was then employed to isolate SUMO1/SUMO2 conjugates, with the target, and possibly the modified lysine(s), subsequently identified by tandem mass spectrometry (MS) (Miller et al., 2010; Rytz et al., 2016).
As a first test of this strategy, we examined the SUMOylation patterns before and after a brief heat stress in mutants attenuating the ligases SIZ1 and MMS21, which have been linked to stress protection (Miura et al., 2005, 2007b; Catala et al., 2007; Park et al., 2011) and DNA endoreduplication/repair (Huang et al., 2009b; Ishida et al., 2009), respectively. Both ligases contain the essential SIZ/PIAS-REALLY INTERSTING NEW GENE (SP-RING) domain that docks with the SUMO-E2 intermediate (Bernier-Villamor et al., 2002). Whereas MMS21 is devoid of other recognizable features, SIZ1 includes signature Scaffold Attachment Factor-A/B/Acinus-PIAS (SAP), plant homeodomain (PHD), and proline-isoleucine-isoleucine-threonine (PIIT) motifs, which are followed by a pair of SIMs (Figure 1A). Only two substrates for MMS21 have been described thus far: DPa1 and BRAHMA (Liu et al., 2016; Zhang et al., 2017). By contrast, SIZ1 drives much of stress-induced SUMOylation by SUMO1 and SUMO2 (Miura et al., 2005; Saracco et al., 2007) and has been connected genetically and/or biochemically to the modification of a number of Arabidopsis proteins, including PHR1, GLOBAL TRANSCRIPTION FACTOR GROUP E3 (GTE3), HSFA2, MYB30, CMT3, FLD, SNF-RELATED KINASE1, CONSTITUTIVE PHOTOMORPHOGENESIS1, and NIA1/2 (Miura et al., 2005; Garcia-Dominguez et al., 2008; Jin et al., 2008; Cohen-Peer et al., 2010; Park et al., 2011; Zheng et al., 2012; Kim et al., 2015; Crozet et al., 2016; Lin et al., 2016).
Figure 1.
Genetic and Phenotypic Description of the siz1 and mms21 Mutants.
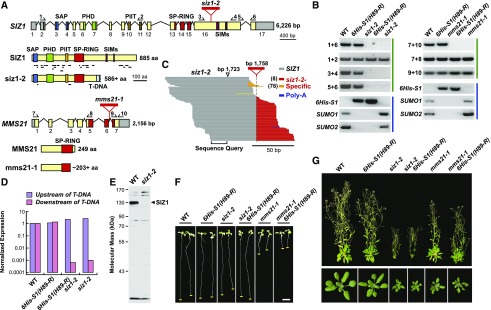
(A) Organization of the SIZ1 and MMS21 genes and proteins. The SAP, PHD, PIIT, SP-RING, and SIM domains are highlighted in blue, green, orange, red, and purple, respectively. Untranslated regions and introns are shown as gray boxes and lines, respectively. Exons are numbered. The red triangles show the positions of the T-DNA insertions. The lines underneath the wild-type and mutant SIZ1 proteins locate the peptides identified during our MS analysis of SUMO1/SUMO2 conjugates. The arrows locate the primers used for RT- PCR in (B). The amino acid (aa) sequence lengths of the siz1-2 and mms21-1 polypeptides that match their wild-type counterparts are shown.
(B) PCR analysis of plants containing wild-type and mutant versions of SIZ1 and MMS21. Panels identified by the green lines represent RT-PCR of the siz1-2 and mms21-1 transcripts. Panels identified by the blue lines represent genomic PCR analyses demonstrating the presence of the 6His-SUMO1(H89-R) transgene and absence of an intact SUMO1 and SUMO2 genes in the respective genotypes.
(C) Alignment of 111 transcripts from siz1-2 plants generated by RNA-seq around the predicted T-DNA insertion site. SIZ1 and T-DNA-related sequences are colored in gray and red/orange, respectively. A poly(A) tract is indicated in blue. The most common junction between the SIZ1 and T-DNA sequences identified the T-DNA insertion site at 1758 bp from the ATG translation start codon (red) with a less prevalent junction upstream (orange). The previously reported insertion site at 1723 bp is also shown (Miura et al., 2005).
(D) Quantification by RT-PCR of SIZ1 transcript levels in wild-type and siz1-2 plants show in (C), using primers that probed the SIZ1 locus either upstream or downstream of the T-DNA insertion. The values were normalized to those for ACT2 and represented as a ratio to the wild type.
(E) Immunoblot detection of SIZ1 protein in 8-d-old unstressed wild-type and siz1-2 seedlings, using anti-SIZ1 antibodies. The arrowhead locates the predicted full-length SIZ1 protein.
(F) and (G) Representative 8-d-old wild-type, siz1-2, and mms21-1 plants with or without the SUMO conjugate purification background [6His-S1(H89-R) sumo1-1 sumo2-1].
(F) Phenotype of young seedlings. Root tips are highlighted by the yellow line. Bar = 0.5 cm.
(G) Plants grown for 20 d (bottom) and 40 d (top) in a long-day photoperiod.
Using improved MS instrumentation, we increased the collection of proteins modified by SUMO1/SUMO2 in Arabidopsis to over a thousand, along with the identification of SUMO1/SUMO2 attachment sites for a subset. Although no targets could be definitively assigned by label-free quantification to MMS21, at least 105 targets were assigned with high confidence to SIZ1, especially after heat stress. Most of these SIZ1-dependent substrates reside in the nucleus and include well-known transcription factors, coactivators/repressors, and chromatin modifiers, as well as many proteins involved in abiotic and biotic stress defense. SIZ1 itself is a prominent SUMO target, but analysis of a SIZ1 mutant immune to SUMOylation suggests that this addition is not physiologically relevant and likely reflects off-target transfer by the conjugation machinery. This deep catalog of SIZ1-dependent SUMO1/SUMO2 targets now provides a framework to better understand how these isoforms and the SIZ1 ligase contribute to plant stress protection.
RESULTS
Development of the siz1-2 and mms21-1 SUMO Conjugate Purification Lines
As a first step toward defining the substrates of SIZ1 and MMS21, we introgressed the 6His-SUMO1(H89-R) sumo1-1 sumo2-1 purification background into the previously described siz1-2 and mms21-1 alleles generated by T-DNA insertional mutagenesis. The T-DNA in siz1-2 was predicted to interrupt the 16th exon at nucleotide 1723 downstream of the ATG start codon (Miura et al., 2005). If transcribed, the resulting mRNA would encode a SIZ1 polypeptide that includes the SAP, PHD, PIIT, and SP-RING domains required for full SIZ1 activity in vitro and in vivo (Garcia-Dominguez et al., 2008; Cheong et al., 2009) but would be missing a substantial portion of the C-terminal half that includes the pair of predicted SIMs (Figure 1A).
RT-PCR analysis of homozygous siz1-2 seedling amplified SIZ1 transcripts 5′ to the insertion site but none 3′, thus roughly supporting the predicted position of the T-DNA (Figure 1B). However, fine mapping by RNA-seq revealed that the siz1-2 insertion site is actually 35 bp downstream at nucleotide 1758, which often generated transcripts encoding the first 586 residues of the SIZ1 polypeptide, followed in-frame by sequence derived from the T-DNA (Figure 1C). The most common siz1-2 mRNAs contained at least 12 additional codons, suggesting that a sizable non-SIZ1 segment follows the truncated siz1-2 polypeptide. As quantified by real-time PCR, these 5′ siz1-2 transcripts accumulate to levels comparable to the full-length SIZ1 transcript in the wild type, suggesting that a partially functional SIZ1 could be synthesized (Figure 1D).
Despite this, attempts to detect the siz1-2 protein by immunoblot analysis failed. Whereas full-length SIZ1 polypeptide was easily detected in wild-type seedling extracts with anti-SIZ1 antibodies, no smaller fragment(s) with an expected mass of ∼67 kD could be assigned to the siz1-2 truncation in mutant seedlings (Figure 1E). However, we detected several siz1-2 peptides during our MS analysis of SUMOylated proteins from the mutant, indicating that the truncated protein does accumulate, albeit at very low levels (Figure 1A). Taken together, the siz1-2 allele should be classified as an attenuated mutant (and not a null) whose protein product might still bind the SUMO-E2 donor and possibly its substrates and/or direct nonspecific SUMOylation but is present at substantially reduced levels.
The T-DNA sequence in the mms21-1 allele (also called hpy2-2; Ishida et al., 2009) is located within the 6th intron that separates the codons for the SP-RING domain (Huang et al., 2009b) (Figure 1A). RT-PCR analysis of homozygous plants found transcripts both upstream and downstream of the T-DNA insertion site but failed to detect transcripts encompassing the full MMS21 coding sequence (Figure 1B). As the resulting polypeptide would be missing part of the SP-RING domain essential for SUMO-E2 binding and subsequent transfer (Cheong et al., 2009; Yunus and Lima, 2009), we considered it likely that mms21-1 is a functionally null allele.
Based on the widely separated chromosomal positions of SUMO1, SUMO2, and MMS21, we predicted that creating the homozygous mms21-1 6His-SUMO1(H89-R) sumo1-1 sumo2-1 line would be straightforward; indeed, generating the quadruple homozygous mutant fit Mendelian segregation ratios in self crosses. [At present, we do not know the insertion position of the 6His-SUMO1(H89-R) transgene.] However, creating similar purification lines harboring siz1-2 was expected to be more difficult given that the SIZ1 and SUMO2 loci are physically linked, being only 1.9 Mb away from each other on chromosome 5. Here, our screen of over 90 offspring from a quadruple heterozygous line identified nine individuals that recombined between the siz1-2 and sumo2-1 alleles, one of which was homozygous for sumo1-1 and sumo2-1, heterozygous for siz1-2, and contained the 6His-SUMO1(H89-R) transgene. Subsequent selfing of this individual generated a line that was homozygous at all four positions [siz1-2, 6His-SUMO1(H89-R), sumo1-1, and sumo2-1], which was confirmed by genomic PCR of its progeny (Figure 1B).
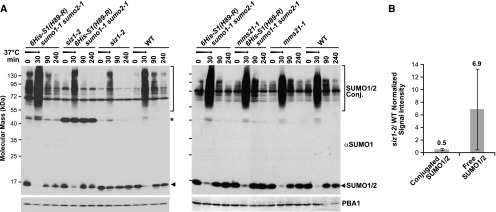
As shown in Figures 1F and 1G, the introgressed lines retained the phenotypes of the siz1-2 and mms21-1 parents, which included smaller leaves and dwarfed stature for siz1-2 plants, and short roots, elongated leaves, and fasciated stems for mms21-1 plants (Miura et al., 2005; Saracco et al., 2007; Huang et al., 2009b; Ishida et al., 2009). When subjected to heat stress (30 min at 37°C), wild-type Arabidopsis seedlings rapidly accumulate high molecular mass SUMO1/SUMO2 conjugates (>50 kD) with a commensurate depletion of the free SUMO1/SUMO2 pool, a response that is reversed upon return to the normal 22°C growth temperature (Figure 2A; Kurepa et al., 2003; Saracco et al., 2007). This accumulation pattern was preserved in mms21-1 seedlings but noticeably dampened in the siz1-2 seedlings [with or without 6His-SUMO1(H89-R) sumo1-1 sumo2-1], as previously reported (Saracco et al., 2007). Quantitative immunoblotting measured a 2-fold decrease in SUMO conjugate levels, concomitant with a 7-fold retention of the free SUMO pool, in siz1-2 seedlings versus wild type after the heat stress (Figure 2B).
Figure 2.
SUMOylation Profile of Wild-Type, siz1-2, and mms21-1 Plants before and after Heat Stress.
(A) Immunoblot analysis of 8-d-old seedlings heat stressed for 30 min at 37°C (arrow), returned to the normal growth temperature of 22°C, and collected at the indicated times. The germplasm included the siz1-2 and mms21-1 mutations by themselves or combined with the SUMO1/2-conjugate purification background [6His-S1(H89-R) sumo1-1 sumo2-1]. The membrane was probed with either anti-SUMO1 or anti-PBA1 antibodies (loading control). High molecular mass SUMO1 and SUMO2 conjugates and free SUMO1/SUMO2 are indicated by the brackets and arrowheads, respectively. Asterisk identified an unknown species abundant in siz1-2 (6His-S1(H89-R) sumo1-1 sumo2-1] seedlings.
(B) Comparison of SUMO1 and SUMO2 conjugate levels in wild-type versus siz1-2 seedlings by quantitative immunoblotting as in (A). Immunoblot signals generated for SUMO conjugates and free SUMO1/SUMO2 from heat-stressed (30 min at 37°C) seedlings were visualized using IRDye 800CW or IRDye 680RD goat anti-rabbit secondary antibodies and normalized to those for PBA1. Each bar represents the average of three biological replicates of independently grown seedlings (±sd). The average ratio of the wild type versus siz1-2 for conjugated and free SUMO is indicated above each bar.
Purification and MS Analysis of SUMO Conjugates in siz1-2 and mms21-1 Seedlings
Using the purification strategy developed by Miller et al. (2010), we generated SUMO1 conjugate-enriched fractions based on the 6His-SUMO1(H89-R) sumo1-1 sumo2-1 background from 8-d-old wild-type, siz1-2, and mms21-1 seedlings either before or after a 30-min heat stress at 37°C. Both the three-step affinity protocol (Ni-NTA, anti-SUMO1 antibody, and Ni-NTA chromatography) and the addition of strong denaturants to many buffers enabled stringent purification with minimal background, as can be seen by the absence of SUMO1 and its conjugates, and protein contaminants when wild-type seedlings without the 6His-SUMO1(H89-R) transgene were used instead (Supplemental Figure 1).
After trypsinization, the peptide pools were separated by reverse-phase liquid chromatography and sequenced by tandem MS using LTQ Orbitrap Velos and Q-Exactive mass spectrometers in the electrospray ionization (ESI) mode. The MS2 spectra were then searched against the Arabidopsis Col-0 proteome database available in TAIR version 10 (http://www.arabidopsis.org) to identify possible SUMO1 or SUMO2 conjugates, using a ≤1% false discovery rate (FDR) cutoff. Those proteins routinely identified in wild-type plants were then subtracted from the search output as likely contaminants (Supplemental Data Sets 1 and 2). The collective MS analysis involved 29 biological replicates from unstressed or heat-stressed samples for wild-type, 6His-SUMO1(H89-R), siz1-2 6His-SUMO1(H89-R), and mms21-1 6His-SUMO1(H89-R) seedlings, with most supported by two technical replicates (Supplemental Data Set 2). In general, strong overlaps in protein identifications were seen among replicates, especially for the heat stress data sets that had substantially more total peptide spectral matches (PSMs), in line with the greater abundance of conjugates measured immunologically (Figure 2; Supplemental Figure 2). Subsequent label-free quantification of SUMO1/SUMO2 conjugate levels, based on distributed normalized spectral abundance factor (dNSAF) values calculated from PSM counts and adjusted based on protein length and shared peptides using Morpheus Spectral Counter (Zhang et al., 2010; Gemperline et al., 2016), also showed high correlations among both technical and biological replicates (R2 values of 0.97–0.99 and 0.5–0.7, respectively; Supplemental Figures 2A and 2B).
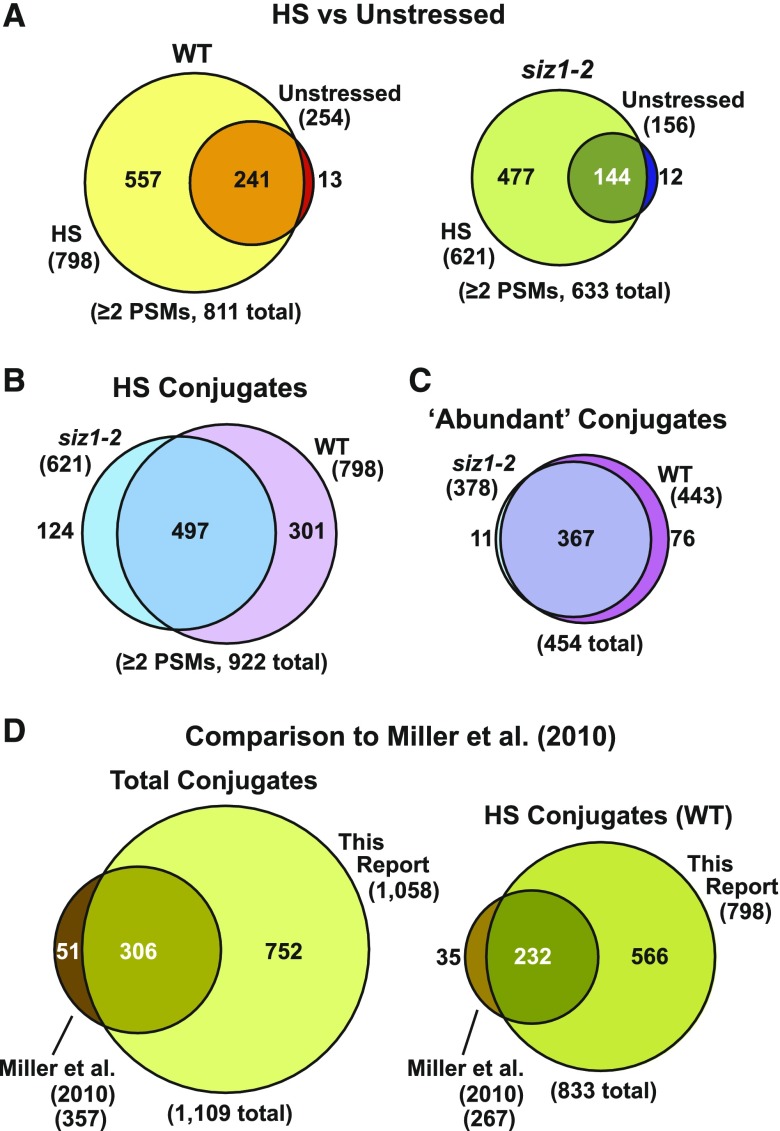
When all the data sets were combined and culled for contaminants, we identified over 1400 potential SUMO1/SUMO2 targets with high statistical support (≤1% FDR). For improved confidence, a collection of 1058 targets detected by at least two PSMs was used for subsequent data set comparisons (Supplemental Data Set 3). This conservative list represents a 3-fold increase in SUMO substrates over that previously determined by Miller et al. (2010) and is likely derived from the use of more advanced Orbitrap mass spectrometers in this study. Remarkably, over 85% of the 357 previously known SUMO1/SUMO2 targets were identified here, thus providing confidence in the methodology (Figure 3D).
Figure 3.
Venn Diagrams Showing the Distribution of SUMOylated Proteins Purified from Wild-Type and siz1-2 Seedlings before and after Heat Stress.
Eight-day-old green seedlings were either kept at 22°C or heat stressed for 30 min at 37°C before tissue collection and SUMO1/SUMO2 conjugate purification.
(A) Overlaps of all SUMO1/SUMO2 conjugates detected in the wild-type or siz1-2 background exposed to heat stress (HS) versus control conditions (unstressed).
(B) Comparison of the total collection of SUMO1/SUMO2 conjugates in wild-type and siz1-2 seedlings after heat stress.
(C) Comparison of the abundant SUMO1/SUMO2 conjugates in wild-type or siz1-2 seedlings after heat stress. Abundant conjugates refer to those detected in three or more biological replicates in either wild-type or siz1-2 seedlings.
(D) Comparisons of the SUMO1/SUMO2 conjugates identified in this study with those previously described by Miller et al. (2010). The left diagram includes all SUMO1/SUMO2 conjugates detected in unstressed samples as well as samples exposed to heat and hydrogen peroxide stress. The right panel includes SUMO1/SUMO2 conjugates that were detected only in heat-stressed samples.
Identification of SIZ1 SUMOylation Targets
As expected based on the immunoblot detection of SUMO1 and SUMO2 conjugates (see Figure 2), we detected only a small pool of adducts in unstressed Arabidopsis seedlings, but their numbers dramatically increased upon heat stress. This rise was caused by the appearance of additional substrates, many of which were at low abundance before the stress based on PSMs and then rapidly rose above our detection threshold upon heat treatment. For example, whereas only 254 and 156 substrates were independently detected in wild-type and siz1-2 seedlings grown at 22°C, respectively, these numbers rose to 798 and 621 in seedlings exposed to the 30-min heat stress at 37°C (Figure 3A). Direct comparisons of the SUMOylomes from wild-type and siz1-2 seedlings after heat stress revealed a substantial loss of conjugates in the siz1-2 background, supporting the role of SIZ1 in modifying a large set of proteins. When considering all SUMO1/SUMO2 conjugates found by two or more PSMs in heat-stressed samples, 301 proteins were absent in the mutant compared with 124 added from a total list of 922 conjugates (Figure 3B). Moreover, when comparing just the 454 conjugates considered to be “abundant” based on their detection in at least three of the five data sets for wild type or siz1-2, 76 conjugates were missing from the siz1-2 seedlings with just 11 added (Figure 3C).
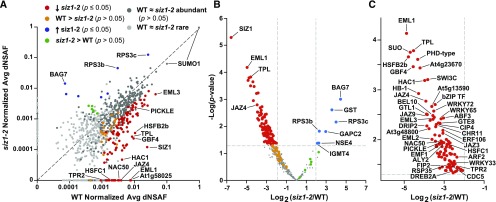
To better evaluate the changes in SUMO1/SUMO2 conjugate patterns in siz1-2 versus the wild type, we quantified the abundance of individual substrates using dNSAF (Zhang et al., 2010; Gemperline et al., 2016). To control for sample-to-sample variations, the dNSAF values in each data set were normalized to those obtained for all peptides derived from SUMO1, based on our observations that the total SUMO1/SUMO2 pool remains unchanged in abundance during this short heat stress, and is equally purified whether in its free or conjugated state (Miller et al., 2010, 2013). When the normalized dNSAF values in the wild type versus siz1-2 plants were plotted for individual SUMO1/SUMO2 targets, we found large deviations from a 1:1 ratio, with many proteins underrepresented or absent in the siz1-2 background both before and after the heat stress (Figure 4A; Supplemental Figures 2 to 4).
Figure 4.
Changes in the SUMO1/SUMO2 Conjugate Accumulation Patterns during Heat Stress in siz1-2 Versus Wild-Type Seedlings.
SUMO1/SUMO2 conjugates detected with at least two PSMs per biological replicate of independently grown seedlings were quantified based on their dNSAF values, which were then normalized based the dNSAF values for SUMO1.
(A) Average normalized dNSAF values of 922 SUMOylated proteins in heat-stressed siz1-2 versus wild-type seedlings (see Figure 3B). Each data point represents the average of five biological replicates of independently grown seedlings. Light-gray points are conjugates considered to be “rare” by their detection in less than three biological replicates in both backgrounds (siz1-2 and/or the wild type). Dark-gray points are conjugates considered to be “abundant” by their detection in three or more biological replicates in either background (siz1-2 and/or the wild type). Proteins with a significant decrease or increase in SUMOylation in the siz1-2 mutant compared with the wild type (P value ≤ 0.05) are highlighted in red and blue, respectively. SUMO targets identified in all wild-type biological replicates and never or only once in the siz1-2 mutant (the wild type > siz1-2), but were above the significance threshold of P value > 0.05, are in orange. Proteins detected in all siz1-2 biological replicates and never or only once in the wild type (siz1-2 > the wild type), but were above the significance threshold (P value > 0.05), are in green. The dashed line represents the theoretical situation where conjugate abundance in the wild type and siz1-2 is equal. Note that two dNSAF values are assigned to SUMO1 by Morpheus Spectral Counter.
(B) Volcano plot of the P value for individual SUMO1/SUMO2 conjugates versus the log2 fold change in wild-type versus siz1-2 seedlings. Missing values were imputed for each biological replicate. The color scheme is the same as in (A). The horizontal dashed line highlights a P value = 0.05. The vertical dashed lines highlight a 4-fold increase or decrease.
(C) Expanded view of (B) highlighting the proteins with a significant reduction of SUMOylation in the siz1-2 mutant. Notable proteins are indicated in (A) to (C).
To further analyze the data, these normalized dNSAF values were processed by the linear models for microarray data (LIMMA) statistical algorithm, which calculated moderate P values for each target (Ritchie et al., 2015). To limit the extent of imputation for substrates with null dNSAF values, we focused only on the 454 SUMO1/SUMO2 substrates considered to be abundant. When illustrated by a volcano plot, 112 proteins were found to have a significant change in SUMOylation in the siz1-2 mutant compared with the wild type (P value ≤0.05) (Figures 4B and 4C). The SUMOylated forms of 18 proteins were not detected and 87 targets showed significantly decreased SUMOylation, while seven proteins had increased SUMOylation in siz1-2. Of note is a second collection of SUMO1/SUMO2 targets (29 total) that were almost always detected in one genotype but consistently undetected among replicates for the other genotype, but remained below the significance threshold (i.e., P values > 0.05: seen on the x and y axis in Figure 4A), and a third collection whose significance did not achieve the P value < 0.05 cutoff (Figure 4B); both might represent additional SIZ1-influenced targets whose scoring was challenged by low abundance and/or poor MS detection. Further comparisons revealed that many of the 105 proteins identified as being significantly less SUMOylated in the siz1-2 background after heat stress were already less prevalent or absent in the data sets before the stress (Supplemental Figures 3 and 4), implying that these targets are modified by SIZ1 under both physiological conditions. Heat stress exacerbated this trend and added more targets to the list.
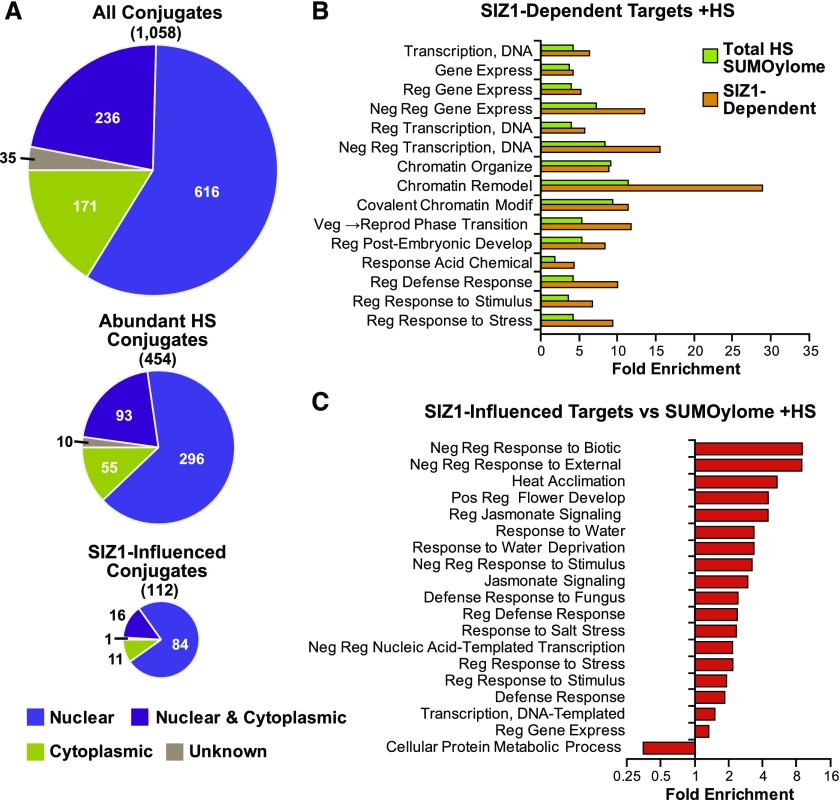
SIZ1-Dependent Conjugates Are Enriched in Stress Regulators
As observed previously in more limited data sets (Miller et al., 2010, 2013), our total Arabidopsis SUMOylome (1058 proteins) was substantially enriched in proteins with predicted or known nuclear locations (80%) based on the Gene Ontology (GO) database (Figure 5A). This nuclear preference was even larger for the 105 SIZ1-dependent targets (89%), which was consistent with the nuclear localization of this ligase (Miura et al., 2005). The remaining targets were considered to be cytoplasmic or undefined. When analyzed for GO functional enrichment using the PANTHER database (Thomas et al., 2003), the whole SUMOylome had a significant focus not only on transcription, DNA repair, chromatin organization, and epigenetic regulatory processes, but also on RNA-related processes, including regulation of RNA splicing, RNA-directed DNA methylation and gene silencing, and mRNA processing (Miller et al., 2010, 2013; Supplemental Figure 5A). Protein interaction analysis of the 454 abundant SUMO1/SUMO2 targets using Cytoscape generated a tightly clustered interaction network with few distinct hubs. The SIZ1-dependent targets of SUMO1/SUMO2 were dispersed throughout the network, suggesting that SIZ1 exerts widespread control (Supplemental Figure 5B).
Figure 5.
Localization and Functional Enrichments of SUMOylated Proteins from Wild-Type and siz1-2 Seedlings.
(A) Pie charts illustrating the known or predicted localization of individual SUMO1/SUMO2 conjugates. Top: All 1058 SUMO1/SUMO2 conjugates detected in wild-type, siz1-2, and mms21-1 plants ± heat stress (HS) at 37°C for 30 min. Middle: The 454 abundant SUMO1/SUMO2 conjugates detection in three or more biological replicates in the siz1-2 and/or wild-type backgrounds upon heat stress. Bottom: The 112 SUMO1/SUMO2 conjugates that were significantly increased/decreased in siz1-2 versus wild-type plants after HS.
(B) GO functional enrichment of all 922 SUMO1/SUMO2 conjugates that appeared during HS in wild-type and siz1-2 seedlings (green) and the 112 SUMO1/SUMO2 conjugates that accumulated during HS and appear to be SIZ1-influenced (orange).
(C) GO functional enrichment of the SIZ1-dependent SUMO1/SUMO2 targets compared with the total collection of 922 SUMO1/SUMO2 targets identified in the wild type and siz1-2 after HS.
Strikingly, a different emphasis of GO categories was seen for the SIZ1-dependent SUMO1/SUMO2 targets, with more enrichment of activities associated with transcription, negative regulation of transcription and chromatin remodeling, and less enrichment of RNA-related processes (Figure 5B). Such a redistribution of GO categories implies that SIZ1 controls only a subset of events directed by SUMO1/SUMO2. Further comparisons showed that SIZ1-dependent targets are specifically focused on abiotic and biotic stress responses, such as heat acclimation, response to drought, hormone signaling, and defense responses (Figure 5C). Taken together, we hypothesize that increased SUMOylation by SIZ1 during stress helps regulate the expression of factors related to defense and hormone signaling by repressing/activating transcription specifically and by altering chromatin access more generally.
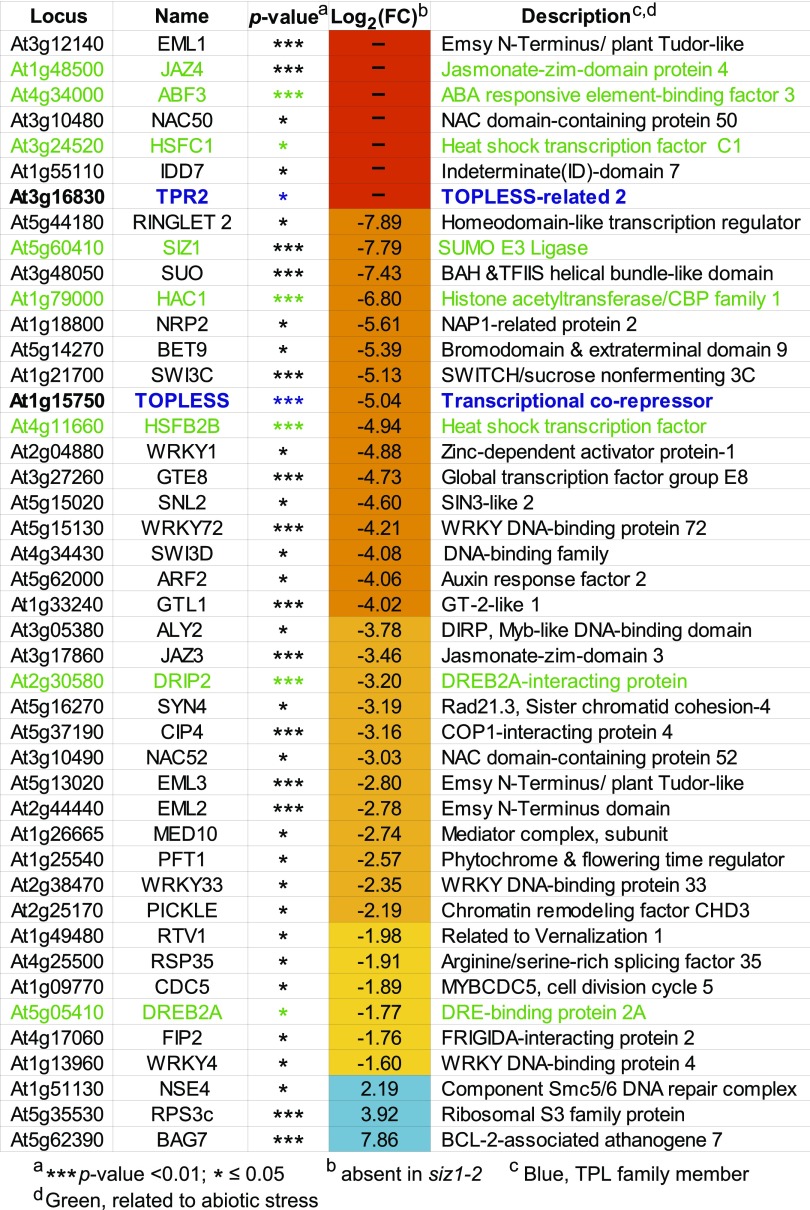
In agreement with this scenario, the list of 105 proteins whose SUMOylation was aided by SIZ1 are enriched in stress-related transcriptional regulators. Included are TPL and TPL-RELATED2 (TPR2), which reside within a more stress-focused subclade of the Groucho/Tup-type corepressor family (Liu and Karmarkar, 2008), along with several of their interacting transcription factors, including AUXIN-RESPONSE FACTOR2 (ARF2), NAM/ATAF1,2/CUC2 52 (NAC52), EMBRYONIC FLOWER1 (EMF1), NAC50, and HSFB2b (Causier et al., 2012) (Figures 4 and 6). TPR1, TPR3, and TPR4 also appeared to be less SUMOylated in the siz1-2 background, but not to levels considered significant by LIMMA (P value > 0.05). However, not all members of the Groucho-Tup corepressor family were affected by SIZ1, as there was no drop in the SUMOylation status of two members of a second, more development-focused subclade containing LEUNIG and LEUNIG HOMOLOG and their interactor SEUSS, in the siz1-2 plants, suggesting that the interactions of SIZ1 with TPL and TPR2 are specific.
Figure 6.
List of Notable Arabidopsis SUMOylation Targets Whose Modification Is Influenced by SIZ1.
TAIR locus identifier, target name, P value, log2 fold change in siz1-2 versus wild-type plants [log2(FC)], and a brief description are listed. The SUMO1/SUMO2 targets absent in the siz1-2 mutant are indicated by a dash. Targets highlighted in blue or green are members of the TPL family or related to abiotic stress responses, respectively.
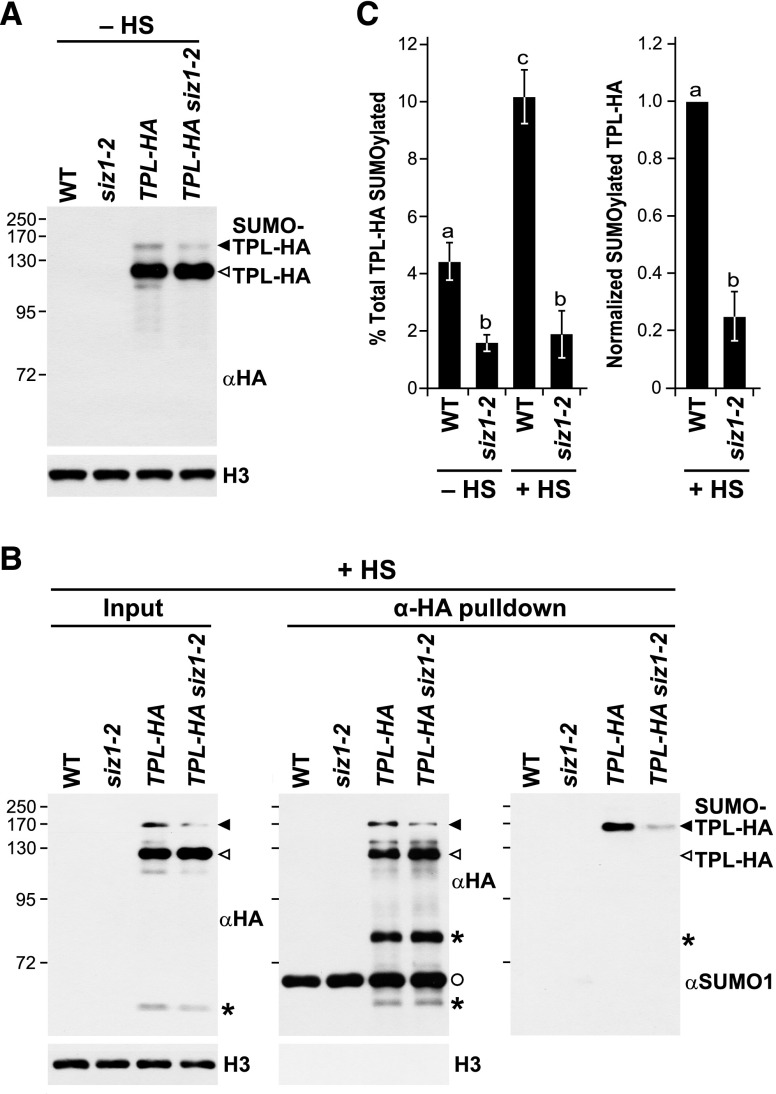
To further validate the connection between SIZ1 and TPL, we directly assessed the SUMOylation status of a HA-tagged version of TPL in siz1-2 plants. As shown in Figure 7A, two forms of TPL could be detected immunologically in unstressed and stressed TPL-HA plants, one at the expected mass for unmodified TPL (125 kD) and another at ∼140 kD that likely represents form(s) modified by SUMO1 and/or SUMO2 (Miller et al., 2010). Subsequent immunoprecipitations with anti-HA antibodies enriched for both species, with the higher mass version(s) also recognized by anti-SUMO1 antibodies (Figure 7B). Consistent with SIZ1 helping generate these SUMOylated species, their abundances as detected immunologically were reduced substantially in TPL-HA siz1-2 plants. Quantification of the immunoblots revealed a 3-fold difference in conjugated TPL in unstressed wild-type versus siz1-2 plants, which increased further to a 6-fold difference after the 30-min heat stress (Figure 7C).
Figure 7.
Inactivation of SIZ1 Suppresses the SUMOylation of TPL.
(A) Detection of SUMOylated TPL in total protein extracts from unstressed (−HS) 8-d-old wild-type, siz1-2, TPL-HA, and TPL-HA siz1-2 seedlings. Immunoblots were performed with anti-HA antibodies using anti-histone H3 antibodies to confirm near equal protein loading. Unmodified and SUMO1/2-modified forms of TPL-HA in (A) and (B) are indicated by the open and closed arrowheads, respectively.
(B) Immunoprecipitation of TPL protein with anti-HA antibodies and subsequent immunoblotting with anti-HA and anti-SUMO1 antibodies from heat stressed (+HS) seedlings. Total protein extracts before (input) and after immunoprecipitation (eluate) are compared. Eight-day-old wild-type, siz1-2, TPL-HA, and TPL-HA siz1-2 seedlings were exposed to a 30 min 37°C heat stress. The asterisks identify an unknown species that could be a breakdown product of TPL, while the circle identifies the eluted anti-HA antibody. Immunoblotting with anti-H3 antibodies was used as a loading control for the inputs and as a judge of enrichment for the eluates.
(C) Quantification of SUMOylated TPL-HA levels isolated from seedlings unstressed (−HS) or exposed to a 30-min 37°C heat stress (+HS). Bars show the levels of conjugated TPL relative to either total TPL-HA purified (left) or relative to that in the wild type (right). Each bar represents the mean of three biological replicates of independently grown plants per genotype (±sd). The letters indicate averages that are statistically significantly different from each other (P value < 0.05).
Other SIZ1-influenced transcription factors include HSFB2b and HSFC1 linked to heat stress; ARF2 linked to auxin signaling; ABA BINDING FACTOR3 (ABF3) linked to ABA signaling; JASMONATE-ZIM DOMAIN3 (JAZ3), JAZ4, and JAZ6 linked to jasmonate signaling; EMSY N TERMINUS/PLANT TUDOR-LIKE1 (EML1), EML2, and EML3 linked to plant defense; and the WRKY DNA BINDING PROTEIN1 (WRKY1), WRKY4, WRKY33, and WRKY72 transcription factors, some of which have been connected to stress protection (Figures 4 and 6). Moreover, both the stress-related transcription factor DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A) and the ubiquitin-protein ligase DREB2A-INTERACTING PROTEIN2 (DRIP2) that directs DREB2A turnover (Qin et al., 2008) were prominent SIZ1 targets. Additionally, a number of the SIZ1-dependent targets are known to associate together in multisubunit complexes, suggesting a broad SUMOylation of all factors by SIZ1. Examples include the aforementioned TPL complexes containing JAZ3/JAZ4/JAZ6, ARF2, and NAC regulators (Causier et al., 2012), components of the stress-sensitive Mediator (MED) transcriptional regulatory complex (MED10 and PHYTOCHROME AND FLOWERING TIME1; Bäckström et al., 2007), and the SWI-SNF chromatin remodeling complex (SWI3C, SWI3D, CHROMATIN REMODELING FACTOR11 (CHR11), CHR17, and PICKLE; Kwon and Wagner, 2007) (Figures 4 and 6).
We also detected seven SUMO targets whose SUMOylation levels were significantly higher in the siz1-2 background. Interestingly, most were not transcription factors or chromatin regulators, but instead included an O-methyltransferase (IGMT4/OMT; At1g21130), a GST (At4g19880), two isoforms of the small ribosomal subunit protein S3 (RPS3b and RPS3c), and glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2) (Figures 4A, 4B, and 6). Intriguingly, the cochaperone BAG7, which participates in the unfolded protein response and was recently shown to be SUMOylated during heat stress (Williams et al., 2010; Li et al., 2017) was also in this group. Its relative abundance in the siz1-2 SUMOylome increased over 200-fold compared with the wild type after 30 min at 37°C to easily become the most abundant SUMO1/SUMO2 conjugate in siz1-2 seedlings subjected to heat stress (Figures 4 and 6). The SUMOylation status of NON-SMC ELEMENT4 (NSE4) also increased in the siz1-2 plants. NSE4 is a part of the SMC5-SMC6 DNA repair complex that coincidently includes MMS21 (Xu et al., 2013), suggesting that its SUMOylation could be driven by this ligase. Notably, we also detected MMS21 in our siz1-2 SUMOylome data sets after heat stress, but not in those from the wild type or mms21-1; this species could reflect auto-SUMOylation of the ligase (see below).
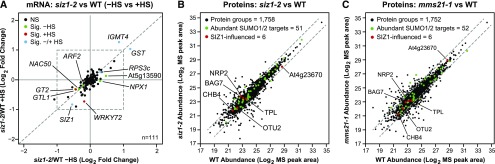
Changes in SUMOylation Levels Were Not Caused by Changes in Target Protein Abundance
It was remotely possible that the strong decreases (or increases) in SUMOylation seen here for individual SUMO1/SUMO2 targets actually reflected substantial changes in the proteome of siz1-2 seedlings, and not specific changes in SUMOylation, given the phenotypic differences between siz1-2 and the wild type (Miura et al., 2005; Saracco et al., 2007; Figures 1F and 1G). To test this possibility, we analyzed at a global level both the mRNA and protein abundances of the 112 SIZ1-influenced targets. Transcriptome comparisons of siz1-2 and wild-type seedlings by RNA-seq indicated that neither the genotype (and resulting phenotype) nor this short heat stress (30 min) had a substantial influence on the abundance of the corresponding mRNA for these targets (Figure 8A; Supplemental Data Set 4). In fact, only 11 of the 112 mRNAs displayed even a modest significant change in abundance (P value <0.05), with only three showing a >2-fold change before or after the brief heat stress. As expected, the SIZ1 mRNA was in the former group consistent with the effects of the siz1-2 allele on its transcript levels (see Figure 1D).
Figure 8.
The siz1-2 and mms21-1 Mutations Do Not Substantially Alter the mRNA and Protein Abundances of SUMO1/SUMO2 Targets Selectively Modified by SIZ1.
(A) Scatterplot comparing the transcript abundance for 112 SIZ1-influenced SUMO1/SUMO2 targets both before and 30 min after a 37°C heat stress (HS). mRNA levels for the 112 targets significantly influenced by SIZ1 (see Figure 4) were determined by RNA-seq of total seedling RNA. The log2 fold change of siz1-2 versus the wild type was compared without or with HS. The dashed box delineates a twofold change in mRNA abundance. Only three transcripts achieve a >2-fold difference in expression, whereas only nine others had slightly significant changes in expression (±HS) below this 2-fold change.
(B) Total proteome analysis comparing unstressed siz1-2 (right) and mms21 seedlings (left) with wild-type seedlings. Total protein from 8-d-old seedlings was trypsinized and subjected to tandem LC-MS. Protein abundances as determined by MS1 peak areas were plotted for the mutants versus the wild type. Protein groups (including isoforms) detected from the complete Arabidopsis proteome (1758 for siz1-2 versus the wild type and 1752 for mms21 versus the wild type), members of the abundant SUMO1/SUMO2 target list (454 total), and members of the SIZ1-influenced target list (112 total) are indicated by the black, green, and red dots, respectively. Dashed lines represent 2-fold changes up or down in protein abundance. Notable SUMO1/SUMO2 targets in (A) and (B) that are influenced by SIZ1 are indicated (see Figure 6).
We next compared the total proteomes of unstressed wild-type and siz1-2 seedlings by tandem MS using the precursor ion intensity of the MS1 scans for quantification. Altogether, 1758 Arabidopsis proteins could be reproducibly identified and quantified in both samples by our liquid chromatography-mass spectrometry (LC-MS) regime analyzed in triplicate, 51 of which were described here as targets of SUMO1 and six as SIZ1-influenced (Supplemental Data Set 5). Comparisons of the total proteomes indicated that the siz1-2 mutation again had little influence, with only 144 individual proteins (8% of the total) showing a 2-fold or greater change in abundance in these 8-d-old seedlings (Figure 8B). This percentage was smaller than those obtained when comparing biological replicates to each other (18–26%), suggesting that much of the variation is related to reproducibility in the MS profiles among replicates. Likewise, levels of the SUMO1/SUMO2 targets were not discernably altered. In fact, both the abundant targets and the SIZ1-influenced targets each showed a <2-fold deviation from the wild type (Figure 8B). Even though only a small portion of the SUMOylated proteins were assayed here (likely due to the low levels of most targets), these proteomic results, combined with the transcriptomic data, strongly suggest that changes in SUMOylation status in the siz1-2 background were not caused by changes in protein abundance.
Attempt to Identify MMS21 SUMOylation Targets
Using the same strategy that cataloged SIZ1-influenced targets, we attempted to identify those influenced by MMS21 by comparing the SUMOylomes of wild-type and mms21-1 seedlings. Both normal and heat stress conditions were analyzed, with the hopes that the stress might boost the MMS21 target(s) to detectable levels, even though the kinetics of heat-induced SUMOylation were unaffected by the mms21-1 mutation (Figure 2A). When we compared the data sets (Supplemental Data Sets 2 and 3), the SUMOylation status of few proteins appeared to be significantly altered in the mms21-1 seedlings with or without heat stress (Supplemental Figures 3, 4, and 6). When considering all conjugates found in heat-stressed wild-type and mms21-1 samples (with ≥2 PSMs), 205 proteins were absent and 126 conjugates were unique in the mms21-1 mutant from a total list of 924 conjugates. However, when compared by dNSAF values, few proteins deviated from the 1:1 ratio in mms21-1 versus the wild type (Supplemental Figure 6B). We did identify 19 proteins that were present in the wild-type data sets but absent in the mms21-1 data sets for seedlings grown under nonstressed conditions (e.g., SKI-INTERACTING PROTEIN, DOF ZINC FINGER PROTEIN2, JAZ12, and HEAT SHOCK COGNATE70; Supplemental Figure 3B). Unfortunately, using the criteria that MMS21-influenced targets should be SUMOylated in wild-type and siz1-2 seedlings, but not in both unstressed and heat-stressed mms21-1 seedlings, none were obvious MMS21 targets. One noteworthy possibility was NSE4 (see above). It matched most criteria by being present in the data sets from heat-stressed wild-type and siz1-2 seedlings but absent in those from mms21-1 seedlings (Supplemental Figure 6B).
As with the siz1-2 background, we also attempted to determine by tandem MS if the mms21-1 mutation alters either the total proteome or the abundance of SUMO1/SUMO2 targets. In total, 1752 Arabidopsis proteins could be reproducibly compared in our LC-MS profile, 52 of which were shown here to be modified by SUMO1/SUMO2 (Supplemental Data Set 5). Comparisons to the wild type indicated that the mms21-1 mutation also had little effect on the total proteome, with only 240 individual proteins (13.6% of total) showing a 2-fold or more change in abundance with much of this variation again related to technical reproducibility (Figure 8C). Importantly, none of the abundant SUMO1/SUMO2 targets (52 detected) were discernably affected.
Analysis of SUMO Footprints
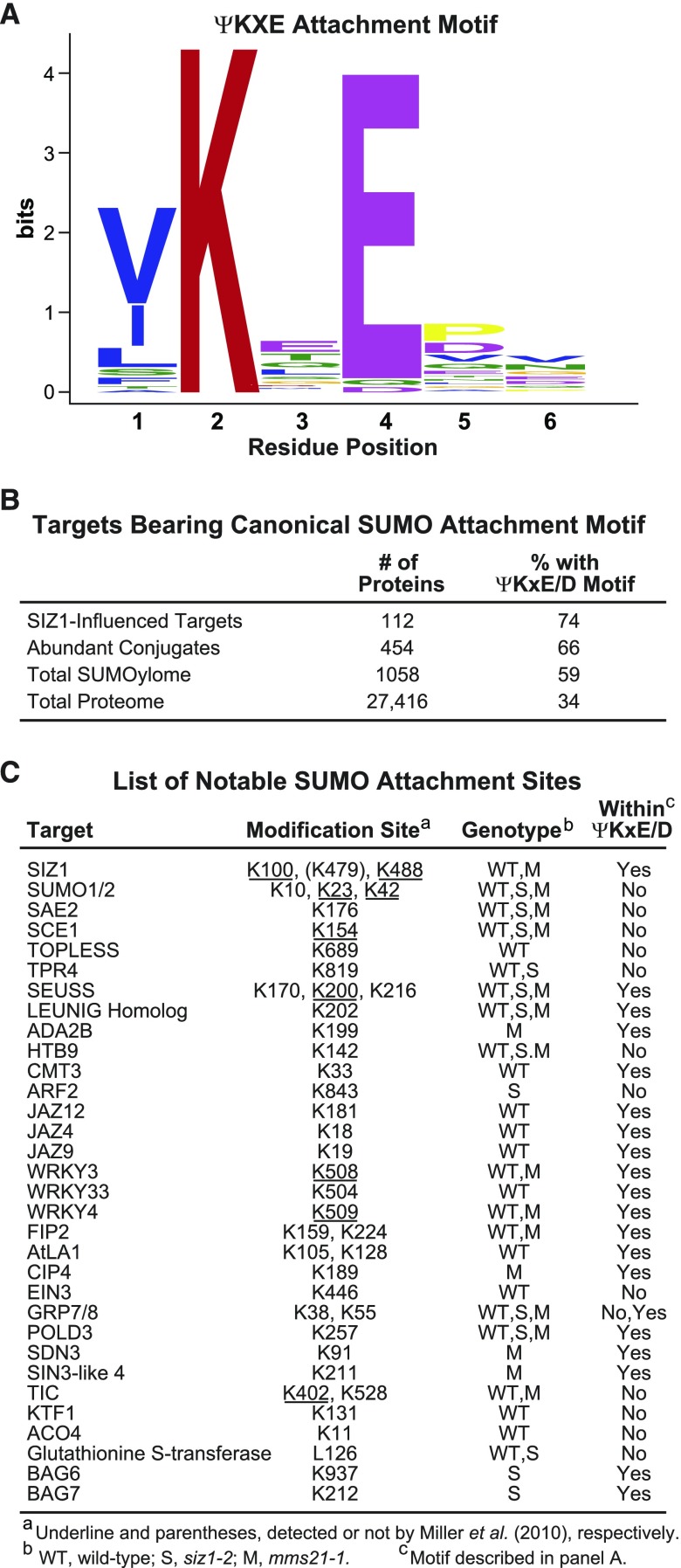
The H89-R substitution in the tagged SUMO1 used here enables detection of SUMOylation sites (“footprints”), which are seen by tandem MS analysis of the trypsinized preparations as a SUMO remnant (QTGG; +326 D) isopeptide linked to the affected lysine in combination with missed trypsin cleavage at that site (Miller et al., 2010; Hendriks and Vertegaal, 2016). From searches of all data sets generated here, we identified 68 SUMO1/SUMO2 modification sites on 53 proteins (Supplemental Data Set 6), with the list encompassing most of the few footprints detected previously (Miller et al., 2010). Motif analysis around the modified lysine using MEME identified a consensus SUMO1/SUMO2 attachment sequence in 44 of the 71 sites (Figure 9A) that strongly matched the ψ-K-x-E/D SUMOylation motif (where ψ represents a hydrophobic amino acid) prevalent in yeast and animal SUMO targets (Rodriguez et al., 2001; Hendriks and Vertegaal, 2016). However, the remaining 24 sites (35%) were unrelated to this motif or among themselves, indicating that noncanonical sites are also common.
Figure 9.
Motif Analysis of SUMO Attachment Sites.
(A) The consensus SUMO1/SUMO2 attachment motif identified by the MEME Suite from the collection of peptides identified through MS analysis to bear a SUMO footprint.
(B) Enrichment of proteins with the canonical ψKxE/D SUMO attachment motif from either SIZ1-influenced SUMOylation targets, the 454 abundant conjugates identified in siz1-2 and/or the wild type during heat stress, the total SUMOylome identified in this study, as well as the complete annotated Arabidopsis proteome. The total number of proteins in each category and the percentage bearing the ψKxE/D motif are indicated.
(C) List of notable SUMO1/SUMO2 attachment sites identified in this study. The sites underlined were detected both here and by Miller et al. (2010). The modification site K479 on SIZ1 (in parenthesis) was identified only by Miller et al. (2010). The genotypes in which the attachment site was identified are shown (WT, wild type; S, siz1-2; M, mms21-1). Match of the SUMO attachment sites with the ψKxE/D motif is indicated.
Interestingly, when we searched the catalogs of SUMO targets generated here using the high probability feature in GPS-SUMO (Zhao et al., 2014), one or more copies of this consensus sequence were detected in 74%, 66%, and 59% of the proteins present in the SIZ1-dependent, abundant, and total SUMOylome lists, respectively, compared with 34% of all proteins in the total Arabidopsis proteome (Figure 9B). For the SIZ1-directed targets, 14 of the 21 mapped attachment sites on 18 targets involved the canonical ψ-K-x-E/D motif, with the remainder having alternative sequences (Supplemental Data Set 6).
SUMO1/SUMO2 attachment sites were mapped for a number of physiologically important Arabidopsis proteins, including TPL and its relative TPR4, several JAZ and WRKY transcription factors, the BAG6/BAG7 cochaperones, and several components of the SUMOylation cascade (SAE2, SCE1, and SIZ1; Figure 9C). Notably, the SUMOylation site identified here for BAG7 at K212 was previously reported to be critical for conferring heat tolerance (Li et al., 2017). We also detected SUMO1 attached to itself via noncanonical linkages, thus confirming the assembly of poly-SUMO chains in plants. These footprints included the previously mapped positions in SUMO1 at K23 and K42 (Miller et al., 2010), as well as at K10, which was predicted previously based on in vitro conjugation assays using Arabidopsis components (Colby et al., 2006). We also searched our SUMOylome data sets for ubiquitylation sites via footprints containing ubiquitin remnants after trypsin cleavage (GG; +114 D). Ubiquitin modifications of SUMO1 at K23 and K42 were detected, as was the presence of polyubiquitins linked internally through K48 linkages, thus providing further evidence that some SUMOylated proteins in Arabidopsis become substrates for ubiquitylation (Miller et al., 2010, 2013).
We note that our MS analysis, like those described previously (Miller et al., 2010, 2013), failed to find any peptides related to either SUMO3 or SUMO5. While their low levels of expression might hinder detection, their absence could indicate that these isoforms have separate sets of targets and ligases and do not assemble into mixed SUMO chains along with SUMO1 or SUMO2.
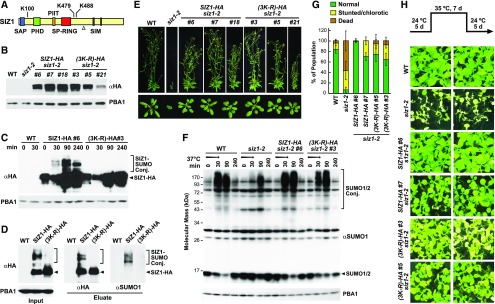
SIZ1 Is SUMOylated
Like Miller et al. (2010), we detected SUMO1 bound to SIZ1 both before and after heat stress. The prior study mapped SUMOylation sites at K100, K479, and K488, while we detected attachment sites only at K100 and K488 (Figures 9C and 10A). This SUMOylation could reflect inadvertent modification of this ligase upon association with the high-energy SUMO-E2 intermediate or might reflect a novel mechanism to control SIZ1 activity. To examine the latter possibility, we attempted to rescue the siz1-2 mutant with a 3xHA-tagged version in which all three modified lysines were substituted for arginines [(3K-R)-HA], thus making it immune to SUMOylation at these positions (Figure 10B). An untagged version of the 3K-R mutant retained its ability to interact with the E2 SCE1 based on yeast two-hybrid assays, implying that the mutant protein is catalytically active (Supplemental Figure 7).
Figure 10.
SUMOylation of SIZ1 at K100, K479, and K488 Does Not Alter SIZ1 Activity nor Its Phenotypic Functions.
(A) Organization of the SIZ1 protein. The SAP, PHD, PIIT, SP-RING, and SIM domains are highlighted in blue, green, orange, red, and purple, respectively. Positions of the modified lysines are indicated. The open triangle marks the termination in siz1-2 protein sequence generated by the T-DNA insertion.
(B) Immunoblot detection of SIZ1 protein in wild-type and siz1-2 seedlings or in a collection of siz1-2 seedlings independently transformed with transgenes encoding HA-tagged SIZ1 or SIZ1 variant in which the lysines at positions K100, K479, and K488 were substituted for arginines [(3K-R)-HA]. The membrane was probed with either anti-HA or anti-PBA1 antibodies (loading control).
(C) Accumulation of SUMOylated forms of SIZ1 during heat stress. Seven-day-old wild-type, SIZ1-HA siz1-2, and (3K-R)-HA siz1-2 seedlings were exposed to heat stress at 37°C for 30 min; total lysates were probed for SIZ1-SUMO1/SUMO2 conjugates by immunoblotting with anti-HA antibodies using anti-PBA1 antibodies as loading control. Unmodified SIZ1-HA and possible SUMO1/SUMO2 conjugates are highlighted by the arrowheads and brackets, respectively.
(D) Detection of SUMO1/2-SIZ1 conjugates by immunoprecipitation. Seven-day-old wild-type, SIZ1-HA siz1-2, and (3K-R)-HA siz1-2 seedlings were exposed to 37°C for 30 min as in (C). SIZ1 protein was isolated by immunoprecipitation with anti-HA antibodies; the eluate was then subjected to immunoblot analysis with anti-HA and anti-SUMO1 antibodies. The left panel shows the levels of HA-tagged SUMO1 before enrichment (Input). Unmodified SIZ1-HA and possible SUMO1 conjugates are highlighted by the arrowheads and brackets, respectively.
(E) Representative wild type, siz1-2, and SIZ1-HA or (K3-R)-HA complemented siz1-2 plants described in (B) were grown for 40 d (top) and 20 d (bottom) in a long-day photoperiod.
(F) Heat stress-induced SUMOylation of 7-d-old wild-type, siz1-2, SIZ1-HA siz1-2, and (3K-R)-HA siz1-2 seedlings. Seedlings were exposed to 37°C for 30 min (arrow) before return to a normal growth temperature of 22°C. Total lysates were probed with either anti-SUMO1 or anti-PBA1 antibodies (loading control). Free SUMO1/SUMO2 is indicated by the arrowhead.
(G) and (H) SUMOylation of SIZ1 at K100, K479, and K488 is not essential for thermotolerance to moderately high temperatures. Diagram of the heat treatment and recovery time course is shown in (H).
(G) Quantification of seedling phenotype after the heat tolerance assay. Each bar represents the mean of four biological replicates (±sd) analyzing at least 25 seedlings each.
(H) Representative wild-type, siz1-2, SIZ1-HA, or (K3-R)-HA complemented siz1-2 seedlings subjected to the temperature treatment. The plants were photographed after the 5-d recovery. Shown are two biological replicates, each consisting of 40 seedlings.
When introduced into siz1-2 plants, the (3K-R)-HA variant lost the ability to be SUMOylated, especially under heat stress. Whereas the accumulation of higher molecular mass forms of SIZ1, potentially representing species modified with SUMO1/SUMO2, were easily seen in SIZ1-HA siz1-2 plants exposed to a 30-min stress at 37 °C, none were evident in (3K-R)-HA siz1-2 plants (Figure 10C). To confirm that these species were indeed SUMOylated forms of SIZ1, we immunoprecipitated SIZ1 from heat-stressed seedlings with anti-HA antibodies and subsequently immunoblotted the samples with anti-SUMO1 antibodies. As shown in Figure 10D, immunoprecipitated SUMO-SIZ1 species of higher mass were readily detected.
To examine the physiological consequences of this block in SUMOylation, we phenotypically examined multiple independent transformations of siz1-2 rescued with SIZ1-HA or (3K-R)-HA. Strikingly, both transgenes fully rescued the siz1-2 phenotype, which restored leaf/rosette size and inflorescence morphology to those seen with the wild type (Figure 10E). Subsequent immunoblot analysis of seedlings exposed to heat stress as well as to stress elicited by ethanol and hydrogen peroxide showed that the (K3-R)-HA variant reestablished the stress-induced profile of SUMOylation (Figure 10F; Supplemental Figure 8). This SUMOylation further confirmed that the (3K-R)-HA protein is catalytically active and discounted a role for auto-SUMOylation in controlling SIZ1 function or Arabidopsis physiology.
Given the importance of SIZ1 to heat tolerance (Kurepa et al., 2003; Yoo et al., 2006; Castro et al., 2012), we compared the sensitivity of siz1-2 and rescued SIZ1-HA and (3K-R)-HA plants in assays that measure thermotolerance to moderately high temperatures (Yeh et al., 2012; Wu et al., 2013). Here, the seedlings were grown for 5 d at 24°C, incubated for 7 d at 35°C and then returned to 24°C for 5 d before seedling viability was measured. Whereas wild-type plants easily survived this treatment based on renewed growth and retained chlorophyll levels, siz1-2 seedlings were highly sensitive based on failed regrowth and/or strong chlorosis by the end of the treatment (Figures 10G and 10H). Complementation with both the SIZ1-HA and (3K-R)-HA transgenes rescued this heat sensitivity, implying that SIZ1 SUMOylation does not alter its activity (Figures 10G and 10H). A slight sensitivity to the prolonged 35°C treatment was seen for the (3K-R)-HA siz1-2 lines, but whether this reflected a slightly dampened SUMOylation activity of the (3K-R)-HA protein or differences in expression levels was not clear.
DISCUSSION
SUMO is emerging as a crucial posttranslational modifier in plants with important roles under normal physiological conditions, during genotoxic stress, and in response to biotic and abiotic challenges (Miura et al., 2007a; Castro et al., 2012; Park and Yun, 2013). Previous studies identified a large suite of conjugates for SUMO1/SUMO2 in whole Arabidopsis seedlings (Miller et al., 2010, 2013), and we have substantially expanded this list here through the use of improved proteomic instrumentation to place it as one of the more pervasive modifications within plant proteomes. Accordingly, the number of high-confidence SUMO targets in this plant (1058 total) now rivals those seen in human cells (Golebiowski et al., 2009; Seifert et al., 2015; Hendriks and Vertegaal, 2016) and other animals such as Caenorhabditis elegans (Kaminsky et al., 2009) and Drosophila melanogaster (Handu et al., 2015) and is larger than the list from the yeast Saccharomyces cerevisiae (Wohlschlegel et al., 2004). Moreover, we expect that the number of SUMO targets will expand further in Arabidopsis as the SUMO3 and SUMO5 isoforms and additional stress conditions, specific tissues, and individual cellular compartments (e.g., the nucleus) are examined in greater depth via proteomics. Like the situation in yeast and animals, the majority of targets reside in the nucleus, with many participating in important functions related to DNA and RNA, including transcription, DNA replication and repair, DNA/nucleosome modification, nucleocytoplasmic transport, chromatin accessibility, epigenetic regulation, and RNA processing, stability, and export. Collectively, the proteomic data implicate SUMO as a key regulator of chromatin maintenance, gene expression, and RNA dynamics.
Here, we attempted to define the catalog of substrates modified by the Arabidopsis SUMO ligases SIZ1 and MMS21. Both the observation that SIZ1 directs much of heat stress-induced modification by SUMO1/SUMO2 (Miura et al., 2005; Saracco et al., 2007), and the expanding list of known SIZ1 targets (e.g., PHR1, GTE3, HSFA2, MYB30, CMT3, and NIA1/NIA2; Miura et al., 2005; Garcia-Dominguez et al., 2008; Cohen-Peer et al., 2010; Park et al., 2011; Zheng et al., 2012; Kim et al., 2015) suggested that this ligase influences a wide array of Arabidopsis proteins, while the limited effect of MMS21 on SUMOylation implied a much smaller subset. These possibilities were confirmed here through MS comparisons of SUMOylated proteins from wild-type, siz1-2, and mms21-1 seedlings. The SUMOylation state of numerous nucleus-enriched proteins was significantly dampened or eliminated in the siz1-2 background, with more such proteins likely to be described as we compare the wild-type and siz1-2 SUMOylomes in greater depth. siz1-2 plants have a heightened sensitivity to prolonged heat stress (Yoo et al., 2006; Figures 10G and 10H), implying that one or more of these SIZ1-influenced targets are crucial for robust thermotolerance.
Importantly, both transcriptomic and proteomic studies are consistent with changes in SUMOylation status underpinning our observations with SIZ1, as opposed to altered accumulation of the parent proteins. In fact, the total proteome in the absence of heat stress was not appreciably altered in the siz1-2 or mms21 backgrounds, indicating that the phenotypic differences seen in young seedlings between the mutants and the wild type are not generated by drastic changes in protein profile and abundance.
In agreement with the role of SIZ1 in stress protection, the catalog of SIZ1-dependent targets included key transcriptional regulators involved in heat, salt, and drought tolerance, and regulation of responses to the hormones, auxin, ABA, and jasmonate, all three of which have roles in stress protection. Notable was the modification of multiple members of the TPL corepressor family that interact with numerous transcription factors connected to stress, including those within the ARF, JAZ, ABF, and NAC families, who themselves are SUMOylated by SIZ1. Also of interest are DREB2A and its ubiquitin ligase DRIP2 that help mediate protection against drought (Qin et al., 2008). siz1-2 plants display a constitutive systemic acquired resistance response that helps promote pathogen defense through elevated production of the stress hormone salicylic acid (Lee et al., 2007). At least some of this activity could be explained by the role of SIZ1 in SUMOylating INDETERMINATE DOMAIN7 and members of the EML and WRKY transcription factor families that are important for biotic stress protection (Zheng et al., 2006; Lai et al., 2008; Tsuchiya and Eulgem, 2011).
The SIZ1-dependent SUMOylation of multiple components of the SWI-SNF chromatin remodeling complex (SWI3C, SWI3D, CHR11, CHR17, and PICKLE) was also seen here. SWI3C and SWI3D are two of the four SWI3 subunits in Arabidopsis that make up part of the core particle, with CHR11, CHR17, and PICKLE interacting with this core to direct the recruitment of the SWI-SNF complex to genes (Kwon and Wagner, 2007; Gentry and Hennig, 2014). Taken together, we speculate that SUMO1/SUMO2 modification of this complex by SIZ1 provides a crucial mechanism for controlling chromatin remodeling and transcriptional regulation.
All the siz1 mutant alleles described thus far contain a T-DNA insertion downstream of the critical SAP, PHD, PIIT, and SP-RING domains (siz1-1, siz1-2, and siz1-3; Miura et al., 2005). Evidence presented here for siz1-2 implies that all three mutant proteins could be expressed (albeit at low levels) and retained at least partial functionality based on our detection of the siz1-2 protein bearing one or more SUMOs. Consequently, a full appreciation of SIZ1 function might eventually require the creation of true null alleles.
By contrast, our MS analyses indicated that the SUMOylation status of few, if any, proteins is altered in the mms21-1 background. One possibility is that the expression and/or activity of MMS21 are tissue specific (e.g., meristems; Liu et al., 2016; Zhang et al., 2017); thus, its targets such as DPa1 and BRAHMA are underrepresented when the proteome of whole seedling is analyzed. Another is that MMS21 has only a few low-abundance targets. In support of this idea, yeast MMS21 physically interacts with Smc5-Smc6 DNA repair complex, which might confine its substrates to this particle (Bermúdez-López et al., 2015). NSE4, which was identified here as an Arabidopsis SUMO1 conjugate, is part of this complex along with MMS21.
Both transcriptomic studies on SUMOylation mutants and chromatin binding studies with anti-SUMO antibodies in animals and yeast are consistent with SUMO becoming localized to the promoters of active genes where it represses transcription upon stress, possibly to prevent detrimental hyperactivation of stress responses (Neyret-Kahn et al., 2013; Ng et al., 2015; Niskanen et al., 2015; Seifert et al., 2015). Certainly, the SIZ1-influenced SUMOylation of the TPL and chromatin remodeling SWI-SNF complexes as well as various negative transcriptional repressors, especially during heat stress, is consistent with this protective function.
One of the more intriguing SUMO1/SUMO2 targets is BAG7, which becomes substantially more SUMOylated in siz1-2 plants upon heat stress. Prior studies by Li et al. (2017) showed that SUMOylation of this endoplasmic reticulum-bound cochaperone promotes its release and/or movement into the nucleus, where it binds to the transcriptional regulator WRKY29 in a SUMOylation-dependent manner. Ultimately, the SUMO-BAG7/WRKY29 complex helps stimulate the unfolded protein response elicited when the endoplasmic reticulum hyperaccumulates misfolded polypeptides. Presumably, the elevated SUMOylation of BAG7 seen here in unstressed and heat-stressed siz1-2 plants reflects attempts to mitigate proteotoxic stress upon eliminating a SIZ1-mediated protective response by constitutively activating the unfolded protein response as an alternative. One of the SUMOylation sites in BAG7 that direct this activation (K212; Li et al., 2017) was also identified here empirically during our searches for SUMO footprints.
A surprising feature of the SUMOylation system compared with that involved in ubiquitylation is that so few SUMO ligases are likely involved. Only four have been identified thus far in Arabidopsis through sequence comparisons and experimental testing (SIZ1, MMS21, PIAL1, and PIAL2; Huang et al., 2009b; Ishida et al., 2009; Tomanov et al., 2014) with as few as 15 present in humans (Jentsch and Psakhye, 2013). Surprisingly, there are more deSUMOylating proteases to remove the SUMO moiety in Arabidopsis than there are known ligases (Colby et al., 2006; Mukhopadhyay and Dasso, 2007). If SIZ1 modifies a moderate fraction of targets (105 identified with high confidence), MMS21 likely has just a few, and the targets of PIAL1 and PIAL2 are also likely limited based on the reasonably normal phenotype of double null mutants, it remains unclear which activities are responsible for modifying the remaining targets, which likely number in the hundreds or more. Certainly, it is possible that additional ligases remain to be discovered in Arabidopsis, but sequence searches have not yet uncovered other proteins with obvious SP-RING domains (Novatchkova et al., 2012; T.C. Rytz and R.D. Vierstra, unpublished data), suggesting that if they exist, they would employ unique domains to bind the SUMO-E2 intermediate. Alternatively, the SCE1 E2 could be responsible possibly through direct interactions with specific substrates either before or after additional posttranslational modifications such as phosphorylation (Bernier-Villamor et al., 2002; Anckar and Sistonen, 2007). SCE1 is encoded by a single essential gene in Arabidopsis (Saracco et al., 2007), thus ruling out the possibility of using E2 isoform diversity to expand target preferences in this species. SCE1 could recognize targets based on the canonical ψKxE/D SUMO conjugation motif, but how it would recognize the myriad of targets that are modified at noncanonical linkage sites is unclear.
Even for SIZ1, it is puzzling how this ligase selectively modifies such a large collection of targets, which do not appear to have common recognition sequences or even SUMOylation sites, with both canonical (ψ-K-x-E/D) and noncanonical sites evident. Possible mechanisms include the selection of targets based on their assembly into multisubunit complexes, and/or their common locations/compartments. In line with “guilt by association” being a key determinant, Psakhye and Jentsch (2012) proposed the “SUMO spray” hypothesis to help explain why a single human ligase might direct SUMOylation of multiple proteins required for DNA maintenance and repair. Once bound to the DNA repair complex, the ligase appears to indiscriminately spray the complex with SUMOs, which then strengthens the interactions among subunits and improves DNA repair efficiency synergistically. That SIZ1 contains two SIMs within its C-terminal half could provide an attractive mechanism whereby addition of a single SUMO1/SUMO2 to a complex would recruit SIZ1, which could then swivel to modify nearby proteins in a relatively nonspecific manner.
An intriguing feature of the heat-induced SUMOylation response is its upregulation within minutes (Kurepa et al., 2003; Golebiowski et al., 2009), implying that the response chain is short and likely posttranslational. Given that some of the more abundant SUMOylated proteins seen in Arabidopsis (Miller et al., 2010, 2013; this report) and human cells (Golebiowski et al., 2009) during thermal stress are components of the SUMO conjugation machinery (MMS21, PIAL2, SAE2, SCE1, and SIZ1), one attractive possibility was that the process is controlled by auto-SUMOylation of this machinery to either accelerate SUMOylation during the induction phase or to suppress it once a sufficient level of conjugation is reached. Given that SIZ1 is responsible for most of the stress-induced modification by SUMO1 and SUMO2 (Miura et al., 2005; Yoo et al., 2006; Saracco et al., 2007; this report), its modification at the mapped sites K100, K479, and K488 could provide the most direct effect. However, our observations that a 3K-R mutant of SIZ1, which is blocked in this stress-induced SUMOylation, easily restored the upregulation of SUMOylation during heat, ethanol, and peroxide stress, and rescued the abnormal growth and heat hypersensitivity of siz1-2 plants indicate that this modification has little influence on SUMOylation driven by SIZ1. As a consequence, other mechanisms for this stress control of SUMOylation must be entertained. One more mundane explanation is that SUMOylation of SIZ1 (and possibly other components of the conjugation machinery) reflects off-target modification by the ligase as it shuttles SUMO1/SUMO2 onto targets, using the high-energy, thioester-linked SUMO-E2 intermediate as the donor.
In sum, this work provides the deepest Arabidopsis catalog of SUMOylated proteins to date, thus offering a rich resource to discover the functions of this posttranslational modification under both physiological and stress conditions in plants. In particular, the expanding list of SUMO1/SUMO2 attachment sites now provides a strategy to discern the function(s) of SUMO addition through the use of arginine substitutions at the modified lysine(s), the potential of which was demonstrated by studies with SIZ1. The large list of targets assigned to SIZ1 now creates a platform to help understand how this ligase plays such profound roles in the protection of plants against various abiotic and biotic challenges.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Col-0 was used as the wild-type genetic background for all germplasm. The SUMO-conjugate purification line [6His-SUMO1(H89R) sumo1-1 sumo2-1], as described by Miller et al. (2010), was introgressed into the siz1-2 (SALK_065397) (Miura et al., 2005) and mms21-1 (hyp2-2, SAIL_77_G06) (Huang et al., 2009b; Ishida et al., 2009) mutants by crossing. Quadruple homozygous lines were identified in the F2 or F3 generations by glufosinate (Sigma-Aldrich) and kanamycin resistance linked to the sumo1-1 and the 6His-SUMO1(H89R) loci, respectively, and by genomic PCR for all loci. Important primers used in this study for both cloning and genotyping are listed in Supplemental Table 1.
The SIZ1-HA rescue lines were created by transforming homozygous siz1-2 plants with a transgene containing the full-length SIZ1 coding sequence followed by the coding sequence for three HA tags (YPYDVPDYASL) linked in tandem and a stop codon, whose expression was driven by the Arabidopsis UBQ10 promoter. The SIZ1 coding sequence (excluding the stop codon) was PCR amplified from cDNA and inserted into the pDONR221p5p2 vector (Thermo Fischer Scientific) at the KpnI/AscI cloning sites along with a 3xHA-tag coding sequence. Lysine-to-arginine codon substitutions within SIZ1 at residues K100, K479, and K498 [(3K-R)-HA] were generated by sequential site-directed mutagenesis of the cDNA in pDONR221p5p2. The UBQ10 promoter was PCR-amplified and inserted into the pMDC99 plant transformation vector at the KpnI/AscI cloning sites upstream of the attB1 site, as described (Suttangkakul et al., 2011). A multisite Gateway LR recombination reaction (Thermo Fisher Scientific) combined the SIZ1-HA construction in the pMDC99-pUBQ10 destination vector. Following transformation by the floral dip method (Saracco et al., 2007), F1 seedlings harboring the SIZ1-HA and (3K-R)-HA transgenes were identified by hygromycin resistance followed by genomic PCR. Homozygous siz1-2 plants expressing the transgenes were selected in the F2 generation by genomic PCR and by the absence of the siz1-2 phenotype. The siz1-2 mutation was introgressed into TPL-HA plants in the Landsberg erecta (Ler) background (p35S:TPL-3HA tpl1 tpl-2; Szemenyei et al., 2008) by crossing. Plants homozygous for the siz1-2 allele and expressing the TPL-HA transgene were identified by genomic PCR and resistance to glufosinate.
Unless otherwise noted, seeds were surface sterilized with bleach and stratified in water at 4°C in the dark for 2 d before sowing. For phenotypic studies, plants were grown at 21°C on soil under long-day photoperiods provided by fluorescence bulbs (long day: 16-h light at 125–150 μmol/m2/s, 8-h dark). For the analysis and purification of SUMO conjugates and TPL-HA immunoprecipitations, seedlings (>100) were grown for 8 d at 22°C under continuous 75 μmol/m2/s fluorescent light on solid Gamborg’s B-5 Basal Medium (GM; Sigma-Aldrich) supplemented with 2% sucrose and containing a 0.8% agar base that was topped with 0.1% agar in GM. For all RNA-seq analyses and SIZ1-HA immunoblot and immunoprecipitation assays, seedlings (>200) were grown under continuous illumination with 75 μmol/m2/s fluorescent light for 7 d in 50 mL liquid cultures containing GM supplemented with 2% sucrose. For the heat stress, the plates or cultures were incubated at 37°C for 30 min in a circulating water bath. For the ethanol and hydrogen peroxide treatments, 10% ethanol or 50 mM hydrogen peroxide (final concentration) was added to 50-mL liquid cultures. At the indicated times, the seedlings (>200) were harvested and frozen to liquid nitrogen temperatures. For the thermotolerance to moderately high temperatures assay, 25 seedlings were germinated on solid Murashige and Skoog medium grown under continuous 75 μmol/m2/s fluorescent light at the indicated temperature regimes (Wu et al., 2013). Each genotype was tested simultaneously on the same plate.
Genomic, RT-PCR, qPCR, and RNA-Seq Analyses
Genomic PCR, RT-PCR, and qPCR analyses employed the oligonucleotide primers described in Supplemental Table 1 with the primers for sumo1-1 available in Saracco et al. (2007). RNA was extracted from 8-d-old seedlings using the RNeasy Plant Mini Kit (Qiagen) followed by first-strand synthesis with oligo(dT)20 primers using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). cDNA and genomic DNA were amplified using EconoTaq Plus Green 2X Master Mix (Lucigen). qPCR was performed with the Bio-Rad CFX Connect Real-Time System together with the LightCycler 480 SYBR Green I Master Mix (Roche); transcript abundance was normalized to that generated with ACT2 based on the comparative threshold method (Pfaffl, 2001).
The RNA-seq data sets for SIZ1 were prepared from total RNA isolated from 7-d-old wild-type and siz1-2 seedlings (>200) grown as above at 22°C or subjected to a 30-min heat stress at 37°C plus 30 min recovery at 24°C. For each condition, three biological replicates were analyzed that contained independently grown wild-type and siz1-2 seedlings. TruSeq mRNA libraries were generated by the University of Wisconsin Gene Expression Center with two of the three biological replicates prepared to maintain strand information; the libraries then were sequenced using the Illumina HiSeq 2000 platform with 2 × 100-bp paired-end reads. The resulting fastq sequence files were manually searched for reads containing the SIZ1 query sequence 5′-CCAACGGCATGGAACTTGAT-3′ or its reverse complement 5′-ATCAAGTTCCATGCCGTTGG-3′, which correspond to the sequence immediately upstream of the T-DNA insertion site reported for siz1-2 (Miura et al., 2005). For total transcriptome analysis, total reads following removal of adapter sequences and low-quality reads/bases with Trimmomatic v0.33 (Bolger et al., 2014) were compared against the Arabidopsis Col-0 reference database in TAIR (http://www.arabidopsis.org) using RSEM v1.2.21 (Li and Dewey, 2011) together with Bowtie2 (Langmead and Salzberg, 2012). Differentially expressed genes were identified with EbSeq v1.12.0 (Leng et al., 2013) using pairwise comparisons between the wild type and siz1-2 under similar growth conditions.
Immunoblot Analyses
Immunodetection of SUMO1/SUMO2 conjugates used frozen tissue pulverized at liquid nitrogen temperatures, mixed with 2 volumes per gram fresh weight (µL/mg) of twice-strength SDS-PAGE sample buffer, heated to 95°C for 5 min, and clarified at 16,000g. The clarified extracts were subjected to SDS-PAGE and transferred onto Immobilon-P PVDF membranes (EMD-Millipore). The membranes were blocked with nonfat dry milk in PBS (12 mM Na2HPO4, pH 7.4, 2.7 mM KCl, and 137 mM NaCl) and probed with rabbit anti-SUMO1 antibodies (Kurepa et al., 2003). Rabbit antibodies against the proteasome subunit PBA1 were used as the loading control (Yang et al., 2004). For the detection of SIZ1, immunoblot analysis was performed as above using anti-SIZ1 antibodies (Miller et al., 2013). For analysis of SIZ1-HA and TPL-HA, the membranes were probed with rabbit polyclonal anti-HA antibodies (Sigma-Aldrich; product number H6908) using the signals from anti-histone H3 (Abcam; product number AB1791) as a control. SIZ1-HA was isolated as described by Miller et al. (2010) using EZview red anti-HA affinity gel (Sigma-Aldrich) for immunoprecipitation.
The relative abundance of SUMO conjugates in siz1-2 and wild-type plants was quantified by immunoblot analysis of a dilution series of the clarified crude extracts with anti-SUMO1 antibodies followed by IRDye 800CW goat anti-rabbit antibodies (LI-COR) and imaged using the 800-nm channel on the LI-COR Odyssey FC fluorimager. Three biological replicates of independently grown plants were used for each dilution series. The signal intensities for free SUMO and the smear of SUMO conjugates at the top of immunoblots were quantified by the LI-COR imaging software and normalized to the signals obtained with anti-PBA1 antibodies in combination with the IRDye 680RD goat anti-rabbit antibodies (LI-COR) detected at 700 nm. The normalized signal intensities of each dilution series were averaged per biological replicate.
Affinity Purification of SUMO Conjugates
SUMO1/SUMO2 conjugates were enriched from 6His-SUMO1(H89R) sumo1-1 sumo2-1 plants using the three-step protocol developed by Miller et al. (2010) with slight modifications (Rytz et al., 2016). Approximately 45 g of frozen tissue was pulverized at liquid nitrogen temperatures and resuspended for 1 h at 55°C in 90 mL of extraction buffer (EXB; 100 mM Na2HPO4, 10 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 10 mM iodoacetamide [IAA]) containing 7 M guanidine-HCl with 10 mM sodium metabisulfate, and 2 mM PMSF added just before use and the pH readjusted to 8.0. The extract was filtered through two layers of Miracloth (EMD Millipore), clarified by centrifugation at 15,000g, and incubated overnight at 4°C with Ni-NTA resin (Qiagen) (0.75 mL resin/5 g of tissue) after addition of imidazole to 10 mM. The Ni-NTA beads were washed sequentially with 10 column volumes of EXB containing 6 M guanidine-HCl, 0.25% Triton X-100, and 10 mM imidazole (pH 8.0), 10 column volumes of EXB containing 8 M urea, 0.25% Triton X-100, and 10 mM imidazole (pH 6.8), and 15 column volumes of EXB containing 8 M urea, 0.25% Triton X-100, and 10 mM imidazole (pH 8.0). SUMO conjugates were eluted with five column volumes of elution buffer (ELB; 350 mM imidazole, 100 mM Na2HPO4, 10 mM Tris-HCl, and 10 mM IAA, pH 8.0). The eluant was concentrated by ultrafiltration with a 10-kD molecular mass cutoff filter (Amicon Ultra-4; EMD Millipore or Vivaspin 6; GE Healthcare Life Sciences).
After two exchanges into ELB without imidazole and reconcentration, samples were renatured by adding dropwise to 25 volumes of ice-cold 0.5× RIPA buffer (100 mM Na2HPO4, pH 7.4, 10 mM Tris-HCl, 200 mM NaCl, 2.5% Nonidet P-40, 1.25% sodium deoxycholate, 0.25% SDS, 10 mM IAA, and 1 mM PMSF). The renatured samples were incubated overnight at 4°C with 0.5 mg of affinity-purified anti-SUMO1 antibodies bound to 500 μL AffiGel 10 beads (Bio-Rad). Beads were washed with 10 column volumes of 0.5× RIPA buffer followed by 100 column volumes of 50 mM NaHPO4 (pH 7.4), 100 mM NaCl, 10 mM IAA, and 1 mM PMSF. SUMO conjugates were eluted by first incubating the beads for 20 min at 65°C with 1 column volume of 1% SDS and 5% 2-mercaptoethanol, and subsequently washing with 10 column volumes of ELB containing 8 M urea (pH 8.0). The eluates were pooled and mixed with 350 μL of Ni-NTA resin for 4 h at 22°C, the beads were washed with 70 mL of ELB containing 8 M urea and 10 mM imidazole (pH 8.0), and the bound conjugates were eluted with six consecutive 500 μL washes of ELB (without IAA) containing 6 M urea and 300 mM imidazole (pH 8.0). The final elute was concentrated to 100 μL by ultrafiltration as above.
For the purification of conjugates from unstressed seedlings, three biological replicates, consisting of independently grown wild-type, 6His-SUMO1(H89-R) sumo1-1 sumo2-1, siz1-2 6His-SUMO1(H89-R) sumo1-1 sumo2-1, and mms21-1 6His-SUMO1(H89-R) sumo1-1 sumo2-1 plants were used. For the heat-stressed samples, five biological replicates of independently grown wild-type, 6His-SUMO1(H89-R) sumo1-1 sumo2-1, and siz1-2 6His-SUMO1(H89-R) sumo1-1 sumo2-1 seedlings were used for the purifications, whereas two biological replicates were used for mms21-1 6His-SUMO1(H89-R) sumo1-1 sumo2-1 seedlings. For four of the five heat-stressed wild-type, 6His-SUMO1(H89-R) sumo1-1 sumo2-1, and siz1-2 6His-SUMO1(H89-R) sumo1-1 sumo2-1 biological replicates, as well as both heat-stressed mms21-1 6His-SUMO1(H89-R) sumo1-1 sumo2-1 biological replicates were analyzed by two LC-MS runs to generate two technical replicates for each.
Tandem Mass Spectrometry
SUMO1/SUMO2 conjugate preparations were reduced for 1 h at 22°C with 10 mM DTT, followed by alkylation with 20 mM IAA for 1 h (Miller et al., 2010; Rytz et al., 2016). The reaction was quenched with 20 mM DTT and diluted with 25 mM ammonium bicarbonate. Trypsin (Trypsin Gold, MS grade; Promega) was added at a 1:20 protease-to-sample weight ratio and incubated at 37°C for 18 h. The digests were acidified with 0.5% trifluoroacetic acid and desalted with OMIX C18 pipette tips (Agilent Technologies), using 75% acetonitrile and 1% formic acid for elution. The samples were vacuum dried, resuspended in 5% acetonitrile and 0.1% formic acid, and subjected to tandem LC-MS using either LTQ Orbitrap Velos (Thermo Fisher Scientific) or Q Exactive Plus (Thermo Fisher Scientific) mass spectrometers operated in the positive ESI mode.
For the LTQ Orbitrap Velos ESI-MS, the tryptic peptides were separated on a 50 × 365-μm fused silica capillary microcolumn packed with 20 cm of 1.7-μm diameter, 130-Å pore size, C18 beads (Waters BEH), with an emitter tip pulled to ∼1 µm. Peptides were eluted over 120 min at a flow rate of 300 nL/min with a linear gradient of 2 to 30% acetonitrile in 0.1% formic acid. Full mass scans were performed in the FT Orbitrap at 300 to 1500 m/z at a resolution of 60,000, followed by 10 MS/MS high-energy collision-induced dissociation (HCD) scans of the 10 highest intensity parent ions at 42% normalized collision energy, 7500 resolution, and a mass range starting at 100 m/z. Dynamic exclusion was set to a repeat count of two over a duration of 30 s and an exclusion window of 120 s.
For the Q-Exactive ESI-MS, the tryptic peptides were separated by nanoscale liquid chromatography (LC) using a Dionex Ultimate 3000 Rapid Separation LC system (Thermo Fisher Scientific) equipped with a 75 µm × 2-cm Acclaim PepMap 100 guard column followed by a 75 µm × 15-cm analytical Acclaim PepMap RSLC C18 column (2-μm particle size, 100-Å pore size; Thermo Fisher Scientific). The peptides were eluted over 120 min at a flow rate of 250 nL/min with a linear gradient of 1.6 to 32% acetonitrile in 0.1% formic acid. Data-dependent acquisition of full MS scans within a mass range of 380 to 1500 m/z at a resolution of 70,000 was performed, with the automatic gain control (AGC) target set to 3 × 106, and the maximum fill time set to 200 ms. HCD fragmentation of the top 15 most intense peaks was performed with a normalized collision energy of 28, with an AGC target of 2 × 105 counts and an isolation window of 3.0 m/z, excluding precursors that had an unassigned, +1, +7, +8, or >+8 charge state. MS2 scans were conducted at a resolution of 17,500, with an AGC target of 2 × 105 and a maximum fill time of 100 ms. Dynamic exclusion was performed with a repeat count of 2 and a duration of 20 s, while the minimum MS ion count for triggering MS2 was set to 4 × 103 counts.
MS Data Analysis
The MS2 spectra were searched using MORPHEUS version 160 (Wenger and Coon, 2013) against the Arabidopsis protein database (TAIR10_PEP_20101214_UPDATED; http://www.arabidopsis.org) with the addition of the SUMO1(H89-R) sequence along with common contaminants (e.g., trypsin and human keratin). The default search parameters were set to a precursor mass tolerance of 2.100 D, product mass tolerance of 0.010 D, maximum FDR of 1%, fixed carbamidomethylation of cysteines, and variable methionine oxidation, along with a maximum of two missed trypsin cleavages.
To provide label-free quantification based on dNSAF values (Zhang et al., 2010), the data sets were filtered through Morpheus Spectral Counter (Gemperline et al., 2016). Background proteins identified by MS analysis from four biological replicates of wild-type plants were classified as contaminants and removed from the SUMO1/SUMO2 conjugate data sets. For the heat stress data sets, only proteins identified by two or more PSMs per biological replicate were included in the final analyses; for the unstressed data sets, all targets were considered due to low protein abundance. All values from the technical replicates were averaged for each biological replicate. dNSAF values for SUMOylated proteins in each biological replicate were normalized based on the dNSAF value for SUMO1. Morpheus assigned the SUMO1 peptides to either SUMO1 or SUMO2 when filtered through the Arabidopsis proteome, thus generating two dNSAF values, which we then combined for normalization.
For the statistical analyses of SUMO1SUMO2 conjugates, missing dNSAF values among biological replicates were imputed using PERSEUS (Tyanova et al., 2016; http://www.perseus-framework.org) with standard settings and applied to each biological replicate separately. To reduce the imputation frequencies, the data set was limited to SUMO targets detected in at least three biological replicates in either background. Before imputation, dNSAF values were transformed by taking the log2 value of the fraction multiplied by 1e10 (x = log2(dNSAF*1e10)). The LIMMA statistical package in R (Ritchie et al., 2015) was used to calculate significant differences between SUMO conjugate profiles from wild-type and siz1-2 plants by adaptation of the source code published by Kammers et al. (2015) to determine the moderate P value. Fold enrichments of specific GO functions were obtained using the PANTHER database (Thomas et al., 2003) with either the default Arabidopsis proteome or the identified SUMOylome serving as the background for enrichment. GO localizations were predicted with DAVID (Huang et al., 2009a). The SUMO conjugate interactome was generated by STRING v10 (Szklarczyk et al., 2015) using coexpression, experimentally determined interactions, database annotations, and automated text mining, and was visualized using Cytoscape version 3.4.0 (Shannon et al., 2003). Proportional Venn diagrams were generated using Vennerable (https://r-forge.r-project.org/projects/vennerable/). Heats maps were visualized using gplots (https://CRAN.R-project.org/package=gplots) and RColorBrewer (https://CRAN.R-project.org/package =RColorBrewer).
SUMO and ubiquitin footprints were identified through Proteome Discoverer (version 2.0.0.802; Thermo Fisher Scientific) by searching the TAIR10 protein database using the variable modification of lysine residues by either SUMO (Glu-Thr-Gly-Gly, +349.149 m/z and pyroGlu-Thr-Gly-Gly, +326.123 m/z [loss of water] or +325.139 m/z [loss of amine]) or ubiquitin (Gly-Gly, +114.043 m/z). Peptides were assigned using SEQUEST HT (Thermo Fisher Scientific), with search parameters set to assume trypsin digestion with a maximum of two missed cleavages, a minimum peptide length of 6, precursor mass tolerances of 10 ppm, and fragment mass tolerances of 0.02 D. Carbamidomethylation of cysteine was specified as a static modification, while oxidation of methionine and N-terminal acetylation were specified as dynamic modifications. The target FDR of ≤1% (strict) was used as validation for PSMs and peptides. Proteins that contained similar peptides and which could not be differentiated based on the MS/MS analysis alone were grouped to satisfy the principles of parsimony. MEME Suite 4.11.4 (Bailey et al., 2009) was used to identify the SUMO binding ψKxE/D motif, whereas the prevalence of these sites were predicted by GPS-SUMO v1.0.1 (Zhao et al., 2014).
MS Analysis of Total Arabidopsis Proteome
For the total proteome analysis, frozen tissue was pulverized at liquid nitrogen temperatures and incubated for 1 h at 55°C in EXB (100 μL of EXB per 60 µg of tissue). After clarification, soluble protein was precipitated using methanol/chloroform and the pellet was washed with acetone. After air-drying, the pellet was reconstituted in 100 μL of 8 M urea and 10 mM DTT and allowed to reduce for 1 h. The samples were alkylated for 1 h with 20 mM IAA, quenched using 20 mM DTT for 5 min, and then diluted with 900 μL of 25 mM ammonium bicarbonate to reduce the urea concentration to below 1.5 M. After overnight trypsinization at 37°C as above, the peptides were acidified with 0.5% trifluoroacetic acid and desalted as above with a 100 μL OMIX C18 pipette tip (Agilent Technologies), vacuum dried, and resuspended in 20 μL of 5% acetonitrile and 0.1% acetic acid.
MS analysis was as above using a Q-Exactive ESI-MS in combination with an Acclaim PepMap RSLC C18 column. HCD fragmentation of the top 15 strongest peaks was performed with a normalized collision energy of 28, an intensity threshold of 4 × 104 counts, and an isolation window of 3.0 m/z, excluding precursors that had an unassigned, +1, +7, +8, or >+8 charge state. MS/MS scans were conducted at a resolution of 17,500, with an AGC target of 2 × 105 and a maximum fill time of 100 msec. Dynamic exclusion was performed with a repeat count of 2 and an exclusion duration of 30 s, while the minimum MS ion count for triggering MS/MS was set to 4 × 103 counts. The resulting MS/MS spectra from each sample analyzed in quadruplicate were searched against the Arabidopsis proteome database by Proteome Discoverer (Thermo Fisher Scientific) and assigned by SEQUEST HT (Eng et al., 1994) assuming a maximum of two missed trypsin cleavages, a minimum peptide length of 6, precursor mass tolerances of 10 ppm, fragment mass tolerances of 0.02 D, and a the target FDR of either 0.01 (high confidence) or 0.05 (medium confidence). Label-free quantification was performed as previously described (Silva et al., 2006) in Proteome Discoverer with a minimum Quan value threshold set to 0.0001 using unique peptides, and “3 Top N” peptides used for area calculation. Protein abundances were normalized based on the least variable proteins (sd/average) using histone abundance as an internal control. Average protein abundances were generated from three biological replicates consisting of independently grown wild-type, siz1-2, and mms21-1 seedlings. Only proteins detected with a protein FDR confidence level of medium or high and by at least five PSMs were considered for the analysis.
Anti-HA Antibody Immunoprecipitations of TPL
For analysis of TPL SUMOylation, seedlings of the wild type (Ler) and siz1-2, with or without the TPL-HA transgene, were plate-grown, subjected to heat stress, and harvested as described above. For each genotype and condition, three biological replicates of independently grown seedlings were used. Samples were homogenized at liquid nitrogen temperatures and extracted into 2 mL/g of cold immunoprecipitation buffer (IPB; 50 mM HEPES, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10% [v/v] glycerol, with 20 mM ATP, 10 mM IAA, 10 mM sodium metabisulphite, 10 mM N-ethylmaleimide, 5 mM DTT, 2 mM PMSF, 6 μM chymostatin, and 1× plant protease inhibitor cocktail [Sigma-Aldrich] added immediately before use). All subsequent manipulations were conducted at 4°C. Extracts were clarified by centrifugation at 16,000g for 10 min, and equal volumes of supernatant (800 μL) were incubated for 2 h with 50 μL EZview red anti-HA affinity gel (100 μL of a 50% slurry; Sigma-Aldrich), preequilibrated in IPB. Beads were collected by centrifugation and washed five times with IPB, and bound proteins were eluted at 95°C in 100 μL of 2× SDS-PAGE sample buffer. Samples were then analyzed by SDS-PAGE and immunoblotting as above. Densitometric quantification of the TPL-HA blots was performed using TotalLab software (Nonlinear Dynamics), with at least three different exposures of the same blot measured to ensure that the exposures were within the linear range of the film. Statistical analysis employed one-way ANOVA to determine the presence of significant differences, followed by Tukey’s post-hoc tests to identify significantly different data points.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the ProQuest Two-Hybrid System (Life Technologies). Pairwise gene combinations in pDEST22 and pDEST32 (or empty vector controls) were cotransformed into the Saccharomyces cerevisiae strain MaV203. Protein-protein interactions were identified by growth at 30°C on medium lacking leucine, tryptophan, and histidine and containing 25 mM 3-amino-1,2,4-triazole.
Accession Numbers
Protein identifiers and the corresponding gene accession numbers for the catalog of Arabidopsis SUMOylation targets identified here can be found in the Supplemental Data Sets. The raw sequence files for the MS data sets are available in the ProteomeXchange database under the submission numbers PXD007054 and PXD009274 within the PRIDE repository (http://www.proteomexchange.org/). RNA-seq files for the transcriptome analysis are available at the NCBI Sequence Read Archive database under the submission number SRP134263 (https://www.ncbi.nlm.nih.gov/sra/SRP134263).
Supplemental Data
Supplemental Figure 1. Affinity purification of SUMOylated proteins from 6His-S1(H89-R) sumo1-1 sumo2-1 seedlings either wild-type or mutant for the SUMO ligases SIZ1 and MMS21.
Supplemental Figure 2. Reproducibility between technical and biological replicates for wild-type, siz1-2, and mms21-1 seedlings.
Supplemental Figure 3. Comparison of SUMO1/SUMO2 conjugate abundance under unstressed conditions in siz1-2 and mms21-1 versus wild-type seedlings.
Supplemental Figure 4. Heat map comparisons of SUMO1/SUMO2 conjugates in siz1-2 and mms21-1 seedlings before and after heat stress.
Supplemental Figure 5. Functional enrichments and interactome analysis of SUMOylated proteins from wild-type and siz1-2 seedlings.
Supplemental Figure 6. MS analysis of SUMOylated proteins purified from the mms21-1 mutant background upon heat stress.
Supplemental Figure 7. The K3-R mutant of SIZ1 retains its interaction with SCE1.
Supplemental Figure 8. SIZ1 modification by SUMO1/SUMO2 upon treatment with ethanol and hydrogen peroxide requires K100, K479, and K488.
Supplemental Table 1. Oligonucleotide primers used in this study.
Supplemental Data Set 1. List of contaminants that copurify with SUMO1/SUMO2 conjugates.
Supplemental Data Set 2. List of SUMO1/SUMO2 conjugates that accumulate in wild-type, siz1-2, and mms21-1 seedlings before and after heat stress.
Supplemental Data Set 3. List of high-confidence SUMO1/SUMO2 conjugates identified by two or more PSMs.
Supplemental Data Set 4. Transcriptome profile of the SIZ1-dependent SUMO1/SUMO2 targets.
Supplemental Data Set 5. Profile of the total proteome and SUMO1/SUMO2 targets detected by MS.
Supplemental Data Set 6. List of Arabidopsis proteins with mapped SUMO footprints.
Supplemental File 1. Statistical analysis of the MS data sets.
Acknowledgments
We thank Joseph M. Walker and Samuel L. York for technical assistance and David C. Gemperline for help with data analysis. This work was supported by a grant from the NSF-Plant Genome Research Program to R.D.V. (IOS-1546862), a NIH NRSA Ruth L. Kirschstein postdoctoral fellowship (5-F32-GM103161) to R.C.A., and graduate fellowships from the NIH-sponsored UW Genetics Training Program (5-T32-GM007133) and the Department of Biology at Washington University in St. Louis to T.C.R. and M.J.M. MS analyses conducted at the University of Wisconsin were supported by grants from NIH/NHGRI (1P50HG004952) and NIH-NIGMS (P01GM081629) to M.S. and L.M.S. Y.-t.J. and Y.-y.C. were supported by the Academia Sinica/Thematic Research Project (AS-102-TP-B05).
AUTHOR CONTRIBUTIONS
M.J.M. and T.C.R developed the affinity purification strategy. T.C.R. created the siz1-2 and mms21-1 germplasm, performed the phenotypic analyses, and generated the SUMO conjugate preparations for MS analysis. F.M., M.S., and L.M.S. developed the SUMOylome data sets by LC-ESI-MS. T.C.R. performed the MS data analyses. M.J.M. created and analyzed the SIZ1-HA and (3K-R)-HA lines. R.C.A. conducted the RNA-seq studies on siz1-2. R.S.M. performed the immunoprecipitation studies for SUMOylated TPL. Y.-t.J. and Y.-y.C. performed the thermotolerance assays. T.C.R., M.J.M., and R.D.V. proposed the research, analyzed the data, and wrote the article with advice from the other authors.
References
- Aguilar-Martinez E., Chen X., Webber A., Mould A.P., Seifert A., Hay R.T., Sharrocks A.D. (2015). Screen for multi-SUMO-binding proteins reveals a multi-SIM-binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proc. Natl. Acad. Sci. USA 112: E4854–E4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J., Sistonen L. (2007). SUMO: getting it on. Biochem. Soc. Trans. 35: 1409–1413. [DOI] [PubMed] [Google Scholar]
- Augustine R.C., York S.L., Rytz T.C., Vierstra R.D. (2016). Defining the SUMO system is maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol. 171: 2191–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-López M., Pociño-Merino I., Sánchez H., Bueno A., Guasch C., Almedawar S., Bru-Virgili S., Garí E., Wyman C., Reverter D., Colomina N., Torres-Rosell J. (2015). ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol. 13: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro P.H., Tavares R.M., Bejarano E.R., Azevedo H. (2012). SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 69: 3269–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong M.S., Park H.C., Hong M.J., Lee J., Choi W., Jin J.B., Bohnert H.J., Lee S.Y., Bressan R.A., Yun D.J. (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 151: 1930–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Peer R., Schuster S., Meiri D., Breiman A., Avni A. (2010). Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol. Biol. 74: 33–45. [DOI] [PubMed] [Google Scholar]
- Colby T., Matthäi A., Boeckelmann A., Stuible H.P. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Price G., O’Donnell E., Schwessinger B., Dominy P., Sadanandom A. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., Tonelli C., Napier R., Hedden P., Bennett M., Sadanandom A. (2014). Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Crozet P., Margalha L., Butowt R., Fernandes N., Elias C.A., Orosa B., Tomanov K., Teige M., Bachmair A., Sadanandom A., Baena-González E. (2016). SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 85: 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N., Coupland G. (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 107: 17415–17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M., March-Diaz R., Reyes J.C. (2008). The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. 283: 21469–21477. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- Gemperline D.C., Scalf M., Smith L.M., Vierstra R.D. (2016). Morpheus Spectral Counter: A computational tool for label-free quantitative mass spectrometry using the Morpheus search engine. Proteomics 16: 920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry M., Hennig L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321: 40–46. [DOI] [PubMed] [Google Scholar]
- Golebiowski F., Matic I., Tatham M.H., Cole C., Yin Y., Nakamura A., Cox J., Barton G.J., Mann M., Hay R.T. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2: ra24. [DOI] [PubMed] [Google Scholar]
- Hammoudi V., Vlachakis G., Schranz M.E., van den Burg H.A. (2016). Whole-genome duplications followed by tandem duplications drive diversification of the protein modifier SUMO in angiosperms. New Phytol. 211: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handu M., Kaduskar B., Ravindranathan R., Soory A., Giri R., Elango V.B., Gowda H., Ratnaparkhi G.S. (2015). SUMO-enriched proteome for Drosophila innate immune response. G3 (Bethesda) 5: 2137–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T. (2013). Decoding the SUMO signal. Biochem. Soc. Trans. 41: 463–473. [DOI] [PubMed] [Google Scholar]
- Hendriks I.A., Vertegaal A.C. (2016). A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17: 581–595. [DOI] [PubMed] [Google Scholar]
- Hickey C.M., Wilson N.R., Hochstrasser M. (2012). Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., Shi S., Wang J., Wang Y., Xie Q., Yang C. (2009b). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60: 666–678. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Psakhye I. (2013). Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47: 167–186. [DOI] [PubMed] [Google Scholar]
- Jin J.B., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A.D., Gygi S.P., Broday L. (2009). SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev. Cell 17: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammers K., Cole R.N., Tiengwe C., Ruczinski I. (2015). Detecting significant changes in protein abundance. EuPA Open Proteom. 7: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Han Y.J., Kim S.I., Song J.T., Seo H.S. (2015). Arabidopsis CMT3 activity is positively regulated by AtSIZ1-mediated sumoylation. Plant Sci. 239: 209–215. [DOI] [PubMed] [Google Scholar]
- Kurepa J., Walker J.M., Smalle J., Gosink M.M., Davis S.J., Durham T.L., Sung D.Y., Vierstra R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278: 6862–6872. [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Wagner D. (2007). Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 23: 403–412. [DOI] [PubMed] [Google Scholar]
- Lai Z., Vinod K., Zheng Z., Fan B., Chen Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49: 79–90. [DOI] [PubMed] [Google Scholar]
- Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., Kendziorski C. (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29: 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Williams B., Dickman M. (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214: 695–705. [DOI] [PubMed] [Google Scholar]
- Lin X.-L., Niu D., Hu Z.-L., Kim D.H., Jin Y.H., Cai B., Liu P., Miura K., Yun D.-J., Kim W.-Y., Lin R., Jin J.B. (2016). An Arabidopsis SUMO ligase SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet. 12: e1006016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13: 137–144. [DOI] [PubMed] [Google Scholar]
- Liu Y., Lai J., Yu M., Wang F., Zhang J., Jiang J., Hu H., Wu Q., Lu G., Xu P., Yang C. (2016). The Arabidopsis SUMO ligase AtMMS21 dissociates the E2Fa/Dpa complex in cell cycle progresssion. Plant Cell 28: 2225–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Scalf M., Rytz T.C., Hubler S.L., Smith L.M., Vierstra R.D. (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., Yun D.J., Hasegawa P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102: 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Hasegawa P.M. (2007a). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10: 495–502. [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007b). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Dasso M. (2007). Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32: 286–295. [DOI] [PubMed] [Google Scholar]
- Neyret-Kahn H., Benhamed M., Ye T., Le Gras S., Cossec J.C., Lapaquette P., Bischof O., Ouspenskaia M., Dasso M., Seeler J., Davidson I., Dejean A. (2013). Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 23: 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C.H., Akhter A., Yurko N., Burgener J.M., Rosonina E., Manley J.L. (2015). Sumoylation controls the timing of Tup1-mediated transcriptional deactivation. Nat. Commun. 6: 6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niskanen E.A., Malinen M., Sutinen P., Toropainen S., Paakinaho V., Vihervaara A., Joutsen J., Kaikkonen M.U., Sistonen L., Palvimo J.J. (2015). Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 16: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M., Tomanov K., Hofmann K., Stuible H.P., Bachmair A. (2012). Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytol. 195: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Yun D.J. (2013). New insights into the role of the small ubiquitin-like modifier (SUMO) in plants. Int. Rev. Cell Mol. Biol. 300: 161–209. [DOI] [PubMed] [Google Scholar]
- Park B.S., Song J.T., Seo H.S. (2011). Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I., Jentsch S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., Hay R.T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276: 12654–12659. [DOI] [PubMed] [Google Scholar]
- Rytz T.C., Miller M.J., Vierstra R.D. (2016). Purification of SUMO conjugates from Arabidopsis for mass spectrometry. In SUMO: Methods and Protocols, M.S. Rodriguez, ed (New York: Springer Science; ), pp. 257–281. [DOI] [PubMed] [Google Scholar]
- Sadanandom A., Ádám É., Orosa B., Viczián A., Klose C., Zhang C., Josse E.M., Kozma-Bognár L., Nagy F. (2015). SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S.A., Miller M.J., Kurepa J., Vierstra R.D. (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A., Schofield P., Barton G.J., Hay R.T. (2015). Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci. Signal. 8: rs7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P.C., Geromanos S.J. (2006). Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5: 144–156. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K., Zeschmann A., Hermkes R., Eifler K., Ziba I., Grieco M., Novatchkova M., Hofmann K., Hesse H., Bachmair A. (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26: 4547–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2011). EMSY-like genes are required for full RPP7-mediated race-specific immunity and basal defense in Arabidopsis. Mol. Plant Microbe Interact. 24: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Kini R.K., Schuurink R.C., Takken F.L. (2010). Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22: 1998–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wenger C.D., Coon J.J. (2013). A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. J. Proteome Res. 12: 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Britt R., Dickman M.B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 107: 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R. III (2004). Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279: 45662–45668. [DOI] [PubMed] [Google Scholar]
- Wu T.Y., Juan Y.T., Hsu Y.H., Wu S.H., Liao H.T., Fung R.W.M., Charng Y.Y. (2013). Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 161: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Yuan D., Liu M., Li C., Liu Y., Zhang S., Yao N., Yang C. (2013). AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 161: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Fu H., Walker J., Papa C.M., Smalle J., Ju Y.M., Vierstra R.D. (2004). Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279: 6401–6413. [DOI] [PubMed] [Google Scholar]
- Yeh C.H., Kaplinsky N.J., Hu C., Charng Y.Y. (2012). Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci. 195: 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.Y., Miura K., Jin J.B., Lee J., Park H.C., Salt D.E., Yun D.J., Bressan R.A., Hasegawa P.M. (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 142: 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus A.A., Lima C.D. (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2017). A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant Physiol. 173: 1574–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wen Z., Washburn M.P., Florens L. (2010). Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82: 2272–2281. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42: W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K.S., Guo Y. (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605. [DOI] [PubMed] [Google Scholar]