The N6-methyladenosine RNA binding proteins ECT2, ECT3, and ECT4 are required for the correct timing of leaf formation and correct leaf and trichome morphogenesis.
Abstract
Methylation of N6-adenosine (m6A) in mRNA is an important posttranscriptional gene regulatory mechanism in eukaryotes. m6A provides a binding site for effector proteins (“readers”) that influence pre-mRNA splicing, mRNA degradation, or translational efficiency. YT521-B homology (YTH) domain proteins are important m6A readers with established functions in animals. Plants contain more YTH domain proteins than other eukaryotes, but their biological importance remains unknown. Here, we show that the cytoplasmic Arabidopsis thaliana YTH domain proteins EVOLUTIONARILY CONSERVED C-TERMINAL REGION2/3 (ECT2/3) are required for the correct timing of leaf formation and for normal leaf morphology. These functions depend fully on intact m6A binding sites of ECT2 and ECT3, indicating that they function as m6A readers. Mutation of the close ECT2 homolog, ECT4, enhances the delayed leaf emergence and leaf morphology defects of ect2/ect3 mutants, and all three ECT proteins are expressed at leaf formation sites in the shoot apex of young seedlings and in the division zone of developing leaves. ECT2 and ECT3 are also highly expressed at early stages of trichome development and are required for trichome morphology, as previously reported for m6A itself. Overall, our study establishes the relevance of a cytoplasmic m6A-YTH regulatory module in the timing and execution of plant organogenesis.
INTRODUCTION
Covalent modification of mRNA is now recognized as a crucial component of posttranscriptional gene regulation in eukaryotes. Methylation of the N6 position of adenosine (m6A) is the most common internal modification in eukaryotic mRNA. Recent transcriptome-wide maps of m6A in human cells identified one m6A peak per 2000 nucleotides on average (Dominissini et al., 2012), and m6A is present in ∼0.5 to 1% of all adenosines in mRNA isolated from sporulating yeast (Bodi et al., 2010) and from different tissues of Arabidopsis thaliana (Zhong et al., 2008). N6-adenosine methylation is catalyzed by a multicomponent enzyme (an m6A “writer”) that methylates similar consensus sites in mammals, plants, and yeast [(R)RACH, R=A/G, H=A/C/U] and whose core components are broadly conserved (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2013; Liu et al., 2014; Shen et al., 2016). Biochemical and genetic studies in mammalian cells and plants have shown that the methyltransferase complex consists of a catalytic heterodimer (METTL3/METTL14) that associates with a number of additional factors required for methylation, including the proteins WTAP and KIAA1429/Virilizer (Zhong et al., 2008; Liu et al., 2014; Schwartz et al., 2014). The biological importance of m6A modification is highlighted by the finding that knockout mutations in homologs of all core methylase components (METTL3, METTL14, WTAP, and KIAA1429/Virilizer) are embryonically lethal in plants (Zhong et al., 2008; Shen et al., 2016; Růžička et al., 2017), as has also been shown for METTL3 and WTAP in mammals and insects (Fukusumi et al., 2008; Hongay and Orr-Weaver, 2011; Geula et al., 2015).
Although m6A does not alter thymidine incorporation by reverse transcription of m6A-sites, it does inhibit A-U base pairing in vitro (Kierzek and Kierzek, 2003; Li et al., 2015). Perhaps as a consequence of impaired A-U pairing, it also influences the formation of secondary RNA structure and, consequently, alters the accessibility of binding sites for RNA binding proteins in cells (Liu et al., 2015). In mammalian cells, such “m6A switches” appear to be particularly prevalent in noncoding RNA and in introns of pre-mRNA (Liu et al., 2015). Nonetheless, the majority of known functional consequences of m6A modification depends on specific interactions with different classes of m6A binding proteins (“m6A readers”) that play roles in distinct steps of mRNA function, including pre-mRNA splicing, mRNA degradation, translation efficiency, and the mode of translation initiation (Wang et al., 2014, 2015; Meyer et al., 2015; Xiao et al., 2016). These molecular functions manifest themselves in fundamental biological processes. For example, in mammalian cells, m6A is important for adaptation to heat shock because it mediates cap-independent translation initiation of mRNAs translated during the heat shock response via direct binding of the translation initiation factor eIF3 (Meyer et al., 2015). It is also becoming clear that m6A has a general function in facilitating transitions between distinct developmental stages, such as the maternal-zygotic transition in zebra fish, oocyte maturation and the endothelial-to-hematopoietic transition in mammals, and onset of the meiotic program in yeast (Shah and Clancy, 1992; Clancy et al., 2002; Ivanova et al., 2017; Zhang et al., 2017; Zhao et al., 2017). In addition, m6A is essential for mammalian stem cell differentiation (Batista et al., 2014; Geula et al., 2015).
In some cases, the functions of m6A in mediating developmental transitions involve a specific family of m6A binding proteins that contain a so-called YT521-B homology (YTH) domain (Stoilov et al., 2002; Schwartz et al., 2013; Ivanova et al., 2017; Zhang et al., 2017; Zhao et al., 2017). The YTH domain specifically recognizes the methyl group in m6A via an aromatic cage formed by three highly conserved aromatic residues, most often tryptophan, and affinity for m6A-containing oligonucleotides is typically in the low micromolar range (Li et al., 2014b; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014). It is important to note, however, that the mere presence of a YTH domain does not necessarily imply m6A binding specificity. For example, the fission yeast (Schizosaccharomyces pombe) YTH domain protein Mmi1 recognizes a specific nucleotide sequence but does not bind to m6A, consistent with amino acid changes in several residues implicated in m6A-RNA binding in this protein (Wang et al., 2016). Two distinct phylogenetic subfamilies of YTH domain proteins can be distinguished, as exemplified by the mammalian YTHDF and YTHDC classes (Patil et al., 2018). YTHDF domain proteins are modular and contain an N-terminal intrinsically disordered region (IDR) followed by a C-terminal YTH domain (Patil et al., 2018). Studies on human YTHDF2 suggest that the two modules function independently of each other such that the YTH domain selects target mRNAs based on specific m6A binding, while the N-terminal IDR can function as a regulatory unit (Wang et al., 2014). It cannot be excluded, however, that the IDR also participates in RNA binding in vivo given the relatively low affinity of isolated YTH domains for RNA. YTHDF2 is required for the degradation of translationally repressed mRNA in cytoplasmic processing (P)-bodies in which many mRNA decay components are concentrated, and artificial tethering of the N-terminal IDR of human YTHDF2 is sufficient to confer localization of tethered mRNA to P-bodies by mechanisms that may include direct recruitment of the CCR4-NOT deadenylase complex (Wang et al., 2014; Du et al., 2016).
In plants, m6A modification of mRNA is of key importance in development, as null alleles of MTA (AT4G10760, METTL3 homolog), METHYLTRANSFERASE B (MTB; AT4G09980, METTL14 homolog), FIP37 (AT3G54170, WTAP homolog), and VIRILIZER (VIR, AT3G05680) are embryonically lethal, and reduced postembryonic expression of these factors results in dramatically reduced m6A levels in mRNA and produces clear developmental phenotypes (Zhong et al., 2008; Bodi et al., 2012; Shen et al., 2016; Růžička et al., 2017). These phenotypes include increased numbers of trichome branches (Bodi et al., 2012), as well as defective leaf initiation coincident with overproliferation of the vegetative shoot apical meristem (Shen et al., 2016). Consistent with these genetic observations, transcriptome-wide m6A mapping has uncovered thousands of m6A sites in Arabidopsis mRNAs (Luo et al., 2014; Shen et al., 2016), and two target mRNAs encoding transcription factors essential for meristem maintenance, WUSCHEL (WUS; AT2G17950) and SHOOT MERISTEMLESS (STM; AT1G62360), have been identified and validated (Shen et al., 2016). In plants that are postembryonically depleted of FIP37, the lack of m6A marks on WUS and STM mRNA coincides with the expansion of their domain of expression and overproliferation of the shoot apical meristem (Shen et al., 2016), suggesting that m6A plays a fundamental role in meristem function. However, the mechanistic underpinnings of these important roles of m6A in plant biology remain unclear. Most importantly, the possible role of YTH domain proteins as m6A readers in plants has not been clarified.
Several YTH domain proteins are encoded in plant genomes (Li et al., 2014a), most belonging to an expanded family of proteins called EVOLUTIONARILY CONSERVED C-TERMINAL REGION1-11 (ECT1-11) in Arabidopsis. ECT1 (AT3G03950) and ECT2 (AT3G13460) interact with CBL-INTERACTING PROTEIN KINASE1 (AT3G17150) (Ok et al., 2005), and several ECT proteins were identified in an mRNA-protein interactome study, suggesting that they have the capacity to bind to mRNA in vivo (Reichel et al., 2016). Consistent with this conclusion, ECT4 (AT1G5550) was found to bind to single-stranded RNA in vitro (Li et al., 2014a). Nonetheless, biological functions and additional biochemical properties of plant YTH-domain proteins, including their potential binding to m6A, remain unknown.
Here, we show that the YTH-domain proteins ECT2 and ECT3 (AT5G61020) are bona fide m6A readers that depend on intact m6A binding sites to control the timing of leaf emergence and define normal leaf morphology. A third YTH-domain protein, ECT4, is also involved in these processes, but its role can only be observed in the absence of both ECT2 and ECT3. Consistent with their functions in leaf formation, all three ECT genes are expressed at the shoot apex in young seedlings and in the cell division zone of developing leaves. The three ECT proteins are cytoplasmic, sometimes forming distinct cytoplasmic foci, suggesting that they mainly interact with mature mRNA. These results demonstrate the crucial importance of a specific set of plant YTH-domain proteins in mediating m6A-dependent effects on the timing and execution of plant development.
RESULTS
Phylogeny and Expression of the ECT Gene Family
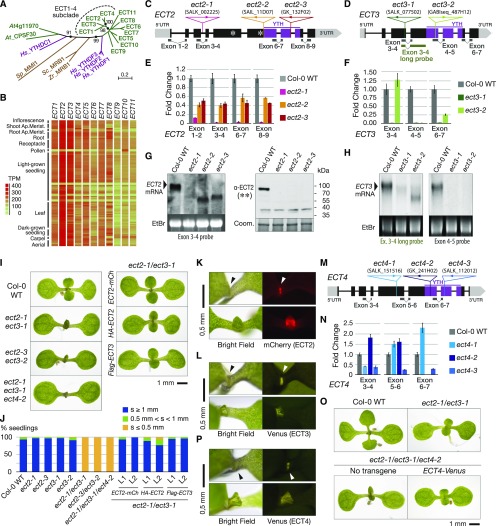
To study potential m6A binding proteins in plants, we first performed phylogenetic analysis of the 13 previously identified Arabidopsis YTH domain-containing proteins (Li et al., 2014a). These 13 proteins fell into the two known major clades: 11 members in the YTHDF clade (ECT1-11) and two members in the YTHDC clade (Figure 1A; Supplemental File 1). ECT1-11 all have an N-terminal region of variable length, and some members have small extensions C-terminal to the YTH domain (Li et al., 2014a). One of the two YTHDC-type proteins is a subunit of the Arabidopsis Cleavage and Polyadenylation Stimulatory Factor (CPSF30; AT1G30460) (Fray and Simpson, 2015). We next used the gene expression information in Araport 11 (Cheng et al., 2017) to survey expression levels and patterns of the ECT gene family. These analyses showed that ECT2 and ECT3 are the most highly and most widely expressed members of the family (Figure 1B). Together with the finding that ECT2 and ECT3 belong to the same phylogenetic subclade of YTHDF-type proteins (Figure 1A; 51% identity), this motivated our initial focus on ECT2 and ECT3 for further study.
Figure 1.
Timing of Leaf Formation Is Controlled by ECT Proteins.
(A) Phylogenetic relationship of Arabidopsis YTH domain proteins (green). The ECT1-4 subclade is highlighted. Human YTHDFs and YTHDC1 (magenta) and three yeast YTH domain proteins (brown) are included as a reference. Hs, Homo sapiens; Zr, Zygosaccharomyces rouxii; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the nodes (marked with black circles) that are relevant for this study. The length of the branches represents the evolutionary distance in number of amino acid substitutions per site.
(B) RNA-seq expression map of ECT genes extracted from ARAPORT (Cheng et al., 2017). Shading is a log2 scale of transcripts per million (TPM).
(C) and (D) Schematic representation of the ECT2 (C) and ECT3 (D) loci and their associated T-DNA insertion lines. Exons are depicted as boxes and introns as lines. Amplicons and probes used for qPCR and RNA gel blot are represented under the gene diagrams. Asterisks on ECT2 indicate the position of the peptides used to raise the antibody used in (G).
(E) and (F) qPCR analysis of ECT2 (E) and ECT3 (F) expression levels in T-DNA insertion alleles, using the amplicons shown in (C) and (D), respectively. Error bars represent the se in three technical replicates.
(G) RNA (left panel) and protein (right panel) blots of total RNA or protein purified from inflorescences of the indicated genotypes. Ethidium bromide (EtBr) and Coomassie blue (Coom.) staining are used as RNA and protein loading controls, respectively. The positions of the probe and the epitopes (**) are shown in (C).
(H) RNA gel blots of total RNA purified from seedlings of the indicated genotypes. Ethidium bromide (EtBr) staining is used as a loading control. Probes refer to (D).
(I) Eight-day-old seedlings of Col-0 wild type, ect2/ect3 double mutants with or without ECT2-mCherry, 3xHA-ECT2, or FLAG-ECT3 transgenes, and the triple mutant ect2-1/ect3-1/ect4-2.
(J) Quantification of the length of the first true leaves at 8 d after germination in the indicated genotypes. Only seedlings with cotyledons of at least 2.5 mm were considered. Between 50 and 100 seedlings were analyzed in all cases, and two independent stable lines (L1 and L2) were analyzed for each transgene. s, size of first true leaves.
(K) Fluorescence microscopy of ECT2-mCherry in ect2-1 ECT2-mCherry seedlings at 4 (upper panel) and 6 (lower panel) d after germination.
(L) Same as in (K) for Venus fluorescence in 4-d-old (upper panel) and 5-d-old (lower panel) ect3-2 ECT3-Venus seedlings. In (K) and (L), arrowheads point to fluorescence detected at sites of leaf formation.
(M) Schematic representation of the ECT4 locus and its associated T-DNA insertion lines, using the same symbols as in (C) and (D).
(N) qPCR analysis of ECT4 expression levels in seedlings of ect4 T-DNA insertion alleles, using the amplicons shown in (M). Error bars represent the se in three technical replicates.
(O) Ten-day-old seedlings of Col-0 wild type, ect2-1/ect3-1, and ect2-1/ect3-1/ect4-2 mutants with or without ECT4-Venus.
(P) Fluorescence microscopy of ECT4-Venus in 4-d-old (upper panel) and 8-d-old (lower panel) ect2-1/ect3-1/ect4-2 ECT4-Venus seedlings. Arrowheads denote detection of fluorescence at the shoot apex and emerging leaves as in (K) and (L).
Single Knockout Mutants in ECT2 and ECT3 Do Not Exhibit Obvious Developmental Phenotypes
We first obtained T-DNA insertion mutants in ECT2 and ECT3 (Figures 1C and 1D) and used quantitative RT-PCR and RNA gel blots to analyze mRNAs produced from the ECT2 and ECT3 genes. None of the three insertion mutants in ECT2 or the two insertion mutants in ECT3 produced detectable levels of full-length mRNA (Figures 1E to 1H), suggesting that all analyzed ect2 and ect3 mutant alleles were null. For ect2 mutant alleles, we corroborated this conclusion by raising functional antibodies recognizing two peptide epitopes in the N-terminal region of the ECT2 protein (Figure 1C). These antibodies clearly detected a protein of apparent molecular mass of 100 kD in wild-type lysates, while no protein was detected in lysates prepared from ect2-1, ect2-2, or ect2-3 mutants (Figure 1G, right panel). We note that the apparent molecular mass of ECT2 as judged by its migration in a denaturing polyacrylamide gel is higher than the expected 72.4 kD, probably because of the long N-terminal region predicted to be intrinsically disordered (Brocca et al., 2009). We conclude from these analyses that our set of ect2 and ect3 insertion mutants represent strong loss-of-function, if not null, alleles and that they are therefore appropriate for phenotypic analyses. Nonetheless, our initial inspection of growth and developmental phenotypes of ect2 and ect3 mutants did not reveal obvious differences from wild type (Supplemental Figure 1).
Timing of Leaf Formation Is Redundantly Controlled by ECT2 and ECT3
We next constructed ect2/ect3 double mutants to uncover possible genetic redundancy between the two ECT genes. Two independent sets of double null mutants were generated (ect2-1/ect3-1 and ect2-3/ect3-2) to establish causal relationships between any observed phenotypes and simultaneous loss of ECT2 and ECT3 function. Both ect2-1/ect3-1 and ect2-3/ect3-2 showed a delay in the emergence of the first true leaves. This phenotype was particularly easy to observe at the time of emergence of the first true leaves in the wild type ∼7 to 8 d after germination (Figure 1I; Supplemental Figure 1). To quantify the phenotype, we sorted 8-d-old seedlings into categories according to the size (s/mm) of true leaves (s ≥ 1; 0.5 < s < 1; s ≤ 0.5). This analysis showed that the phenotype was fully penetrant in both double mutant allele combinations (Figure 1J). It also allowed us to obtain formal statistical proof that while the distributions of true leaf sizes are not significantly different between the wild type and single mutants (P = 0.50, Fisher’s exact test), both ect2/ect3 double mutants have significantly smaller true leaves than the wild type at 8 d post germination (P = 2.2 × 10−16, Fisher’s exact test). Thus, ECT2 and ECT3 are required for the correct timing of leaf formation. In addition, transgenic expression of ECT2-mCherry, 3xHA-ECT2, and FLAG-ECT3 restored the delayed leaf emergence in ect2-1/ect3-1 (Figures 1I and 1J). All of these constructs used endogenous ECT promoters to drive expression, and for ECT2-mCherry and 3xHA-ECT2, we verified that protein levels were comparable to those of endogenous ECT2 (Supplemental Figure 2). These observations further confirm the redundant control of the timing of leaf formation by ECT2 and ECT3 and validate the functionality of the tagged proteins.
ECT2 and ECT3 Are Highly Expressed at Sites of Leaf Formation
To study in detail the expression of ECT2 and ECT3, we analyzed ECT2-mCherry and ECT3-Venus protein fusions under the control of the corresponding endogenous ECT2/ECT3 promoters. Similar to ECT2-mCherry, ECT3-Venus was suitable for detailed expression analyses, because the construct complemented ect2/ect3 double mutants, and the lines accumulated an intact fusion protein (Supplemental Figures 1 and 2). In young seedlings, ECT2-mCherry and ECT3-Venus showed strong fluorescence signals at the shoot apex, clearly above the background fluorescence of nontransgenic controls. Gratifyingly, the signal was particularly distinct at the sites of leaf formation and in emerging leaves (Figures 1K and 1L), thus providing clear coherence between the expression pattern and the importance of ECT2 and ECT3 in the timing of leaf formation, as revealed by genetic analysis.
Mutation of ECT4 Enhances Delayed Leaf Emergence in ect2/ect3
Because of the close sequence similarity between ECT2 and ECT4 (72% amino acid identity; Figure 1A) and because ECT4 promoter:GUS fusions were reported to show activity in emerging leaves (Li et al., 2014a), we also included the ect4-2 insertion mutant (GK-241H02) in our analysis (Figure 1M). ect4-2 is probably a null allele, as judged by the lack of ECT4 mRNA 3′ to the insertion site in the region encoding the YTH domain (Figures 1M and 1N). ect4-2 single and both ect2-1/ect4-2 and ect3-2/ect4-2 double mutant seedlings showed normal shoot development, as in the case for ect2 and ect3 single mutants (Supplemental Figure 1). In contrast, ect2-1/ect3-1/ect4-2 triple mutants showed a consistent delay of leaf formation similar to that of ect2/ect3 double mutants (Figures 1I and 1J), but the phenotype was more readily observable because the cotyledons of ect2-1/ect3-1/ect4-2 appeared more widely separated and the emergence of the first true leaves was slightly more delayed than in ect2/ect3 double mutants (Figure 1I). The enhancement of delayed leaf emergence was caused by ECT4 mutation because expression of an ECT4-Venus fusion in the ect2-1/ect3-1/ect4-2 background resulted in seedlings exhibiting the weaker ect2/ect3 double mutant phenotype (Figure 1O; Supplemental Figure 2). Analyses of ECT4-Venus fluorescence in these transgenic lines showed that ECT4 is also specifically expressed at sites of leaf formation in young seedlings (Figure 1P). Taken together, the expression of ECT2/3/4 at sites of leaf formation in young seedlings and the complete penetrance of delayed leaf emergence phenotypes in ect2/ect3 and ect2/ect3/ect4 mutants indicate that ECT2, ECT3, and ECT4 constitute a component required for the genetic control of the timing of postembryonic leaf formation.
m6A Binding Sites Are Required for ECT2 and ECT3 Function in Leaf Formation
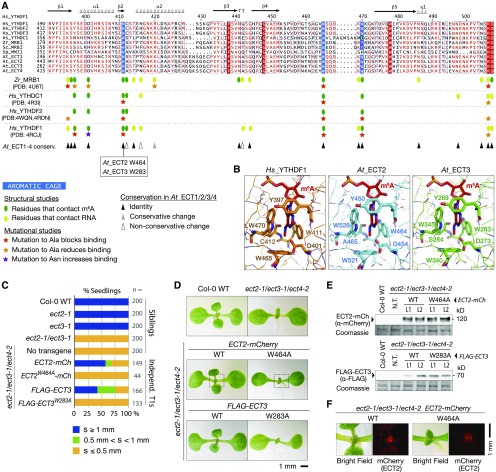
While the analyses of knockout alleles clearly established the importance of ECT2/3/4 in the timing of leaf emergence, they did not show whether binding to m6A-containing mRNA may underlie their biological role. To answer this key question, we first examined the patterns of conservation of the YTH domains in detail to determine whether they may possess m6A binding activity. Of 20 amino acid residues shown by structural analyses to be involved in RNA and m6A binding (Li et al., 2014b; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014, 2015), 16 were identical between ECT2/ECT3/ECT4 and human YTHDF proteins, and an additional residue had a conservative change (Figure 2A; Supplemental File 1). Of particular interest, the invariant residues included the tryptophans that form the aromatic cage essential for high-affinity binding to m6A-containing RNA (Figure 2A) (Fray and Simpson, 2015), suggesting that ECT2, ECT3, and ECT4 bind m6A. To further explore this property, we modeled the 3D structures of the YTH domains of ECT2 and ECT3 using the fully automated homology-modeling server SWISS-MODEL (Biasini et al., 2014). The crystal structure of the YTH domain of human YTHDF1 in complex with a 5mer m6A-containing RNA was the best template for both ECT2 and ECT3. Both models had excellent quality estimates (see Methods). The models showed that the structural arrangement of the m6A recognition pocket is nearly perfectly conserved in ECT2 and ECT3, and the only variable amino acid (Cys-412 in YTHDF1, Ala-465 in ECT2, and Ser-284 in ECT3) contacts m6A via backbone rather than side chain atoms (Figure 2B). In particular, no amino acid side chains in ECT2 or ECT3 sterically clash with the methyladenosine ligand, suggesting that the pocket is available for m6A binding (Figure 2B). Thus, the patterns of amino acid conservation and homology models strongly support m6A binding by ECT2 and ECT3. In human YTHDF1/2 and YTHDC1, mutation of the tryptophan residue corresponding to Trp-464 in ECT2 (ECT2W464) and Trp-283 in ECT3 (ECT3W283) to alanine is sufficient to fully abrogate the increased affinity to m6A-containing RNA over unmodified RNA (Li et al., 2014b; Xu et al., 2014, 2015; Zhu et al., 2014) (Figure 2A). Importantly, circular dichroism spectroscopy of this Trp-Ala mutant in human YTHDF2 has verified that it has secondary structure composition similar to the wild type, suggesting that it maintains the structural integrity of the YTH domain (Zhu et al., 2014). For these reasons, we constructed ECT2W464A-mCherry and FLAG-ECT3W283A m6A binding site mutants and scored their ability to rescue the leaf initiation defect in ect2-1/ect3-1/ect4-2 triple mutants compared with the corresponding wild-type constructs. Since a certain percentage of stable transgenic lines will always be noncomplementing, even when the protein encoded by the transgene is functional, we first determined complementation frequencies in large numbers of primary transformants using the same quantitative criteria as we used to describe the delayed leaf emergence in ect2/ect3 mutants (Figure 1J). By these criteria, roughly 70% of primary transformants obtained with wild-type transgenes rescued the seedling phenotype of ect2/ect3/ect4, while rescue was not observed in any primary transformants expressing the m6A binding site mutants (Figures 2C and 2D). This highly significant difference (P = 2.2 × 10−16, Fisher’s exact test) demonstrates that the aromatic cage mutants of ECT2 and ECT3 do not have biological function. To rule out trivial reasons for loss of function, such as protein instability in vivo, we isolated and characterized individual wild-type and mutant lines. Independent lines expressing wild-type ECT2-mCherry or FLAG-ECT3 fully rescued the ect2/ect3/ect4 seedling phenotype, in contrast to ECT2W464A-mCherry and FLAG-ECT3W283A lines that expressed levels of protein comparable to the wild-type fusions (Figure 2E). For the ECT2-mCherry fusions, we also verified that both wild-type ECT2-mCherry and ECT2W464A-mCherry were expressed in similar patterns at the shoot apex (Figure 2F). Taken together, these observations argue that loss of protein activity rather than misexpression or instability is the cause of noncomplementation of aromatic cage mutants of ECT2 and ECT3. Thus, intact m6A binding sites are required for the in vivo functions of ECT2 and ECT3.
Figure 2.
m6A Binding Sites Are Required for the in Vivo Function of ECT2 and ECT3.
(A) Multiple sequence alignment of part of the YTH domain of the proteins described in Figure 1A. The secondary structure elements of Hs_YTHDF1 are indicated, and amino acids are colored according to level of sequence conservation: red letters, similar residues; red boxes, identical residues (ESPript; Robert and Gouet, 2014). Amino acids that form the methyl-interacting aromatic cage are highlighted in blue. Oval marks and stars summarize the main finding of previous structural and mutational studies of the YTH domain of Zr_MRB1, Hs_YTHDC1, and Hs_YTHDF1/2 as indicated (Luo and Tong, 2014; Xu et al., 2014, 2015; Li et al., 2014b; Zhu et al., 2014). Triangles indicate the degree of conservation of the studied residues in ECT1/2/3/4. PDB, Protein Data Bank (IDs).
(B) Experimentally determined structure of the YTH domain of human YTHDF1 in complex with m6A RNA (PDB: 4RCJ), and models of the YTH domains of ECT2 and ECT3 generated using the homology-modeling server SWISS-MODEL. Residues forming contacts with m6A are highlighted.
(C) Percentages of primary transgenic lines of the indicated genotypes in three categories defined by the length of the first true leaves at 8 d after germination. s, size of first true leaves. Only seedlings with cotyledons longer than 2.5 mm were considered.
(D) Phenotypes of 9-d-old seedlings of the indicated genotypes. Dotted lines delimit the areas analyzed in (F).
(E) Protein blot analyses of wild type and mutant ECT2-mCherry (top) and FLAG-ECT3 (bottom). Two lines (L1 and L2) of each kind with comparable expression levels are shown. Coomassie staining is used as a loading control. N.T., no transgene.
(F) Expression of the wild type and mutant ECT2-mCherry at the shoot apex detected by fluorescence microscopy (mCherry).
Leaf Morphogenesis Requires ECT2, ECT3, and ECT4
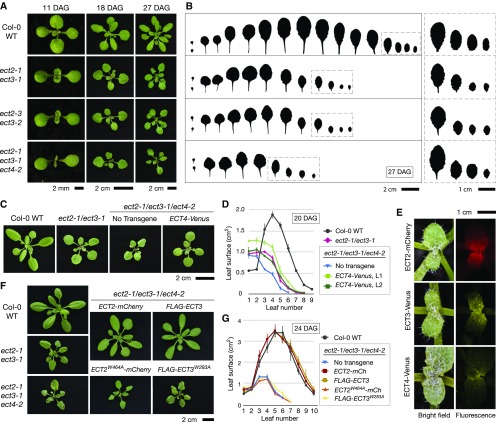
We next investigated whether the ECT genes might be involved in the definition of leaf shape in addition to the timing of leaf emergence. Indeed, leaves of ect2-1/ect3-1 and ect2-3/ect3-2 mutants exhibited an altered, triangular shape (Figures 3A and 3B), demonstrating that ECT2 and ECT3 control leaf development in an apparently redundant manner. In addition, the rosettes of double mutants were smaller than those of the wild type due to delayed leaf formation throughout development (Figures 3A and 3B). No leaf formation and morphology phenotypes could be observed in ect2/ect4 or ect3/ect4 double mutants (Supplemental Figure 1), but those phenotypes were strongly enhanced in ect2-1/ect3-1/ect4-2 triple mutants compared with ect2/ect3 double mutants: The overall rosette size was clearly smaller (Figures 3A and 3B), the serrations of young leaves were more pronounced (Figure 3B, right panel), and the triangular shape of, in particular, the first adult leaves was exaggerated compared with ect2/ect3 double mutants (Figures 3A and 3B). To confirm that mutation of ECT4 caused the enhancement of ect2/ect3 phenotypes, we used the transgenic lines expressing ECT4-Venus in the ect2/ect3/ect4 mutant background. Several independent lines showed restoration of ect2/ect3/ect4 leaf morphology and rosette size to an ect2/ect3-like phenotype based on visual inspection (Figure 3C). We also quantified this complementation by measuring leaf surface areas of individual leaves. These measurements confirmed that the expression of ECT4-Venus in ect2/ect3/ect4 restored leaf surface areas to the level observed in ect2/ect3 (Figure 3D) and thereby demonstrate a role for ECT4 in leaf development together with ECT2 and ECT3. We next used the fluorescent protein fusions to examine the expression of ECT2, ECT3, and ECT4 in developing leaves. All three genes were expressed in the proximal leaf parts in which cell division occurs, although the expression of ECT4 was weaker and more restricted to the proximal edge of the emerging leaves (Figure 3E). These expression patterns are consistent with the role of ECT2, ECT3, and ECT4 in leaf development.
Figure 3.
Leaf Morphogenesis Requires ECT2, ECT3, and ECT4.
(A) Seedlings and young rosettes of the indicated genotypes at three different time points. All plants were germinated directly on soil. DAG, days after germination.
(B) Leaf profiles of the plants in (A) at 27 DAG. Note the abnormal shape and delayed development in ect2-1/ect3-1, ect2-3/ect3-2, and ect2-1/ect3-1/ect4-2. The four younger leaves of all plants are magnified on the right side (dashed squares) to show the different margins.
(C) and (D) Partial restoration of ect2-1/ect3-1/ect4-2 rosette phenotypes (20 DAG) by expression of transgenic ECT4-Venus, as indicated by overall rosette phenotype (C) or measured by average leaf surface areas (D). T2 plants of two independent transgenic lines (L1 and L2) are analyzed. Dots indicate the average area of every type of leaf among three to five plants grown in parallel. Lines connect dots to facilitate reading. Error bars indicate se.
(E) Fluorescence microscopy of the second pair of true leaves of plants expressing ECT2-mCherry, ECT3-Venus, or ECT4-Venus as indicated.
(F) and (G) Complementation of ect2-1/ect3-1/ect4-2 rosette phenotypes (24 DAG) by expression of transgenic ECT2-mCherry or FLAG-ECT3, but not of their corresponding m6A binding site mutants. Data were obtained and represented as in (C) and (D), except that the plants are older and averages of transgenic lines are calculated from independent T1 plants.
The delayed formation, triangular shape, and serrations at the bases of leaves of ect2-1/ect3-1/ect4-2 bear remarkable resemblance to those of transgenic lines exhibiting weak silencing of the m6A methyltransferase subunit MTA by the expression of a complementary artificial miRNA (Shen et al., 2016). This observation strongly suggests that the leaf morphology defect of ect2/ect3/ect4 involves m6A and that ECT2/ECT3/ECT4 are the main effectors of m6A modifications implicated in the definition of leaf morphology. To prove this point rigorously, we used the characterized transgenic lines expressing wild-type and aromatic cage mutants in the ect2/ect3/ect4 background. In contrast to the wild-type fusions, ECT2W464A-mCherry and FLAG-ECT3W283A failed to complement ect2/ect3/ect4, as assessed by visual inspection (Figure 3F) and by measurement of leaf surface areas of individual leaves produced during rosette development (Figure 3G). We conclude that ECT2, ECT3, and ECT4 play overlapping roles in the control of leaf morphology and that at least ECT2 and ECT3 rely on intact m6A binding sites for this function.
Trichome Branching Is Stochastically Affected in the Absence of ECT2 and ECT3
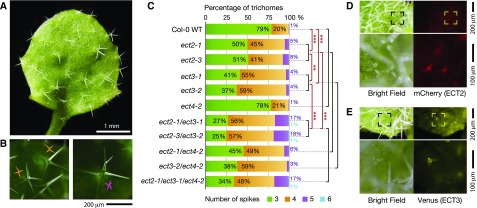
Since transgenic lines depleted of m6A during postembryonic development exhibit an increased number of trichome branches (Bodi et al., 2012), we also examined rosette leaves of ect2-1/ect3-1 mutants for aberrant trichome branching. We observed a striking increase in the number of spikes of the majority of trichomes (Figures 4A and 4B). ect2-3/ect3-2 exhibited a very similar trichome branching phenotype, demonstrating that aberrant trichome branching was due to the simultaneous loss of ECT2 and ECT3 function. To quantify more precisely the contribution of each gene to defective trichome branching, we counted the number of spikes on roughly 1000 trichomes per mutant line and compared the branching pattern of wild-type Col-0 and of ect2/ect3 double mutants to that of the single mutants used to construct them: ect2-1, ect2-3, ect3-1, and ect3-2. We also included ect4-2, ect2-1/ect4-2, ect3-2/ect4-2, and ect2-1/ect3-1/ect4-2 to test any potential involvement of ECT4 in trichome branching (Figure 4C). First, we did a likelihood ratio test for the null hypothesis that different mutant alleles of the same gene, or allele combinations of the same genes, produce the same branching patterns. This null hypothesis was accepted (likelihood ratio = 1.6841, df = 3, P = 0.6405). We next queried the data for significantly different branching patterns. These analyses showed that single mutation of either ECT2 or ECT3 caused significantly increased numbers of trichome branches (Figure 4C). The stronger effect of ect3 compared with ect2 (60% versus 50% trichomes with more than three branches) was also statistically significant, and the effect was significantly more pronounced in ect2/ect3 double mutants than in either single mutant (Figure 4C). In contrast, ect4-2 did not show any significant difference from the wild type, nor did mutation of ECT4 significantly alter the trichome branching pattern of ect2, ect3, or ect2/ect3 mutants (Figure 4C). We conclude that ECT3 and ECT2 are major effectors of the previously described role of m6A in trichome morphogenesis (Bodi et al., 2012) and that the effect of ECT4, if any, in this process cannot be observed in the genetic backgrounds used here. We also note that contrary to leaf initiation, the trichome branching defect in ect2/ect3 mutants is stochastic, implying that trichome morphogenesis can be correctly completed in the absence of a functional m6A-ECT2/ECT3 axis, but it is less likely to do so than in the wild type. Finally, we reinspected ECT2-mCherry and ECT3-Venus expression in young leaves too see if trichome expression could be detected. Mature trichomes did not display fluorescence signal, but several highly fluorescent dots were noted at the bases of leaves (Figure 3E). Careful inspection of such dots showed that they are in fact young trichomes at an early stage of development (Figures 4D and 4E) in which the branching pattern is defined (Hülskamp et al., 1994). Thus, ECT2 and ECT3 are highly expressed in young trichomes, and each protein plays necessary, if perhaps partly overlapping, roles in trichome development.
Figure 4.
Aberrant Trichome Morphology in the Absence of ECT2 and ECT3.
(A) Young leaf with several trichomes showing an increased number of branches. The picture shown is of an ect2-1/ect3-1/ect4-2 triple mutant, but ect2/ect3 double mutants exhibit the same branching pattern.
(B) Examples of trichomes with aberrant branching. Left, four-spiked trichomes marked with orange symbols; right, five-spiked trichome marked with a purple symbol.
(C) Branching pattern sorted by number of spikes in the indicated genotypes. Branches were counted on at least 150 trichomes on each of at least 6 plants for each genotype (n = ∼1000). Seven observations of two-spiked trichomes and one observation of a seven-spiked trichome were removed from the total of >12,000 trichomes counted for simplicity. Percentages corresponding to three and four spikes are overlaid on the green and orange colored bars, while percentages of five and six spikes, if any, are indicated to the right of the bar in purple and cyan numbers, respectively. Data were fitted to a proportional odds model in R for statistical analyses as described in the Methods. Asterisks indicate Bonferroni-corrected P values: **P < 0.01 and ***P < 0.001. Black bars indicate no significant difference.
(D) and (E) Fluorescence microscopy images of young trichomes in plants expressing ECT2-mCherry (D) or ECT3-Venus (E). Framed areas on upper panels are magnified below.
ECT2, ECT3, and ECT4 Are Enriched in the Cytoplasm
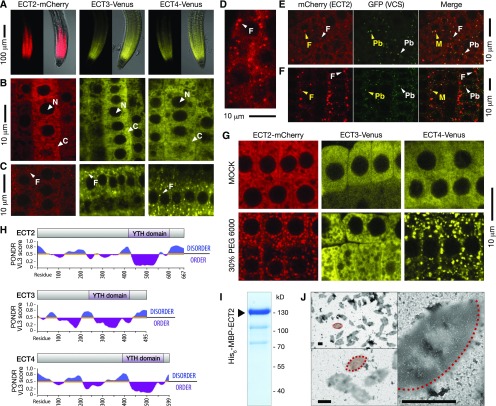
YTH domain proteins may exert different functions in gene expression in the nucleus and in the cytoplasm. For example, mRNA processing, mRNA export, and even epigenetic regulation may be controlled by nuclear YTH domain proteins in animals, while cytoplasmic YTH domain proteins may control the translation or stability of mature mRNA (Patil et al., 2018). To obtain first insights into how ECT2, ECT3, and ECT4 control plant gene expression, we analyzed their subcellular localization using our established fluorescent protein fusions. We noticed that all three proteins were well expressed in root meristems (Figure 5A) and used these cells to analyze subcellular localization because their small vacuoles and lack of chlorophyll facilitate observations by fluorescence microscopy. For all three proteins, fluorescence was detected only in the cytoplasm (Figure 5B). Thus, similar to animal proteins in the YTHDF clade (Wang et al., 2014, 2015; Li et al., 2017), ECT2, ECT3, and ECT4 are cytoplasmic proteins.
Figure 5.
ECT2, ECT3, and ECT4 Are Cytoplasmic and May Aggregate in Granules.
(A) Confocal images of mCherry or Venus fluorescence in root tips grown inside MS-agar of ect2-1 ECT2-mCherry, ect3-2 ECT3-Venus, and ect2-1/ect3-1/ect4-2 ECT4-Venus. Left panels display only fluorescence, and right panels show an overlay of fluorescence over bright field images.
(B) and (C) Fluorescence of root tips of the plants described in (A) at higher magnification. N, nucleus; C, cytoplasm; F, foci.
(D) Confocal image of ECT2-mCherry-expressing roots grown on the agar surface.
(E) and (F) Confocal images of mCherry and GFP fluorescence in root tips grown inside MS-agar of plants coexpressing VCS-GFP and ECT2-mCherry. Unstressed roots (E) and roots stressed (F) by partial dehydration in 0.8% noble agar for 10 h. Arrowheads point to different kinds of cytoplasmic bodies: F, ECT2-mCherry Foci; Pb, P-body; M, merge. Yellow coloring indicates rare colocalization.
(G) Confocal images of root tips of the plants described in (A) after treatment with 30% PEG6000 in MS, or mock treatment (MS), for 1 h.
(H) Intrinsic disorder prediction of ECT2/3/4 by PONDR-VL3 (Predictor of Naturally Disordered Regions-VL3) scores (Peng et al., 2005). PONDR-VL3 values increase with the predicted increase in disorder.
(I) Coomassie-stained SDS-PAGE gel of Ni2+-NTA-affinity purified His6-MBP-ECT2.
(J) Negative stain transmission electron microscopy of His6-MBP-ECT2 shown in (I). Examples of eye-shaped electron dense assemblages are outlined and shaded in red (only half-outlined at the highest magnification). Bars = 1 μm.
ECT2, ECT3, and ECT4 May Localize to Cytoplasmic Foci
In cells of roots grown inside Murashige and Skoog (MS)-agar medium, fluorescence was detected in a mostly diffuse, but somewhat heterogeneous, pattern (Figure 5B). Occasionally, some roots tips had foci in most cells (Figure 5C). For ECT2-mCherry, this pattern became prevalent when roots were grown on the surface of the medium (Figure 5D). To allow an assessment of potential localization to cytoplasmic P-bodies, we expressed the ECT2-mCherry fusions in plants containing GFP-tagged VARICOSE (VCS; AT3G13300) (Roux et al., 2015), an essential subunit of the decapping complex and a well-known P-body component (Goeres et al., 2007). Some mCherry fluorescence overlapped with VCS-GFP in P-bodies, but the ECT2 granules were bigger and the majority of the signal did not colocalize (Figure 5E). We speculated that drought stress could be the reason for the enhanced formation of foci in roots grown on the agar surface. Indeed, when roots embedded in 0.8% noble agar between a glass slide and its cover slip were left for 10 h at room temperature to allow for partial dehydration of the agar, most of the diffuse ECT2-mCherry signal was replaced by bigger and brighter foci (Figure 5F), suggesting that ECT2 aggregates in foci upon stress. Time-lapse imaging showed that these foci were dynamic, and occasionally, two distinct foci would form contacts and eventually merge (Supplemental Movie 1). To test whether osmotic stress can indeed induce the formation of foci, we exposed roots of transgenic ECT2-mCherry, ECT3-Venus, and ECT4-Venus lines to osmotic shock by treatment with 30% PEG6000 for 1 h (Xiong et al., 1999) and examined the subcellular localization of the fusion proteins. Cytoplasmic foci were observed for ECT2-mCherry and ECT4-Venus, but not for ECT3-Venus (Figure 5G). However, the foci did not form in every root tip every time, and we often saw some degree of condensation of the signal to a more granular texture, an effect that was also observed for ECT3-Venus (Figure 5G). We conclude that at least ECT2 and ECT4 may localize to foci upon stress. While we did not observe such localization of ECT3, we do not exclude the possibility that it also has the ability to relocalize to foci, although it appears to do so less readily than ECT2 and ECT4.
Pure ECT2 Can Form Regularly Sized Assemblages in Vitro
As in mammalian YTHDF proteins, the N-terminal part of ECT2, ECT3, and ECT4 is predicted to be intrinsically disordered (Figure 5H). It is an important biophysical property of many IDRs, in particular in RNA binding proteins that they are able to form assemblages with hydrogel-like properties in vitro (Kato et al., 2012). Such assemblages may underlie subcellular localization to membraneless bodies in vivo and are visible in vitro by electron microscopy (Banani et al., 2016). To examine whether ECT2 might possess such biophysical properties, we expressed full-length ECT2 fused to His6-MBP in Escherichia coli. The protein was purified by immobilized Ni2+-affinity chromatography and analyzed by negative stain electron microscopy (Figures 5I and 5J). This analysis showed the occurrence of regularly sized and shaped electron-dense assemblages, similar to what has been observed with other IDR-containing RNA binding proteins in vitro (Kato et al., 2012). We stress that these analyses show that ECT2 has the ability to form regular assemblages in vitro, but our experiments do not define the precise biochemical conditions (salt, protein concentration, temperature) under which these assemblages form.
DISCUSSION
m6A Dependence of ECT2/ECT3/ECT4 Function in Plant Developmental Timing and Morphogenesis
Our demonstration that the loss of ECT2, ECT3, and ECT4 causes delayed leaf initiation and defective leaf and trichome morphogenesis establishes the crucial importance of these YTH domain proteins in plant development. Three sets of observations argue that this biological importance is m6A-dependent. First, homology modeling strongly indicates the existence of intact m6A binding sites in ECT2 and ECT3, consistent with the nearly perfect conservation of residues implicated in m6A-RNA binding. Second, point mutants in ECT2 and ECT3 in an aromatic cage residue crucial for m6A recognition completely lose biological function. These observations indicate that specific m6A binding activity is required for their in vivo functions. Third, at least two developmental phenotypes of ect2/ect3 and ect2/ect3/ect4 are remarkably similar to those described for plants subjected to postembryonic knockdown of RNA methyltransferase subunits: defective trichome morphogenesis with increased numbers of branches (Bodi et al., 2012) and, in particular, delayed rosette development and triangular leaf shape with increased serrations (Shen et al., 2016). This equivalence between knockdown of m6A methyltransferase subunits and mutation of ECT2/ECT3/ECT4 suggest that the regulatory functions of m6A require ECT2/ECT3/ECT4. Taken together, these observations establish ECT2/ECT3/ECT4 as m6A readers and thereby introduce the m6A-ECT2/ECT3/ECT4 regulatory module as an important player in plant development. We note that the delayed leaf formation observed in ect2/ect3/ect4 here is reminiscent of the delayed larval development in zebrafish knockout mutants of YTHDF2 (Zhao et al., 2017), suggesting that control of developmental timing may be a recurrent function of eukaryotic YTHDF proteins.
Molecular Basis of the Biological Functions of the m6A-ECT2/ECT3/ECT4 Module
Although the goal of this study was limited to establishing biological importance of plant YTH domain proteins and their relation to m6A, it is of interest consider which molecular mechanisms may underlie their biological functions. Four observations suggest that similar to YTHDF2 in animals, ECT2/ECT3/ECT4 may accelerate target mRNA degradation to exert their developmental functions. First, postembryonic loss of m6A in FIP37-suppressed lines led to reduced decay rates of m6A-modified WUS and STM mRNAs (Shen et al., 2016), suggesting that m6A modification in the shoot apical meristem causes accelerated mRNA decay. Since ect2-1/ect3-1/ect4-2 triple mutant leaves phenocopy those of transgenic lines with weakly silenced MTA expression (Shen et al., 2016), the same molecular defects are likely to result from reduced m6A levels and the loss of ECT2/ECT3/ECT4 in leaf development. Second, ECT2, ECT3, and ECT4 are cytoplasmic, strongly suggesting that they exert their functions through effects on the fate of mature mRNA rather than mRNA processing. Thus, the subcellular localization of ECT2/ECT3/ECT4 is consistent with effects on mRNA stability but clearly cannot be taken as evidence for such a function in and of itself. Third, the observation that leaf initiation is delayed, but not abrogated, in ect2-1/ect3-1 and ect2-1/ect3-1/ect4-2 mutants is consistent with a function in accelerated target mRNA decay because leaf initiation from the periphery of the SAM requires repression of the Class I KNOX family of transcription factors (Hay and Tsiantis, 2010) that includes STM encoded by an m6A-containing mRNA. Fourth, studies of m6A in mammalian neural progenitor cells showed its relevance for correctly timed cell cycle exit through degradation of cell cycle-related mRNAs (Yoon et al., 2017). A similar principle may underlie the aberrant trichome morphology phenotype observed here because the number of endoreplication cycles in the trichome controls branch number (Schnittger and Hülskamp, 2002). Thus, continued endoreplication as a result of delayed cell cycle exit could cause increased trichome branching in ect2/ect3 mutants. Clear answers to whether functions of ECT2/ECT3/ECT4 in the accelerated decay of key developmental regulators underlie the leaf and trichome development phenotypes of the triple knockout mutant must await identification of m6A-modified target mRNAs bound directly by these proteins, as well as comparative analysis of their accumulation over time in meristems, leaf primordia, and trichomes of wild-type and mutant seedlings. In this way, this study opens up rich avenues of research to clearly define how m6A sculpts plant development.
Redundant Functions of ECT2, ECT3, and ECT4?
Since multiple ect knockouts were required to reveal the implication of an m6A-YTH module in leaf development, it is tempting to conclude that ECT2, ECT3, and ECT4 perform redundant functions in wild-type plants. We hesitate to suggest such a model for two reasons. First, knockout mutants are imperfect tests of gene function because of unnatural compensatory functions in vivo. For example, the closely related yeast MAP kinases Fus3 and Kss1 perform entirely different functions in controlling mating and virulence programs in wild-type cells (Schwartz and Madhani, 2004). Nonetheless, double knockout mutants are required to observe fully defective mating (Elion et al., 1991) because Kss1 adopts an unnatural function in the mating pathway in the absence of Fus3. Second, the analyses of trichome phenotypes in ect mutants show that knockout of single ECT genes can be sufficient to obtain an observable defect. Again, transcriptome-wide views of target mRNAs bound directly by the different ECT proteins will be crucial for understanding their potential functional overlap or specialization.
Properties of the N- and C-Terminal IDRs of Plant YTHDF Proteins: Subcellular Localization and Biophysical Properties
Although our study shows that ECT2, ECT3, and ECT4 are cytoplasmic, a previous study of the subcellular localization of the close ECT3 homolog, ECT1 (55% identity), concluded that it is a nuclear protein and that this nuclear localization is driven by its YTH domain (Ok et al., 2005). Since the YTH domains of ECT1 and ECT3 are very closely related (86% similar; 75% identical), the reason for this difference may be found in the properties of their N-terminal and C-terminal IDRs. Remarkably, we found that ECT2 and ECT4 in particular have the capacity to form cytoplasmic foci in vivo and that the pure, full-length ECT2 protein is capable of forming assemblages of roughly constant size and shape in vitro. Thus, biophysical properties intrinsic to the ECT2 protein itself may contribute to its ability to form such cytoplasmic foci in vivo. It is likely that the IDRs of ECT2, perhaps in particular the long N-terminal IDR, contribute to these remarkable properties because several RNA helicases and other RNA binding proteins that may accumulate in membraneless foci in vivo typically contain low-complexity regions able to drive hydrogel formation in vitro (Beckham et al., 2008; Kato et al., 2012; Lin et al., 2015; Banani et al., 2016). We also note that tethering of the IDR of human YTHDF2 to mRNA is sufficient for its localization to P-bodies (Wang et al., 2014), although it is unknown whether pure YTHDF2 is able to form assemblages in vitro similar to the ones we describe here for ECT2.
METHODS
Phylogenetic Analysis
Sequences of selected YTH-domain-containing proteins were downloaded from UniProt (Apweiler et al., 2004) (www.uniprot.org) and aligned using Clustal Omega (Sievers et al., 2011) with the following parameters: program, clustalo; version, 1.2.4; output guide tree, false; output distance matrix, false; dealign input sequences, false; mBed-like clustering guide tree, true; mBed-like clustering iteration, true; number of iterations, 0; maximum guide tree iterations, −1; maximum HMM iterations, −1; output alignment format, fa; output order, input; sequence type, protein. The secondary structure from the YTH domain of human YTHDF1 (Xu et al., 2015) (Protein Data Bank accession number 4RCJ) was annotated in the alignment using ESPript (Robert and Gouet, 2014). Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). The phylogenetic tree was generated from the alignment described above (Supplemental File 1) using the neighbor-joining method (Saitou and Nei, 1987). Values of the bootstrap test (Felsenstein, 1985) were inferred from 1000 replicates. Branches corresponding to partitions reproduced in <50% of the bootstrap replicates are collapsed. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 20 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 155 positions in the final data set.
Homology Modeling
The 3D structures of the YTH domains of ECT2 and ECT3 were modeled using the fully automated homology-modeling server SWISS-MODEL (Biasini et al., 2014). The crystal structure of the YTH domain of YTHDF1 in complex with RNA (Protein Data Bank entry 4RCJ) was the best template for structure modeling of both ECT2 and ECT3. Sequence identity to the template was 54% for ECT2 and 57% for ECT3. Quality estimates of the models by QMEAN Z-scores were as follows: QMEAN Z-score(ECT2) = −0.50, QMEAN Z-score(ECT3) = −1.35. Z-scores close to 0 indicate that global structural features are comparable to what is expected from experimentally determined structures of similar size, while Z-scores lower than −4.0 are indicative of a low-quality model (Biasini et al., 2014).
Plant Material and Growth Conditions
All lines used in this study are in the Arabidopsis thaliana Col-0 ecotype background, including ect2-1 (SALK_002225), ect2-2 (SAIL_11_D07), ect2-3 (GK_132_F02), ect3-1 (SALKseq_63401: SALK T-DNA NextG Seq for SALK_077502), ect3-2 (GABIseq487H12.1), ect4-1 (SALK_151516), ect4-2 (GK-241H02), and ect4-3 (SALK_112012). T-DNA insertion lines were obtained from Nottingham Arabidopsis Stock Centre. VCS-GFP plants were described by Roux et al. (2015). Seeds were surface-sterilized by immersion in 70% ethanol for 2 min followed by incubation in 1.5% hypochlorite, 0.05% Tween 20 for 10 min, and immediately rinsed with water. Sterilized seeds were stratified for 2 to 5 d at 4°C in darkness to obtain synchronous germination. Seedlings grown on MS-agar medium (4.4 g/L MS salt mixture, 10 g/L sucrose, and 8 g/L agar), pH 5.7, for 10 d were transferred to soil and maintained in Percival incubators with a 16-h-light/8-h-dark supplemental light cycle. Alternatively, surface-sterilized seeds were sown directly on soil when indicated. Philips fluorescent tubes (TL-D 90 De Luxe 36W) were used as the light source. The plants received light intensities of ∼70 μmol m−2 s−1 during in vitro growth and ∼100 μmol m−2 s−1 in Percival incubators.
Generation of 3xHA-ECT2, ECT2-mCherry, FLAG-ECT3, ECT3-Venus, and ECT4-Venus Transgenic Lines
The coding sequences of ECT2 (AT3G13460), ECT3 (AT5G61020), or ECT4 (AT1G55500), their upstream regulatory elements (ECT2P, ECT3P, or ECT4P), and downstream terminators (ECT2T, ECT3T, or ECT4T) were amplified from genomic DNA of Col-0 wild-type inflorescences by PCR using USER-compatible primers and KAPA HiFi Hotstart Uracil+ ReadyMix. Genomic DNA template for PCR was prepared as described by Arribas-Hernández et al. (2016).The primers were designed to create overhangs compatible with either the PacI USER cassette present in pCAMBIA3300U/2300U (pCAMBIA3300/2300 with a double PacI USER cassette inserted between the PstI-XmaI sites at the multiple cloning site; Nour-Eldin et al., 2006) or with the flanking sequences of the tags of choice (3xHA, mCherry, FLAG, or Venus). The 3xHA, mCherry, and Venus fragments were amplified by PCR from pSLF173 (Forsburg and Sherman, 1997), pCAMBIA-mCherry, and Venus-FANCD2 (Pedersen et al., 2015), respectively, using the same USER-compatible methodology. The FLAG sequence (DYKDDDDK) was obtained from synthetic 5′P-oligonucleotides. A list of constructs with the sequences of the oligonucleotides used in their production can be found in Supplemental Data Set 1. To obtain the constructs, their fragments were combined and inserted into pCAMBIA3300U by USER cloning (Bitinaite and Nichols, 2009) except for ECT3-Venus, which was constructed using pCAMBIA2300U. In all cases, kanamycin-resistant colonies were inspected by restriction digestion analysis and sequencing. Of note, manipulation of FLAG-ECT3 and ECT3-Venus was extraordinarily difficult due to unusually high rates of PCR-derived mutations clustered in the same region encoding the end of the IDR and the beginning of the YTH domain. In addition, the plasmids often rearranged during amplification in Escherichia coli, probably due to repetitive sequences located at the end of the IDR.
Arabidopsis stable transgenic lines were generated by floral dip transformation (Clough and Bent, 1998) with Agrobacterium tumefaciens GV3101. Selection of primary transformants (T1) produced with pCAMBIA3300U was done on soil by spraying seedlings with 0.2 g/L BASTA 10 d after germination or on MS-agar plates supplemented with glufosinate ammonium (Fluka; 7.5 mg/L). Alternatively, T1 seeds transformed with pCAMBIA2300U were selected on plates supplemented with kanamycin (50 mg/L). Segregation studies of T2 populations were performed on plates in the same manner. For every construct, at least three independent lines with a single T-DNA insertion and comparable expression patterns were selected in T2.
Transgenic lines coexpressing ECT2-mCherry and VCS-GFP were obtained by direct transformation of pCAMBIA3300U-ECT2-mCherry into VCS-GFP-expressing plants.
Site-Directed Mutagenesis
Mutation of the first tryptophan of the aromatic cage of ECT2 and ECT3 to alanine (ECT2-W464A and ECT3-W283A) in pCAMBIA3300U-ECT2-mCherry and -FLAG-ECT3 was performed by QuickChange site-directed mutagenesis (Agilent Technologies) following the manufacturer’s instructions. Phusion High-Fidelity DNA Polymerase (NEB) was used for PCR, and NEB DH 5-alpha Competent E. coli (High Efficiency) cells were used for chemical transformation. Primer sequences to produce and detect the mutations are detailed in Supplemental Data Set 1.
Fluorescence Microscopy
Imaging of whole seedlings and leaves was done using a Leica MZ16 F stereomicroscope mounted with a Sony a6000 camera. Roots were imaged with a Zeiss LSM700 confocal microscope. mCherry fluorescence was excited using a 555-nm laser, and GFP and Venus with a 488-nm laser. Emitted light was captured by the filter configuration preprogrammed for mCherry, GFP, and ZsYellow with the microscope software. Time-lapse images were taken every 30 s. For osmotic stress treatment, seedlings were transferred to filter paper soaked in 30% polyethylene glycol molecular weight 6000 (PEG6000) in MS and placed in the light at room temperature for one hour as described by Xiong et al. (1999).
Phenotypic Characterization
Images of seedlings, rosettes, and trichomes were acquired with a Leica MZ16 F stereomicroscope mounted with a Sony a6000 camera for specimens smaller than 2 cm or with a Canon EOS 1100 D camera when larger.
Pictograms of detached leaves were obtained from photographs using the tool “Adjust/Threshold” of the image processing package FIJI (Schindelin et al., 2012). The area of every leaf (including petiole) was determined with the same software applying “Analyze Particles” to pictograms of three to five plants for each genotype.
Quantification of trichome branching was done manually by counting the number of spikes in all trichomes of two to three half leaves of at least six plants per genotype, producing a data set of roughly 1000 trichome counts for each of the 11 genotypes analyzed. The plants were grown in parallel and the quantification started using the biggest leaves of 20-d-old plants. As the counting process spanned 4 d, we counted the bigger plants first (Col-0 wild type, single mutants, and ect4-containing double mutants). The smaller ect2/ect3 and ect2/ect3/ect4 plants were characterized last, allowing the leaves to mature to more comparable sizes. For statistical analyses, we applied a proportional odds model for ordinal regression using the ordinal package in the R software system. The response variable (number of trichome spikes) was on an ordinal scale from three to six spikes. Proportional odds ordinal regression was done with random effect of individual plants. The random effect was used to correct for multiple measurements on the same plants. P values were Bonferroni corrected for multiple testing.
Recombinant Protein Expression and Purification
cDNA encoding residues 1 to 667 (full coding sequence) of Arabidopsis ECT2 (AT3G13460.1) was cloned into pDEST-HisMBP (Nallamsetty et al., 2005) from the clone U15252 from the ABRC. The expression of His6-MBP-ECT2 (His6-MBP-ECT2) was induced by the addition of 1 mM IPTG to E. coli BL21 (DE3) codon plus (RIL) cells grown at 37°C in Luria-Bertani medium (100 μg/mL ampicillin and 33 μg/mL chloramphenicol) to OD600 ≈ 0.5. Cultures were grown overnight at 16°C, and pelleted bacteria were resuspended in IMAC binding buffer (20 mM Tris HCl, pH 8.0, 300 mM NaCl, and 10 mM imidazole) supplemented with EDTA-free protease inhibitor (cOmplete; Roche) and 1 mM TCEP. Cells were lysed using a French press, and the crude lysate was cleared by centrifugation at 12,500g, filtered through a 0.45-μm membrane, and applied to a gravity-flow chromatography column of Ni2+-NTA resin (Macherey-Nagel). IMAC washing buffer (IMAC binding buffer with 20 mM imidazole) was used for intermediate washing steps. Bound protein was eluted with IMAC elution buffer (IMAC binding buffer with 300 mM imidazole).
Electron Microscopy
Ten-microliter droplets of 0.5 mg/mL Ni2+-NTA-purified His6-MBP-ECT2 (in 25 mM Tris-HCl, pH 7.5, and 300 mM NaCl) were spotted onto Parafilm. Transmission electron microscopy grids were applied on top of the droplets (carbon-coated copper grids, 200 mesh; Electron Microscopy Sciences) for 10 min at room temperature. To remove the excess of adsorbed protein, the grids were washed for 10 s with water, and 10 μL of 2% uranyl acetate droplets were applied for 2 min for negative staining. Residual uranyl acetate was removed with filter paper before analysis of the grids under a JEOL 1400 transmission electron microscope.
Genotyping
Genomic DNA from young leaves or inflorescences was extracted as described by Arribas-Hernández et al. (2016). A detailed list of T-DNA lines with the corresponding PCR primers for genotyping and the observed lengths of the obtained amplicons can be found in Supplemental Data Set 1.
Quantitative RT-PCR
cDNA was obtained by reverse transcription of DNase-treated total RNA from inflorescences (ect2 alleles) or seedlings (ect3 and ect4 alleles) by TRIzol extraction (Arribas-Hernández et al., 2016). Briefly, 1 μg of RNA was added to a total volume of 10 μL reaction containing 1 μL (1 unit) of DNase I, 0.2 μL (0.2 units) of Ribolock, and 1× DNase buffer (Thermo Scientific). After 30 min at 37°C, the reaction was stopped with 1 μL of 50 mM EDTA and 10 min incubation at 65°C. Four microliters of the reaction containing DNA-free RNA was mixed with 0.5 μg (1 μL) of oligo(dT)18 (Thermo Scientific), denatured for 5 min at 65°C, and added to a total of 20 μL of first-strand cDNA synthesis reaction containing 1 unit of reverse transcriptase, 0.5 units of Ribolock, 4 mM dNTP mix, and 1× RT buffer (Thermo Scientific). The RT reaction proceeded at 42°C for 1 h, followed by 10 min at 70°C to deactivate the enzyme.
Quantitative RT-PCR (qPCR) reactions consisted of 5 μL of Maxima SYBR Green qPCR Master Mix (2×) (Thermo Scientific) mixed with 0.5 μL of the cDNA prepared as described above and the appropriate primers (Supplemental Data Set 1) in 0.3 μM final concentration each, to reach a total volume of 10 μL. The reactions were monitored in real time in a Bio-Rad CFX Connect thermal cycler, and expression analysis was performed following the ΔΔCT method (Pfaffl, 2001). Fold changes of gene expression in the different genotypes were calculated relative to Col-0 wild type using ACTIN2 (AT3G18780) for normalization of cDNA input. In all cases, means and standard errors were calculated based on CT values of three technical replicates. Primers were designed with the Universal Probe Library (Roche) online resource, and their sequences can be found in Supplemental Data Set 1.
Protein Gel Blotting
Protein blots from plant extracts were performed as described by Arribas-Hernández et al. (2016). We used commercially available antibodies against FLAG (Sigma-Aldrich A8592; 1:750 dilution), HA (Abnova 12CA5; 1:2000 dilution), mCherry (Abcam ab183628; 1:1000 dilution), and Venus (Sicgen AB2166-100; 1:1000 dilution). The anti-ECT2 antibody was affinity-purified by Biogenes from sera collected from rabbits immunized with a 1:1 mix of the peptides C+KGNLDDSLEVKE-NH2 and C+QDPRYAYEGYYAPVPW-NH2, and diluted 1:500. Peptides were synthesized by Schafer-N.
mRNA Gel Blot Analysis
Twenty-five micrograms of total RNA extracted from inflorescences or seedlings as described by Arribas-Hernández et al. (2016) and dissolved in loading buffer (1× HEPES [20 mM HEPES, 1 mM EDTA, and 17 mM KOH, pH 7.8], 45% formamide, 16% formaldehyde, 16% ethidium bromide, and bromophenol blue) was denatured by incubation at 65°C for 10 min and loaded into a 1% agarose gel prepared with 16% formaldehyde 1× HEPES buffer. The RNA was separated during 4 h of electrophoresis at 120 V in 1× HEPES buffer and blotted overnight onto an Amersham Hybond-NX nylon membrane (GE Healthcare Life Sciences) by the capillary motion of 20× SSC buffer (3 M NaCl and 300 mM sodium citrate, pH 7.0) absorbed by Whatman 3MM paper and additional paper towels with a weight on top. After blotting, the RNA was UV-cross-linked to the membrane. The membrane was then soaked and incubated with PerfectHyb Plus Hybridization buffer (Sigma-Aldrich) at 65°C for 20 min before the addition of radioactively labeled ECT2 or ECT3 probes (primers LA355-356, LA391-MH35, and LA755-756; Supplemental Data Set 1) for hybridization overnight at 65°C with constant rotation. The probe was synthesized using the Prime-a-Gene (Promega) kit and denatured (5 min at 95°C) before incubation with the membrane. After hybridization and washing (3 washes of 20 min with 2× SSC 0.1% SDS at 65°C), the membrane was exposed to a phosphor imager screen for image analysis.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MTA (AT4G10760), MTB (AT4G09980), FIP37 (AT3G54170), VIR (AT3G05680), ECT2 (AT3G13460), ECT3 (AT5G61020), ECT4 (AT1G55500), VCS (AT3G13300), WUS (AT2G17950), and STM (AT1G62360).
Supplemental Data
Supplemental Figure 1. Control of the timing of leaf organogenesis and morphogenesis by ECT proteins.
Supplemental Figure 2. Expression of ECT2-mCherry, 3xHA-ECT2, FLAG-ECT3, ECT3-Venus, and ECT4-Venus.
Supplemental Movie 1. Confocal time-lapse imaging of the movement of foci containing ECT2-mCherry.
Supplemental Data Set 1. Oligonucleotides used in this study.
Supplemental File 1. Alignment in FASTA format used to produce the phylogenetic tree shown in Figure 1 and the ESPript representation shown in Figure 2.
Acknowledgments
This work was supported by a Hallas Møller Stipend from the Novo Nordisk Foundation (HM-2010), a project grant from the Lundbeck Foundation (R83-A7859), Starting and Consolidator Grants from the European Research Council (MICROMECCA 282460 and PATHORISC 726417), and equipment grants from Brdr Hartmann Fonden, Augustinus Fonden, and Carlsberg Fondet, all to P.B. We thank Sara Simonini for her helpful suggestions and comments on this manuscript. We thank Lena Bjørn Johansson, Mathias Tankmar, and Emilie Oksbjerg for their help with experimental procedures. We also thank Morten Nørholm for the gift of pCAMBIA3300U/2300U plasmids, Vibe H. Østergaard for Venus-FANCD2, and Bo Markussen for help with statistical analysis.
AUTHOR CONTRIBUTIONS
L.A.-H. and P.B. designed the study. L.A.-H., M.H.H., and C.P. did phylogenetic analysis of ECT proteins and expressed ECT2. C.P. did protein structure modeling. L.A.-H. and M.H.H. characterized T-DNA insertion lines and constructed double and triple mutants. L.A.-H. constructed and characterized all ECT transgenic lines. L.A.-H. and S.B. did confocal microscopy. S.E. did electron microscopy analyses. L.A.-H., S.B., and P.B. analyzed data. P.B. and L.A.-H. wrote the manuscript with input from all authors.
Footnotes
Articles can be viewed without a subscription.
References
- Apweiler R., et al. (2004). UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernández L., Marchais A., Poulsen C., Haase B., Hauptmann J., Benes V., Meister G., Brodersen P. (2016). The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K. (2016). Compositional control of phase-separated cellular bodies. Cell 166: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J., et al. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C., Hilliker A., Cziko A.M., Noueiry A., Ramaswami M., Parker R. (2008). The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 19: 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaite J., Nichols N.M. (2009). DNA cloning and engineering by uracil excision. Curr. Protoc. Mol. Biol. 3: 21. [DOI] [PubMed] [Google Scholar]
- Bodi Z., Button J.D., Grierson D., Fray R.G. (2010). Yeast targets for mRNA methylation. Nucleic Acids Res. 38: 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z., Zhong S., Mehra S., Song J., Graham N., Li H., May S., Fray R.G. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocca S., Samalíková M., Uversky V.N., Lotti M., Vanoni M., Alberghina L., Grandori R. (2009). Order propensity of an intrinsically disordered protein, the cyclin-dependent-kinase inhibitor Sic1. Proteins 76: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Clancy M.J., Shambaugh M.E., Timpte C.S., Bokar J.A. (2002). Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. (2016). YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7: 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A., Brill J.A., Fink G.R. (1991). Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harb. Symp. Quant. Biol. 56: 41–49. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L., Sherman D.A. (1997). General purpose tagging vectors for fission yeast. Gene 191: 191–195. [DOI] [PubMed] [Google Scholar]
- Fray R.G., Simpson G.G. (2015). The Arabidopsis epitranscriptome. Curr. Opin. Plant Biol. 27: 17–21. [DOI] [PubMed] [Google Scholar]
- Fukusumi Y., Naruse C., Asano M. (2008). Wtap is required for differentiation of endoderm and mesoderm in the mouse embryo. Dev. Dyn. 237: 618–629. [DOI] [PubMed] [Google Scholar]
- Geula S., et al. (2015). m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006. [DOI] [PubMed] [Google Scholar]
- Goeres D.C., Van Norman J.M., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Hongay C.F., Orr-Weaver T.L. (2011). Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl. Acad. Sci. USA 108: 14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M., Misŕa S., Jürgens G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566. [DOI] [PubMed] [Google Scholar]
- Ivanova I., Much C., Di Giacomo M., Azzi C., Morgan M., Moreira P.N., Monahan J., Carrieri C., Enright A.J., O’Carroll D. (2017). The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67: 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E., Kierzek R. (2003). The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31: 4472–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27: 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang H., Hong Y., Huang L., Li X., Zhang Y., Ouyang Z., Song F. (2014a). Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Report. 32: 1169–1186. [Google Scholar]
- Li F., Zhao D., Wu J., Shi Y. (2014b). Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 24: 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Zhang Z., Zamudio A.V., Zhao J.C. (2015). Genome-wide detection of high abundance N6-methyladenosine sites by microarray. RNA 21: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., Parker R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60: 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10: 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518: 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Tong L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA 111: 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.Z., MacQueen A., Zheng G., Duan H., Dore L.C., Lu Z., Liu J., Chen K., Jia G., Bergelson J., He C. (2014). Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S., Austin B.P., Penrose K.J., Waugh D.S. (2005). Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 14: 2964–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ok S.H., Jeong H.J., Bae J.M., Shin J.S., Luan S., Kim K.N. (2005). Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol. 139: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D.P., Pickering B.F., Jaffrey S.R. (2018). Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen R.T., Kruse T., Nilsson J., Oestergaard V.H., Lisby M. (2015). TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J. Cell Biol. 210: 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. (2005). Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 3: 35–60. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M., Liao Y., Rettel M., Ragan C., Evers M., Alleaume A.-M., Horos R., Hentze M.W., Preiss T., Millar A.A. (2016). In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 28: 2435–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42: W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M.E., Rasmussen M.W., Palma K., Lolle S., Regué À.M., Bethke G., Glazebrook J., Zhang W., Sieburth L., Larsen M.R., Mundy J., Petersen M. (2015). The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J. 34: 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A., Hülskamp M. (2002). Trichome morphogenesis: a cell-cycle perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., et al. (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.A., Madhani H.D. (2004). Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38: 725–748. [DOI] [PubMed] [Google Scholar]
- Shah J.C., Clancy M.J. (1992). IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Liang Z., Gu X., Chen Y., Teo Z.W., Hou X., Cai W.M., Dedon P.C., Liu L., Yu H. (2016). N(6)-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 38: 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P., Rafalska I., Stamm S. (2002). YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27: 495–497. [DOI] [PubMed] [Google Scholar]
- Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42: 13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhu Y., Bao H., Jiang Y., Xu C., Wu J., Shi Y. (2016). A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res. 44: 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., et al. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61: 507–519. [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J.-K. (1999). HOS5-a negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. Plant J. 19: 569–578. [DOI] [PubMed] [Google Scholar]
- Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10: 927–929. [DOI] [PubMed] [Google Scholar]
- Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. (2015). Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290: 24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.-J., et al. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., et al. (2017). m6A modulates haematopoietic stem and progenitor cell specification. Nature 549: 273–276. [DOI] [PubMed] [Google Scholar]
- Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C. (2017). m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542: 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y. (2014). Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24: 1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, Bryson V., Vogel H.J., eds (New York: Academic Press; ), pp. 97–166. [Google Scholar]