The receptor-like cytoplasmic kinase STRK1 positively regulates rice salt and oxidative stress tolerance through maintaining H2O2 homeostasis by phosphorylating CatC mainly at Tyr-210.
Abstract
Salt stress can significantly affect plant growth and agricultural productivity. Receptor-like kinases (RLKs) are believed to play essential roles in plant growth, development, and responses to abiotic stresses. Here, we identify a receptor-like cytoplasmic kinase, salt tolerance receptor-like cytoplasmic kinase 1 (STRK1), from rice (Oryza sativa) that positively regulates salt and oxidative stress tolerance. Our results show that STRK1 anchors and interacts with CatC at the plasma membrane via palmitoylation. CatC is phosphorylated mainly at Tyr-210 and is activated by STRK1. The phosphorylation mimic form CatCY210D exhibits higher catalase activity both in vitro and in planta, and salt stress enhances STRK1-mediated tyrosine phosphorylation on CatC. Compared with wild-type plants, STRK1-overexpressing plants exhibited higher catalase activity and lower accumulation of H2O2 as well as higher tolerance to salt and oxidative stress. Our findings demonstrate that STRK1 improves salt and oxidative tolerance by phosphorylating and activating CatC and thereby regulating H2O2 homeostasis. Moreover, overexpression of STRK1 in rice not only improved growth at the seedling stage but also markedly limited the grain yield loss under salt stress conditions. Together, these results offer an opportunity to improve rice grain yield under salt stress.
INTRODUCTION
Adverse environmental stresses, such as drought and salinity, can seriously affect plant growth and crop production. In response to environmental cues, plants have evolved a variety of physiological and biochemical mechanisms to cope with adverse conditions during their growth and development (Boyer, 1982). Signal transduction mediated by membrane-anchored receptor-like kinases (RLKs) is an important regulatory mechanism by which plants respond to different environmental stresses and developmental cues (Kim et al., 2011; Chen et al., 2013).
RLKs constitute a large gene family, with over 610 genes in Arabidopsis thaliana and over 1131 in rice (Oryza sativa) (Shiu et al., 2004). Structurally, RLKs typically include an extracellular domain, a transmembrane domain, and an intracellular Ser/Thr protein kinase domain (Torii, 2000). Mechanistically, the extracellular domains of RLKs perceive a wide range of extracellular signals or stimuli, resulting in homodimerization or heterodimerization followed by autophosphorylation of the intracellular kinase domains and then activation of downstream signaling factors by transphosphorylation (Osakabe et al., 2013). However, one of the RLK families lacks an extracellular domain and includes only an intracellular kinase domain or only the transmembrane domain with an intracellular kinase domain. This family was thus designated as the receptor-like cytoplasmic kinases (RLCKs) (Shiu et al., 2004). There are 200 RLCK genes in Arabidopsis and 379 in rice (Vij et al., 2008). So far, only a few RLCKs have been shown to be involved in abiotic stress responses. Overexpression of OsRLCK253 in Arabidopsis improved tolerance to water deficit and salt stress through its interaction with stress-associated proteins (OsSAP1/11) (Giri et al., 2011). Heterologous expression of the receptor-like cytoplasmic kinase GsRLCK, identified from Glycine soja, improved tolerance of Arabidopsis plants to salt and drought stresses with increased expression levels of stress-induced genes (Sun et al., 2013). A rice receptor-like cytoplasmic kinase, GUDK, mediates drought stress signaling through phosphorylation and activation of OsAP37, leading to transcriptional activation of stress-induced genes, which confers drought tolerance and improves grain yield in rice (Ramegowda et al., 2014). Recently, a cold-activated receptor-like cytoplasmic kinase, CRPK1, was reported to phosphorylate 14-3-3 proteins and induce their nuclear import for fine-tuning C-repeat binding factor signaling during the cold response in Arabidopsis (Liu et al., 2017).
Reactive oxygen species (ROS), such as superoxide anion radical (·O2−), hydroxyl radical (·OH), and hydrogen peroxide (H2O2), serve as vital signaling molecules in many biological processes, including biotic and abiotic stress tolerance (Mittler et al., 2011; Schippers et al., 2012; Schmidt et al., 2013). Salt-induced ROS, mainly in the form of H2O2, emerge within several minutes of the applied stress, which depends on the activity of NADPH oxidases that contributes to salt or drought stress tolerance in plants (Hong et al., 2009; Ma et al., 2012; Wang et al., 2016). SALT-RESPONSIVE ERF1 involving H2O2-dependent molecular signaling cascade is required for the regulation of the initial transcriptional response during salt stress in rice (Schmidt et al., 2013). However, excessive accumulation of ROS is potentially harmful to cells and causes oxidative damage to proteins, DNA, and lipids (Apel and Hirt, 2004). Production of ROS is enhanced in plants suffering from diverse abiotic stresses, such as drought, salt, and temperature (Mittler et al., 2004). Plants have evolved efficient enzymatic and nonenzymatic detoxification mechanisms to scavenge ROS (Xu et al., 2013). Among the ROS-scavenging enzymes, catalase (CAT) is a strong antioxidant enzyme that catalyzes the decomposition of H2O2 to water and oxygen and thus plays a critical role in plant responses to abiotic stresses (Mhamdi et al., 2010). Resistance to abiotic stresses is greatly enhanced in transgenic plants overexpressing the CAT gene (Shikanai et al., 1998; Miyagawa et al., 2000; Polidoros et al., 2001; Moriwaki et al., 2007). In rice, CatA, CatB, and CatC are involved in environmental stress responses, and overexpression of CatA and CatC conferred tolerance to drought stress in transgenic rice (Joo et al., 2014).
Phosphorylation has been demonstrated as an important posttranslational modification in many RLKs and RLCKs to regulate diverse signaling pathways (Shiu et al., 2004; Macho et al., 2015). Plant RLKs and RLCKs are traditionally classified as serine/threonine kinases (Shiu and Bleecker, 2001), but recent work has revealed that tyrosine phosphorylation is also crucial for the activation of RLK/RLCK-mediated signaling in plants (Macho et al., 2015). Well-studied examples of RLK/RLCK-mediated signaling pathways are the steroid hormone brassinosteroid (BR) signaling pathway, which promotes plant growth (Zhu et al., 2013), and the initiation of immune signaling triggered by plant pattern-recognition receptors (Boller and Felix, 2009). BRASSINOSTEROID-INSENSITIVE1 (BRI1) and BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), the receptor and coreceptor of BR, are dual-specificity kinases, and tyrosine phosphorylation plays a prominent role in the perception of BR and subsequent signal transduction. For example, phosphorylation of a specific tyrosine residue (Tyr-211) occurs in BKI1, a kinase inhibitor of BRI1, in response to BR perception, which releases BKI1 into the cytosol and enables in turn formation of an active signaling complex (Wang et al., 2014). In the innate immune signaling pathway, two RLKs, EF-TU RECEPTOR (EFR) and BAK1, interact with an RLCK BOTRYTIS-INDUCED KINASE1 (BIK1) to initiate plant immune responses to bacterial elongation factor Tu (EF-Tu; or elf18) (Macho and Zipfel, 2014). Tyr-836 of EFR is phosphorylated in vivo after elf18 perception, which is required for the activation of EFR and downstream immune responses (Macho et al., 2014). BIK1 is autophosphorylated and phosphorylated by BAK1 at multiple tyrosine residues in addition to serine/threonine residues. Notably, several BIK1 tyrosine residues are required for the BIK1-mediated phosphorylation of substrates in vitro and for BIK1-dependent immune responses in planta (Lin et al., 2014).
CAT activity was reported to be activated by protein kinase through phosphorylation (Kumar et al., 2010; Rafikov et al., 2014; Zou et al., 2015). Ser-167 of CAT is phosphorylated by protein kinase Cδ (PKCδ) in response to endothelin 1 in humans, which increases CAT activity and decreases cellular H2O2 levels (Kumar et al., 2010; Rafikov et al., 2014). An Arabidopsis calcium-dependent protein kinase, CPK8, was reported to mediate drought stress signaling through phosphorylation at Ser-261 and activation of CAT3, which plays an important role in maintaining H2O2 homeostasis (Zou et al., 2015). So far, although several RLKs/RLCKs such as the CrRLK1Ls (Boisson-Dernier et al., 2013) and FERONIA (Duan et al., 2014) have been reported to regulate H2O2 homeostasis, there is no report of RLKs/RLCKs being involved in the regulation of H2O2 homeostasis and improvement of abiotic tolerance by tyrosine phosphorylation on CAT. In this study, we characterized a novel rice receptor-like cytoplasmic kinase, STRK1 (salt tolerance receptor-like cytoplasmic kinase 1), which activates CatC activity mainly through phosphorylation at Tyr-210 of CatC to regulate H2O2 homeostasis and improve salt tolerance. Notably, overexpression of STRK1 in rice significantly improved the tolerance to salt and oxidative stresses and increased grain yield.
RESULTS
An RLCK, STRK1, Positively Regulates Salt Tolerance in Rice
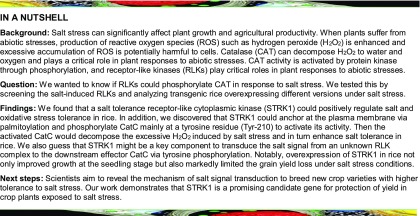
To identify genes that contribute to salt stress tolerance, we compared transcript profiles of rice RLKs based on chip data (Tyagi et al., 2007; Vij et al., 2008), and partial salt-induced RLKs were selected and further verified by a real-time PCR analysis in rice (O. sativa cv Kitaake) under salt treatment. The transcription of six candidate RLK/RLCK genes, Os01g05960, Os04g45730, Os06g07070, Os07g03810, Os08g03020, and Os09g39930, were found to be induced by salt stress (Supplemental Figure 1). To study the function of these RLKs in plant responses to salt, the transgenic rice plants overexpressing each RLK/RLCK were generated by introducing the pCAMBIA1301GW (Zhou et al., 2014, 2015) vector that harbored either a fragment of the extracellular and transmembrane domain coding sequence of RLK or a full-length coding sequence of RLCK to generate the dominant-negative or gain-of-function transgenic plants, respectively (Supplemental Figure 2A). T2 seedlings of transgenic lines were used to identify the phenotypes under salt stress conditions. Among them, an RLCK gene, Os04g45730, with a gene size of 1744 bp and encoding a protein of 361 amino acids, was found to improve significantly the salt tolerance in transgenic rice plants (Figures 1A and 1B; Supplemental Figure 2B). Thus, the RLCK gene Os04g45730 was referred to as STRK1.
Figure 1.
STRK1 Positively Regulates Salt Tolerance in Rice at the Seedling Stage.
(A) Phenotypic comparison of seedlings grown under salt stress at the seedling stage. For STRK1-OE transgenic plants, 15-d-old seedlings were treated with 140 mM NaCl for 10 d and then recovered for 6 d. For STRK1-RNAi transgenic plants, 15-d-old seedlings were treated with 140 mM NaCl for 8 d and then recovered for 6 d.
(B) and (C) Survival rates of transgenic and wild-type plants in (A) after 6 d of recovery. Forty plants in each line were used for survival rate analysis.
(D) Chlorophyll content in leaves of 15-d-old plants treated with 100 mM NaCl for 7 d.
(E) MDA concentrations in leaves of 15-d-old plants after 100 mM NaCl treatment for 24 h.
(F) Relative ion leakage in leaves of 15-d-old plants after 100 mM NaCl treatment for 24 h. For all panels, OE indicates STRK1-overexpressing plants; Ri indicates STRK1-RNAi plants; numbers indicate different transgenic lines; data in (B) to (F) are presented as mean ± sd (n = 3, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
To completely investigate the function of STRK1, the RNAi of STRK1 transgenic rice plants was also generated (Supplemental Figures 2C and 2D). To study whether the expression of homologous genes of STRK1 were affected in STRK1-RNAi plants, the expression of OsRLCK1 (Os02g43290) and OsRLCK2 (Os03g02190), which are homologous to STRK1 (Supplemental Figures 3A and 3B and Supplemental Data Set 1), was determined in STRK1-RNAi plants using RT-qPCR. Compared with wild-type plants, only the expression of STRK1 was dramatically reduced in STRK1-RNAi plants, but no obvious change was observed for OsRLCK1 and OsRLCK2 (Supplemental Figure 3C). These results indicated that the STRK1-RNAi plants were genuine STRK1 knockdown transgenic plants.
To further verify the role of STRK1 on salt tolerance in rice, the seedlings of wild-type, STRK1-overexpressing, and STRK1-RNAi transgenic plants were treated with salt stress (140 mM NaCl). Without NaCl treatment, no obvious difference was observed between the wild-type and all transgenic plants (Figure 1A). After 10 d of NaCl treatment, the STRK1-overexpressing plants were more robust than the wild-type plants. Conversely, after 8 d of NaCl treatment, the STRK1-RNAi plants were more wilted compared with the wild-type plants (Figure 1A). Furthermore, after treatment with NaCl for 10 d and recovery by regular hydroponic culture for 6 d, the survival rate of STRK1-overexpressing plants ranged from 50 to 75%, whereas that of wild-type plants was only 11 to 17% (Figure 1B). Additionally, after treatment with NaCl for 8 d and recovery for 6 d, the survival rate of wild-type plants was 20 to 28%, whereas that of STRK1-RNAi plants was only 3 to 9% (Figure 1C). Together, these observations suggested that STRK1 confers tolerance to salt stress in rice at the seedling stage.
To elucidate the physiological changes in transgenic plants under salt stress, 15-d-old seedlings were exposed to 100 mM NaCl for 7 d. As expected, compared with the wild-type plants, both the relative shoot fresh weight (FW) and dry weight were significantly higher in STRK1-overexpressing plants but markedly lower in STRK1-RNAi plants (Supplemental Figures 4A and 4B). Similarly, the chlorophyll content was also significantly higher in STRK1-overexpressing plants but markedly lower in STRK1-RNAi plants under salt stress (Figure 1D). However, no significant difference in chlorophyll content was observed between transgenic and wild-type plants under normal growth conditions. Malondialdehyde (MDA) is an indicator of oxidative attack on membrane lipids and ion leakage reflects membrane injury (Ouyang et al., 2010). Under normal growth conditions, no significant differences in MDA content and relative ion leakage were detected between transgenic and wild-type plants. Compared with wild-type plants, salt stress resulted in a significant decrease in both MDA accumulation and relative ion leakage in STRK1-overexpressing plants but a dramatic increase in STRK1-RNAi plants (Figures 1E and 1F). Additionally, the ability to avoid Na+ accumulation and maintain a low Na+/K+ ratio contributes to salt tolerance in plants (Zhu, 2003). The Na+ and K+ contents in seedlings treated with or without 100 mM NaCl for 5 d were therefore detected. Under both normal and salt stress growth conditions, the STRK1-overexpressing line (OE18) could avoid Na+ accumulation and maintain a low Na+/K+ ratio. By contrast, compared with the wild-type plants, the STRK1-RNAi line (Ri11) accumulated more Na+, leading to a higher Na+/K+ ratio under salt stress (Supplemental Figures 4C and 4D). Taken together, these results suggest that STRK1 might positively regulate the tolerance to salt stress via the avoidance of Na+ accumulation in plants and relief of the membrane damage caused by salt stress.
STRK1 Functions at the Plasma Membrane by Palmitoyl Anchors
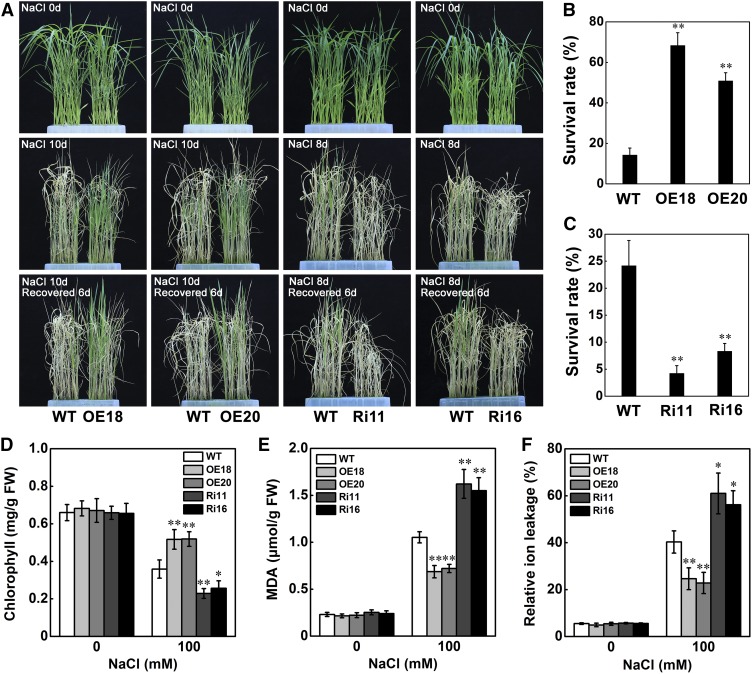
STRK1 belongs to a member of the OsRLCK XIII subfamily (Shiu et al., 2004) and contains an intracellular kinase domain (Vij et al., 2008). To investigate the subcellular localization of STRK1, a STRK1-YFP fusion protein was transiently expressed in rice protoplasts and the subcellular distribution of STRK1-YFP was examined using confocal microscopy. The STRK1-YFP fusion protein was found to specifically localize at the plasma membrane (Figure 2A). Some previous studies reported that RLCKs could be anchored to the plasma membrane by forming a complex with RLK (Tanaka et al., 2012; Yamaguchi et al., 2013) or N-terminal putative palmitoylation and/or myristoylation motifs (Veronese et al., 2006; Tang et al., 2008; Kim et al., 2011). Particularly, protein S-palmitoylation, the covalent lipid modification of the side chain of cysteine residues with the 16-carbon fatty acid palmitate, is the most common acylation of proteins in eukaryotic cells (Aicart-Ramos et al., 2011). Because STRK1 has three cysteine residues in its N-terminal region predicted to be palmitoylation sites (Figure 2A; Supplemental Figure 5), STRK1 may be palmitoylated and subsequently anchored to the plasma membrane. As expected, substitution of these cysteine residues with alanine (STRK1-C5,10,14A-YFP) resulted in cytoplasmic localization of STRK1 in rice protoplasts (Figure 2A). Meanwhile, the same subcellular distribution of fusion proteins was observed in the root cells or protoplasts of overexpressing STRK1-YFP or STRK1-C5,10,14A-YFP transgenic seedlings, respectively (Supplemental Figures 6A, 6B, and 6D). To further verify this hypothesis, the subcellular localization of STRK1-YFP upon treatment with hydroxylamine (NH2OH), a protein de-S-palmitoylation reagent (Duan et al., 2017), was also detected in the root cells of transgenic seedlings overexpressing STRK1-YFP. After NH2OH treatment, STRK1-YFP was distributed in the cytoplasm but not at the plasma membrane, which was consistent with the localization pattern of STRK1-C5,10,14A-YFP (Supplemental Figures 6B and 6C). Together, these results suggested that STRK1 is actually anchored to the plasma membrane by palmitoylation. Furthermore, compared with wild-type plants, the salt tolerance of overexpressing STRK1-C5,10,15A-YFP transgenic plants disappeared under NaCl treatment, whereas plants overexpressing STRK1-YFP remained salt tolerant and had higher survival rates and chlorophyll contents (Supplemental Figures 7 and 8). These results suggested that plasma membrane localization is required for STRK1 to function in salt signal transduction.
Figure 2.
Subcellular Localization and Expression Pattern of STRK1.
(A) Plasma membrane localization of STRK1-YFP anchored by palmitoylation. Amino acid sequence of the N-terminal region of STRK1 showed the three cysteine residues (bold) as putative palmitoylation sites. Confocal images showed localization of STRK1-YFP or STRK1-C5,10,14A-YFP in rice protoplasts. Bar = 10 µm.
(B) to (H) STRK1 promoter-GUS expression patterns in transgenic rice plants. GUS expression was observed in young root ([B] and [C]), leaf vein (D), 4-d-old seedling (E), stem (F), leaf sheath (G), and young spikelet (H).
(I) and (J) Relative mRNA levels of STRK1 by RT-qPCR in the root (I) and shoot (J) of three-leaf stage rice seedlings treated with 150 mM NaCl and 1% (v/v) H2O2. Data are presented as mean ± sd (n = 3).
Additionally, the tissue expression pattern of STRK1 was detected by transforming rice plants with a GUS construct driven by the STRK1 promoter. The results of GUS staining indicated that STRK1 was mainly expressed in the young root, leaf vein, 4-d-old seedling, stem, leaf sheath, and young spikelet, with the highest expression level in the young root (Figures 2B to 2H), which was further confirmed by RT-qPCR (Supplemental Figure 9). Since STRK1 positively regulated salt tolerance and reduced the oxidative damage caused by salt stress (Figure 1), the expression of STRK1 was further examined in rice seedlings under salt or oxidative stress treatments. The results showed that STRK1 was apparently induced by NaCl and H2O2 in both shoots and roots of seedlings (Figures 2I and 2J), suggesting that STRK1 is involved in the response to both salt and oxidative stress.
STRK1 Interacts with CatC in Vitro and in Vivo
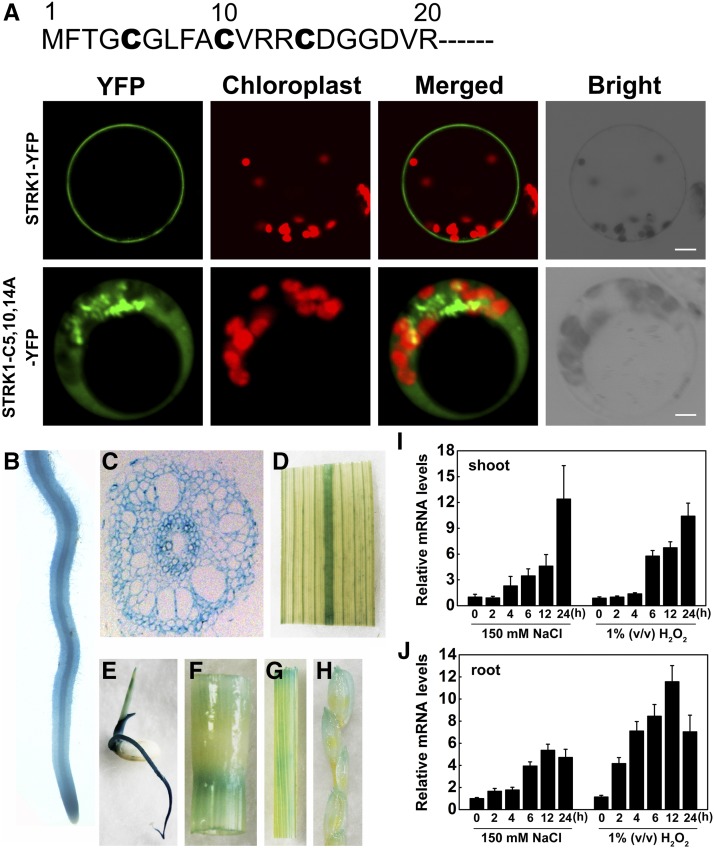
To gain insight into the mechanism by which STRK1 modulates salt tolerance in plants, a yeast two-hybrid screen was performed to identify STRK1-interacting proteins. The full-length STRK1 was fused to the Gal4 DNA binding domain of a bait vector (BD-STRK1). Several interacting clones were isolated and identified to correspond to the catalase domain containing protein (CatC). To verify this interaction, full-length CatC was introduced into the Gal4 activation domain of prey vector (AD-CatC). The bait and prey vectors were cotransformed into yeast and the protein-protein interaction was reconstructed (Figure 3A). CatC is one of three genes in the rice CAT family (CatA, CatB, and CatC) (Morita et al., 1994), and CatC shares a high level of amino acid identity with CatA and CatB (Supplemental Figure 10). Thus, the interaction between STRK1 and the other two CATs, CatA and CatB, was also examined. STRK1 could also interact with CatA and CatB (Supplemental Figure 11A). In addition, the yeast two-hybrid assay also showed that STRK1 interacted with itself (Supplemental Figure 11A), indicating that STRK1 could form homodimers or homomultimers under these conditions. Recently, Zhang et al. (2016) reported that CatA localizes mainly to the cytoplasm, but CatB and CatC localize mainly to peroxisomes. The bimolecular fluorescence complementation (BiFC) assay was used to further confirm the interaction between STRK1 and CATs in planta. The BiFC assays showed that STRK1 could specifically interact with CATs and itself, and the STRK1/CATs complex was localized to the plasma membrane (Figure 3B; Supplemental Figure 11B). Strikingly, similar to the STRK1-CATs interaction in rice, an Arabidopsis calcium-dependent protein kinase, CPK8, was also reported to physically interact with CAT3 at the plasma membrane (Zou et al., 2015), which implies that the kinase-CATs interaction at the plasma membrane might be a basic mechanism of catalase activation. Meanwhile, the STRK1-CatC interaction was determined to be the strongest of the STRK1-CATs interactions (Supplemental Figure 12), so we focused our analysis on the STRK1-CatC interaction for the rest of this study.
Figure 3.
STRK1 Physically Interacts with CatC.
(A) Yeast two-hybrid assay of STRK1 interaction with CatC. STRK1 (bait) and CatC (prey) were introduced into yeast cells as indicated and grown on selection medium minus leucine and tryptophan (-LW) or leucine, tryptophan, histidine, and adenine (-LWHA). Empty vectors pGBKT7 and pGADT7 were used as controls.
(B) BiFC assay for interaction between STRK1 and CatC in Arabidopsis protoplasts. Full-length STRK1 protein was fused to C-terminal CFP (STRK1-cCFP), and full-length CatC was fused to N-terminal Venus (nVenus-CatC). The expression of nVenus/STRK1-cCFP was used as control. Before imaging, the protoplasts were treated with FM4-64 (2 μM) for 5 min to show the plasma membrane. Bar = 10 µm.
(C) Pull-down assay for interaction between STRK1 and CatC. Purified His-TF-CatC was used to copurify STRK1-FLAG from OE18 seedling protein extracts (Input) following incubation with Ni-NTA agarose (see Methods). The flow-through was collected after centrifugation (Flow) and two washes (Wash1 and Wash2), and the eluted fractions (Elute) were separated by 10% SDS-PAGE and immunoblotted with anti-FLAG antibodies. His-TF was used as control.
(D) Co-IP assay for interaction between STRK1 and CatC in N. benthamiana leaves. Protein extracts (Input) were immunoprecipitated with anti-FLAG antibody (IP). Immunoblots were developed with anti-FLAG antibody to detect STRK1 and with anti-GFP to detect CatC.
To further confirm the interaction between STRK1 and CatC, the purified recombinant His-TF-CatC and isolated proteins from the OE18 line were subjected to a pull-down assay. As expected, STRK1-FLAG was pulled down by His-TF-CatC but not by His-TF (Figure 3C). Subsequently, the in vivo interaction between STRK1 and CatC was confirmed by a coimmunoprecipitation (co-IP) assay. When STRK1-FLAG and CatC-GFP fusion proteins were transiently coexpressed in tobacco (Nicotiana benthamiana) leaf cells, CatC-GFP fusion protein could be coimmunoprecipitated with STRK1-FLAG (Figure 3D). Taken together, these results indicate that STRK1 physically interacts with CatC.
STRK1 Phosphorylates CatC Mainly at Tyr-210 and Regulates Its Activity
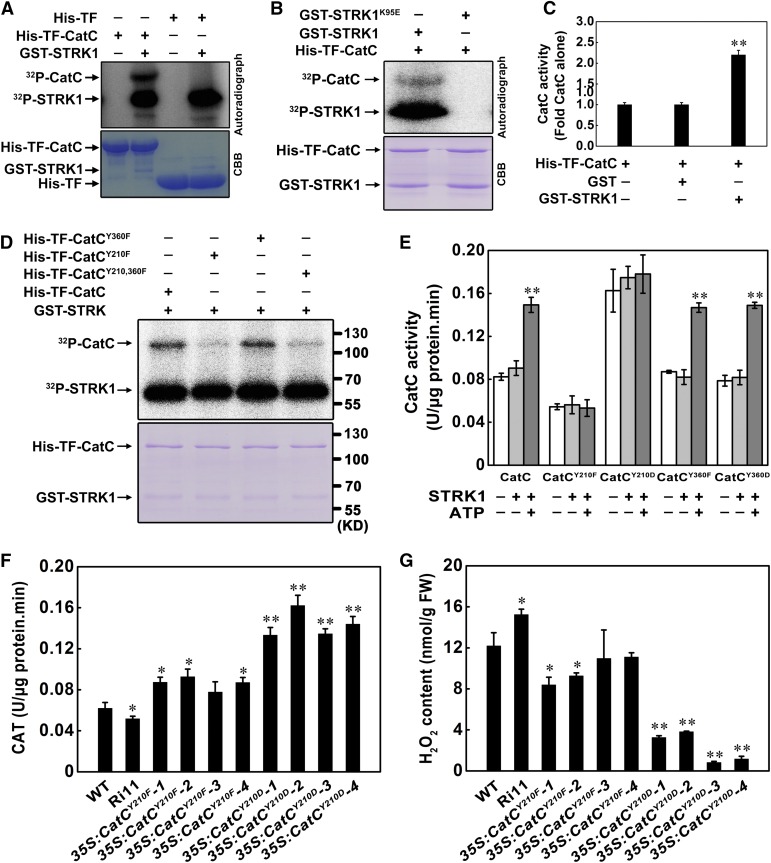
To determine whether STRK1 is a functional protein kinase, a recombinant fusion protein with glutathione S-transferase (GST-STRK1) was expressed in Escherichia coli and purified. In in vitro kinase assays, strong autophosphorylation of STRK1 was detected and the myelin basic protein (MBP) as an in vitro substrate was phosphorylated by STRK1 (Supplemental Figure 13), indicating that STRK1 is an active kinase. To investigate whether CatC could be phosphorylated by STRK1, GST-STRK1 was incubated with His-TF-CatC and tested for protein kinase activity. The results showed that STRK1 could phosphorylate CatC in vitro (Figure 4A), indicating that CatC is a candidate substrate of STRK1. Additionally, to determine whether the phosphorylation of CatC is indeed dependent on the kinase activity of STRK1, the conserved Lys-95 (Supplemental Figure 5), which is required for the activities of other kinases (Li and Nam, 2002; Liu et al., 2017), was mutated to Glu to generate a kinase-dead form of STRK1 (STRK1K95E). STRK1K95E failed to show either autophosphorylation or phosphorylation of CatC (Figure 4B). This result suggests that the kinase activity of STRK1 is essential for its function in salt signal transduction. Previous studies have shown that the catalase activity is regulated by phosphorylation in humans (Kumar et al., 2010; Rafikov et al., 2014) and in Arabidopsis (Zou et al., 2015). Thus, we hypothesized that STRK1 could modulate CatC activity through phosphorylation. As expected, the CatC activity in the presence of STRK1 was ∼2.5-fold of that in the absence of STRK1 (Figure 4C), indicating that CatC activity might be enhanced by STRK1 through phosphorylation.
Figure 4.
STRK1 Phosphorylates CatC Mainly at Tyr-210 and Activates Its Activity.
(A) STRK1 phosphorylates CatC in vitro. His-TF or His-TF-CatC was incubated with GST-STRK1 and [γ-32P]ATP. Upper panel shows autoradiography and bottom panel Coomassie brilliant blue (CBB) staining of the gel.
(B) Phosphorylation of CatC is indeed dependent on the kinase activity of STRK1. His-TF-CatC was incubated with GST-STRK1, GST-STRK1K95E and 32P-γ-ATP. GST-STRK1K95E is the kinase dead form of STRK1. Upper panel shows autoradiography and bottom panel Coomassie brilliant blue (CBB) staining of the gel.
(C) STRK1 stimulates CatC activity in vitro. Recombinant His-TF-CatC expressed in E. coli was purified, and the CatC activity was measured with GST or GST-STRK1 in the presence of ATP. Data are presented as mean ± sd (n = 3, **P ≤ 0.01, Student’s t test).
(D) An equal amount of His-TF fused CatC, CatCY210F, CatCY360F, and CatCY210,360F was incubated with GST-STRK1 and [γ-32P]ATP. Upper panel shows autoradiography and bottom panel Coomassie brilliant blue staining of the gel.
(E) Phosphorylation of CatC at Tyr-210 is essential for CatC activity. CatCY210Fand CatCY360F, recombinant CatC with Tyr-210 and Tyr-360 mutated to Phe, CatCY210D and CatCY360D, and recombinant CatC with Tyr-210 and Tyr-360 mutated to Asp. Each data point represents the mean ± se (n = 6). Asterisk indicates significant difference relative to CatC activity in the absence of STRK1 and ATP (Student’s t test, **P < 0.01).
(F) and (G) CAT activity (F) and H2O2 contents (G) in transgenic seedlings (T0) overexpressing CatCY210F and CatCY210D in the Ri11 background, a STRK1 knockdown mutant. The Kitaake (WT) and Ri11 seedlings regenerated from the corresponding calli were used as control. Data are presented as mean ± sd (n = 3, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
To investigate the potential phosphorylation sites of CatC by STRK1, the His-TF-CatC incubated with GST-STRK1 was used for mass spectrometry. The results of mass spectrometry showed five phospho-serines (Ser-9, Ser-10, Ser-11, Ser-18, and Ser-205), four phospho-threonines (Thr-19, Thr-209, Thr-211, and Thr-292), and two phospho-tyrosines (Tyr-210 and Tyr-360) were identified in CatC (Supplemental Figure 14A). Therefore, we replaced these Ser (S) and Thr (T) residues of CatC with an Ala (A) residue, and the Tyr(Y) residues with a Phe (F) residue, producing His-TF-CatCS9,10,11,18,205A, His-TF-CatCT19,209,211,292A, and His-TF-CatCY210,360F, to mimic nonphosphorylation. In vitro phosphorylation assays showed that STRK1-mediated phosphorylation of CatC was largely abolished when only containing two tyrosine point mutations. However, the phosphorylation of CatC was not obviously affected when these five serines and four threonines were mutated to Ala (Supplemental Figure 14B). These results indicate that STRK1 might phosphorylate CatC at both or either of Tyr-210 and Tyr-360. Subsequently, the Tyr-210 and Tyr-360 residues of CatC were separately mutated to Phe (His-TF-CatCY210F and His-TF-CatCY360F). Then, almost no phosphorylation of His-TF-CatCY210F was observed, which was similar to the double point mutants His-TF-CatCY210,360F, whereas another single point mutant protein His-TF-CatCY360F showed no obvious effect on STRK1-mediated phosphorylation compared with the wild-type His-TF-CatC (Figure 4D). These results further suggest that Tyr-210 of CatC is the major phosphorylated residue recognized by STRK1. Strikingly, the sequence alignment showed that the motif containing Tyr-210 is highly conserved in the rice CAT family (CatA, CatB, and CatC; Supplemental Figure 10A) and Arabidopsis CAT3 (Supplemental Figure 15), implying that the residue Tyr-210 plays an essential role in regulating CAT activity.
To study the significance of phosphorylated Tyr-210 of CatC in vitro and in planta, the Tyr-210 and Tyr-360 residues of CatC were separately mutated to Phe (F) and Asp (D), to mimic nonphosphorylation (CatCY210F and CatCY360F) and phosphorylation (CatCY210D and CatCY360D), respectively. Then, the catalytic activities of mutant CatC were analyzed in the presence of STRK1. As expected, the results showed that STRK1 significantly enhanced CatC activity in the presence of ATP. The catalase activity of CatCY210F could not be stimulated by STRK1, while the mutated CatCY210D showed higher catalase activity even in the absence of STRK1 (Figure 4E). As a control, similar to the wild-type CatC, the mutations of Tyr-360 (CatCY360F and CatCY360D) did not affect STRK1-activated CatC activity, whose catalase activities were only enhanced in the presence of STRK1 and ATP (Figure 4E). Furthermore, CatCY210F and CatCY210D were overexpressed in the Ri11 line, a STRK1 knockdown mutant, respectively (Supplemental Figure 16), and the CAT activity and H2O2 concentration were measured under normal growth condition. As expected, compared with the control Ri11 plants, CatCY210D significantly enhanced the CAT activity with the lowest H2O2 accumulation in transgenic plants, while CatCY210F only moderately increased the CAT activity with a lower H2O2 accumulation (Figures 4F and 4G). These results indicate that CatCY210D has a stronger ability than CatCY210F to rescue the CAT activity in STRK1-RNAi plants, which means that, even if the stress signal transduction via upstream phosphorylation was disrupted in STRK1-RNAi plants, the mimic phosphorylation form CatCY210D could still enhance the capacity of scavenging and detoxification of ROS. It should be noted that, as CatCY210F still had partial CAT activity and was overexpressed (Figure 4E; Supplemental Figure 16B), the CAT activity in CatCY210F transgenic plants was also higher than that in the wild-type Kitaake plants but lower than that in CatCY210D transgenic plants under normal growth conditions (Figure 4F). Together, these results demonstrate that the mimic phosphorylation form CatCY210D exhibits higher catalase activity both in vitro and in planta, and Tyr-210 in CatC is an essential residue for CatC activation by STRK1.
Salt Stress Enhances STRK1 Tyrosine Phosphorylation on CatC
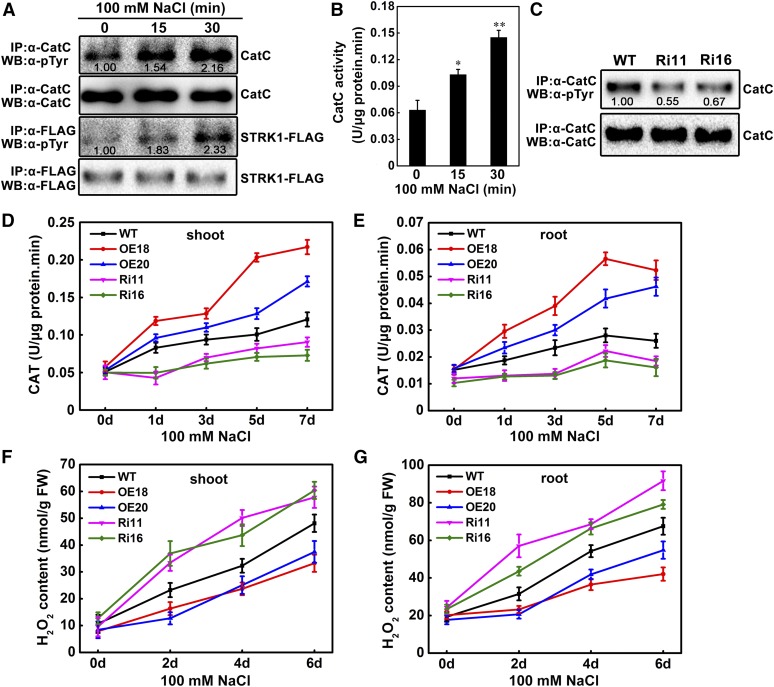
To determine the in planta tyrosine phosphorylation states of CatC and STRK1 in response to salt stress, the fusion protein STRK1-FLAG and endogenous CatC were immunoprecipitated by anti-FLAG and anti-CatC antibodies, respectively, from OE18 seedlings after a short period of NaCl treatment, and the tyrosine phosphorylation of STRK1-FLAG and CatC was analyzed with an anti-phospho-tyrosine antibody. As shown in Figure 5A, endogenous CatC was also tyrosine phosphorylated in planta, and the signal of phospho-tyrosine could be obviously detected in CatC even without NaCl treatment. Particularly, the tyrosine phosphorylation level of CatC significantly increased with the time of NaCl treatment (Figure 5A). As mentioned above, although two phospho-tyrosines (Tyr-210 and Tyr-360) were identified in CatC, only Tyr-210 could be phosphorylated by STRK1 (Supplemental Figure 14A; Figure 4). Thus, we can speculate that the basic signal of phospho-tyrosine under non-salt stress might mainly come from the phosphorylated Tyr-360, and the increased signals might result from the phosphorylated Tyr-210 in response to salt stress. Furthermore, similar to CatC, the tyrosine phosphorylation level of STRK1 also increased with the time of NaCl treatment (Figure 5A). Combined with the previous results showing that the transcriptional level of STRK1 was induced by NaCl (Figures 2I and 2J), we can also speculate that salt stress not only induces the transcription of STRK1 but also stimulates the phosphorylation of STRK1, which finally enhances the phosphorylation of CatC. Additionally, the catalase activity of immunoprecipitated endogenous CatC was analyzed. As shown in Figure 5B, the CatC activity also increased significantly with the time of NaCl treatment, indicating that the activity of CatC is positively correlated with the level of tyrosine phosphorylated CatC. These results suggest that salt stress enhances STRK1 tyrosine phosphorylation on CatC and in turn stimulates CatC activity. Meanwhile, the tyrosine phosphorylation level of endogenous CatC in STRK1-RNAi plants under normal growth condition was also analyzed. As expected, compared with the wild-type plants, the signal of phospho-tyrosine decreased ∼40% in the endogenous CatC of STRK1-RNAi plants (Figure 5C), indicating that the disruption of STRK1 can actually decrease the tyrosine phosphorylation of its substrate CatC.
Figure 5.
Salt Stress Induces STRK1 Tyrosine Phosphorylation on CatC.
(A) NaCl treatment enhances STRK1 tyrosine phosphorylation on CatC. STRK1-FLAG and CatC proteins were immunoprecipitated from OE18 seedling after treatment with NaCl for the indicated periods and then subjected to an immunoblot assay. Anti-phospho-tyrosine (α-pTyr) antibody was used to probe for STRK1 and CatC phosphorylation. Immunoprecipitated endogenous CatC and STRK1-FLAG were analyzed by immunoblotting with anti-CatC and anti-FLAG antibodies, respectively. The relative intensities of tyrosine phosphorylation of CatC and STRK1-FLAG without NaCl treatment (0 min) were set to 1.00.
(B) The CatC activity is associated with its tyrosine phosphorylation levels. The phosphorylated CatC proteins in (A) were used to determine the catalase activity. Data are presented as mean ± sd (n = 3, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
(C) Knockdown of STRK1 reduces the tyrosine phosphorylation level of CatC. Endogenous CatC proteins were immunoprecipitated from Ri11 and Ri16, the STRK1 knockdown mutants, and the wild-type seedlings were grown under normal growth conditions and then detected by immunoblotting. Anti-phospho-tyrosine (α-pTyr) antibody was used to probe for CatC phosphorylation, and anti-CatC antibody was used to show the immunoprecipitated CatC. The relative intensity of tyrosine phosphorylation of CatC in wild-type seedlings was set to 1.00.
(D) and (E) CAT activity in rice shoots (D) and roots (E) of seedlings grown under normal conditions or after 100 mM NaCl stress for the indicated periods. Data are presented as mean ± sd (n = 3).
(F) and (G) H2O2 contents in rice shoots (F) and roots (G) of seedlings grown under normal conditions or after 100 mM NaCl stress for the indicated periods. Data are presented as mean ± sd (n = 3).
To further investigate the effect of STRK1 on catalase activity in planta, the CAT activity and H2O2 concentration were measured in shoots and roots of transgenic and wild-type plants under normal growth and salt stress conditions. Under salt stress, the CAT activity was significantly enhanced and the H2O2 accumulation was obviously decreased in shoots and roots of STRK1-overexpressing plants compared with wild-type plants. By contrast, the CAT activity was apparently reduced and the H2O2 accumulation was markedly increased in shoots and roots of salt-treated STRK1-RNAi plants compared with wild-type plants (Figures 5D to 5G). Under normal growth conditions, both CAT activity and H2O2 concentration showed no significant difference in either shoots or roots between transgenic and wild-type plants. These results further support the hypothesis that STRK1 improves salt tolerance through regulating H2O2 homeostasis by modulating CAT activity.
STRK1 Also Positively Regulates Oxidative Tolerance in Rice
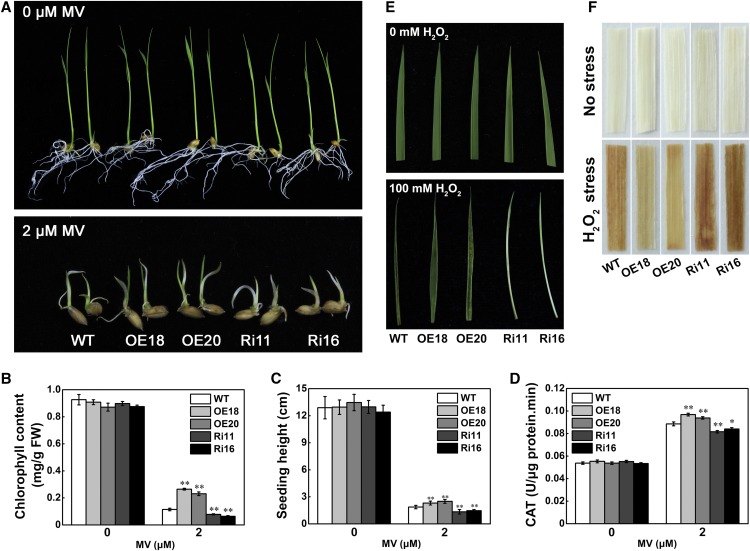
As shown previously, the expression of STRK1 was induced by H2O2 (Figures 2I and 2J) and H2O2 homeostasis was maintained through regulation of CAT activity by STRK1 in response to salt stress (Figures 5D to 5G), which might contribute to improved tolerance to other oxidative stresses. To test this, the response of STRK1 transgenic plants to oxidative stress was investigated by application of methylviologen (MV), a well-known oxidative stress inducer in plants. Under normal conditions, there was no significant difference in growth among the transgenic and wild-type seedlings (Figure 6A). By contrast, after 6 d MV (2 µM) treatment, STRK1-overexpressing seedlings were more green than the wild type, and STRK1-RNAi seedlings exhibited an etiolated and dwarf phenotype (Figure 6A). Then, the chlorophyll content, seedling height, and CAT activity in these seedlings were determined. Under normal conditions, no significant differences in chlorophyll content, seedling height and CAT activity were observed among the transgenic and wild-type seedlings (Figures 6B to 6D). On the contrary, under MV stress, STRK1-overexpressing plants showed higher chlorophyll content, seedling height, and CAT activity compared with wild-type plants, whereas the STRK1-RNAi plants exhibited the opposite results. In another oxidative stress test, three-leaf stage seedlings of wild-type and transgenic plants were treated with 100 mM H2O2. After 2 d H2O2 treatment, severe necrosis and leaf rolling were observed in STRK1-RNAi plants (Figure 6E). It should be noted that, although no obvious chlorosis or damage was detected in the leaves of STRK1-overexpressing and wild-type plants, wild-type plants exhibited stronger leaf rolling than STRK1-overexpressing plants after H2O2 treatment. The accumulation of H2O2 was further determined by 3,3′-diaminobenzidine (DAB) staining. After 1 d H2O2 treatment, compared with the wild type, less accumulation of H2O2 was observed in STRK1-overexpressing leaves but greater accumulation of H2O2 in STRK1-RNAi leaves (Figure 6F). Under normal growth conditions, no obvious staining was observed in all transgenic and wild-type plants. These results indicated that overexpression of STRK1 in rice can increase oxidative stress tolerance resulting from an enhanced ROS-scavenging ability.
Figure 6.
STRK1 Positively Regulates Oxidative Tolerance in Rice.
(A) Phenotypic comparison of rice plants subjected to MV stress. The germinated seeds were transplanted into either 0.5× MS medium or 0.5× MS medium supplemented with 2 μM MV for 6 d.
(B) to (D) Chlorophyll content in leaves (B), seedling height (C), and CAT activity in leaves (D) of STRK1 transgenic and wild-type plants under normal and MV stress conditions. Data are presented as mean ± sd (n = 3, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
(E) Leaf phenotype of STRK1 transgenic and wild-type plants at the three-leaf stage under normal conditions or after 100 mM H2O2 stress for 2 d.
(F) DAB staining for H2O2 in leaves from unstressed and H2O2-stressed STRK1 transgenic and wild-type plants for 1 d.
STRK1 Improves the Grain Yield of Rice under Salt Stress
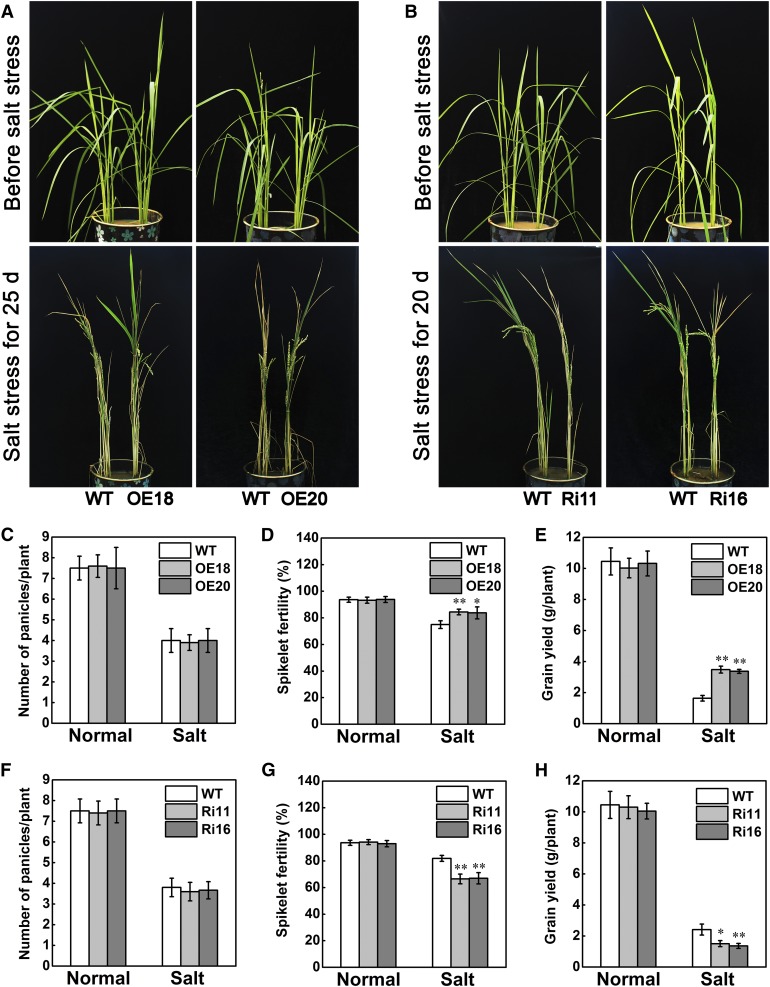
As rice is an economically important cereal crop, its grain yield is the focus of breeding. Although STRK1 has been verified to significantly improve growth under salt stress and positively regulate salt tolerance at the seedling stage (Figure 1), it is worth investigating whether STRK1 can also improve agronomic traits, especially grain yield, under salt stress and enhance salt tolerance at the reproductive stage in rice. Therefore, the STRK1 transgenic plants were also tested for salt tolerance at the reproductive stage when grown in plastic buckets. To obtain more apparent phenotypes between transgenic and wild-type plants, 20 and 25 d salt treatments were applied for STRK1-RNAi and STRK1-overexpressing plants as well as their corresponding wild-type plants, respectively. After salt treatments, STRK1-overexpressing plants had more green leaves than wild-type plants (Figure 7A). By contrast, STRK1-RNAi plants had more withered leaves than wild-type plants (Figure 7B). These results indicated that STRK1 could also significantly improve growth under salt stress at the reproductive stage. Meanwhile, compared with their corresponding wild-type plants, no significant differences were observed in panicle numbers of STRK1-overexpressing and STRK1-RNAi plants under both normal growth and salt stress conditions (Figures 7C and 7F). Under salt stress conditions, significantly higher spikelet fertility was observed in STRK1-overexpressing plants (Figure 7D), but markedly lower spikelet fertility in STRK1-RNAi plants (Figure 7G). There was no difference in spikelet fertilities among STRK1-overexpressing, STRK1-RNAi, and wild-type plants under normal growth conditions (Figures 7D and 7G). Furthermore, under salt stress conditions, the grain yields of STRK1-overexpressing plants were significantly higher than those of wild-type plants (Figure 7E). By contrast, the grain yields of STRK1-RNAi plants were markedly lower than those of wild-type plants under salt stress conditions (Figure 7H). Similar to the spikelet fertilities, there was no obvious difference in grain yield between wild-type and transgenic plants under normal growth conditions (Figures 7E and 7H). These results indicated that STRK1 could also positively regulate salt tolerance at the reproductive stages and limit the grain yield loss of rice under salt stress.
Figure 7.
STRK1 Improves the Grain Yield of Rice under Salt Stress at the Reproductive Stage.
(A) and (B) Phenotypic comparison of rice plants under salt stress. Salt stress of plants was initiated at the panicle development stage (top) by exposure to 1% NaCl as indicated (down), and then the plants were recovered with irrigation for 10 d and harvested.
(C) to (E) Panicle number (C), spikelet fertility (D), and grain yield (E) of STRK1-overexpressing and wild-type plants in (A) after 10 d of recovery. Data are presented as mean ± sd (n = 20,*P ≤ 0.05, **P ≤ 0.01, Student’s t test).
(F) to (H) Panicle number (F), spikelet fertility (G), and grain yield (H) of STRK1-RNAi and wild-type plants in (B) after 10 d of recovery. Data are presented as mean ± sd (n = 20,*P ≤ 0.05, **P ≤ 0.01, Student’s t test).
DISCUSSION
STRK1 Positively Regulates Salt Tolerance in Rice
Salinity not only inhibits plant growth but also reduces the crop productivity at reproductive stages, so the sustained growth and yield under salt stress conditions will be the main criterion for developing salt-tolerant crop plants (Zeng and Shannon, 2000; Zhu, 2001). In this study, we have identified a novel salt stress-induced RLCK gene, STRK1, from rice and showed that STRK1 was involved in the response to salt stress in rice. As STRK1 expression is induced by salt (Figure 2I; Supplemental Figure 1), it was a possibility that this gene was involved in the response to salt stress. Indeed, at the seedling stage, STRK1-overexpressing plants exhibited tolerance to salt stress, whereas STRK1-RNAi plants were sensitive to salt stress (Figure 1), which indicates that STRK1 positively regulates salt tolerance in rice. The ability to avoid Na+ accumulation and maintain a low Na+/K+ ratio contributes to salt tolerance in plants (Zhu, 2003). It is well-known that the salt overly sensitive (SOS) pathway plays a critical role in sodium ion homeostasis during salt stress (Zhu, 2003; Tan et al., 2016). In Arabidopsis, the SOS pathway consists of at least three major components: SOS1, SOS2, and SOS3, which have been identified as a plasma membrane-localized Na+/H+ antiporter, a Ser/Thr protein kinase, and a calcium binding protein, respectively (Guo et al., 2001; Zhu, 2003). Among them, the SOS1 phosphorylated by SOS2 can transport cytosol-accumulated Na+ out of the cell, thereby enhancing salt tolerance (Quintero et al., 2011). Interestingly, in addition to an enhanced capacity of scavenging and detoxification of ROS, STRK1-overexpressing plants exhibited another important aspect of salt tolerance with a low Na+/K+ ratio (Supplemental Figures 4C and 4D), meaning that STRK1 might partially be involved in the SOS pathway to regulate the tolerance to salt stress via the avoidance of Na+ accumulation in plants. This possibility merits further investigation.
In rice, the spikelet fertility rate is highly correlated with grain yield under stress conditions (Flowers, 2004). For example, the reduced grain yield of gudk mutant plants was mainly due to lower spikelet fertility under drought stress (Ramegowda et al., 2014). In this study, the spikelet fertility rates were significantly higher in STRK1-overexpressing plants but markedly lower in STRK1-RNAi plants than those in the corresponding wild-type plants (Figures 6D and 6G), which resulted in the increased and reduced grain yield in STRK1-overexpressing and STRK1-RNAi plants, respectively (Figures 7E and 7H). It is noteworthy that there was no significant difference in panicle numbers between transgenic and wild-type plants under both normal growth and salt stress conditions (Figures 7C and 7F). According to the development of inflorescences and spikelets (Ikeda et al., 2004), most panicles (inflorescences) developed or started to develop when the rice plants began to suffer from salt stress (Figures 7A and 7B). Thus, the panicle numbers were hardly affected by salt treatment. Together, these results suggest that STRK1 is a promising candidate gene for yield improvement in rice under salt stress conditions.
STRK1 Regulates H2O2 Homeostasis by Activating CatC
Although ROS can serve as vital signaling molecules in many biological processes, excessive accumulation of ROS in living plant cells results in cellular damage by oxidative stress (Apel and Hirt, 2004; Mittler et al., 2004; Schippers et al., 2012; Schmidt et al., 2013). Thus, the cytoplasmic concentration of ROS must be strictly regulated. Abiotic stress induces ROS production in plant cells, so a stronger ROS scavenging capacity often confers the higher tolerance to abiotic stresses in plants (Apel and Hirt, 2004). For instance, overexpression of OsSIK2 in rice enhanced plant tolerance to high salinity and drought stress through scavenging and detoxification of ROS (Chen et al., 2013). OsCPK12-overexpressing rice plants improved tolerance to salt stress by reducing ROS accumulation (Asano et al., 2012). The ospp18 mutant exhibited an increased sensitivity to drought and oxidative stresses through the reduced activity of ROS scavenging enzymes (You et al., 2014). Strikingly, plants have evolved efficient nonenzymatic and enzymatic detoxification mechanisms that scavenge ROS. Nonenzymatic antioxidants include the major cellular redox buffers, glutathione and ascorbate, as well as flavonoids, alkaloids, tocopherol, and carotenoids. Enzymatic ROS-scavenging mechanisms in plants include CAT (EC 1.11.1.6), superoxide dismutase (SOD; EC 1.15.1.1), ascorbate peroxidase (APX; EC 1.11.1.11), and glutathione peroxidase (EC 1.11.1.7) (Apel and Hirt, 2004; Mittler et al., 2004). Among them, CAT is a key H2O2-scavenging enzyme that plays an important role in protecting plant cells against stresses (Xu et al., 2013).
Previous studies revealed that CAT activity is regulated by phosphorylation in humans (Kumar et al., 2010; Rafikov et al., 2014) and Arabidopsis (Zou et al., 2015). PKCδ phosphorylated CAT at Ser-167 in response to endothelin 1 and increased CAT activity, leading to lower cellular H2O2 accumulation in humans (Kumar et al., 2010; Rafikov et al., 2014). CPK8 can specifically phosphorylate CAT3 at Ser-261 and regulate its activity to maintain H2O2 homeostasis in response to drought stress in Arabidopsis (Zou et al., 2015). In the this study, STRK1 phosphorylated CatC at Tyr-210 and stimulated its activity to regulate H2O2 homeostasis and improve salt tolerance in rice (Figures 1 and 4). Especially, the mutated CatCY210D exhibits higher catalase activity both in vitro and in planta (Figures 4E and 4F), which indicates that Tyr-210 in CatC is an essential residue for CatC activation by STRK1. In addition to the two reported phosphorylation sites, Ser-167 in human CAT and Ser-261 in Arabidopsis CAT3, the Tyr-210 in rice CatC is the third and only tyrosine residue identified to date to be phosphorylated by a definite protein kinase in catalases. As the CAT family contains CatA, CatB, and CatC in rice (Morita et al., 1994), the activity of CAT measured in planta may be the sum of activity of CAT proteins. Although we focused on the analysis of the STRK1-CatC interaction in this study, all CAT proteins were found to interact with STRK1 (Figure 3; Supplemental Figures 11 and 12). Consequently, STRK1 probably stimulates the activities of CatA and CatB as well as CatC, which will require further investigation. The increased CAT activity would in turn result in H2O2 degradation and enhance the tolerance to salt stress. These results suggested that STRK1 mediates salt tolerance through activating CatC activity and maintaining H2O2 homeostasis in rice.
Tolerant species have an excellent capacity for protecting themselves from salt-induced oxidative stress through mechanisms of antioxidant protection (Azevedo-Neto et al., 2006). The transgenic Nicotiana benthamiana plants heterologously expressing GhWRKY39-1 exhibited improved tolerance to high salt and oxidative stress resulted from an increased capacity for ROS scavenging (Shi et al., 2014). Overexpression of HSFA4A in Arabidopsis reduced H2O2 accumulation and lipid peroxidation, which resulted in the enhancement of salt and oxidative stress tolerance (Pérez-Salamó et al., 2014). In this study, induction of STRK1 upon H2O2 treatment suggests that it functions in oxidative stress adaptation. MDA, an important intermediate in ROS scavenging, is toxic to plant cells if it accumulates excessively, so it is often used as an indicator of oxidative attack on membrane lipids (Apel and Hirt, 2004; Mittler et al., 2004). Thus, the reduced MDA contents in STRK1-overexpressing plants (Figure 1E) indicated that less oxidative damage occurred in the cells than in those of the wild type during salt stress treatment. Relevant to these findings, the STRK1-overexpressing seedlings exhibited higher chlorophyll content, seedling height, and CAT activity as well as less accumulation of H2O2 than wild-type plants under oxidative stress (Figure 6). By contrast, STRK1-RNAi seedlings showed reduced tolerance to oxidative stress. These results confirmed that the function of STRK1 in salt tolerance is associated with the increase of antioxidation ability.
STRK1 Might Act as a Key Component of Receptor Kinase Complex-Mediated Salt or Oxidative Stress Signaling
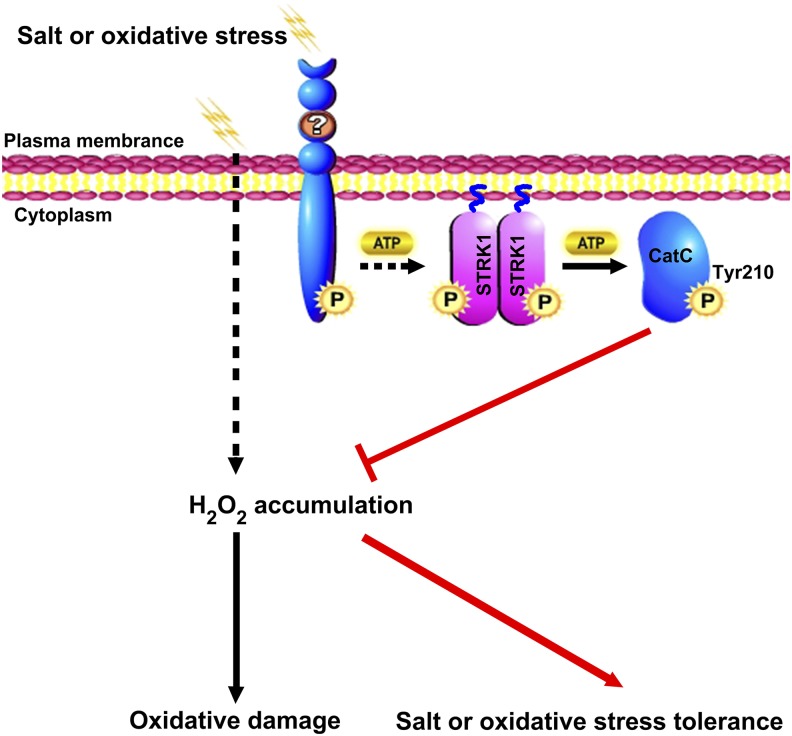
Typically, RLKs are transmembrane proteins that perceive the signals through their extracellular domains and propagate the signals via their intracellular kinase domains (Shiu et al., 2004). Protein phosphorylation cascades via RLKs plays important roles in plant responses to abiotic stress. For instance, a rice leucine-rich repeat RLK, OsSIK1, mediates tolerance to drought and salt stress in rice through activating the antioxidative system (Ouyang et al., 2010). A rice lectin RLK, SIT1, mediates salt stress signal transduction from the cell surface to intracellular MAPK3/6 modules (Li et al., 2014). So far, some RLCKs have also been reported to be involved in abiotic stress responses (Giri et al., 2011; Sun et al., 2013; Ramegowda et al., 2014; Liu et al., 2017). Similar to RLKs, although some RLCKs lack the extracellular and transmembrane proteins, they are potentially anchored to the plasma membrane through formation of a complex with RLK (Tanaka et al., 2012; Yamaguchi et al., 2013) or N-terminal putative palmitoylation and/or myristoylation motifs (Veronese et al., 2006; Tang et al., 2008; Kim et al., 2011; Li et al., 2016). Membrane-anchored proteins have various functions, including cellular regulation, signal transduction, and translocation (Podell and Gribskov, 2004). In this study, STRK1, a member of the RLCK XIII family in rice, can be anchored to the plasma membrane by palmitoylation (Figure 2A; Supplemental Figure 6). Moreover, the plasma membrane localization is required for STRK1 to function in salt signal transduction (Supplemental Figure 7). Localization of STRK1 to the plasma membrane implies that STRK1 may act as an early component of the plant salt stress response, either directly in salt stress signal recognition or early in the signaling cascade. However, as STRK1 lacks both an extracellular domain and a transmembrane domain, it cannot directly sense the extracellular signal of salt stress. Previous studies showed that RLCKs often functionally and physically associate with RLKs as a complex that relays intracellular signaling via transphosphorylation events (Lin et al., 2013). BIK1, a plant RLCK that is anchored to the plasma membrane by myristoylation, is rapidly phosphorylated upon flagellin perception in a FLAGELLIN-SENSING2- and BAK1-dependent manner to regulate plant innate immunity (Veronese et al., 2006; Lu et al., 2010). Constitutive differential growth 1, a member of the RLCK VIIc family that is anchored to the plasma membrane by palmitoylation, can be activated by BRI1 transphosphorylation to transduce the BR signal from BRI1 receptor kinase to the BSU1 phosphatase and BIN2 kinase (Kim et al., 2011). Thus, it is possible that there is a RLK that directly senses and transmits the extracellular signal of salt or oxidative stress to STRK1 through phosphorylation. As a consequence, the phosphorylated STRK1 phosphorylates CatC and stimulates its activity to maintain H2O2 homeostasis in response to salt or oxidative stress. Based on this hypothesis, we propose a working model of STRK1 under early events of salt or oxidative stress signaling (Figure 8).
Figure 8.
A Proposed Model for the Role of STRK1 in Regulating Salt or Oxidative Stress Tolerance.
Salt or oxidative stress induces H2O2 accumulation, which causes oxidative damage to the cell. In response to salt or oxidative stress, the extracellular signals are transmitted from an unknown RLK to STRK1, a membrane-anchored RLCK by palmitoylation, through phosphorylation. The phosphorylated STRK1 stimulates CatC activity via phosphorylation (P) mainly at Tyr-210, which in turn inhibits H2O2 accumulation and, ultimately, increases the salt or oxidative stress tolerance in plants. The question mark indicates an unknown RLK. The wavy lines indicate the palmitoyl groups. The step marked with a black dashed line with an arrow between the unknown RLK and STRK1 is currently unknown.
Recently, tyrosine phosphorylation has been revealed to be an essential regulatory mechanism in the initiation and transduction of RLK/RLCK-mediated BR and innate immune signaling (Macho et al., 2015). For instance, two RLKs, EFR and BAK1, the receptor and coreceptor for a bacterial elongation factor Tu (EF-Tu; or elf18) are phosphorylated at Tyr-836 and Tyr-610, respectively, to initiate plant immune responses after elf18 perception. Subsequently, a plant RLCK BIK1 is rapidly autophosphorylated and phosphorylated by BAK1 at multiple tyrosine residues, including Tyr-23, Tyr-150, Tyr-168, Tyr-214, Tyr-234, Tyr-243, and Tyr-250, in addition to serine/threonine residues to positively regulate plant innate immunity (Veronese et al., 2006; Lu et al., 2010). It should be noted that phosphorylation of these particular, distinct tyrosine residues is required for activation of EFR and downstream immunity to the phytopathogenic bacterium Pseudomonas syringae. Interestingly, a tyrosine phosphatase, HopAO1, secreted by the pathogen P. syringae, can reduce EFR tyrosine phosphorylation and prevent subsequent immune responses (Macho et al., 2014). In this study, CatC was phosphorylated by STRK1 at Tyr-210 (Figure 4), and both STRK1 and CatC tyrosine phosphorylation were promptly increased in response to salt stress (Figure 5A), which means that a tyrosine phosphorylation cascade might exist in the initiation and transduction of RLK/RLCK-mediated salt or oxidative stress signaling. We propose that, similar to BIK1 in innate immune signaling, STRK1 might act as a key component that transduces the signal from an unknown receptor kinase complex to the downstream effector CatC via tyrosine phosphorylation to maintain H2O2 homeostasis in response to salt or oxidative stress (Figure 8). However, this hypothesis remains to be tested.
In summary, the results presented in this study demonstrate that STRK1 functions in maintaining H2O2 homeostasis in response to salt or oxidative stress by phosphorylating CatC at Tyr-210 and regulating its activity. The significantly improved growth and yield under salt stress of STRK1-overexpressing rice plants indicates that STRK1 is a promising candidate gene for maintaining yield in crop plants exposed to salt stress.
METHODS
Plant Materials and Stress Treatments
Fifteen-day-old rice seedlings (Oryza sativa cv Kitaake) were treated with 150 mM NaCl for various time periods and then an RT-qPCR analysis was performed to investigate the gene expression of RLKs selected based on chip data (Tyagi et al., 2007; Vij et al., 2008) (primers shown in Supplemental Table 1). Constructs of overexpression for these candidate RLK/RLCK genes were made and introduced into Kitaake via Agrobacterium tumefaciens-mediated transformation as described previously (Lin et al., 2009; primers shown in Supplemental Table 1). All plants were grown in a greenhouse with white fluorescent light provided at a photon flux density of 300 to 350 μmol m−2 s−1 and with a 16-h-light/8-h-dark cycle at 30°C. The relative humidity in the greenhouse was maintained at 60 to 70%.
The T2 generation of transgenic rice plants was used to investigate the stress response. For salt stress at the seedling stage, 15-d-old seedlings (40 plants each genotype) were transferred to a hydroponic culture solution (Ren et al., 2005) containing 140 mM NaCl. To obtain more apparent phenotypes between transgenic and wild-type plants, 8- and 10-d NaCl treatments were applied for STRK1-RNAi and STRK1-overexpressing seedlings as well as their corresponding wild-type seedlings, respectively. Then, the seedlings were transferred to a normal hydroponic culture solution to recover for 6 d and the survival rates were measured. Meanwhile, all leaves of 15-d-old seedlings (40 plants each genotype) treated with 100 mM NaCl for 7 d were used for the measurement of chlorophyll content, and those treated with 100 mM NaCl for 24 h were used for the measurements of MDA concentration and relative ion leakage. The salt stress at the reproductive stage was performed in plastic buckets. One wild-type plant and one positive transgenic plant each were planted per plastic bucket (20 plants each genotype). Plants were exposed to 1% NaCl at the panicle development stage. Then, NaCl solution was removed from the plastic bucket, and the plants were recovered with irrigation for 10 d and harvested. The panicle number, spikelet fertility, and grain yield were measured. For the MV treatment, germinated seeds (30 seeds per genotype) were transplanted onto hydroponic culture solution supplemented with 2 μM MV, and after 6 d, the chlorophyll content, CAT activity in the leaves, and seedling height were measured. For the H2O2 treatment, three-leaf stage seedlings (30 plants each genotype) were submerged into 100 mM H2O2 solution. After 1 d, the leaves were stained with DAB following the previously described method (Ouyang et al., 2010). Each data point represents the average of three replicates. Each stress experiment was repeated three times, and the results were consistent. The result from one set of experiments is presented here.
To study the significance of phosphorylated Tyr-210 of CatC in planta, the coding sequence of mutated CatC (CatCY210F and CatCY210D) was generated by site-directed mutagenesis and subcloned into the pCAMBIA2300 vector (primers listed in Supplemental Table 2). The mutated CatC (CatCY210F and CatCY210D) sequences were overexpressed in Ri11, a STRK1-RNAi line, via Agrobacterium-mediated transformation as described previously (Lin et al., 2009). The seedlings (T0) regenerated from the positive transgenic calli were used to measure the CAT activity and H2O2 content. The Kitaake (wild type) and Ri11 seedlings regenerated from the corresponding calli were used as controls.
RT-qPCR Analysis
To measure the transcript level of STRK1 under various stresses, Kitaake rice seedlings at the three-leaf stage were treated with salt stress (150 mM NaCl) or oxidative stress (1% [v/v] H2O2). After treatment, the shoots and roots were sampled at 0, 2, 4, 6, 12, and 24 h and used for total RNA isolation using TRIzol reagent (Invitrogen). The synthesis of first-stand cDNAs and real-time PCR analysis was performed as described previously (Li et al., 2016). Rice Actin1 was used as an internal control.
Physiological Measurements
Total chlorophyll content was measured according to the method described previously (Ouyang et al., 2010) with slight modification. The leaf samples (150 mg) were ground in liquid nitrogen and then transferred to a 10-mL falcon tube. Then, 2.5 mL of 80% acetone was added to the tube, and the samples were mixed thoroughly and incubated in darkness overnight at 4°C. The mixtures were centrifuged at 5000g for 15 min at 4°C. Supernatant was transferred to a colorimetric tube for absorbance measurement at 663 and 645 nm with a spectrophotometer (Shimadzu). The total chlorophyll content was calculated and expressed as mg g−1 FW.
The MDA content was determined as previously described (Ouyang et al., 2010) with slight modification. Briefly, ∼0.5 g of rice leaves was homogenized in 5 mL of 10% trichloroacetic acid and centrifuged at 5000g for 10 min at 4°C. Two milliliters of the supernatant was reacted with 2 mL of 0.6% (w/v) thiobarbituric acid (made in 10% trichloroacetic acid). The mixture was boiled for 15 min and centrifuged at 12,000g at 4°C for 10 min. The absorbance of the supernatant was read at 450, 532, and 600 nm. The MDA content was estimated using the extinction coefficient of 155 (nmol/L/cm) and expressed as μmol g−1 FW.
The relative ion leakage was measured according to the method described previously (Cao et al., 2007).
The CAT activity was determined by measuring the rate of decomposed H2O2 according to the method described previously (Ouyang et al., 2010). The reaction mixture (2 mL) contained 50 mM phosphate buffer (pH 7.0), 10 mM H2O2, and 10 μL enzyme extract. One unit of CAT activity was defined as 0.01 absorbance decrease per minute at 240 nm. The enzyme activity was expressed as U min−1 µg−1 protein. The concentration of H2O2 was determined using commercial kits (Beyotime) according to the manufacturer’s instructions.
The relative ion accumulation of Na+ and K+ was measured according to the method described previously (Schmidt et al., 2013).
For the above parameters, each data point represents the average of three replicates. Three experiments were performed, and the results were consistent. The result from one set of experiments is presented here.
Subcellular Localization Analysis
For detection of the subcellular localization of STRK1, the coding region of STRK1 or the mutational coding sequence of STRK1 were amplified and respectively subcloned into the pCAMBIA1300-YFP vector, in which the YFP-coding sequence was fused in frame to the 3′ end of the STRK1 gene sequence. Primers are listed in Supplemental Table 2. The plasmid constructs were transformed into rice protoplasts and calli as described previously (Lin et al., 2009; Lv et al., 2014). The fluorescence was then observed with a confocal laser scanning microscope (Olympus FV1000).
BiFC Assays
For generation of the BiFC vectors, the coding sequence of STRK1 was cloned into pE3449 vector, resulting in STRK1-cCFP, and the coding regions of STRK1, CatA, CatB, and CatC were respectively cloned into pE3228 vector, resulting in nVenus-STRK1, nVenus-CatA, nVenus-CatB, and nVenus-CatC. Primers used are listed in Supplemental Table 2. Transient Arabidopsis thaliana protoplast expression was performed using the PEG-mediated transformation method described previously (Yoo et al., 2007). Fluorescence was visualized in Arabidopsis protoplasts by a confocal laser scanning microscope (Olympus FV1000).
Coimmunoprecipitation Assays
For co-IP assays, the constructs Ubi:STRK1-FLAG and 35S:CatC-GFP were generated as described previously (Earley et al., 2006; Zhou et al., 2014, 2015). Then two constructs were transiently expressed in 3-week-old Nicotiana benthamiana leaves by Agrobacterium infiltration. The co-IP assay was performed as described previously (Feng et al., 2008). Anti-FLAG beads (Sigma-Aldrich) were used to immunoprecipitate protein complexes. The immunoprecipitated proteins were subsequently released by boiling in 2.5× SDS sample buffer and subjected to 10% SDS-PAGE. Immunoblot analyses using anti-GFP (Abmart; catalog no. M20004) and anti-FLAG (Abmart; catalog no. M20008) antibodies were then performed.
The in vivo pull-down assay was performed as described previously (Wang et al., 2013). CatC was expressed in Escherichia coli using the pCold-TF expression system according to the manufacturer’s instructions (Takara; catalog no. 3365). His-TF-CatC fusion protein was affinity purified using Ni-NTA agarose beads (Invitrogen). Total plant proteins (1 mg) of the STRK1-overexpressing line (OE18) were incubated with 10 μg of purified His-TF-CatC protein bound to Ni-NTA agarose overnight at 4°C on a rotary shaker.
Yeast Two-Hybrid Assays
Yeast two-hybrid analysis was performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech) according to the manufacturer’s instructions. Full-length STRK1 was cloned into the pGBKT7 vector, and then the construct was transformed into the yeast strain Y2HGold. The transformed Y2HGold yeast strain was fused with the Y187 yeast strain containing a rice cDNA library. Medium lacking Leu-Trp-His-Ade was used for selection. Positive clones were selected for sequencing.
The interactions between STRK1 and CATs or STRK1 were verified by yeast two-hybrid assays as described previously (Jing et al., 2015). The coding region of STRK1, CatA, CatB, and CatC were cloned into pGADT7 prey plasmid and cotransformed into the yeast strain AH109 with STRK1 bait, respectively. Primers used are listed in Supplemental Table 2. Medium lacking Leu-Trp-His-Ade was used for selection. The β-galactosidase analyses were performed according to the manufacturer’s recommendations (Clontech), using chlorophenol red-β-d-galactopyranoside as the substrate.
Promoter-GUS Analysis
For the promoter-GUS reporter assay, the STRK1 promoter (1771-bp DNA fragment upstream of the translation start site) was amplified from rice genomic DNA by PCR with a pair of primers, STRK1-ProF/R (Supplemental Table 2). The amplified STRK1 promoter was digested with HindIII and NcoI and then cloned into the corresponding restriction site of the pCAMBIA1301 vector with a GUS reporter gene to generate pCAMBIA1301-STRK1pro:GUS. The primer sequences are listed in Supplemental Table 2. The construct was transformed into rice (O. sativa cv Kitaake) by Agrobacterium-mediated transformation as described previously (Lin et al., 2009). Histochemical GUS staining was performed according to the method described previously (Asano et al., 2012).
Kinase Assay
The STRK1 expressed in E. coli as GST fusions using pGEX4T-1 (Amersham Biosciences) was purified with glutathione agarose beads (Roche) and subjected to an in vitro kinase assay as described previously (Tanaka et al., 2012).
For the in vitro phosphorylation assay of the MBP (Sigma-Aldrich) and rice CatC by recombinant STRK1, 10 µg of MBP, His-TF-CatC, His-TF-CatCS9,10,11,18,205A, His-TF-CatCT19,209,211,292A, His-TF-CatCY210,360F, His-TF-CatCY210F, and His-TF-CatCY360F expressed and purified from E. coli were incubated respectively with 0.5 µg of GST-STRK1 in kinase reaction buffer (50 mM HEPES, pH 7.4, 50 mM NaCl, 0.1% Triton X-100, 10% glycerol, 0.1 mM Na3VO4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 8 µCi [γ-32P]ATP) for 1.5 h at 30°C. Kinase reactions were terminated by adding an appropriate volume of 2× SDS sample buffer. The phosphorylated proteins were analyzed by 10% SDS-PAGE and autoradiography.
For immunocomplex kinase assays, the OE18 transgenic seedlings expressing STRK1-FLAG were treated with NaCl for the indicated time period. Next, the STRK1-FLAG and endogenous CatC were immunoprecipitated by anti-FLAG beads (Sigma-Aldrich) and anti-CatC antibody (Abmart; its specificity shown in Supplemental Figure 17) as previously described (Li et al., 2014). The tyrosine phosphorylation of STRK1-FLAG and CatC were analyzed by immunoblot using anti-phospho-tyrosine antibody (Abcam; catalog no. ab17302). Meanwhile, the catalase activity of CatC was measured as described previously (Ouyang et al., 2010).
Phylogenetic Analysis
The amino acid sequences of STRK1 and its homologs (Supplemental Data Set 1) were aligned using ClustalX2, and the resulting alignment was used to construct a phylogenetic tree using MEGA 5 software with the neighbor-joining method. Bootstrap values were derived from 1000 replications and are shown at branch nodes.
Accession Numbers
Sequence data from this article can be found in the Michigan State University Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu) (Ouyang et al., 2007) using the Michigan State University accession numbers: STRK1 (Os04g45730), CatA (Os02g02400), CatB (Os06g51150), CatC (Os03g03910), and Actin1 (Os03g50890). The RLK genes include LOC_Os01g12720, LOC_Os01g49920, LOC_Os03g17300, LOC_Os04g44910, LOC_Os05g03620, LOC_Os05g24010, LOC_Os07g03810, LOC_Os08g03020, and LOC_Os11g40970 in Supplemental Figure 1. The RLCK genes include LOC_Os01g05960, LOC_Os04g45730, LOC_Os06g07070, LOC_Os07g48730, LOC_Os08g28710, and LOC_Os09g39930 in Supplemental Figure 1.
Supplemental Data
Supplemental Figure 1. The expression profiles of RLKs genes under salt stress.
Supplemental Figure 2. Diagrammatic representation of the vector constructs and expression analysis of STRK1 in transgenic plants.
Supplemental Figure 3. Sequence similarity matrix of STRK1.
Supplemental Figure 4. Effect of STRK1 on biomass reduction and ion accumulation under salt stress.
Supplemental Figure 5. Schematic overview of the STRK1 protein.
Supplemental Figure 6. Subcellular localization of STRK1 in transgenic rice plants.
Supplemental Figure 7. Mislocalized STRK1 failed to activate salt stress signaling.
Supplemental Figure 8. Chlorophyll content in the leaves of wild-type and transgenic rice seedlings treated with 100 mM NaCl for 7 d.
Supplemental Figure 9. RT-qPCR analyses of STRK1 expression in young root, stem, leaf, leaf sheath, and young spikelet from rice.
Supplemental Figure 10. Sequence similarity matrix of CAT family members in rice.
Supplemental Figure 11. CAT family members CatA and CatB also interact with STRK1.
Supplemental Figure 12. β-Galactosidase assay showing interaction of STRK1 with members of the rice CAT family in yeast cells.
Supplemental Figure 13. The GST-STRK1 shows strong autophosphorylation and substrate (MBP) phosphorylation.
Supplemental Figure 14. Preliminary identification of phosphorylation sites of CatC recognized by STRK1.
Supplemental Figure 15. Phosphorylation sites of CAT proteins identified to date to be phosphorylated by definite protein kinases.
Supplemental Figure 16. Relative expression levels of STRK1 and CatC in transgenic seedlings (T0) overexpressing CatCY210F and CatCY210D under the Ri11 background, a STRK1 knockdown mutant, as determined by RT-qPCR.
Supplemental Figure 17. Specificity verification of a polyclonal anti-CatC antibody.
Supplemental Table 1. Primers for analysis of the candidate RLK/RLCK genes induced by salt stress.
Supplemental Table 2. The other primers used in this study.
Supplemental Data Set 1. The amino acid sequences of STRK1 and its homologs used for constructing a phylogenetic tree.
Acknowledgments
This work was supported by the National Science Foundation of China (31170172 to J.-Z.L. and 31571635 to X.-M.L.), by Hunan Provincial Natural Science Foundation of China (2017JJ2042 to D.-Y.T.), by Planned Science and Technology Project of Hunan Province (2017WK2012 to X.-M.L.), and by Planned Science and Technology Project of Changsha City (kq1701028 to J.-Z.L.). We thank Chentao Lin (University of California) for guidance in the initial work. We thank Hang He and Shoucheng Liu (Peking University, China) for guidance in the analysis of RLK transcript profile based on ChIP data. We thank Zecheng Zuo (Jilin University, China) for guidance in mass spectrometry. We thank Jia Li (Lanzhou University, China), Vivian F. Irish (Yale University), and Yan Guo (China Agricultural University, China) for critically reading the manuscript and constructive suggestions.
AUTHOR CONTRIBUTIONS
J.-Z.L. and X.-M.L. conceived the project and designed the research. Y.-B.Z. and C.L. designed the research, performed the experiments, and analyzed the data. D.-Y.T. and L.Y. analyzed the CAT activity and H2O2 concentration. X.-D.T. constructed the transgenic rice plants. D.W., Y.-Z.Y., J.-S.G., X.-Y.Z., L.-G.L., F.Y., J.-L.L., L.-L.L., and Y.-H.Z. partially participated in the experiments and analyzed the data. Y.-B.Z., J.-Z.L., and X.-M.L. wrote the article.
References
- Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I. (2011). Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808: 2981–2994. [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Asano T., Hayashi N., Kobayashi M., Aoki N., Miyao A., Mitsuhara I., Ichikawa H., Komatsu S., Hirochika H., Kikuchi S., Ohsugi R. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69: 26–36. [DOI] [PubMed] [Google Scholar]
- Azevedo-Neto A.D., Prisco J.T., Enéas-Filho J., Abreu C.E.B., Gomes-Filho E. (2006). Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Bot. 56: 87–94. [Google Scholar]
- Boisson-Dernier A., Lituiev D.S., Nestorova A., Franck C.M., Thirugnanarajah S., Grossniklaus U. (2013). ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 11: e1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boyer J.S. (1982). Plant productivity and environment. Science 218: 443–448. [DOI] [PubMed] [Google Scholar]
- Cao W.-H., Liu J., He X.-J., Mu R.-L., Zhou H.-L., Chen S.-Y., Zhang J.-S. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-J., et al. (2013). An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 163: 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M., Zhang R., Zhu F., Zhang Z., Gou L., Wen J., Dong J., Wang T. (2017). A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase/expression during drought stress. Plant Cell 29: 1748–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Kita D., Johnson E.A., Aggarwal M., Gates L., Wu H.M., Cheung A.Y. (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 5: 3129. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T.J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55: 307–319. [DOI] [PubMed] [Google Scholar]
- Giri J., Vij S., Dansana P.K., Tyagi A.K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191: 721–732. [DOI] [PubMed] [Google Scholar]
- Guo Y., Halfter U., Ishitani M., Zhu J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C.Y., Chao Y.Y., Yang M.Y., Cheng S.Y., Cho S.C., Kao C.H. (2009). NaCl-induced expression of glutathione reductase in roots of rice (Oryza sativa L.) seedlings is mediated through hydrogen peroxide but not abscisic acid. Plant Soil 320: 103–115. [Google Scholar]
- Ikeda K., Sunohara H., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54: 147–156. [Google Scholar]
- Jing H., Yang X., Zhang J., Liu X., Zheng H., Dong G., Nian J., Feng J., Xia B., Qian Q., Li J., Zuo J. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6: 7395. [DOI] [PubMed] [Google Scholar]
- Joo J., Lee Y.H., Song S.I. (2014). Rice CatA, CatB, and CatC are involved in environmental stress response, root growth, and photorespiration, respectively. J. Plant Biol. 57: 375–382. [Google Scholar]
- Kim T.-W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sud N., Fonseca F.V., Hou Y., Black S.M. (2010). Shear stress stimulates nitric oxide signaling in pulmonary arterial endothelial cells via a reduction in catalase activity: role of protein kinase C δ. Am. J. Physiol. Lung Cell. Mol. Physiol. 298: L105–L116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li C.H., Wang G., Zhao J.-L., Zhang L.-Q., Ai L.-F., Han Y.-F., Sun D.-Y., Zhang S.-W., Sun Y. (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26: 2538–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lin J., Li L., Peng Y., Wang W., Zhou Y., Tang D., Zhao X., Yu F., Liu X. (2016). DHHC-cysteine-rich domain S-acyltransferase protein family in rice: organization, phylogenetic relationship and expression pattern during development and stress. Plant Syst. Evol. 302: 1405–1417. [Google Scholar]
- Lin J., Zhou B., Yang Y., Mei J., Zhao X., Guo X., Huang X., Tang D., Liu X. (2009). Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of indica rice. Plant Cell Rep. 28: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Lin W., Ma X., Shan L., He P. (2013). Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 55: 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Li B., Lu D., Chen S., Zhu N., He P., Shan L. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 111: 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jia Y., Ding Y., Shi Y., Li Z., Guo Y., Gong Z., Yang S. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q., Zhong Y., Wang Y., Wang Z., Zhang L., Shi J., Wu Z., Liu Y., Mao C., Yi K., Wu P. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang H., Sun L., Jiao Y., Zhang G., Miao C., Hao F. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63: 305–317. [DOI] [PubMed] [Google Scholar]
- Macho A.P., et al. (2014). A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54: 263–272. [DOI] [PubMed] [Google Scholar]
- Macho A.P., Lozano-Durán R., Zipfel C. (2015). Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 20: 269–272. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61: 4197–4220. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: the new wave? Trends Plant Sci. 16: 300–309. [DOI] [PubMed] [Google Scholar]
- Miyagawa Y., Tamoi M., Shigeoka S. (2000). Evaluation of the defense system in chloroplasts to photooxidative stress caused by paraquat using transgenic tobacco plants expressing catalase from Escherichia coli. Plant Cell Physiol. 41: 311–320. [DOI] [PubMed] [Google Scholar]
- Morita S., Tasaka M., Fujisawa H., Ushimaru T., Tsuji H. (1994). A cDNA clone encoding a rice catalase isozyme. Plant Physiol. 105: 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki T., Yamamoto Y., Aida T., Funahashi T., Shishido T., Asada M., Prodhan S.H., Komamine A., Motohashi T. (2007). Overexpression of the Escherichia coli catalase gene, katE, enhances tolerance to salinity stress in the transgenic indica rice cultivar, BR5. Plant Biotechnol. Rep. 2: 41–46. [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S.P. (2013). Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64: 445–458. [DOI] [PubMed] [Google Scholar]
- Ouyang S., et al. (2007). The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 35: D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S.-Q., Liu Y.-F., Liu P., Lei G., He S.-J., Ma B., Zhang W.-K., Zhang J.-S., Chen S.-Y. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62: 316–329. [DOI] [PubMed] [Google Scholar]
- Pérez-Salamó I., et al. (2014). The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 165: 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S., Gribskov M. (2004). Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidoros A.N., Mylona P.V., Scandalios J.G. (2001). Transgenic tobacco plants expressing the maize Cat2 gene have altered catalase levels that affect plant-pathogen interactions and resistance to oxidative stress. Transgenic Res. 10: 555–569. [DOI] [PubMed] [Google Scholar]
- Quintero F.J., Martinez-Atienza J., Villalta I., Jiang X., Kim W.-Y., Ali Z., Fujii H., Mendoza I., Yun D.-J., Zhu J.-K., Pardo J.M. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 108: 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafikov R., Kumar S., Aggarwal S., Hou Y., Kangath A., Pardo D., Fineman J.R., Black S.M. (2014). Endothelin-1 stimulates catalase activity through the PKCδ-mediated phosphorylation of serine 167. Free Radic. Biol. Med. 67: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda V., Basu S., Krishnan A., Pereira A. (2014). Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 166: 1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z.-H., Gao J.P., Li L.G., Cai X.L., Huang W., Chao D.Y., Zhu M.Z., Wang Z.Y., Luan S., Lin H.X. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Schippers J.H.M., Nguyen H.M., Lu D., Schmidt R., Mueller-Roeber B. (2012). ROS homeostasis during development: an evolutionary conserved strategy. Cell. Mol. Life Sci. 69: 3245–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Mieulet D., Hubberten H.M., Obata T., Hoefgen R., Fernie A.R., Fisahn J., San Segundo B., Guiderdoni E., Schippers J.H.M., Mueller-Roeber B. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 2115–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Hao L., Li J., Liu D., Guo X., Li H. (2014). The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep. 33: 483–498. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Takeda T., Yamauchi H., Sano S., Tomizawa K.-I., Yokota A., Shigeoka S. (1998). Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett. 428: 47–51. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.-H., Mayer K.F.X., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Sun M., Luo X., Ding X., Cai H., Bai X., Liu X., Zhu Y. (2013). A Glycine soja ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses. Planta 237: 1527–1545. [DOI] [PubMed] [Google Scholar]
- Tan T., Cai J., Zhan E., Yang Y., Zhao J., Guo Y., Zhou H. (2016). Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol. Biol. 92: 391–400. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Osakabe Y., Katsura S., Mizuno S., Maruyama K., Kusakabe K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70: 599–613. [DOI] [PubMed] [Google Scholar]
- Tang W., Kim T.-W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.-Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U. (2000). Receptor kinase activation and signal transduction in plants: an emerging picture. Curr. Opin. Plant Biol. 3: 361–367. [DOI] [PubMed] [Google Scholar]
- Tyagi A.K., Kapoor S., Khurana J.P., Ray S. (2007). Expression data for stress treatment in rice seedlings. NCBI, http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi.
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R.A., Hirt H., Mengiste T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij S., Giri J., Dansana P.K., Kapoor S., Tyagi A.K. (2008). The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 1: 732–750. [DOI] [PubMed] [Google Scholar]
- Wang J., Yu Y., Zhang Z., Quan R., Zhang H., Ma L., Deng X.W., Huang R. (2013). Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiang J., Wang J., Chen L., Fan S.L., Wu J.W., Wang X., Wang Z.X. (2014). Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 24: 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang M.M., Wang Y.J., Gao Y.T., Li R., Wang G.F., Li W.Q., Liu W.T., Chen K.M. (2016). The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought-stress response in rice. Physiol. Plant. 156: 421–443. [DOI] [PubMed] [Google Scholar]
- Xu J., Duan X., Yang J., Beeching J.R., Zhang P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161: 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., et al. (2013). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- You J., Zong W., Hu H., Li X., Xiao J., Xiong L. (2014). A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166: 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Shannon M.C. (2000). Salinity effects on seedling growth and yield components of rice. Crop Sci. 40: 996–1003. [Google Scholar]
- Zhang Z., Xu Y., Xie Z., Li X., He Z.H., Peng X.X. (2016). Association-dissociation of glycolate oxidase with catalase in rice: A potential switch to modulate intracellular H2O2 levels. Mol. Plant 9: 737–748. [DOI] [PubMed] [Google Scholar]
- Zhou Y., et al. (2014). Over-expression of a fungal NADP(H)-dependent glutamate dehydrogenase PcGDH improves nitrogen assimilation and growth quality in rice. Mol. Breed. 34: 335–349. [Google Scholar]
- Zhou Y., Zhang C., Lin J., Yang Y., Peng Y., Tang D., Zhao X., Zhu Y., Liu X. (2015). Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta 241: 727–740. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4: 401–406. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6: 441–445. [DOI] [PubMed] [Google Scholar]
- Zhu J.Y., Sae-Seaw J., Wang Z.Y. (2013). Brassinosteroid signalling. Development 140: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.J., Li X.D., Ratnasekera D., Wang C., Liu W.-X., Song L.-F., Zhang W.-Z., Wu W.-H. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27: 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]