The story behind m6A (methylation of the N6 position of adenosine), the most common internal mRNA modification in eukaryotes, has long been a source of intrigue. This epitranscriptomic mark is deposited at specific mRNA sequences by m6A writers and removed by m6A erasers. The m6A marks recruit and anchor m6A binding proteins (readers) that function in processes such as pre-mRNA splicing, mRNA degradation, and translation, thus regulating gene expression. The best-characterized m6A readers, YTH domain proteins, contain a highly conserved aromatic cage that recognizes m6A. YTH proteins function in fundamental processes ranging from stem cell fate regulation in vertebrates to sex determination in fruit fly (Drosophila melanogaster), but whether these or similar proteins act as m6A readers in plants has been a mystery.
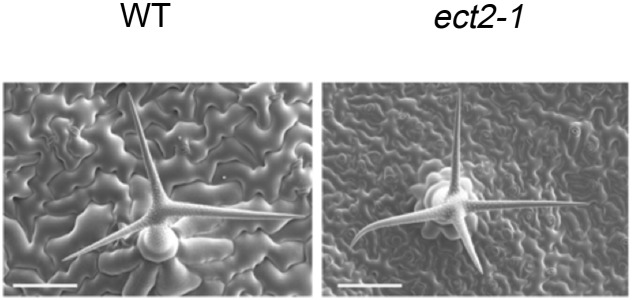
Arabidopsis thaliana contains 13 YTH proteins with highly conserved C termini, including EVOLUTIONARILY CONSERVED C-TERMINAL REGION1 (ECT1) to ECT11. The observation that ECT2 is highly expressed in Arabidopsis, especially in rapidly growing tissues, prompted Wei et al. (2018) to focus on ECT2 as a possible m6A reader. Cryo-scanning electron microscopy showed that ect2 mutants have increased trichome branching compared with the wild type (see figure). ECT2 binds strongly to m6A residues in mRNA, whereas ECT2 lacking the YTH domain does not. Complementation experiments confirmed the role of ECT2 as an m6A reader that functions in trichome development. Transcriptome-wide analysis to identify ECT2-RNA interaction sites using a new formaldehyde cross-linking/immunoprecipitation method uncovered a plant-specific m6A motif in the 3′ untranslated regions of target genes that is recognized by ECT2, which was confirmed to be methylated in vitro. While the human m6A reader, YTHDF2, promotes mRNA degradation, ECT2 appears to support m6A-mediated mRNA stability in the cytoplasm. Indeed, ECT2 bound to three trichome morphogenesis-related transcripts with m6A modifications, and disrupting ECT2 accelerated their degradation and altered trichome branching.
Cryo-scanning electron microscopy of trichome morphology in wild-type (WT) and ect2 plants. Trichomes are from the third and fourth leaves of 3-week-old Arabidopsis plants. Bars = 100 μm. (Reprinted from Wei et al. [2018], Figure 2A.)
Scutenaire et al. (2018) took the long view before honing in on ECT2. Evolutionary analysis of YTH domain proteins suggested that all Viridiplantae YTH domains display a canonical aromatic cage and thus carry bona fide m6A binding pockets. However, unlike their animal counterparts, it appears that the functions of plant YTH proteins are not fully redundant, as YTH domains from different clades are predicted to have different m6A binding affinities, a feature conserved across evolution. Epifluorescence imaging suggested that the trichome branching defect in ect2 mutants is related to an excessive number of endoreduplication cycles, a phenotype also observed in hypomethylated plants. Like many mRNA binding proteins, ECT2 relocates from the cytoplasm to stress granules upon exposure to heat stress. The authors demonstrated that ECT2 bind to m6A-RNA in vivo; this property requires a functional aromatic cage.
Since ECT3 is also highly expressed in Arabidopsis and belongs to the same phylogenetic subclade as ECT2, Arribas-Hernández et al. (2018) focused their analysis on both proteins. They showed that in addition to ECT2, ECT3 is necessary for trichome branching and that ECT2 and ECT3 are only expressed during early stages of trichome development. In addition, the ect2 ect3 double mutants displayed a compelling new phenotype: delayed first true leaf emergence. The mutation of ECT4, a close homolog of ECT2, enhanced this phenotype, suggesting that ECT2, ECT3, and ECT4 control the timing of postembryonic leaf formation. These proteins, and the m6A binding sites in ECT2 and ECT3, are also required for proper leaf morphology.
The story of m6A readers in plants is nowhere near complete. Additional studies will no doubt leave their mark on plant biology.
Footnotes
Articles can be viewed without a subscription.
References
- Arribas-Hernández L., Bressendorff S., Hansen M.H., Poulsen C., Erdmann S., Brodersen P. (2018). An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30: 952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutenaire J., Deragon J.-M., Jean V., Benhamed M., Raynaud C., Favory J.-J., Merret R., Bousquet-Antonelli C. (2018). The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30: 986–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.-H., Song P., Wang Y., Lu Z., Tang Q., Yu Q., Xiao Y., Zhang X., Duan H.-C., Jia G. (2018). The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30: 968–985. [DOI] [PMC free article] [PubMed] [Google Scholar]