Summary
Natural killer (NK) cells express an array of germ‐line encoded receptors that are capable of triggering cytotoxicity. NK cells tend to express many members of a given family of signalling molecules. The presence of many activating receptors and many members of a given family of signalling molecules can enable NK cells to detect different kinds of target cells, and to mount different kinds of responses. This contributes also to the robustness of NK cells responses; cytotoxic functions of NK cells often remain unaffected in the absence of selected signalling molecules. NK cells express many MHC‐I‐specific inhibitory receptors. Signals from MHC‐I‐specific inhibitory receptors tightly control NK cell cytotoxicity and, paradoxically, maintain NK cells in a state of proper responsiveness. This review provides a brief overview of the events that underlie NK cell activation, and how signals from inhibitory receptors intercept NK cell activation to prevent inappropriate triggering of cytotoxicity.
Keywords: activating receptor, cytotoxic synapse, immunoreceptor tyrosine‐based inhibitory motif (ITIM), inhibitory receptor, inhibitory synapse, killer‐cell Ig‐like receptor (KIR), natural killer cell
Introduction
Natural killer (NK) cells are lymphocytes with features of both innate and adaptive immunity. They contribute to immune defense by killing unhealthy cells, secreting soluble factors, and regulating the responses of antigen‐presenting cells and the adaptive T cells. NK cells are the predominant lymphocyte population in the uterus, and play important roles in reproduction.1, 2 Unlike adaptive lymphocytes, wherein receptor diversity is generated by DNA rearrangements, NK cells express an array of germ‐line encoded receptors capable of triggering activation.3, 4, 5, 6, 7, 8 Despite this difference, NK cells do display the features of adaptive immunity.9 NK cells can mount a response against infections, including viral, and register a memory for that response. NK cells can distinguish healthy cells from diseased cells. Understanding how specificity is achieved in the recognition of target cells, and how healthy cells are spared from NK cell attack, are among the major goals of NK cell studies. With a focus on human NK cells, this review examines briefly the molecular events of NK cell cytotoxicity, and how NK cell cytotoxicity is controlled to distinguish healthy cells from diseased ones.
NK cell cytotoxicity
Receptors for NK cell cytotoxicity
Natural killer cells express many receptors that can activate their cytotoxic and secretory functions. These receptors recognize ligands on the surface of infected, transformed or stressed cells. The Fc receptor Fcγ RIIIa (CD16) on NK cells recognizes the Fc portion of antibodies, for example, bound to unhealthy cells, and triggers NK cell activation through a process termed as antibody‐dependent cell‐mediated cytotoxicity.
The natural cytotoxicity receptors (NCR) (NKp30, NKp44 and NKp46) are among the earliest identified NK cell‐activating receptors.10 They are type I transmembrane receptors, which belong to the immunoglobulin superfamily. They are potent inducers of NK cell cytotoxicity, and are important for NK cell‐mediated tumour immunosurveillance. They are expressed largely on NK cells. While NKp46 is specific to NK cells, the other two are expressed also on T cell subsets.11 While NKp46 and NKp30 are expressed on both resting and activated NK cells, expression of NKp44 is induced upon NK cell activation.12, 13, 14, 15 Identification of the cellular ligands of NCRs has been challenging, except the identification of B7‐H6 as an NKp30′s cellular ligand expressed on the surface of tumour cells.16, 17 A number of cellular and viral ligands of these NCRs have been reported, which could trigger or block the receptor.10, 18, 19 For example, a nuclear protein BAT3, a ligand of NKp30, could be released from tumour cells in exosomes or as soluble proteins to activate or inhibit NKp30, respectively. The protein CMV pp65 has been described as a viral ligand of NKp30, which binds and inhibits NKp30.
Natural‐killer group 2, member D (NKG2D) is a homodimeric C‐type lectin‐like receptor, which is expressed on the surface of NK cells and cytotoxic T cells.20 It is a type II transmembrane receptor, belonging to the CD94/NKG2 family.21, 22 NKG2D ligands are many structural homologues of class I MHC, including human ULBPs and MICA/B, and mouse Mult1, H60 and Rae‐1.22, 23 The NKG2D ligands are upregulated in infected, stressed and tumour cells, indicating important roles of NKG2D in immune defense against abnormal cells. Tumour cells have been seen to shed NKG2D ligands, which has important implications in tumour immunosurveillance.24, 25
Natural killer cells express other C‐type lectin‐like activating receptor NKp80, which is a dimeric type II transmembrane receptor. NKp80 is expressed on all NK cells in the peripheral blood. It recognizes a C‐type lectin‐like ligand, activation‐induced C‐type lectin (AICL).26, 27 AICL is upregulated on activated monocytes and NK cells, and NKp80–AICL interaction promotes NK cell‐mediated control of monocytes and autologous NK cells.27
Natural killer cells also express the members of the signalling lymphocytic activation molecule (SLAM) family of receptors. The SLAM receptor family is a group of type I transmembrane receptors, and has six members.7, 18 NK cells express every member except SLAM (CD150, SLAMF1). The other five members are: 2B4 (CD244, SLAMF4), Ly‐9 (CD229, SLAMF3); natural killer, T‐ and B‐cell antigen (NTB‐A) or Ly108 (in mouse) (SLAMF6); CD84 (SLAMF5); and CD2‐like receptor‐activating cytotoxic cells (CRACC) (CD319, SLAMF7). They are expressed on other immune cells too, but not on non‐immune cells. The members of SLAM receptor family promote cell–cell interactions through homophilic binding, i.e. they bind themselves in trans. The only exception is 2B4, which recognizes another Ig‐like molecule CD48 expressed on nearly every haematopoietic cell.28, 29
DNAX accessory molecule‐1 (DNAM‐1) is an Ig‐like domain‐containing receptor.18, 30, 31 It is expressed on both mouse and human NK cells. DNAM‐1 is also expressed on other immune cells, including CD8+ T cells and myeloid cells. DNAM‐1 recognizes CD155 (poliovirus receptor) and CD122 (nectin adhesion molecule),32 both of which are upregulated on cancerous and virus‐infected cells. In human, DNAM‐1 associates, physically and functionally, with the β2‐integrin LFA‐1.33
In addition to the above‐mentioned receptors, NK cells express many other activating receptors, including CD2, CD44, fractalkine receptor, CD27, CD160, CD137, activating Ly49 receptors (in mouse) and activating killer‐cell Ig‐like receptors (in human).
Tyr‐based motifs for NK cell activation
Most activating receptors of NK cells signal through their cytosolic Tyr‐based motifs, and signalling by them initiates with phosphorylation of the key Tyr residues in the motif.18, 34 An assortment of Tyr‐based motifs has been described for NK cell activation. The Fc receptor Fc γ RIIIa (CD16) signals upon association with homo‐ or hetero‐dimer of the immunoreceptor Tyr‐based activation motif (ITAM)‐bearing FcR γ and/or CD3 ζ chains. Similarly, the NCRs NKp30 and NKp46 associate with FcR γ and/or CD3 ζ chains.10 However, the NCR NKp44 associates with the adaptor protein DAP12,35 which is homodimeric, with each DAP12 molecule containing a single ITAM.36, 37 DAP12 is the ITAM‐bearing partner of the activating killer‐cell Ig‐like receptors (KIRs) and CD94‐NKG2 receptors.4, 38 NKG2D associates with the adaptor molecule DAP10, which bears the activating Tyr‐based motif YxxM, which is distinct from the ITAMs.39 The SLAM family of receptors, such as the best‐studied member 2B4, does not require association with a partner chain for the Tyr‐based activation motifs. They rather possess a Tyr‐based motif S/TxYXXL/I, referred to as immunoreceptor Tyr‐based switch motifs (ITSM),40 in their cytosolic tails, which could signal for NK cell activation.29 NKp80 also does not require a partner chain for activation motifs, and possesses a motif corresponding to half of an ITAM.41 Signalling through DNAM‐1 requires phosphorylation of a conserved Tyr (Y319 in mouse and Y322 in human) and a conserved Asn (N321 in mouse and N324 in human) that are present in its cytosolic tail.30
Signalling pathways of NK cell‐activating receptors
Because ITAMs are utilized by many immunoreceptors, including the T cell receptor (TCR), ITAM‐based signalling is among the best‐understood pathways.42 ITAMs are phosphorylated by the Src family kinases. The phosphorylated ITAMs, through SH2 domain‐based interactions, recruit the Tyr kinases ZAP‐70 and Syk, which, in turn, could phosphorylate transmembrane adaptor proteins, leading to recruitment of several signalling molecules, including the phosphoinositide 3‐kinase (PI3K), phospholipase C (PLC)‐γ1 and ‐γ2, and the guanine nucleotide exchange factors Vav‐1, 2, 3. The importance of different isoforms of PLC‐γ and Vav could be different for different ITAM‐bearing receptors.43, 44, 45, 46
Signalling by NKG2D occurs through DAP10, and does not require the Tyr kinases ZAP‐70 or Syk.47, 48, 49 The YxxM motif of DAP10 could be phosphorylated by Src family kinases, and could bind to either PI3K or the cytosolic adaptor protein Grb2. The p85 subunit of PI3K can bind to the cytosolic adaptor proteins CrkL and Grb2, and to the guanine nucleotide exchange factor Vav‐1, leading to the activation of GTPases to promote NK cell‐target cell adhesion and synapse formation.47, 50, 51, 52, 53
2B4, a member of the SLAM family of receptors, is among the best‐understood NK cell‐activating receptors.7 SLAM receptor's ITSMs can be phosphorylated by Src family kinases. Phosphorylated ITSM recruits the SAP family of small cytosolic adaptor proteins, SLAM‐associated protein (SAP), Ewing's sarcoma‐associated transcript‐2 (EAT‐2) or EAT‐2‐related transducer (ERT).54, 55, 56, 57 It appears that both SAP and EAT‐2 contribute to NK cell activation by 2B4. However, NK cell activation by CRACC is SAP‐independent and requires EAT‐2.58 The SAP family adaptors are composed largely of an SH2 domain. SAP could contribute to SLAM receptor‐mediated NK cell activation by preventing the recruitment of SH2‐domain containing inositol 5′ phosphatase‐1 (SHIP‐1).59 The SAP family adaptors could prevent the recruitment of SHIP, the protein Tyr phosphatases SHP‐1/2 and the protein Tyr kinase Csk.60, 61 Further, SAP family adaptors could also recruit specific molecules to elicit signalling by SLAM receptors. For example, SAP promotes activation of mouse NK cells by Fyn‐induced phosphorylation of Vav‐1.59
The conserved Tyr‐ and Asn‐based motif of DNAM‐1 could be phosphorylated by Src family kinases, which, upon phosphorylation, could recruit Grb2.30 In this respect, DNAM‐1 is similar to the YxxM motif of DAP10. The Met residue at the position +3 (relative to the key Tyr residue) of the YxxM motif is essential for the recruitment of the p85 subunit of PI3K. DNAM‐1 lacks this +3 Met residue, but yet recruits p85, seemingly through an indirect mode. The signalling outcomes of such differences in recruiting the same set of molecules by different activation receptors are unclear.
CD2 possesses Pro‐rich sequences in its cytosolic tail, which are seen to interact with the Src family kinase Lck and many adaptors.62 How exactly signalling is achieved by CD2 is unclear.
NK cell cytotoxic synapse
When an NK cell recognizes a sensitive target cell, it makes a cell–cell junction with the target cell. The synaptic cleft formed at the cell–cell junction is mediated by engagements of many transmembrane receptors with their cognate cell surface ligands. This creates a highly organized intercellular junction, called immunological synapse, for vectoral communication between the cells.63 Immunological synapses accumulate and integrate signals from the engaged receptors to determine the signalling outcomes.63, 64
Signalling at NK cell cytotoxic synapses, in response to sensitive target cells, triggers a series of cell biological processes that culminates in cytolytic degranulation towards the target cells and secretion of soluble factors.65 After initial NK cell‐target cell adhesion, the lytic granules of NK cells dock onto the microtubule‐organizing centre (MTOC) in a process termed granule convergence. Then, the MTOC, along with the docked lytic granules, is polarized towards the NK‐target cell synapses. This process is known as granule polarization, and is associated with dramatic cytoskeletal rearrangements, including actin accumulation at the synapses. A fraction of the polarized lytic granules traverses through the actin meshwork66, 67 at the synapse, and exocytose their lytic contents (degranulation) that leads to target cell killing. It appears that the requirement of signal strength is different for different NK cell responses, such as degranulation versus cytokine secretion.3
Natural killer cells, like cytotoxic T cells, require signalling by the β2‐integrin LFA‐1 (αLβ2, CD11a/CD18), for adhesion to target cells and proper cytotoxicity. LFA‐1 recognizes ICAM‐1 on target cells. In primary T cells, LFA‐1 requires activation by inside‐out signals, from TCR or a chemokine receptor, in order to bind ICAM‐1.68, 69 Contrarily, in NK cells, LFA‐1 molecules exit in intermediate conformations, which can bind ICAM‐1 and signal autonomously.70, 71 Therefore, it has been possible to dissect out the signalling properties of LFA‐1 in the absence of other signals. It is seen that LFA‐1 signalling is sufficient to induce signals for actin reorganization and granule polarization in human NK cells.72, 73
After the initial contact of NK cells with the target cells, LFA‐1 appears to initiate the process of synapse formation. Using human NK cells, it is seen that many of the signalling molecules, including PLC‐γ and Syk kinase, that are phosphorylated upon CD16 engagement, are also phosphorylated in response to LFA‐1 engagement.74 Despite this overlap, LFA‐1 signalling fails to induce Ca2+ mobilization, and CD16 signalling, although induces Ca2+ mobilization, cannot lead to granule polarization. A recent study with human NK cells, using proteomics, has identified signals that emerge upon LFA‐1 engagement, and has validated candidate molecules for their roles in granule polarization.75 It was seen that LFA‐1‐induced granule polarization utilizes a conserved set of signalling events, involving an integrin‐linked kinase ILK, Pyk2, Paxillin and RhoGEF7, which are also used to establish cell polarity during migration. Further, using a human NK cell line that has granules pre‐converged at the MTOC, it was seen that Pyk2, Leupaxin, Cdc42 or CLIP‐170 is required, and ILK or RhoGEF7 is not required, for granule convergence.75
LFA‐1 alone fails to induce degranulation. It is possible that LFA‐1‐mediated adhesion and granule polarization are transient processes, unless stabilized by signals from activating receptors and/or not intercepted by inhibitory signals (see below). Experiments involving activation of primary human NK cells by insect cells expressing ligands of individual or combination of receptors (involving NKG2D, DNAM‐1, 2B4, CD2 and LFA‐1) have shown that each activating receptor alone can elicit inside‐out signal for LFA‐1 activation but none, except CD16, could induce degranulation at its own.76 CD16 alone or a synergistic pair of activating receptors could induce degranulation, but without polarization.73 Such unpolarized degranulation is not sufficient for efficient target cell lysis. Polarized degranulation towards a target cell could be achieved when LFA‐1 is engaged together with CD16 or a synergistic receptor pair. Therefore, a collective engagement of LFA‐1 and a synergistic receptor pair (or CD16 alone) represents the minimal requirement to trigger NK cell cytotoxicity.76
Receptor synergy for inducing secretion by NK cells appears more complex.77 For example, in human NK cells, 2B4, which alone cannot induce degranulation, could induce IFN‐γ at its own. However, 2B4 can induce higher IFN‐γ secretion in the presence of a synergistic partner.77 The molecular basis of synergy among NK cell‐activating receptors is not well understood.78, 79 The requirement of synergistic receptor pairs could maintain a high threshold for NK cell activation, and thus could serve as a safeguard mechanism to ensure restrained activation. CD16 induces NK cell activation through its interaction with the Fc portion of antibodies, and thus the adaptive arm of the immune system determines specificity here. This could be the reason why CD16 does not require a synergistic partner for inducing degranulation.
Studying synapses formed between NK cells and target cells, it is seen that adhesion molecules, including LFA‐1, quickly segregate to the periphery of the synapses,80, 81, 82 where they contribute to adhesion as well as activation signals for F‐actin accumulation. Actin polymerization and its synaptic accumulation are necessary for the formation of NK cell activation synapses and NK cell cytotoxicity.72, 80, 83 Similar peripheral distribution of LFA‐1 is seen at T cell immune synapses.84 Perforin is seen to accumulate in the central region of the NK cell activation synapses. It appears that perforin accumulation follows F‐actin and adhesion molecule accumulation at the synapses.80 Further, accumulation of adhesion molecules at the synapses requires actin, but not microtubule functions.65 On the other hand, as expected, perforin accumulation requires microtubule functions.65
While LFA‐1 accumulates at the periphery of the NK cell activation synapses, different distributions of activating receptors are seen. A recent investigation has studied NK cell activation synapses formed by applying human NK cells onto supported lipid bilayer carrying ligands for LFA‐1 and/or NKG2D‐2B4 synergistic pair of activating receptors, as well as those formed between NK cells and target cells.85 It is seen that LFA‐1 controls the distribution of the synergistic pair of the activating receptors. In the absence of LFA‐1 engagement, 2B4 and NKG2D form separate clusters in a non‐concentric manner. However, co‐engagement of LFA‐1 imposes a distinct distribution, wherein 2B4 accumulates at the centre of the synapses, while NKG2D accumulates at the periphery. This spatial segregation of this synergistic receptor pair of NK cells is reminiscent of that seen for TCR‐CD28 and TCR‐CD2 pairs at T cell activation synapses.86, 87 Further, LFA‐1 could control synaptic accumulation of CD16. Thus, it appears that formation of organized receptor–ligand pairs at NK cell activation synapses is dependent on LFA‐1.85 Further, the initial accumulation of LFA‐1 at the periphery of NK cell cytotoxic synapses seems to create a confined central zone for degranulation and membrane retrieval.
Inhibition of NK cell cytotoxicity
Natural killer cells possess a vast array of germ‐line encoded receptors that can trigger cytotoxicity. This immediately raises the questions as to how NK cells achieve specificity in recognition of target cells and how healthy cells are spared from NK cell cytotoxicity. As noted above, individual activating receptors fail in triggering NK cell cytotoxicity, and at least a pair of receptors, that provides synergistic signals, is required for triggering NK cell activation.78, 79 This requirement of synergistic pairs of receptors for NK cell activation could be a safeguard mechanism to avoid unrestrained NK cell cytotoxicity. Further, many of the ligands of NK cell activating receptors are expressed specifically on unhealthy cells, which could ensure specific killing of unhealthy cells. However, ligands of NK cell‐activating receptors can be expressed constitutively on healthy cells. For example, normal human articular chondrocytes express a ligand for NKp44.88 The most important regulation of NK cell activation is provided by MHC‐I‐specific inhibitory receptors.89 The activation of NK cells is tightly controlled by signals from MHC‐I‐specific inhibitory receptors. MHC‐I‐expressing healthy cells, therefore, could avoid NK cell attack by engaging those MHC‐I‐specific inhibitory receptors on NK cells, and thereby triggering inhibition of cytotoxicity. Signalling by inhibitory receptors also provides NK cells a proper responsiveness via a process termed as licensing; NK cells lacking inhibitory receptors are hyporesponsive.90 Aged T cells could acquire these inhibitory receptors, which may dampen T cell responses in aged individuals.91
MHC‐I‐specific receptors for inhibition of NK cell activation
Human NK cells express two major classes of inhibitory receptors, the inhibitory members of KIR and the CD94‐NKG2A heterodimer. While NKG2A is expressed in both mouse and human, KIR is expressed in human and not in mouse. KIR is a type I transmembrane receptor possessing extracellular Ig‐like domains. NKG2A is a type II transmembrane receptor, belonging to the CD94‐NKG2 C‐type lectin family. While KIR recognizes HLA‐C expression, NKG2A recognizes the expression of the non‐classical HLA‐E on other cells. Both these classes of inhibitory receptors signal through immunoreceptor Tyr‐based inhibitory motif (ITIM), which has a consensus amino acid sequence of V/IxYxxL/V.89, 92, 93, 94 Unlike the mouse MHC‐I‐specific Ly49 inhibitory receptors,95 which possess only one ITIM, KIR as well as NKG2A possess two ITIMs that are separated by ~26 amino acid residues. In the case of the mouse Ly49 receptor, it is likely that receptor homo‐dimerization provides two ITIMs.
Immunoreceptor Tyr‐based inhibitory motif‐dependent inhibition was first seen in mouse with the receptor FcγRIIb, which inhibits B cell receptor signalling by recruiting the inositol phosphatase SHIP.96 However, most of the ITIM‐bearing receptors recruit the Tyr phosphatase SHP‐1 or SHP‐2, not SHIP. The ITIM‐dependent inhibition of NK cell activation prefers recruitment of SHP‐1 to the ITIMs.94, 97, 98 SHP‐1 is composed of two tandem SH2 domains followed by a catalytic domain and a C‐terminal tail. SHP‐1 is maintained in an auto‐inhibited conformation in the cytosol, wherein its N‐terminal SH2 domain docks onto its catalytic domain.99 Binding of SHP's tandem SH2 domains with ITIMs can relieve the auto‐inhibited conformation to activate its catalytic activity.100
Signalling pathways of MHC‐I‐specific inhibitory receptors of NK cells
It is not known which kinase phosphorylates ITIMs; the Src family kinases Lck and Fyn are the candidates.94, 101, 102 The two phospho‐ITIMs (pITIM) that are separated by ~26 amino acid residues provide specific binding sites for the two tandem SH2 domains of SHP‐1. These interactions could break the interaction of SHP‐1′s N‐terminal SH2 domain with its catalytic domain, and thus could relieve the auto‐inhibited conformation of SHP‐1. According to the initial view, activated SHP‐1 may dephosphorylate multiple signalling molecules to block activation.89 However, Vav‐1 is the only major substrate identified directly from NK cell inhibition (Fig. 1).103 Vav‐1 is essential for TCR‐mediated signals for Ca2+ mobilization, actin remodelling and synapse formation.104 Therefore, SHP‐1‐mediated dephosphorylation of activating Tyr residues of Vav‐1 could be sufficient, and dephosphorylation of multiple activating molecules may not be necessary for inhibition.
Figure 1.
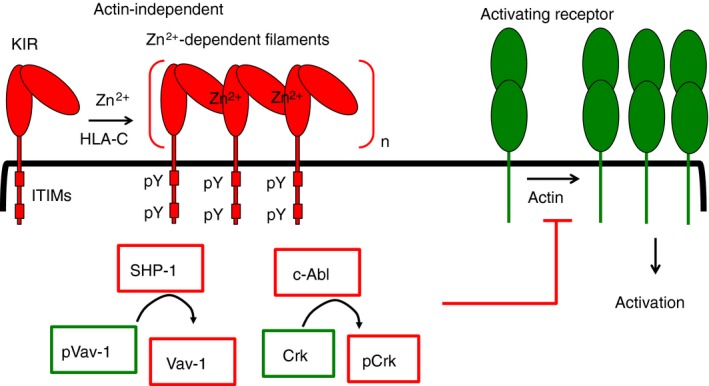
Interception of natural killer (NK) cell activation by killer‐cell Ig‐like receptor (KIR). At the inhibitory synapses formed between KIR+ NK cells and HLA‐C+ target cells, KIR clusters rapidly, in actin‐independent manner. The zinc‐dependent polymerization of KIR into filaments could contribute to the rapid and actin‐independent KIR clustering at these synapses. The Src family kinase Lck and Fyn are candidate kinases for immunoreceptor Tyr‐based inhibitory motif (ITIM) phosphorylation. The protein Tyr phosphates SHP‐1, recruited and activated by its interaction with phospho‐ITIMs (pITIMs), dephosphorylates the guanine nucleotide exchange factor Vav‐1. The c‐Abl kinase is recruited to the inhibitory synapses through an unknown mechanism. The c‐Abl kinase phosphorylates the small adaptor protein Crk, and dissociates it from a signalling complex (not shown here) formed during activation. Vav‐1 dephosphorylation and Crk phosphorylation contribute to blockage of actin‐dependent signals for NK cell activation, and thus could contribute to inhibition of proximal actin‐dependent steps, such as LFA‐1 activation (not shown here) and clustering of activating receptors.
Inhibitory receptors are seen to co‐cluster with activating receptors and invoke inhibition locally81, 105, 106, 107 without impairing the ability of NK cells to respond to other stimuli. This led to the term co‐inhibition, and the inhibitory receptors were considered as co‐receptors. However, it is now clear that the NK cell inhibitory receptors can signal independently.108 During inhibition by ITIM‐bearing receptor of human NK cells, a small adaptor protein Crk becomes phosphorylated and associates with the protein Tyr kinase c‐Abl.108, 109 During activation of human NK cells, Crk is seen to be associated with signalling complexes involving c‐Cbl, C3G and p130CAS. As a result of its phosphorylation during inhibition, Crk dissociates from these complexes. Experiments involving human NK cells on ligand‐attached lipid bilayer have shown that HLA‐E alone on the lipid bilayer can induce Crk phosphorylation, indicating that ITIM‐bearing inhibitory receptors can signal independently.108 The signalling complexes involving Crk, c‐Cbl, C3G and p130CAS are seen to control cytoskeletal‐remodelling.110, 111, 112 Crk is seen to be required for different steps of NK cell cytotoxicity.113 Crk is required for CD16 signalling in human NK cells on Fc‐containing lipid bilayer; silencing of Crk impairs movement of Fc clusters and reduces CD16‐induced Vav‐1 phosphorylation. Co‐engagement of HLA‐E with CD16 induces Crk phosphorylation and prevents actin‐dependent signals in human NK cells.108 A membrane‐target Crk, which lacks the Tyr residues that are substrates of c‐Abl kinase, partially relieves KIR‐mediated inhibition of NK cells.109 These observations suggest that Crk phosphorylation contributes to NK cell inhibition (Fig. 1).
In summary, NK cell inhibitory receptors can signal independently, and they signal through two pathways (Fig. 1). One pathway involves Vav‐1 dephosphorylation by SHP‐1. The other pathway does not involve dephosphorylation of activating molecules. Rather, it involves c‐Abl‐mediated phosphorylation of Crk and its dissociation from signalling complexes formed during activation. Both Vav‐1 dephosphorylation and Crk phosphorylation can contribute to inhibition of actin‐dependent signals of activation.
NK cell Inhibitory synapses
Upon binding to MHC‐I ligands on another cell, inhibitory receptors of an NK cell cluster at the inhibitory synapse formed between the two cell types.114 The amount of KIR accumulated at the synapse is seen to be proportional to the number of KIR molecules engaged with HLA ligands.115 Accordingly, inhibition of killing increases with an increase in the expression level of HLA ligands on the target cells.115 NK cell inhibitory receptors possess unusual features. Clustering of KIR and NKG2A is independent of actin‐related processes and ATP.114, 116 KIR accumulation at inhibitory synapses is independent of ITIM signalling, ICAM‐1‐mediated adhesion, or any other vertebrate molecule on the target cell.116, 117 Interestingly, KIR clustering requires the metal ion Zn2+ (see below).
Upon binding with HLA‐C on target cells, KIR is seen to cluster quickly at the centre of the inhibitory synapses along with some SHP‐1 molecules.107, 114 The centre of the inhibitory synapses also accumulates some GM1 sphingolipid, suggesting accumulation of activating receptors, which do not spatially overlap with KIR clusters.107 At this early time, LFA‐1 localizes at the periphery of the synapses, encircling the centrally accumulated KIR. As the inhibitory synapses further develop, this specific pattern of synaptic organization is disrupted and GM1 or LFA‐1 accumulation is blocked.107
It is seen that the organization of inhibitory synapses depends on the amount of KIR engagement.115 When a lower number of KIR molecules on NK cells is engaged with HLA‐C on target cells, numerous small KIR microclusters are seen at the inhibitory synapses. However, at a higher level of KIR engagement, mixed patterns of KIR clustering are seen, where the majority of synapses form a homogenous large KIR cluster. This phenomenon is independent of actin‐dependent processes. The relative spatial localization of KIR and LFA‐1 at inhibitory synapses is also seen to depend on the level of KIR engagement with HLA‐C.115 The extent of segregation between the KIR and LFA‐1 clusters is seen to be proportional to the number of KIR molecules on NK cells that are engaged with HLA‐C on target cells. The extent of segregation of LFA‐1 and KIR clusters correlates positively with the extent of inhibition of NK cell‐mediated killing of target cells.115 Therefore, it appears that the number of engaged inhibitory receptors controls the synaptic organization as well as inhibitory function of the receptor.
Signalling‐competent NK cell inhibitory synapses can be reconstituted on a planar lipid bilayer carrying only HLA‐E, suggesting that NK cell inhibitory receptors can signal independently.108 On lipid bilayers carrying HLA‐E alone, HLA‐E initially forms a peripheral ring, which rapidly moves towards the centre of the synapse. On lipid bilayers carrying Fc only, Fc initially forms numerous microclusters that move centripetally to accumulate at the centre. On lipid bilayers carrying both HLA‐E and Fc, they are seen to form a few microclusters and to co‐localize to the centre of the synapse. Fewer Fc microclusters are formed in the presence of HLA‐E, but the central accumulation of Fc microclusters is not impaired in the presence of HLA‐E. Therefore, NKG2A engagement prevents formation of CD16 microclusters.108 At the T cell activation synapses too, early TCR microclusters are formed that later converge into a central cluster, and these TCR microclusters are the sites of active signalling.118 Conceivably, CD16 microclusters contribute to signalling, and thus NKG2A‐mediated reduction in the number of Fc microclusters would prevent CD16 signalling.
It appears from these studies that activation and inhibitory receptors initially accumulate at the periphery and later converge at the centre of inhibitory synapses, LFA‐1 appears to encircle the centrally accumulated activation and inhibitory receptors, and inhibitory receptors prevent further accumulation of LFA‐1 and activating receptors.
Lck and Fyn are candidate kinases for ITIM phosphorylation. How the phosphorylation of KIR's ITIMs is initiated and controlled is not understood. A H36A mutation in KIR results in constitutive phosphorylation and self‐association of the receptor.119 It appears that KIR self‐association makes its cytosolic ITIMs inaccessible to phosphatases, which favours ITIM phosphorylation. It is proposed that ITIM phosphorylation is controlled by KIR self‐association, and that His36 acts as a gatekeeper to avoid uncontrolled KIR signalling.119 In the kinetic partitioning model120 of TCR phosphorylation, ITAM phosphorylation is controlled by a size‐based exclusion of the transmembrane phosphatase CD45 from the central synaptic zones. The receptor self‐association model119 is similar, wherein phosphorylation is achieved by protecting the phosphorylated Tyr residues from phosphatases. However, in the receptor self‐association model of KIR phosphorylation, the receptor itself converts into a state that protects ITIM's Tyr residues from local phosphatases.
Killer‐cell Ig‐like receptor phosphorylation is localized to the synaptic region and does not spread to the extra‐synaptic regions of the plasma membrane.105 Thus, NK cell inhibition is local and not cell‐wide. Further, phosphorylated KIR molecules are not distributed uniformly; rather, they are localized in discrete microclusters within the larger KIR cluster at the inhibitory synapses.105 In a recent study with super‐resolution microscopy, it is seen that KIR organizes into nanoclusters, and that SHP‐1 preferentially associates with larger KIR nanoclusters.121 The implication of such distribution of phosphorylated KIR at inhibitory synapses is not understood.
Interception of NK cell activation pathways by signals from inhibitory receptors
As noted above, signals for granule polarization and degranulation can be uncoupled in NK cells.73, 76, 78 LFA‐1 alone can induce granule polarization without degranulation, and CD16 alone (or a synergistic activating receptor pair) can induce degranulation without polarization of granules towards the target cells. While inhibition of degranulation appears to have much more stringent requirements, inhibitory receptors efficiently prevent LFA‐1‐induced granule polarization, indicating that granule polarization is highly sensitive to inhibition.122
Inhibitory signals can intersect NK cell activation at multiple points, including adhesion to target cells, inside‐out signals for LFA‐1 activation, clustering of activating receptors, synaptic accumulation of F‐actin, and Ca2+ flux.65 These steps are actin‐dependent, which can be blocked by Vav‐1 dephosphorylation and/or Crk phosphorylation during inhibition (Fig. 1). The movement of lytic granules along microtubules and their accumulation at the MTOC depend on dynein, not on actin or microtubule reorganization. Interestingly, this granule convergence, which is independent of actin or microtubule reorganization, is not sensitive to signals from these inhibitory receptors.123
Natural killer cell inhibitory receptors have an ability to accumulate rapidly at inhibitory synapses.124 This rapid accumulation at inhibitory synapses, which is independent of actin, may confer to these inhibitory receptors ability of blocking proximal activation signals, such as actin‐dependent adhesion, LFA‐1 activation and clustering of activating receptors, before the full cascade for activation.76, 124, 125 By blocking proximal activation signals, signals from NK cell inhibitory receptors tend to dominate over those from activating receptors. Signalling by chemokine receptor for fractalkine (CX3CL1) is the only example, apart from activation by soluble molecules, which is not sensitive to the signals from inhibitory receptors.126 The signalling properties of the fractalkine receptor, which confer to the receptor this unusual feature, are not understood.
Therefore, control of NK cell cytotoxicity is not a simple balance between signals from activation and inhibitory receptors. Rather, the physiological outcome, of the co‐engagement of NK cell activation and inhibitory receptors, is tilted towards the inhibitory signals. However, it is not clear as to how NK cell inhibitory receptors could achieve a rapid actin‐independent clustering and signalling.
A role of Zn2+ at NK cell inhibitory synapses
The inhibitory function of KIR has long been known to be dependent on Zn2+.127 The primary amino acid sequence of KIR is rich in His residues, including the N‐terminal HExxH zinc‐binding motif, which can bind to Zn2+.128, 129 While the clustering and functioning of KIR at NK cell inhibitory synapses are independent of actin or ATP, they require Zn2+.114 Zn2+ does not contribute to the binding of KIR to HLA‐C.128
How Zn2+ contributes to clustering and signalling of KIR is not understood. Zn2+ induces self‐association of soluble KIR (purified ectodomain of KIR) into filamentous polymers.130 The soluble KIR possesses three Trp residues (at positions 29, 188 and 207) that span the entire length of the protein. The Zn2+‐induced KIR polymerization is coupled with a change in Trp environment, suggesting a Zn2+‐induced conformational change in KIR. Zn2+ treatment of KIR expressing NK cells leads to masking of KIR epitopes for antibody as well as HLA‐C binding, which could arise from Zn2+‐induced conformational change in the receptor and/or receptor polymerization in NK cells. Similar KIR filaments are isolated from the lysate of Zn2+‐treated NK cells. KIR filaments form spontaneously, without addition of exogenous Zn2+, at functional inhibitory synapses formed between KIR expressing NK cells and HLA‐C expressing target cells. Zn2+ treatment of NK cells expressing Venus‐tagged KIR leads to a decrease in the fluorescence anisotropy of Venus, indicating KIR self‐association at the surface of intact NK cells. Two independent mutations in KIR's extracellular portion, which are away from the HLA‐C binding site and do not impair HLA‐C binding, impair Zn2+‐induced polymerization and inhibitory function. These results indicate that KIR undergoes Zn2+‐dependent polymerization during its signalling, and that Zn2+‐dependent polymerization is required for KIR's inhibitory function.130 However, Zn2+ alone is not sufficient, and HLA‐C binding is required, to induce ITIM phosphorylation.
Zn2+‐induced polymerization of KIR represents a new mode of transmembrane receptor signalling, wherein a transmembrane receptor polymerizes into higher‐order assemblies that are much larger than the known dimers and oligomers of transmembrane receptors.130 Zn2+‐dependent KIR polymerization represents a new role for Zn2+ in the regulation of receptor signalling at the plasma membrane. KIR is organized into nanoclusters at the plasma membrane of resting NK cells,121 which could promote rapid Zn2+‐dependent polymerization. Thereby, conceivably, Zn2+‐induced polymerization could contribute to the rapid, actin‐independent KIR clustering and signalling at inhibitory synapses.
Many signalling pathways involve assembly of cytosolic molecules into supramolecular assemblies.131, 132 Zn2+‐dependent KIR polymerization advances this paradigm of signalling by supramolecular assemblies by providing the first example of a transmembrane receptor that signals upon polymerization. Protein assembly is often described by a nucleation‐dependent polymerization (NDP) mechanism,133, 134 wherein the initial steps (nucleation) are slower than the later ones. In the case of KIR, it is conceivable that the proximity of KIR molecules in the nanoclusters121 and involvement of Zn2+ 130 make the nucleation favourable to achieve a rapid polymerization. Assembly of supramolecular signalling complexes via NDP mechanism may invoke new mechanisms of signal amplification, setting threshold and spatio‐temporal regulation in signal transduction.132 While principles of protein polymerization134 predict potential implications of assembly of supramolecular signalling complexes in signal transduction, whether and how a transmembrane receptor can control these assembly reactions in the cytosol are unclear. KIR offers a model system to understand this.
Summary
Natural killer cells possess many receptors that can trigger their cytotoxic functions. Many of these receptors signal through ITAM‐dependent and many through ITAM‐independent pathways. The receptors that signal through ITAM‐dependent pathways associate with ITAM‐containing chains, such as FcRγ, CD3ζ and DAP12. NKG2D signals through its association with DAP10, which possesses the YxxM motif that is distinct from ITAM. SLAM family of receptors signals through their cytosolic ITSM. Utilization of different kinds of signalling pathways could explain why NK cells tend to possess several members of a given family of signalling molecules. These different kinds of activating receptors and many members of a given family of signalling molecules may enable NK cells to detect different kinds of target cells and to mount different kinds of responses. The β2‐integrin LFA‐1, which accumulates at the periphery of cytotoxic synapses, appears to play a pivotal role in organizing the signalling complexes and creating a confined central zone for degranulation at the NK cell cytotoxic synapses. A major goal of current NK cell biology studies is to understand the signalling properties of different activating receptors and how signals from multiple receptors are integrated to mount NK cell responses.
Natural killer cell cytotoxicity is tightly controlled by the dominant inhibition exerted by MHC‐I‐specific inhibitory receptors. The MHC‐I‐specific inhibitory receptors of human NK cells, KIR and NKG2A, signal through their cytosolic ITIMs. Phosphorylated ITIMs recruit and activate the protein Tyr phosphatase SHP‐1, which dephosphorylates the guanine nucleotide exchange factor Vav‐1. During inhibition, the small adaptor protein Crk becomes phosphorylated, dissociates from the signalling complexes formed during activation, and associates with the c‐Abl kinase. The dephosphorylation of Vav‐1 and phosphorylation of Crk contribute to inhibition. Future investigations should focus on how these components of inhibitory signalling pathways operate and coordinate at NK cell inhibitory synapses. Are there more ITIM‐dependent pathways for interception of activation signals and/or for licensing of NK cells? KIR signalling requires its Zn2+‐dependent polymerization at inhibitory synapses. An important future goal would be to elucidate how Zn2+ is delivered at NK cell inhibitory synapses and how Zn2+‐dependent polymers contribute to KIR signalling.
Disclosures
There is no conflict of interest
Acknowledgement
The author thanks Shweta Jain (University of California San Francisco, USA) for her comments.
References
- 1. Moffett‐King A. Natural killer cells and pregnancy. Nat Rev Immunol 2002; 2:656–63. [DOI] [PubMed] [Google Scholar]
- 2. Moffett‐King A, Entrican G, Ellis S, Hutchinson J, Bainbridge D. Natural killer cells and reproduction. Trends Immunol 2002; 23:332–3. [DOI] [PubMed] [Google Scholar]
- 3. Bryceson YT, Chiang SC, Darmanin S, Fauriat C, Schlums H, Theorell J et al Molecular mechanisms of natural killer cell activation. J Innate Immun 2011; 3:216–26. [DOI] [PubMed] [Google Scholar]
- 4. Lanier LL. NK cell recognition. Annu Rev Immunol 2005; 23:225–74. [DOI] [PubMed] [Google Scholar]
- 5. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008; 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tassi I, Klesney‐Tait J, Colonna M. Dissecting natural killer cell activation pathways through analysis of genetic mutations in human and mouse. Immunol Rev 2006; 214:92–105. [DOI] [PubMed] [Google Scholar]
- 7. Veillette A. SLAM‐family receptors: immune regulators with or without SAP‐family adaptors. Cold Spring Harb Perspect Biol 2010; 2:a002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu N, Veillette A. SLAM family receptors in normal immunity and immune pathologies. Curr Opin Immunol 2016; 38:45–51. [DOI] [PubMed] [Google Scholar]
- 9. Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 2016; 16:112–23. [DOI] [PubMed] [Google Scholar]
- 10. Koch J, Steinle A, Watzl C, Mandelboim O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol 2013; 34:182–91. [DOI] [PubMed] [Google Scholar]
- 11. Hudspeth K, Silva‐Santos B, Mavilio D. Natural cytotoxicity receptors: broader expression patterns and functions in innate and adaptive immune cells. Front Immunol 2013; 4:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A et al NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med 1999; 189:787–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L et al Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med 1998; 188:953–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C et al p46, a novel natural killer cell‐specific surface molecule that mediates cell activation. J Exp Med 1997; 186:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E et al NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non‐major histocompatibility complex‐restricted tumor cell lysis. J Exp Med 1998; 187:2065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B et al The B7 family member B7‐H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 2009; 206:1495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaifu T, Escaliere B, Gastinel LN, Vivier E, Baratin M. B7‐H6/NKp30 interaction: a mechanism of alerting NK cells against tumors. Cell Mol Life Sci 2011; 68:3531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 2013; 31:227–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajagopalan S, Long EO. Found: a cellular activating ligand for NKp44. Blood 2013; 122:2921–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL et al Activation of NK cells and T cells by NKG2D, a receptor for stress‐inducible MICA. Science 1999; 285:727–9. [DOI] [PubMed] [Google Scholar]
- 21. Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res 2006; 35:263–78. [DOI] [PubMed] [Google Scholar]
- 22. Cerwenka A, Lanier LL. Ligands for natural killer cell receptors: redundancy or specificity. Immunol Rev 2001; 181:158–69. [DOI] [PubMed] [Google Scholar]
- 23. Lanier LL. NKG2D receptor and its ligands in host defense. Cancer Immunol Res 2015; 3:575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deng W, Gowen BG, Zhang L, Wang L, Lau S, Iannello A et al Antitumor immunity. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science 2015; 348:136–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J, Basher F, Wu JD. NKG2D ligands in tumor immunity: two sides of a coin. Front Immunol 2015; 6:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vitale M, Falco M, Castriconi R, Parolini S, Zambello R, Semenzato G et al Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol 2001; 31:233–42. [DOI] [PubMed] [Google Scholar]
- 27. Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80‐AICL interaction. Nat Immunol 2006; 7:1334–42. [DOI] [PubMed] [Google Scholar]
- 28. Chen R, Relouzat F, Roncagalli R, Aoukaty A, Tan R, Latour S et al Molecular dissection of 2B4 signaling: implications for signal transduction by SLAM‐related receptors. Mol Cell Biol 2004; 24:5144–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veillette A. NK cell regulation by SLAM family receptors and SAP‐related adapters. Immunol Rev 2006; 214:22–34. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Z, Wu N, Lu Y, Davidson D, Colonna M, Veillette A. DNAM‐1 controls NK cell activation via an ITT‐like motif. J Exp Med 2015; 212:2165–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shibuya A, Campbell D, Hannum C, Yssel H, Franz‐Bacon K, McClanahan T et al DNAM‐1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996; 4:573–81. [DOI] [PubMed] [Google Scholar]
- 32. Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B et al Identification of PVR (CD155) and Nectin‐2 (CD112) as cell surface ligands for the human DNAM‐1 (CD226) activating molecule. J Exp Med 2003; 198:557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibuya K, Lanier LL, Phillips JH, Ochs HD, Shimizu K, Nakayama E et al Physical and functional association of LFA‐1 with DNAM‐1 adhesion molecule. Immunity 1999; 11:615–23. [DOI] [PubMed] [Google Scholar]
- 34. Watzl C, Long EO. Signal transduction during activation and inhibition of natural killer cells. Curr Protoc Immunol 2010; Chapter 11: Unit 11 9B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J 2004; 23:255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine‐based activation motif is involved in activating NK cells. Nature 1998; 391:703–7. [DOI] [PubMed] [Google Scholar]
- 37. Tomasello E, Olcese L, Vely F, Geourgeon C, Blery M, Moqrich A et al Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor‐associated protein (KARAP)/DAP‐12. J Biol Chem 1998; 273:34 115–9. [DOI] [PubMed] [Google Scholar]
- 38. Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane‐embedded interaction site. PLoS Biol 2006; 4:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL et al An activating immunoreceptor complex formed by NKG2D and DAP10. Science 1999; 285:730–2. [DOI] [PubMed] [Google Scholar]
- 40. Sidorenko SP, Clark EA. The dual‐function CD150 receptor subfamily: the viral attraction. Nat Immunol 2003; 4:19–24. [DOI] [PubMed] [Google Scholar]
- 41. Dennehy KM, Klimosch SN, Steinle A. Cutting edge: NKp80 uses an atypical hemi‐ITAM to trigger NK cytotoxicity. J Immunol 2011; 186:657–61. [DOI] [PubMed] [Google Scholar]
- 42. Smith‐Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol 2009; 27:591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ting AT, Karnitz LM, Schoon RA, Abraham RT, Leibson PJ. Fc gamma receptor activation induces the tyrosine phosphorylation of both phospholipase C (PLC)‐gamma 1 and PLC‐gamma 2 in natural killer cells. J Exp Med 1992; 176:1751–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Billadeau DD, Brumbaugh KM, Dick CJ, Schoon RA, Bustelo XR, Leibson PJ. The Vav‐Rac1 pathway in cytotoxic lymphocytes regulates the generation of cell‐mediated killing. J Exp Med 1998; 188:549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cella M, Fujikawa K, Tassi I, Kim S, Latinis K, Nishi S et al Differential requirements for Vav proteins in DAP10‐ and ITAM‐mediated NK cell cytotoxicity. J Exp Med 2004; 200:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Upshaw JL, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. The isoforms of phospholipase C‐gamma are differentially used by distinct human NK activating receptors. J Immunol 2005; 175:213–8. [DOI] [PubMed] [Google Scholar]
- 47. Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D‐DAP10 triggers human NK cell‐mediated killing via a Syk‐independent regulatory pathway. Nat Immunol 2003; 4:557–64. [DOI] [PubMed] [Google Scholar]
- 48. Chiesa S, Mingueneau M, Fuseri N, Malissen B, Raulet DH, Malissen M et al Multiplicity and plasticity of natural killer cell signaling pathways. Blood 2006; 107:2364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zompi S, Hamerman JA, Ogasawara K, Schweighoffer E, Tybulewicz VL, Di Santo JP et al NKG2D triggers cytotoxicity in mouse NK cells lacking DAP12 or Syk family kinases. Nat Immunol 2003; 4:565–72. [DOI] [PubMed] [Google Scholar]
- 50. Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol 2002; 3:1150–5. [DOI] [PubMed] [Google Scholar]
- 51. Graham DB, Cella M, Giurisato E, Fujikawa K, Miletic AV, Kloeppel T et al Vav1 controls DAP10‐mediated natural cytotoxicity by regulating actin and microtubule dynamics. J Immunol 2006; 177:2349–55. [DOI] [PubMed] [Google Scholar]
- 52. Upshaw JL, Arneson LN, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. NKG2D‐mediated signaling requires a DAP10‐bound Grb2‐Vav1 intermediate and phosphatidylinositol‐3‐kinase in human natural killer cells. Nat Immunol 2006; 7:524–32. [DOI] [PubMed] [Google Scholar]
- 53. Segovis CM, Schoon RA, Dick CJ, Nacusi LP, Leibson PJ, Billadeau DD. PI3K links NKG2D signaling to a CrkL pathway involved in natural killer cell adhesion, polarity, and granule secretion. J Immunol 2009; 182:6933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL et al Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol 2003; 5:149–54. [DOI] [PubMed] [Google Scholar]
- 55. Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D et al The X‐linked lymphoproliferative‐disease gene product SAP regulates signals induced through the co‐receptor SLAM. Nature 1998; 395:462–9. [DOI] [PubMed] [Google Scholar]
- 56. Veillette A, Dong Z, Perez‐Quintero LA, Zhong MC, Cruz‐Munoz ME. Importance and mechanism of ‘switch’ function of SAP family adapters. Immunol Rev 2009; 232:229–39. [DOI] [PubMed] [Google Scholar]
- 57. Roncagalli R, Taylor JE, Zhang S, Shi X, Chen R, Cruz‐Munoz ME et al Negative regulation of natural killer cell function by EAT‐2, a SAP‐related adaptor. Nat Immunol 2005; 6:1002–10. [DOI] [PubMed] [Google Scholar]
- 58. Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M. Activation of NK cell‐mediated cytotoxicity by a SAP‐independent receptor of the CD2 family. J Immunol 2001; 167:5517–21. [DOI] [PubMed] [Google Scholar]
- 59. Dong Z, Davidson D, Perez‐Quintero LA, Kurosaki T, Swat W, Veillette A. The adaptor SAP controls NK cell activation by regulating the enzymes Vav‐1 and SHIP‐1 and by enhancing conjugates with target cells. Immunity 2012; 36:974–85. [DOI] [PubMed] [Google Scholar]
- 60. Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A. Regulation of SLAM‐mediated signal transduction by SAP, the X‐linked lymphoproliferative gene product. Nat Immunol 2001; 2:681–90. [DOI] [PubMed] [Google Scholar]
- 61. Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood 2005; 105:4722–9. [DOI] [PubMed] [Google Scholar]
- 62. Bell GM, Fargnoli J, Bolen JB, Kish L, Imboden JB. The SH3 domain of p56lck binds to proline‐rich sequences in the cytoplasmic domain of CD2. J Exp Med 1996; 183:169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dustin ML, Groves JT. Receptor signaling clusters in the immune synapse. Annu Rev Biophys 2012; 41:543–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pageon SV, Cordoba SP, Owen DM, Rothery SM, Oszmiana A, Davis DM. Superresolution microscopy reveals nanometer‐scale reorganization of inhibitory natural killer cell receptors upon activation of NKG2D. Sci Signal 2013; 6:ra62. [DOI] [PubMed] [Google Scholar]
- 65. Mace EM, Dongre P, Hsu HT, Sinha P, James AM, Mann SS et al Cell biological steps and checkpoints in accessing NK cell cytotoxicity. Immunol Cell Biol 2014; 92:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rak GD, Mace EM, Banerjee PP, Svitkina T, Orange JS. Natural killer cell lytic granule secretion occurs through a pervasive actin network at the immune synapse. PLoS Biol 2011; 9:e1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brown AC, Oddos S, Dobbie IM, Alakoskela JM, Parton RM, Eissmann P et al Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super‐resolution microscopy. PLoS Biol 2011; 9:e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol 2007; 25:619–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anikeeva N, Somersalo K, Sims TN, Thomas VK, Dustin ML, Sykulev Y. Distinct role of lymphocyte function‐associated antigen‐1 in mediating effective cytolytic activity by cytotoxic T lymphocytes. Proc Natl Acad Sci USA 2005; 102:6437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barber DF, Long EO. Coexpression of CD58 or CD48 with intercellular adhesion molecule 1 on target cells enhances adhesion of resting NK cells. J Immunol 2003; 170:294–9. [DOI] [PubMed] [Google Scholar]
- 71. Urlaub D, Hofer K, Muller ML, Watzl C. LFA‐1 activation in NK cells and their subsets: influence of receptors, maturation, and cytokine stimulation. J Immunol 2017; 198:1944–51. [DOI] [PubMed] [Google Scholar]
- 72. Barber DF, Faure M, Long EO. LFA‐1 contributes an early signal for NK cell cytotoxicity. J Immunol 2004; 173:3653–9. [DOI] [PubMed] [Google Scholar]
- 73. Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med 2005; 202:1001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. March ME, Long EO. beta2 integrin induces TCRzeta‐Syk‐phospholipase C‐gamma phosphorylation and paxillin‐dependent granule polarization in human NK cells. J Immunol 2011; 186:2998–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang M, March ME, Lane WS, Long EO. A signaling network stimulated by beta2 integrin promotes the polarization of lytic granules in cytotoxic cells. Sci Signal 2014; 7:ra96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood 2009; 114:2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK‐cell cytokine and chemokine production by target cell recognition. Blood 2010; 115:2167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim HS, Das A, Gross CC, Bryceson YT, Long EO. Synergistic signals for natural cytotoxicity are required to overcome inhibition by c‐Cbl ubiquitin ligase. Immunity 2010; 32:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim HS, Long EO. Complementary phosphorylation sites in the adaptor protein SLP‐76 promote synergistic activation of natural killer cells. Sci Signal 2012; 5:ra49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Orange JS, Harris KE, Andzelm MM, Valter MM, Geha RS, Strominger JL. The mature activating natural killer cell immunologic synapse is formed in distinct stages. Proc Natl Acad Sci USA 2003; 100:14 151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vyas YM, Mehta KM, Morgan M, Maniar H, Butros L, Jung S et al Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I‐regulated noncytolytic and cytolytic interactions. J Immunol 2001; 167:4358–67. [DOI] [PubMed] [Google Scholar]
- 82. Roda‐Navarro P, Mittelbrunn M, Ortega M, Howie D, Terhorst C, Sanchez‐Madrid F et al Dynamic redistribution of the activating 2B4/SAP complex at the cytotoxic NK cell immune synapse. J Immunol 2004; 173:3640–6. [DOI] [PubMed] [Google Scholar]
- 83. Orange JS, Ramesh N, Remold‐O'Donnell E, Sasahara Y, Koopman L, Byrne M et al Wiskott‐Aldrich syndrome protein is required for NK cell cytotoxicity and colocalizes with actin to NK cell‐activating immunologic synapses. Proc Natl Acad Sci USA 2002; 99:11 351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM et al The immunological synapse: a molecular machine controlling T cell activation. Science 1999; 285:221–7. [DOI] [PubMed] [Google Scholar]
- 85. Liu D, Bryceson YT, Meckel T, Vasiliver‐Shamis G, Dustin ML, Long EO. Integrin‐dependent organization and bidirectional vesicular traffic at cytotoxic immune synapses. Immunity 2009; 31:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kaizuka Y, Douglass AD, Vardhana S, Dustin ML, Vale RD. The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. J Cell Biol 2009; 185:521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yokosuka T, Kobayashi W, Sakata‐Sogawa K, Takamatsu M, Hashimoto‐Tane A, Dustin ML et al Spatiotemporal regulation of T cell costimulation by TCR‐CD28 microclusters and protein kinase C theta translocation. Immunity 2008; 29:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bialoszewska A, Baychelier F, Niderla‐Bielinska J, Czop A, Debre P, Vieillard V et al Constitutive expression of ligand for natural killer cell NKp44 receptor (NKp44L) by normal human articular chondrocytes. Cell Immunol 2013; 285:6–9. [DOI] [PubMed] [Google Scholar]
- 89. Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev 2008; 224:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim S, Poursine‐Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L et al Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 2005; 436:709–13. [DOI] [PubMed] [Google Scholar]
- 91. Abedin S, Michel JJ, Lemster B, Vallejo AN. Diversity of NKR expression in aging T cells and in T cells of the aged: the new frontier into the exploration of protective immunity in the elderly. Exp Gerontol 2005; 40:537–48. [DOI] [PubMed] [Google Scholar]
- 92. Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine‐based inhibition motifs: a quest in the past and future. Immunol Rev 2008; 224:11–43. [DOI] [PubMed] [Google Scholar]
- 93. Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 2008; 8:34–47. [DOI] [PubMed] [Google Scholar]
- 94. Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T et al Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity 1996; 4:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McVicar DW, Burshtyn DN. Intracellular signaling by the killer immunoglobulin‐like receptors and Ly49. Sci STKE 2001; 2001:re1. [DOI] [PubMed] [Google Scholar]
- 96. Ono M, Bolland S, Tempst P, Ravetch JV. Role of the inositol phosphatase SHIP in negative regulation of the immune system by the receptor Fc(gamma)RIIB. Nature 1996; 383:263–6. [DOI] [PubMed] [Google Scholar]
- 97. Olcese L, Lang P, Vely F, Cambiaggi A, Marguet D, Blery M et al Human and mouse killer‐cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol 1996; 156:4531–4. [PubMed] [Google Scholar]
- 98. Burshtyn DN, Yang W, Yi T, Long EO. A novel phosphotyrosine motif with a critical amino acid at position ‐2 for the SH2 domain‐mediated activation of the tyrosine phosphatase SHP‐1. J Biol Chem 1997; 272:13 066–72. [DOI] [PubMed] [Google Scholar]
- 99. Yang J, Liu L, He D, Song X, Liang X, Zhao ZJ et al Crystal structure of human protein‐tyrosine phosphatase SHP‐1. J Biol Chem 2003; 278:6516–20. [DOI] [PubMed] [Google Scholar]
- 100. Hof P, Pluskey S, Dhe‐Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP‐2. Cell 1998; 92:441–50. [DOI] [PubMed] [Google Scholar]
- 101. Binstadt BA, Brumbaugh KM, Dick CJ, Scharenberg AM, Williams BL, Colonna M et al Sequential involvement of Lck and SHP‐1 with MHC‐recognizing receptors on NK cells inhibits FcR‐initiated tyrosine kinase activation. Immunity 1996; 5:629–38. [DOI] [PubMed] [Google Scholar]
- 102. Chan VW, Lowell CA, DeFranco AL. Defective negative regulation of antigen receptor signaling in Lyn‐deficient B lymphocytes. Curr Biol 1998; 8:545–53. [DOI] [PubMed] [Google Scholar]
- 103. Stebbins CC, Watzl C, Billadeau DD, Leibson PJ, Burshtyn DN, Long EO. Vav1 dephosphorylation by the tyrosine phosphatase SHP‐1 as a mechanism for inhibition of cellular cytotoxicity. Mol Cell Biol 2003; 23:6291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tybulewicz VL. Vav‐family proteins in T‐cell signalling. Curr Opin Immunol 2005; 17:267–74. [DOI] [PubMed] [Google Scholar]
- 105. Treanor B, Lanigan PM, Kumar S, Dunsby C, Munro I, Auksorius E et al Microclusters of inhibitory killer immunoglobulin‐like receptor signaling at natural killer cell immunological synapses. J Cell Biol 2006; 174:153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vyas YM, Maniar H, Dupont B. Cutting edge: differential segregation of the SRC homology 2‐containing protein tyrosine phosphatase‐1 within the early NK cell immune synapse distinguishes noncytolytic from cytolytic interactions. J Immunol 2002; 168:3150–4. [DOI] [PubMed] [Google Scholar]
- 107. Vyas YM, Maniar H, Lyddane CE, Sadelain M, Dupont B. Ligand binding to inhibitory killer cell Ig‐like receptors induce colocalization with Src homology domain 2‐containing protein tyrosine phosphatase 1 and interruption of ongoing activation signals. J Immunol 2004; 173:1571–8. [DOI] [PubMed] [Google Scholar]
- 108. Liu D, Peterson ME, Long EO. The adaptor protein Crk controls activation and inhibition of natural killer cells. Immunity 2012; 36:600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Peterson ME, Long EO. Inhibitory receptor signaling via tyrosine phosphorylation of the adaptor Crk. Immunity 2008; 29:578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal 2009; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chodniewicz D, Klemke RL. Regulation of integrin‐mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim Biophys Acta 2004; 1692:63–76. [DOI] [PubMed] [Google Scholar]
- 112. Nakashima N, Rose DW, Xiao S, Egawa K, Martin SS, Haruta T et al The functional role of CrkII in actin cytoskeleton organization and mitogenesis. J Biol Chem 1999; 274:3001–8. [DOI] [PubMed] [Google Scholar]
- 113. Liu D. The adaptor protein Crk in immune response. Immunol Cell Biol 2014; 92:80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Davis DM, Chiu I, Fassett M, Cohen GB, Mandelboim O, Strominger JL. The human natural killer cell immune synapse. Proc Natl Acad Sci USA 1999; 96:15 062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Almeida CR, Davis DM. Segregation of HLA‐C from ICAM‐1 at NK cell immune synapses is controlled by its cell surface density. J Immunol 2006; 177:6904–10. [DOI] [PubMed] [Google Scholar]
- 116. Faure M, Barber DF, Takahashi SM, Jin T, Long EO. Spontaneous clustering and tyrosine phosphorylation of NK cell inhibitory receptor induced by ligand binding. J Immunol 2003; 170:6107–14. [DOI] [PubMed] [Google Scholar]
- 117. Fassett MS, Davis DM, Valter MM, Cohen GB, Strominger JL. Signaling at the inhibitory natural killer cell immune synapse regulates lipid raft polarization but not class I MHC clustering. Proc Natl Acad Sci USA 2001; 98:14 547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor‐proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 2006; 25:117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kumar S, Sarkar P, Sim MJ, Rajagopalan S, Vogel SS, Long EO. A single amino acid change in inhibitory killer cell Ig‐like receptor results in constitutive receptor self‐association and phosphorylation. J Immunol 2015; 194:817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Davis SJ, van der Merwe PA. The kinetic‐segregation model: TCR triggering and beyond. Nat Immunol 2006; 7:803–9. [DOI] [PubMed] [Google Scholar]
- 121. Oszmiana A, Williamson DJ, Cordoba SP, Morgan DJ, Kennedy PR, Stacey K et al The size of activating and inhibitory killer ig‐like receptor nanoclusters is controlled by the transmembrane sequence and affects signaling. Cell Rep 2016; 15:1957–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Das A, Long EO. Lytic granule polarization, rather than degranulation, is the preferred target of inhibitory receptors in NK cells. J Immunol 2010; 185:4698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mentlik AN, Sanborn KB, Holzbaur EL, Orange JS. Rapid lytic granule convergence to the MTOC in natural killer cells is dependent on dynein but not cytolytic commitment. Mol Biol Cell 2010; 21:2241–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Abeyweera TP, Merino E, Huse M. Inhibitory signaling blocks activating receptor clustering and induces cytoskeletal retraction in natural killer cells. J Cell Biol 2011; 192:675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Burshtyn DN, Shin J, Stebbins C, Long EO. Adhesion to target cells is disrupted by the killer cell inhibitory receptor. Curr Biol 2000; 10:777–80. [DOI] [PubMed] [Google Scholar]
- 126. Pallandre JR, Krzewski K, Bedel R, Ryffel B, Caignard A, Rohrlich PS et al Dendritic cell and natural killer cell cross‐talk: a pivotal role of CX3CL1 in NK cytoskeleton organization and activation. Blood 2008; 112:4420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rajagopalan S, Winter CC, Wagtmann N, Long EO. The Ig‐related killer cell inhibitory receptor binds zinc and requires zinc for recognition of HLA‐C on target cells. J Immunol 1995; 155:4143–6. [PubMed] [Google Scholar]
- 128. Rajagopalan S, Long EO. Zinc bound to the killer cell‐inhibitory receptor modulates the negative signal in human NK cells. J Immunol 1998; 161:1299–305. [PubMed] [Google Scholar]
- 129. Vales‐Gomez M, Erskine RA, Deacon MP, Strominger JL, Reyburn HT. The role of zinc in the binding of killer cell Ig‐like receptors to class I MHC proteins. Proc Natl Acad Sci USA 2001; 98:1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kumar S, Rajagopalan S, Sarkar P, Dorward DW, Peterson ME, Liao HS et al Zinc‐induced polymerization of killer‐cell ig‐like receptor into filaments promotes its inhibitory function at cytotoxic immunological synapses. Mol Cell 2016; 62:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol 2014; 14:821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wu H. Higher‐order assemblies in a new paradigm of signal transduction. Cell 2013; 153:287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ferrone FA. Nucleation: the connections between equilibrium and kinetic behavior. Methods Enzymol 2006; 412:285–99. [DOI] [PubMed] [Google Scholar]
- 134. Kumar S, Udgaonkar JB. Mechanisms of amyloid fibril formation by proteins. Curr Sci 2010; 98:639–56. [Google Scholar]


