Summary
Acquisition of T‐cell central tolerance involves distinct pathways of self‐antigen presentation to thymocytes. One pathway termed indirect presentation requires a self‐antigen transfer step from thymic epithelial cells (TECs) to bone marrow‐derived cells before the self‐antigen is presented to thymocytes. The role of indirect presentation in central tolerance is context‐dependent, potentially due to variation in self‐antigen expression, processing and presentation in the thymus. Here, we report experiments in mice in which TECs expressed a membrane‐bound transgenic self‐antigen, hen egg lysozyme (HEL), from either the insulin (insHEL) or thyroglobulin (thyroHEL) promoter. Intrathymic HEL expression was less abundant and more confined to the medulla in insHEL mice compared with thyroHEL mice. When indirect presentation was impaired by generating mice lacking MHC class II expression in bone marrow‐derived antigen‐presenting cells, insHEL‐mediated thymocyte deletion was abolished, whereas thyroHEL‐mediated deletion occurred at a later stage of thymocyte development and Foxp3+ regulatory T‐cell differentiation increased. Indirect presentation increased the strength of T‐cell receptor signalling that both self‐antigens induced in thymocytes, as assessed by Helios expression. Hence, indirect presentation limits the differentiation of naive and regulatory T cells by promoting deletion of self‐reactive thymocytes.
Keywords: central tolerance, indirect presentation, regulatory T cells, T‐cell deletion, T‐cell tolerance
Abbreviations
- Aire
autoimmune regulator
- APC
antigen‐presenting cells
- BM‐APCs
bone marrow‐derived antigen‐presenting cells
- BM
bone marrow
- CD4SP
CD4 single‐positive
- cTEC
cortical thymic epithelial cells
- DAB
3,3′‐diaminobenzidine
- DP
CD4+ CD8+ double‐positive
- EdU
5‐ethynyl‐2′‐deoxyuridine
- HEL
hen egg lysozyme
- insHEL
insulin promoter‐driven hen egg lysozyme transgene
- MHCII
major histocompatibility complex class II
- mTEC
medullary thymic epithelial cells
- Non‐Tg
non‐transgenic
- RFI
relative fluorescence intensity
- RIP‐OVAhi
rat insulin promoter‐deriven ovalbumin
- TCR
T‐cell receptor
- TECs
thymic epithelial cells
- thyroHEL
thyroglobulin promoter‐driven hen egg lysozyme transgene
- Treg
regulatory T
Introduction
During T‐cell development in the thymus, the primary determinant of thymocyte fate is T‐cell antigen receptor (TCR) engagement. Thymocytes that express a TCR with low affinity for a self‐peptide displayed by a major histocompatibility complex (MHC) molecule are induced to differentiate into naive T cells. Several outcomes are possible in thymocytes that receive a strong TCR signal due to expressing a TCR with high affinity for self‐peptide/MHC. Most strongly TCR‐signalled thymocytes are eliminated by apoptosis (called deletion hereafter), but some differentiate into Foxp3+ T regulatory (Treg) cells. One clear difference between these two mechanisms is the stage of thymocyte maturation at which they occur. Although deletion can occur at any stage after the onset of TCR expression,1, 2 Foxp3 up‐regulation occurs predominantly in mature CD4+ CD8– single‐positive (CD4SP) thymocytes.3, 4 To understand how these two mechanisms cooperate to establish self‐tolerance, it is important to elucidate factors that determine the maturation stage at which thymocytes encounter self‐antigens.
Self‐antigen presentation in the thymus is broadly divisible into direct and indirect presentation pathways. Direct presentation is the process whereby an antigen‐presenting cell (APC) expresses a self‐antigen and presents it to thymocytes, whereas indirect presentation involves an intercellular self‐antigen transfer step that precedes self‐antigen presentation to thymocytes. In the classic indirect presentation pathway, self‐antigens are transferred from thymic epithelial cells (TECs) to bone‐marrow‐derived APCs (BM‐APCs).5, 6 For some self‐antigens, TECs7, 8, 9 or BM‐APCs10, 11, 12 can delete thymocytes solely through direct presentation. In contrast, some self‐antigens require indirect presentation to delete thymocytes. This has been demonstrated for CD4+ T cells that recognize the transgenic self‐antigens, rat insulin promoter‐driven ovalbumin (RIP‐OVA)hi 13 and Aire‐GCLG120A,14 or the natural self‐antigens, myelin basic protein15 and interphotoreceptor retinoid‐binding protein.16 Deletion mediated by RIP‐OVAhi and interphotoreceptor retinoid‐binding protein also requires autoimmune regulator (Aire),13, 16 a transcription factor that promotes expression of a broad array of self‐antigens in a subset of medullary thymic epithelial cells (mTECs).17, 18, 19 TCR sequencing data indicate that almost half of the TCRs that require Aire to undergo deletion also require MHC class II (MHCII) expression in BM‐APCs to undergo deletion.20 Hence, a substantial proportion of self‐antigens appears to require indirect presentation to delete high‐affinity CD4+ T cells in the thymus.
Impairment of indirect presentation sometimes causes a small but reproducible increase in the CD4SP thymocyte population size, consistent with a partial defect in thymocyte deletion.13, 14 In these situations it is possible that changes in the maturation stage at which thymocytes engage the deleting self‐antigen have been overlooked. Thymocytes up‐regulate the transcription factor, Helios, in response to strong TCR signalling and down‐regulate Helios in response to weak TCR signalling.21 Helios labelling therefore provides a way to define the maturation stage at which thymocytes receive a strong TCR signal due to self‐antigen engagement.
Here, we examined the role of indirect presentation in the deletion of thymocytes expressing the 3A9 transgenic TCR22 which has high affinity for a peptide derived from hen egg lysozyme (HEL) complexed with the MHCII molecule, I‐Ak.23 Membrane‐bound HEL was expressed in the thymus using a transgene containing either the insulin (insHEL) or thyroglobulin (thyroHEL) promoter.24 3A9 thymocyte deletion by insHEL requires Aire expression in non‐haematopoietic cells, indicating that TECs are involved in the deletion mechanism.25 Aire is also required for thyroHEL to delete 3A9 thymocytes.26 However, it was unknown whether indirect presentation by BM‐APCs is required for insHEL or thyroHEL to delete 3A9 thymocytes. To impair indirect presentation, we made chimeric mice lacking MHCII expression in BM‐APCs. This perturbation abrogated deletion in the insHEL system. In the thyroHEL system, the same perturbation delayed deletion and increased Treg cell differentiation. In both systems, the presence of deleting self‐antigen was associated with increased Helios expression in CD4+ CD8+ double‐positive (DP) thymocytes in a manner that required MHCII expression in BM‐APCs. Hence, indirect presentation promotes self‐antigen recognition in DP thymocytes and limits the differentiation of naive CD4+ T cells and Treg cells.
Materials and methods
Mice
B10.BR (H‐2k), 3A9,22 insHEL and thyroHEL (initially called ILK‐3 and TLK‐2, respectively),24 Bim –/– (Bcl2l1 tm1Ast)27 and H2‐Aa –/– (H2‐Aa tm1Blt)28 mice were bred and housed in the Australian Phenomics Facility, Canberra. Mice were genotyped for transgenes and mutations by PCR and used 7–28 (typically 10–16) weeks after birth. Mice carrying the 3A9 and insHEL or thyroHEL transgene(s) were hemizygous for the transgene. To make chimeras, recipients were irradiated with X‐rays (two doses of 4·5 Gy given 4 hr apart) then injected intravenously with at least 2 × 106 bone marrow cells. They were allowed to reconstitute for 4–6 weeks before analysis. The Animal Experimentation Ethics Committees of the Australian National University or Monash University approved all procedures.
RNA in situ hybridization
HEL mRNA was detected using the RNAscope® 2.0 HD Detection Kit (Brown) (Advanced Cell Diagnostics Inc., Newark, CA, Cat # 320497) according to the manufacturer's instructions.29 Briefly, 5‐μm‐thick formalin‐fixed paraffin‐embedded thymus tissue sections were deparaffinized, incubated with Pretreat 1 reagent (hydrogen peroxide) for 10 min, boiled in Pretreat 2 reagent (epitope retrieval) for 15 min, and treated with Pretreat 3 reagent (protease) for 15 min. Tissue sections were then hybridized with a custom‐made HEL mRNA‐specific probe (Cat # 445141) or a negative control probe targeting the bacterial gene, DapB, for 2 hr at 40° in a HybEZ™ Oven. Signal amplification steps using AMP1 to AMP6 reagents were performed, culminating in the binding of horseradish peroxidase‐labelled probe followed by the chromogenic 3,3′‐diaminobenzidine (DAB) substrates (DAB‐A and DAB‐B). Sections were counterstained with haematoxylin and mounted with xylene‐based mounting medium; photographs were taken using a bright field microscope (Olympus IX71).
Sorting of thymic APC subsets
After removing fat and connective tissue, individual thymi were placed in a Petri dish with 5 ml of warm RPMI‐1640 medium, small incisions were made in the capsule to release thymocytes and tissue pieces were then transferred into one well of a 24‐well plate with 1 ml warm RPMI‐1640. A plastic transfer pipette was used to liberate thymocytes into the supernatant, which was discarded. Larger fragments were digested through a series of incubations in 1 ml of digestion buffer at 37° for 10 min at a time. After each digestion, supernatant was collected and stored on ice. Three incubations were performed using RPMI‐1640 containing 1 mg/ml Collagenase 4 (Worthington Biochemical Corporation, Lakewood, NJ, USA) with 0·2 mg/ml DNAse 1 (Roche, Basel, Switzerland), followed by three incubations in 1 mg/ml Collagenase/Dispase (Roche) with 0·2 mg/ml DNAse 1. The final two digestion fractions were pooled and passed through a 100‐μm mesh. Cells were pelleted, washed in FACS buffer and stained in a 500 μl antibody cocktail containing propidium iodide and the antibodies (BioLegend, San Diego, CA): allophycocyanin‐conjugated anti‐Ly5.1, allophycocyanin‐Cy7‐conjugated anti‐EpCAM, anti‐MHCII eF450, anti‐TCR‐β BV510, phycoerythrin‐Cy7‐conjugated anti‐CD11c and phycoerythrin‐conjugated anti‐CD45.1 (Miltenyi Biotec, Bergisch Gladbach, Germany) plus fluorescein isothiocyanate‐conjugated anti‐UEA‐1 (Vector Laboratories, Burlingame, CA, USA). Cells were washed and sorted directly into 350 μl of RNeasy Lysis Buffer (Qiagen, Hilden, Germany) using the 100 μm nozzle on an Influx 3 cell sorter (Becton Dickinson, Franklin Lakes, NJ). Testes from insHEL mice were digested and lysed using the same protocol excluding the staining and sorting steps.
Real‐time quantitative PCR
RNA was isolated using the RNeasy Micro kit (Qiagen), cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen), PCRs were performed using reagents from the QuantiNova SYBR Green RT‐PCR Kit (Qiagen) on a Rotor‐Gene Q real‐time PCR machine (Qiagen). Primer sequences were: Actb_l, CGTGAAAAGATGACCCAGATCA; Actb_r TGGTACGACCAGAGGCATACAG; HEL_l, GTGTGCCGCAAAATTCGAGA; HEL_r, TGTTGCACAGGTTCCTGGAG. PCR conditions were 95° for 2 min followed by 40 cycles of 95° for 5 seconds and 58° for 10 seconds. PCR amplification of a 10‐fold dilution series of cDNA from insHEL testis tissue, which is known to express HEL,26 was used to verify that cDNA molecules from HEL and the internal control gene Actb were amplified equally efficiently. Relative expression levels of HEL and Actb were calculated using the comparative C T method.30
Flow cytometry
Single‐cell thymocyte suspensions were pelleted by centrifugation and incubated for 30 min at 4° in culture supernatant from the 1G12 hybridoma (specific for TCR3A9) (American Type Culture Collection, Manassas, VA; Cat #CRL‐2827). Then, cells were incubated for 30 min at 4° in FACS buffer (PBS containing 2% v/v heat‐inactivated bovine serum and 0·01% m/v sodium azide) containing assortments of fluorochrome‐conjugated monoclonal antibodies against CD4 (BioLegend, Cat #100430), CD8α (BioLegend, Cat #100765), CD45.1 (BioLegend, Cat #110739), CD45.2 (BioLegend, Cat #109828) or mouse IgG1 (to detect 1G12) (Becton Dickinson, Cat # 560089). After washing in FACS buffer, cells were fixed and permeabilized using the Foxp3/Transcription Factor Staining Buffer Set (Affymetrix eBioscience, Santa Clara, CA), then incubated with antibodies specific for intracellular proteins, Helios (clone 22F6; BioLegend) and Foxp3 (eBioscience, Cat # 11‐5773‐80). Data were acquired with LSRFortessa or LSR II flow cytometers (Becton Dickinson) and analysed using flowjo software (FlowJo LLC, Ashland, OR).
EdU labelling
For 5‐ethynyl‐2′‐deoxyuridine (EdU) labelling, 0·25 mg of EdU was injected intraperitoneally (0·1 ml of a 2·5 mg/ml m/v in DMSO solution) per mouse. After intracellular staining as described above, cells were processed following the manufacturer's instructions (Click‐iT® EdU Flow Cytometry Assay Kit, Thermo Fisher Scientific, Waltham, MA) except that Click‐iT® EdU buffer additive (Component G) was used at one‐fifth of the concentration recommended.
Statistical analysis
graphpad prism version 7 for Mac was used to conduct statistical analyses as described in the figure legends.
Results
Intrathymic HEL expression is less abundant and more restricted to the medulla in insHEL mice compared with thyroHEL mice
To determine the intrathymic location of HEL expression in insHEL and thyroHEL mice, we used in situ hybridization to detect HEL mRNA in thymus sections. Intense focal staining was observed in rare cells in the medulla in both insHEL and thyroHEL thymi (Fig. 1a). In addition, punctate staining of lower intensity was observed in a diffuse pattern throughout the cortex and medulla of thyroHEL but not insHEL thymic sections (Fig. 1a). Next, we measured HEL mRNA abundance in FACS‐sorted thymic APC subsets using real‐time PCR. Although HEL mRNA was undetectable in thymic dendritic cells, HEL mRNA was detected at low levels in two out of three cortical TEC (cTEC) samples from thyroHEL mice (Fig. 1b). The mTEC population was subdivided into MHCIIlo and MHCIIhi subsets. HEL mRNA was detected in five out of six mTEC samples from thyroHEL mice and was detected at a lower level in one out of four mTEC samples from insHEL mice (Fig. 1b). These data indicate that HEL expression is less abundant and more confined to the medulla in insHEL mice compared with thyroHEL mice.
Figure 1.
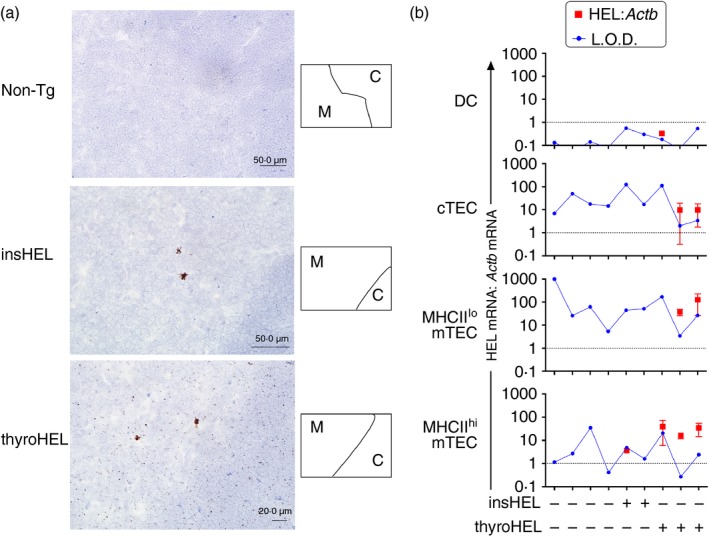
Differential intrathymic hen egg lysozyme (HEL) expression in mice in which HEL is driven by insulin (insHEL) or thyroglobulin (thyroHEL) promoter. (a) HEL mRNA detection using in situ hybridization in thymic tissue sections from mice of the genotypes indicated (left). Cortical (C) and medullary (M) regions were distinguished based on cell density, as shown in the schematics (right). Sections exposed to the negative control probe targeting the bacterial gene, DapB, were indistinguishable from the non‐transgenic (Non‐Tg) section (data not shown). Sections from thyroHEL mice (n = 3 mice) exhibited low‐intensity punctate staining throughout cortical and medullary regions. (b) Quantification of HEL expression in the thymic antigen‐presenting cell (APC) subsets indicated (left). Dendritic cells (DCs; CD45+ CD11c+), cortical thymic epithelial cells (cTECs; CD45– EpCAM+ UEA‐1– Ly51+), MHCIIlo medullary TECs (mTECs; CD45– EpCAM+ UEA‐1+ MHCII–) and MHCIIhi mTECs (CD45– EpCAM+ UEA‐1+ MHCII+) were sorted from individual thymi of mice of the indicated genotypes (bottom) and cDNA was analysed by real‐time PCR. For samples in which a product was detected in the HEL‐specific PCR, red symbols on graphs show HEL expression relative to the internal control gene, Actb, calculated using the comparative C T method. Values were normalized to HEL expression levels in the insHEL testis, which is represented by the dotted line at 1 on the y‐axis. Limits of detection (L.O.D.) were determined based on the maximum C T value for the lowest concentration of insHEL testis cDNA that generated a product in nine out of nine HEL‐specific PCRs. Error bars show the standard deviation of observations from three independent experiments performed on the same panel of cDNA samples.
Both insHEL and thyroHEL delete DP thymocytes albeit to different extents
To define the maturation stages at which insHEL and thyroHEL delete thymocytes, we used flow cytometry to examine thymocytes from 3A9×insHEL or 3A9×thyroHEL double‐transgenic mice and 3A9 single‐transgenic littermates. Thymocytes expressing the transgenic 3A9 TCR (TCR3A9) were detected in all groups except non‐transgenic (Non‐Tg) controls (Fig. 2a). At both DP and CD4SP stages the numbers of TCR3A9+ Foxp3– thymocytes were significantly lower in 3A9×insHEL mice compared with 3A9 mice, while 3A9×thyroHEL mice had even fewer cells than 3A9×insHEL mice (Fig. 2b). Hence insHEL and thyroHEL both delete thymocytes at the DP stage, but more deletion occurs at the DP stage in 3A9×thyroHEL mice than in 3A9×insHEL mice.
Figure 2.
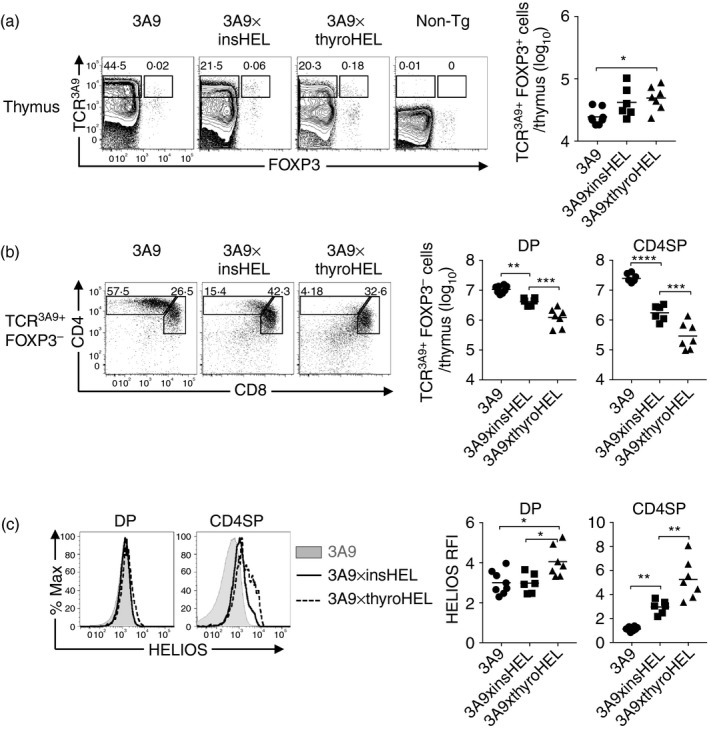
Both insulin promoter‐driven hen egg lysozyme transgene (insHEL) and thyroglobulin promoter‐driven hen egg lysozyme transgene (thyroHEL) delete thymocytes at the double‐positive (DP) stage of development albeit to different extents. (a) Representative flow cytometry plots show expression of the 3A9 TCR (TCR3A9) versus Foxp3 on all thymocytes from 3A9, 3A9×insHEL, 3A9×thyroHEL and non‐transgenic (Non‐Tg) mice with gates enclosing the TCR3A9+ Foxp3– and TCR3A9+ Foxp3+ populations (left). Graph shows the number of TCR3A9+ Foxp3+ cells per thymus (right). (b) TCR3A9+ Foxp3– thymocytes were analysed for CD4/CD8 expression (left). Graphs show the number of TCR3A9+ Foxp3– thymocytes divided into DP and CD4 single‐positive (CD4SP) stages as gated in the plots. (c) Helios expression in TCR3A9+ Foxp3– DP or CD4SP thymocytes from mice of the indicated genotypes (left). Helios relative fluorescence intensities (RFIs) were calculated relative to Non‐Tg DP thymocytes analysed in parallel. (a–c) Data compiled from three independent experiments were analysed using one‐way analysis of variance and Sidak's multiple comparisons test. Lines show the group mean; one symbol represents one mouse; *P < 0·05; **P < 0·01; ***P < 0·001; ****P < 0·0001.
Surprisingly, there was no difference between 3A9 mice and 3A9×insHEL mice in Helios expression within thymocytes at the DP stage (Fig. 2c). We noted that DP thymocytes in 3A9 mice had Helios relative fluorescence intensity values of ~3, which means their Helios expression was three‐fold higher than Non‐Tg DP thymocytes analysed in parallel (Fig. 2c). However, Helios was significantly up‐regulated in DP thymocytes from 3A9×thyroHEL mice (Fig. 2c). Increased deletion at the DP stage in 3A9×thyroHEL mice was therefore associated with increased Helios expression in DP thymocytes. Helios expression in CD4SP cells was greater in 3A9×insHEL mice than in 3A9 mice, an effect mainly attributable to a decrease in Helios expression during the DP to CD4SP transition in 3A9 mice (Fig. 2c).
As strong TCR signalling can also induce Foxp3+ Treg cell differentiation, TCR3A9+ Foxp3+ thymocytes were enumerated. Compared with 3A9 mice, there was a significant increase in TCR3A9+ Foxp3+ thymocytes in 3A9×thyroHEL mice, but not in 3A9×insHEL mice (Fig. 2a).
Helios up‐regulation can be limited by intraclonal competition and by rapid deletion
Intraclonal competition limits Helios expression in CD4SP thymocytes in 3A9×insHEL mice.21 In addition, rapid deletion might limit the extent of Helios up‐regulation. Thymocytes lacking the pro‐apoptotic protein, Bim (encoded by the Bcl2 l11 gene, referred to as Bim hereafter), are able to survive stimuli that normally induce thymocyte deletion.27 To investigate more closely the stage and extent of Helios up‐regulation induced by insHEL and thyroHEL, chimeric mice were generated using a BM mixture composed of 90% Non‐Tg (CD451/2) BM, 5% Bim +/+ 3A9 (CD451/1) BM and 5% Bim –/– 3A9 (CD452/2) BM. This enabled comparison of co‐resident wild‐type and apoptosis‐defective 3A9 TCR transgenic thymocytes at moderate clonal frequencies.
In Non‐Tg hosts, Bim‐deficiency caused a small but statistically significant decrease in CD4SP frequency (Fig. 3b) but there was no effect on Helios expression (Fig. 3c). In insHEL hosts, Bim‐deficiency caused a marked increase in CD4SP frequency (Fig. 3b), consistent with the requirement for Bim in insHEL‐mediated thymocyte deletion.21 DP thymocytes expressed significantly more Helios in insHEL hosts than in Non‐Tg hosts (Fig. 3c). The finding that reducing the frequency of HEL‐specific thymocytes to 10% led to insHEL‐dependent Helios up‐regulation in DP thymocytes suggests that intraclonal competition limits Helios expression at the DP stage in 3A9×insHEL mice, as previously observed at the CD4SP stage.21
Figure 3.
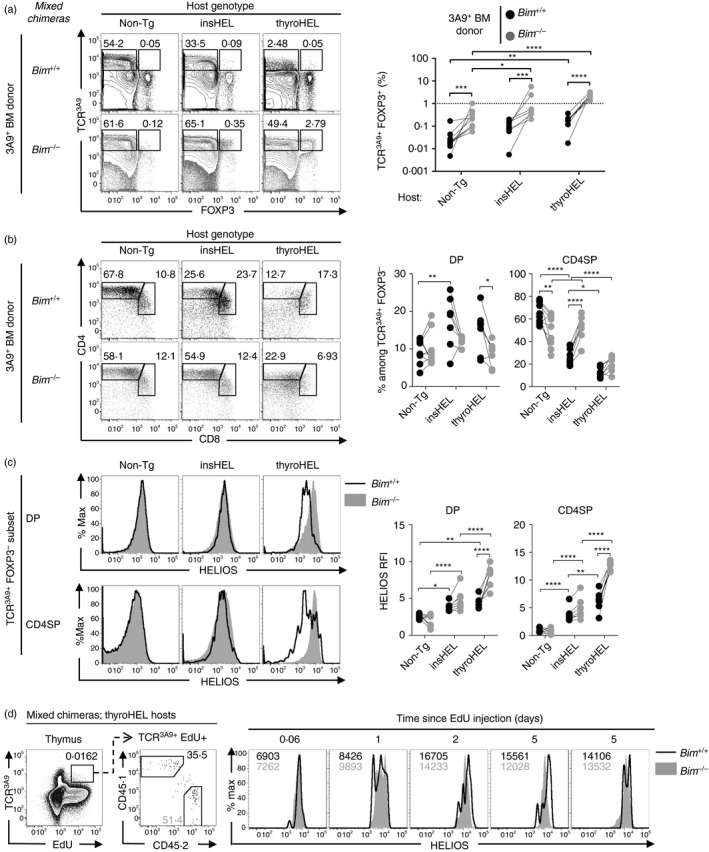
Phenotypes of 3A9 thymocytes rescued from deletion by a defect in apoptosis. Mixed bone marrow (BM) chimeras were generated by reconstituting irradiated non‐transgenic (Non‐Tg) (n = 9), insulin promoter‐driven hen egg lysozyme transgene (insHEL) (n = 7) or thyroglobulin promoter‐driven hen egg lysozyme transgene (thyroHEL) (n = 7) hosts with a cell mixture comprising 90% CD451/2 Bim +/+ Non‐Tg BM, 5% CD451/1 Bim +/+ 3A9 BM and 5% CD452/2 Bim –/– 3A9 BM. (a) Six weeks post‐reconstitution, electronically gated CD451/1 (Bim +/+) and CD452/2 (Bim –/–) thymocyte populations were analysed for expression of TCR3A9 and Foxp3 (left) with a summary graph showing the TCR3A9+ Foxp3+ cell frequencies within the CD451/1 and CD452/2 populations as gated in the plots (right). (b) CD4/CD8 phenotype of Bim +/+ or Bim –/– TCR3A9+ Foxp3– thymocytes (left) and summaries (right) show the double‐positive (DP) and CD4 single‐positive (CD4SP) cell frequencies as gated in the plots. (c) Helios expression on the DP and CD4SP subsets gated in (b) with a summary showing the Helios relative fluorescence intensity (RFI) values relative to Non‐Tg DP thymocytes analysed in parallel (right). (d) ThyroHEL hosts were injected intraperitoneally with EdU at the indicated times before analysis. Representative plots show the gate for the EdU+ TCR3A9+ population (first column) that was divided into CD451/1 (Bim +/+) and CD452/2 (Bim –/–) subsets (second column) that were compared for Helios expression with the mean fluorescence intensity of Bim +/+ and Bim –/– cells shown in black and grey text, respectively (columns 3–7). Each pair of symbols joined by a line in (a–c) and each histogram overlay in (d) represents one mixed chimera. Statistical comparisons used two‐way analysis of variance with Sidak's multiple comparisons test. *P < 0·05; **P < 0·01; ***P < 0·001; ****P < 0·0001.
In thyroHEL hosts, Bim‐deficiency did not affect the CD4SP frequency (Fig. 3b), indicating that thyroHEL prevents naive T‐cell differentiation in a Bim‐independent manner. Notably, in thyroHEL hosts, Bim‐deficiency caused a significant increase in Helios expression (Fig. 3c), which prompted us to formulate two hypotheses. First, Bim‐deficiency may affect the kinetics of Helios up‐regulation induced by thyroHEL. Second, Bim‐deficient thymocytes may survive for a longer period after engaging the thyroHEL self‐antigen and accumulate higher levels of Helios protein over time. To distinguish between these hypotheses, some of the mixed chimeras described above were injected with EdU, a thymidine analogue that is incorporated by DNA‐synthesizing cells.31 This procedure labels immature thymocytes, which can then be tracked as they progress through thymocyte development. Helios expression levels in EdU+ TCR3A9+ thymocytes were lower at < 1 day after EdU injection compared with 2 or 5 days after EdU injection (Fig. 3d), indicating that the older thymocytes had higher Helios expression. Age‐matched Bim +/+ and Bim –/– thymocytes had similar levels of Helios expression at all time‐points (Fig. 3d), consistent with the hypothesis that Bim‐deficient thymocytes survive longer after engaging the thyroHEL self‐antigen and accumulate higher levels of Helios protein over time.
Bim‐deficiency consistently increased Treg cell differentiation in TCR3A9+ Foxp3+ thymocytes (Fig. 3a). In addition, the TCR3A9+ Foxp3+ thymocyte frequencies were usually higher in insHEL and thyroHEL hosts than in Non‐Tg hosts (Fig. 3a). Apoptosis therefore limits Treg cell differentiation in 3A9 thymocytes. Furthermore, inhibiting apoptosis revealed that HEL expression can promote Treg cell differentiation in both insHEL and thyroHEL mice.
Direct presentation by BM‐APCs has no role in deletion mediated by insHEL or thyroHEL
3A9 thymocytes are not deleted when only the BM‐APCs carry the insHEL transgene.32 To test whether BM‐APCs carrying the thyroHEL transgene can mediate deletion through direct presentation, chimeras were made by reconstituting Non‐Tg hosts with BM from Non‐Tg, insHEL or thyroHEL mice that was mixed 9 : 1 with 3A9 BM. Six weeks later, the CD4SP cell frequency among TCR3A9+ Foxp3– thymocytes were similar in all groups (Fig. 4), which shows that neither insHEL BM‐APCs nor thyroHEL BM‐APCs are sufficient to delete 3A9 thymocytes through direct presentation.
Figure 4.
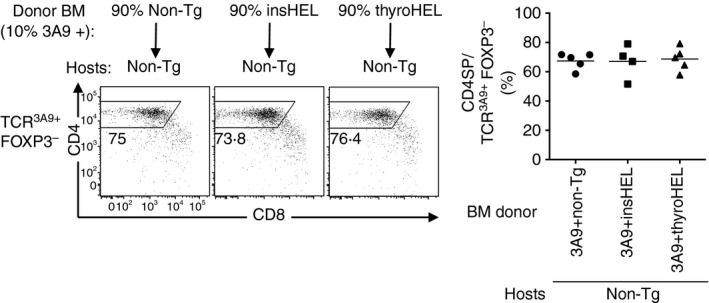
Neither insulin promoter‐driven hen egg lysozyme transgene (insHEL) nor thyroglobulin promoter‐driven hen egg lysozyme transgene (thyroHEL) mediates deletion when only the bone marrow‐derived antigen presenting cells (BM‐APCs) are transgenic. Irradiated non‐transgenic (Non‐Tg) hosts (CD452/2) were reconstituted with Non‐Tg, insHEL or thyroHEL (CD451/1) BM mixed with 3A9 (CD451/2) BM at a ratio of 9 : 1. Six weeks later, CD451/2 TCR3A9+ Foxp3– thymocytes were analysed for CD4/CD8 phenotype (left) and the CD4SP cell percentage was determined (right). Each symbol represents one mouse; each line shows the group mean.
Indirect presentation is essential for insHEL‐mediated deletion
Having excluded a role for direct presentation by BM‐APCs, the remaining pathways by which insHEL and thyroHEL must delete thymocytes include indirect presentation and direct presentation by TECs. To test whether indirect presentation is required, chimeras were made using wild‐type or MHCII‐deficient (H2‐Aa –/–) 3A9 BM. The insHEL and thyroHEL hosts with wild‐type BM‐APCs had similar thymic phenotypes to non‐chimeric 3A9×insHEL and 3A9×thyroHEL mice, respectively (Fig. 5b and 2b, respectively). In contrast, insHEL hosts with MHCII‐deficient BM‐APCs were indistinguishable from Non‐Tg hosts (Fig. 5b,c) demonstrating that indirect presentation is essential for insHEL‐mediated deletion.
Figure 5.
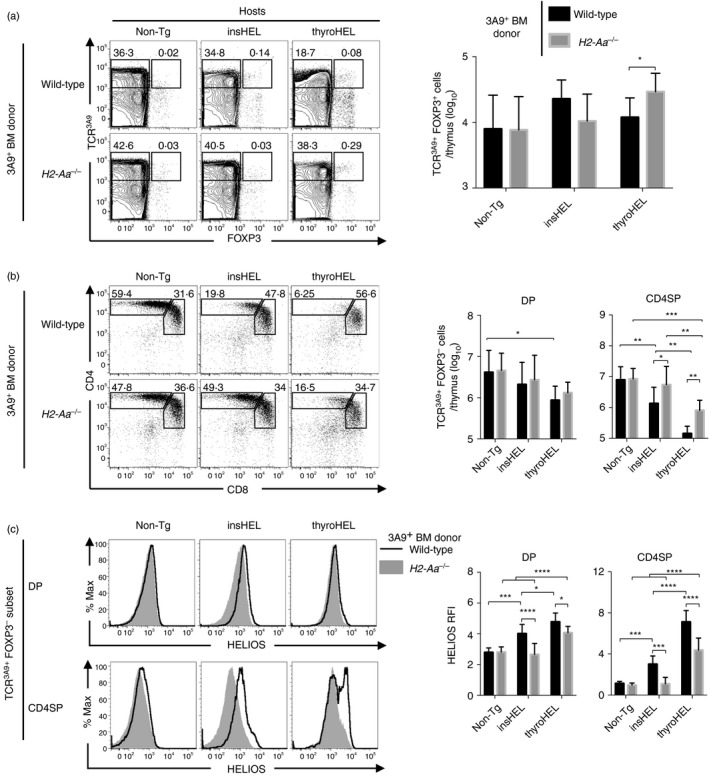
Defective indirect presentation by bone marrow antigen‐presenting cells (BM‐APCs) abrogates insulin promoter‐driven hen egg lysozyme transgene (insHEL)‐mediated deletion and delays thyroglobulin promoter‐driven hen egg lysozyme transgene (thyroHEL)‐mediated deletion. Irradiated none‐transgenic (Non‐Tg), insHEL or thyroHEL hosts were reconstituted with 3A9 BM that was either wild‐type (H2‐Aa +/+) or MHCII‐deficient (H2‐Aa –/–) and analysed 4–6 weeks later. (a) Thymocytes were gated for TCR3A9+ Foxp3+ and TCR3A9+ Foxp3– subsets (left) and the summary shows the number of TCR3A9+ Foxp3+ thymocytes (right). (b) TCR3A9+ Foxp3– thymocytes were analysed for CD4/CD8 expression (left) and summaries show the number of double‐positive (DP) and CD4 single‐positive (CD4SP) cells (right). (c) Helios expression on DP and CD4SP subsets of TCR3A9+ Foxp3– thymocytes (left) with Helios relative fluorescence intensity (RFI) values relative to Non‐Tg DP thymocytes (right). Statistical comparisons used two‐way analysis of variance and Sidak's multiple comparisons test. P‐value symbols: *P < 0·05; **P < 0·01; ***P < 0·001; ****P < 0·0001.
Defective indirect presentation delays deletion and increases Treg cell differentiation mediated by thyroHEL
Compared with thyroHEL hosts with wild‐type BM‐APCs, thyroHEL hosts with MHCII‐deficient BM‐APCs had a larger CD4SP thymocyte population (Fig. 5b), which is indicative of a defect in thymocyte deletion. In thyroHEL hosts, MHCII deficiency in BM‐APCs also decreased Helios expression (Fig. 5c). However, thyroHEL hosts with MHCII‐deficient BM‐APCs had fewer CD4SP thymocytes and higher Helios expression than Non‐Tg hosts (Fig. 5b,c). As these findings show that at least some deletion was occurring, impairing indirect presentation only caused a partial defect in thyroHEL‐mediated deletion. However, thymic TCR3A9+ Foxp3+ cells were 2·5‐fold more numerous in thyroHEL hosts with MHCII‐deficient BM‐APCs compared with thyroHEL hosts with wild‐type BM‐APCs (Fig. 5a). Impairing indirect presentation therefore delayed thyroHEL‐mediated deletion and was associated with an increase in thyroHEL‐mediated Treg differentiation.
Discussion
In the thymus, the indirect presentation pathway is important for the establishment of immunological self‐tolerance by promoting deletion or Treg cell differentiation. The data above show that indirect presentation amplifies TCR signalling and promotes thymocyte deletion mediated by the transgenic self‐antigens, insHEL and thyroHEL. In the insHEL model indirect presentation is absolutely required for deletion. In the thyroHEL model indirect presentation is required to promote deletion at the DP stage and to limit Treg cell differentiation. Indirect presentation therefore limits differentiation of both naive CD4+ T cells and Treg cells by promoting self‐antigen recognition at the DP stage of thymocyte development.
We examined Bim‐deficient thymocytes to rescue thymocytes from deletion so that we could determine the maturation stage at which thymocytes encounter insHEL and thyroHEL by assessing Helios expression. The mixed chimeras made for this purpose revealed that both self‐antigens increased Helios expression in DP thymocytes (Fig. 3c). The experiment also provided unanticipated insights. First, Bim‐deficiency enabled thymocytes encountering thyroHEL to express very high levels of Helios. That Bim‐deficient thymocytes expressed markedly more Helios in response to thyroHEL compared with insHEL is consistent with the evidence of higher intrathymic HEL expression in thyroHEL mice compared with insHEL mice (Fig. 1). Second, Bim‐deficiency had different effects on tolerance induction in the two models. ThyroHEL prevents naive T‐cell development in a Bim‐independent manner, as observed in other models where the deleting self‐antigen is expressed in the thymic cortex and deletion occurs at the DP stage.33, 34, 35 These findings suggest that tolerance induction in the thymic cortex is Bim‐independent. By contrast, Bim is essential for the mechanisms by which insHEL prevents CD4+ T‐cell development21 and RIP‐mOVA prevents CD8+ T‐cell development.35, 36 To our knowledge, no models of thymocyte deletion in the thymic medulla are Bim‐independent. However, defects in medullary deletion caused by Bim‐deficiency appear to be offset by an increase in Treg cell differentiation in CD4+ T cells (Fig. 3) or induction of anergy in CD8+ T cells.35 While Bim‐deficiency is sufficient to precipitate organ‐specific autoimmunity,36 tolerance is induced in substantial numbers of self‐reactive T cells by Bim‐independent mechanisms,33, 34, 35, 37 which may explain why the organ‐specific autoimmunity caused by Bim‐deficiency is relatively mild.
Thymus‐grafting experiments excluded the blood and BM‐APCs as potential sources of the insHEL self‐antigen that deletes 3A9 thymocytes.32 Aire is required for insHEL‐mediated deletion,25, 26 indicating that TECs are involved in the mechanism. However, we found that insHEL TECs were unable to delete 3A9 thymocytes via direct presentation (Fig. 5). These results align insHEL with RIP‐OVAhi13, Aire‐GCLG120A,14 myelin basic protein15 and interphotoreceptor retinoid‐binding protein16 in a class of self‐antigens for which thymocyte deletion requires both TECs and BM‐APCs, but only the BM‐APCs actually present the self‐antigen to CD4+ thymocytes.
At what site and stage do thymocytes engage insHEL? The smaller DP population size in 3A9×insHEL mice compared with 3A9 mice suggests that insHEL is engaged at the DP stage. The insHEL‐mediated Helios induction in DP thymocytes required MHCII expression in BM‐APCs (Fig. 5). Multi‐photon imaging in thymic slices revealed that DP thymocytes localize to the thymic cortex,38, 39 yet insHEL expression was detected only in the medulla (Fig. 1). To reconcile these findings, it is conceivable that migratory DCs transport self‐antigens from the medulla to the cortex. Only ~2% of DCs in the thymic cortex were observed to migrate,40 which may explain why insHEL‐mediated Helios induction in DP thymocytes required conditions of low intraclonal competition (Fig. 3).
The HEL protein concentration in the blood is lower in thyroHEL mice than in insHEL mice (0·5 ng/ml and 1·2 ng/ml, respectively),24 which excludes the blood as a potential source of the thyroHEL self‐antigen that triggers thymocyte deletion. ThyroHEL can delete 3A9 thymocytes through direct presentation by TECs (Fig. 5) but not through direct presentation by BM‐APCs (Fig. 4). However, indirect presentation by BM‐APCs increases thyroHEL‐mediated Helios induction and promotes deletion at the DP stage. This observation extends in vitro evidence that mTEC‐derived self‐antigens can activate T cells more strongly when presented by thymic dendritic cells than by the mTECs themselves.5 ThyroHEL and the transgenic self‐antigen pigeon cytochrome c14, 41 fall within a class of self‐antigens for which direct presentation by TECs can delete thymocytes, but indirect presentation by BM‐APCs enhances deletion at the DP stage.
T‐cell receptor sequencing data indicate that indirect presentation of Aire‐dependent self‐antigens plays a crucial role in promoting Treg cell selection.20 Our results reveal an effect in the opposite direction, that is, indirect presentation can also limit Treg cell selection in thyroHEL mice (Fig. 5). How might indirect presentation limit Treg selection? The ‘avidity hypothesis’ postulates that strong TCR signalling induces deletion whereas intermediate TCR signalling induces Treg cell selection.9, 42 The data above are consistent with the ‘avidity hypothesis’ because a decrease in Helios induction was associated with a shift from deletion towards Treg cell selection in thyroHEL mice. Alternatively, it is also significant that indirect presentation was required for normal deletion at the DP stage in thyroHEL mice. High‐affinity TCR engagement is less likely to induce Treg cell differentiation in a thymocyte at the DP stage than at the CD4SP stage.43, 44 In summary, this study demonstrates that indirect presentation amplifies TCR signalling induced by certain self‐antigens and thereby limits the differentiation of both naive CD4+ T cells and Treg cells in the thymus.
Author contributions
JYY, RCW, AC and DRH performed the experiments; JYY, RCW, AC and SRD analysed data and prepared figures; CCG and SRD designed the study; JYY and SRD drafted the paper and all authors read and edited the paper.
Disclosures
All authors have declared that no conflict of interest exists.
Acknowledgements
We thank the Australian Phenomics Facility Genotyping Team and staff for animal genotyping and husbandry. This research was supported by National Health and Medical Research Council Grants 1107464 to SRD, 1108800 to CCG and SRD, 1016953 and 585490 (Australia Fellowship) to CCG and by the Monash Biomedicine Discovery Institute.
References
- 1. Baldwin KK, Trenchak BP, Altman JD, Davis MM. Negative selection of T cells occurs throughout thymic development. J Immunol 1999; 163:689–98. [PubMed] [Google Scholar]
- 2. Douek DC, Corley KT, Zal T, Mellor A, Dyson PJ, Altmann DM. Negative selection by endogenous antigen and superantigen occurs at multiple thymic sites. Int Immunol 1996; 8:1413–20. [DOI] [PubMed] [Google Scholar]
- 3. Fontenot JD, Dooley JL, Farr AG, Rudensky AY. Developmental regulation of Foxp3 expression during ontogeny. J Exp Med 2005; 202:901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee HM, Hsieh CS. Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. J Immunol 2009; 183:2261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koble C, Kyewski B. The thymic medulla: a unique microenvironment for intercellular self‐antigen transfer. J Exp Med 2009; 206:1505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Millet V, Naquet P, Guinamard RR. Intercellular MHC transfer between thymic epithelial and dendritic cells. Eur J Immunol 2008; 38:1257–63. [DOI] [PubMed] [Google Scholar]
- 7. Klein L, Klein T, Ruther U, Kyewski B. CD4 T cell tolerance to human C‐reactive protein, an inducible serum protein, is mediated by medullary thymic epithelium. J Exp Med 1998; 188:5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aschenbrenner K, D'Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, et al Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol 2007; 8:351–8. [DOI] [PubMed] [Google Scholar]
- 9. Hinterberger M, Aichinger M, da Costa OP, Voehringer D, Hoffmann R, Klein L. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nat Immunol 2010; 11:512–9. [DOI] [PubMed] [Google Scholar]
- 10. Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol 2002; 3:756–63. [DOI] [PubMed] [Google Scholar]
- 11. Gallegos AM, Bevan MJ. Central tolerance to tissue‐specific antigens mediated by direct and indirect antigen presentation. J Exp Med 2004; 200:1039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perchellet A, Brabb T, Goverman JM. Crosspresentation by nonhematopoietic and direct presentation by hematopoietic cells induce central tolerance to myelin basic protein. Proc Natl Acad Sci USA 2008; 105:14040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hubert FX, Kinkel SA, Davey GM, Phipson B, Mueller SN, Liston A, et al Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood 2011; 118:2462–72. [DOI] [PubMed] [Google Scholar]
- 14. Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med 2013; 210:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huseby ES, Sather B, Huseby PG, Goverman J. Age‐dependent T cell tolerance and autoimmunity to myelin basic protein. Immunity 2001; 14:471–81. [DOI] [PubMed] [Google Scholar]
- 16. Taniguchi RT, DeVoss JJ, Moon JJ, Sidney J, Sette A, Jenkins MK, et al Detection of an autoreactive T‐cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)‐mediated selection. Proc Natl Acad Sci USA 2012; 109:7847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, et al Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med 2005; 202:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sansom SN, Shikama‐Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al Population and single‐cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self‐antigen expression in thymic epithelia. Genome Res 2014; 24:1918–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al Projection of an immunological self shadow within the thymus by the aire protein. Science 2002; 298:1395–401. [DOI] [PubMed] [Google Scholar]
- 20. Perry JSA, Lio CJ, Kau AL, Nutsch K, Yang Z, Gordon JI, et al Distinct contributions of Aire and antigen‐presenting‐cell subsets to the generation of self‐tolerance in the thymus. Immunity 2014; 41:414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daley SR, Hu DY, Goodnow CC. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD‐1 or NF‐κB. J Exp Med 2013; 210:269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho WY, Cooke MP, Goodnow CC, Davis MM. Resting and anergic B cells are defective in CD28‐dependent costimulation of naive CD4+ T cells. J Exp Med 1994; 179:1539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen PM, Unanue ER. Differential requirements for antigen processing by macrophages for lysozyme‐specific T cell hybridomas. J Immunol 1984; 132:1077–9. [PubMed] [Google Scholar]
- 24. Akkaraju S, Ho WY, Leong D, Canaan K, Davis MM, Goodnow CC. A range of CD4 T cell tolerance: partial inactivation to organ‐specific antigen allows nondestructive thyroiditis or insulitis. Immunity 1997; 7:255–71. [DOI] [PubMed] [Google Scholar]
- 25. Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ‐specific T cells. Nat Immunol 2003; 4:350–4. [DOI] [PubMed] [Google Scholar]
- 26. Liston A, Gray DH, Lesage S, Fletcher AL, Wilson J, Webster KE, et al Gene dosage–limiting role of Aire in thymic expression, clonal deletion, and organ‐specific autoimmunity. J Exp Med 2004; 200:1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, et al BH3‐only Bcl‐2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 2002; 415:922–6. [DOI] [PubMed] [Google Scholar]
- 28. Kontgen F, Suss G, Stewart C, Steinmetz M, Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int Immunol 1993; 5:957–64. [DOI] [PubMed] [Google Scholar]
- 29. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al RNAscope: a novel in situ RNA analysis platform for formalin‐fixed, paraffin‐embedded tissues. J Mol Diagn 2012; 14:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative CT method. Nat Protoc 2008; 3:1101–8. [DOI] [PubMed] [Google Scholar]
- 31. Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo . Proc Natl Acad Sci USA 2008; 105:2415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liston A, Lesage S, Gray DH, Boyd RL, Goodnow CC. Genetic lesions in T‐cell tolerance and thresholds for autoimmunity. Immunol Rev 2005; 204:87–101. [DOI] [PubMed] [Google Scholar]
- 33. Hu Q, Sader A, Parkman JC, Baldwin TA. Bim‐mediated apoptosis is not necessary for thymic negative selection to ubiquitous self‐antigens. J Immunol 2009; 183:7761–7. [DOI] [PubMed] [Google Scholar]
- 34. Kovalovsky D, Pezzano M, Ortiz BD, Sant'Angelo DB. A novel TCR transgenic model reveals that negative selection involves an immediate, Bim‐dependent pathway and a delayed, Bim‐independent pathway. PLoS ONE 2010; 5:e8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suen AY, Baldwin TA. Proapoptotic protein Bim is differentially required during thymic clonal deletion to ubiquitous versus tissue‐restricted antigens. Proc Natl Acad Sci USA 2012; 109:893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gray DH, Kupresanin F, Berzins SP, Herold MJ, O'Reilly LA, Bouillet P, et al The BH3‐only proteins Bim and Puma cooperate to impose deletional tolerance of organ‐specific antigens. Immunity 2012; 37:451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jorgensen TN, McKee A, Wang M, Kushnir E, White J, Refaeli Y, et al Bim and Bcl‐2 mutually affect the expression of the other in T cells. J Immunol 2007; 179:3417–24. [DOI] [PubMed] [Google Scholar]
- 38. Ehrlich LI, Oh DY, Weissman IL, Lewis RS. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity 2009; 31:986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Halkias J, Melichar HJ, Taylor KT, Ross JO, Yen B, Cooper SB, et al Opposing chemokine gradients control human thymocyte migration in situ . J Clin Invest 2013; 123:2131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ladi E, Schwickert TA, Chtanova T, Chen Y, Herzmark P, Yin X, et al Thymocyte‐dendritic cell interactions near sources of CCR7 ligands in the thymic cortex. J Immunol 2008; 181:7014–23. [DOI] [PubMed] [Google Scholar]
- 41. Oehen S, Feng L, Xia Y, Surh CD, Hedrick SM. Antigen compartmentation and T helper cell tolerance induction. J Exp Med 1996; 183:2617–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014; 14:377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cowan JE, Parnell SM, Nakamura K, Caamano JH, Lane PJ, Jenkinson EJ, et al The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J Exp Med 2013; 210:675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu DY, Yap JY, Wirasinha RC, Howard DR, Goodnow CC, Daley SR. A timeline demarcating two waves of clonal deletion and Foxp3 upregulation during thymocyte development. Immunol Cell Biol 2016; 94:357–66. [DOI] [PubMed] [Google Scholar]


