Abstract
The rust fungi (Pucciniales) are the most speciose natural group of plant pathogens, members of which possess the most complex lifecycles in Fungi. How natural selection works on the Pucciniales has been the subject of several hypotheses in mycology. This study uses molecular age estimation using sequence data from multiple loci, and cophylogeny reconciliation analyses to test hypotheses regarding how the aecial and telial stages in the lifecycle of rust fungi may have differentially impacted their diversification. Molecular age estimates show that the timing of diversification in the Pucciniales correlates with the diversification of their gymnosperm and angiosperm hosts. Host reconciliation analyses suggest that systematic relationships of hosts from the aecial stage of the Pucciniales lifecycle better reflect the systematic relationships among the Pucciniales. The results demonstrate the relative importance of this stage on the overall evolution of the Pucciniales and supports hypotheses made by Leppik over half a century ago. This study represents the first evaluation of how different life stages in the Pucciniales shape the evolution of these fungi.
Key words: Basidiomycota, Biogenic radiation, Biological specialization, Hologenetic ladder hypothesis, Melampsorineae, Phytopathogenic fungi, Pucciniaceae, Pucciniomycotina, Uredinales
Introduction
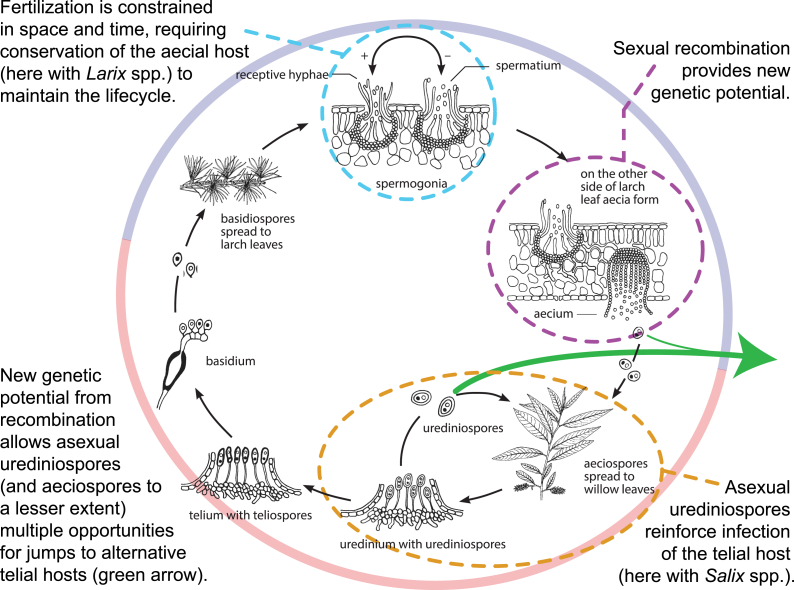
Rust fungi (Pucciniales) are simultaneously the most speciose and the most complex group of plant pathogenic fungi. The majority of Pucciniales require two specific but unrelated plant hosts in order to complete their life cycle (Fig. 1). This heteroecious life cycle can be distilled into two phases, each occurring on its associated host, herein referred to as the aecial and the telial stages/hosts. The aecial stage represents the part of the Pucciniales life cycle where haploid monokaryons (i.e., spermatia) are brought together through fertilization (i.e., plasmogamy) to form a dikaryon. Dikaryotic aeciospores are then formed and function to disperse the dikaryon to the telial host. On the telial host, asexual propagation occurs via production of urediniospores. Ultimately, usually in response to environmental cues, the dikaryon will cease asexual sporulation and form teliospores. It is during this stage that karyogamy, followed by meiosis, takes place. Haploid basidiospores are ultimately produced from germinating teliospores, which carry the new monokaryon back to the aecial host (Fig. 1). Given the complexity of this lifecycle, and the diversity of hosts observed in both the aecial and telial stages, several hypotheses regarding how coevolution in this pathogenic relationship has shaped Pucciniales diversity have been postulated. However, no studies have been performed to effectively test these hypotheses.
Fig. 1.
Alternation of generations, depicted in a species of Melampsora, illustrating stages of the Pucciniales life cycle that may reinforce host association via natural selection. In the outer circle, blue indicates aecial (gametophytic) stage and red indicates telial (sporophytic) stage. Illustration adapted from Merje Toome (2010) with permission.
Several papers by Leppik published between 1953 and 1967 postulated how Pucciniales evolution may have been shaped by their plant hosts. One mechanism, termed the hologenetic ladder hypothesis, envisioned a scenario whereby individual Pucciniales lineages leverage the alternating stages of the life cycle to switch from ancestral hosts to younger, more recently derived hosts (Leppik 1953), thereby allowing speciation through specialization on new hosts as well as an avenue for avoiding co-extinction of lineages adapted to phylogenetically older hosts. In later papers, Leppik discussed the duality of the Pucciniales heteroecious life cycle as an alternation between biological specialization (BSp) and biogenic radiation (BgR). In BSp the fungus is specialized to exploit a very specific host, typically as a result of the gene-for-gene type of interactions proposed by Flor (1956) and (Leppik 1965). The taxonomic identity of this host is typically conserved between related Pucciniales taxa, displaying a strong degree of host fidelity or specialization. The reverse is true for BgR. In this process the pathogens show less fidelity and specificity resulting in numerous potential hosts with a much higher degree of taxonomic variability (Leppik 1967).
Although the trends of BSp and BgR are observed across the Pucciniales lineages, understanding how plant hosts have shaped their evolution in a phylogenetic context remains to be thoroughly examined. In addition to the work of Leppik, other early rust biologists looked for evidence that evaluated the importance of BSp or BgR on rust evolution. The development of molecular phylogenetics has provided tools for testing these hypotheses but is underemployed in urediniology. The most recent studies to analyse the phylogenetic associations between the Pucciniales and their hosts have either examined patterns for a single stage, such as analyses of telial stage/host relationships (e.g., van der Merwe et al. 2008), or examined patterns between Pucciniales and hosts while assuming the same evolutionary forces are acting equally across all stages/hosts in the rust life cycle (e.g., McTaggart et al. 2016). Likewise, published molecular phylogenetic studies that describe the systematic relationships within the Pucciniales have focused on molecular interpretations of these relationships and their impact on taxonomy (Maier et al., 2003, Bauer et al., 2006, van der Merwe et al., 2008, Dixon et al., 2010, Feau et al., 2011); few have speculated on how host associations may have shaped the diversity of the Pucciniales.
Attempting to look for evidence of coevolution between the rust fungi and their hosts is complicated because there can be multiple hosts per fungal species. As a result, different selective pressures may be at work during different phases of the life cycle. Aime (2006) noted a strong association between aecial hosts and deeper phylogenetic nodes for Pucciniales. Following that observation, the present study attempts to deconstruct the coevolutionary signal between the Pucciniales and their hosts by testing assumptions about equality of host conservation across the Pucciniales. For this study, we compiled previously published sequences with ca. 100 newly generated sequences from four target loci to reconstruct the phylogenetic relationships within Pucciniales as well as for their host plants. We used molecular dating to evaluate their aecial and telial host relationships across three evolutionary time scales. We then used reconciliation analysis between the rust and host phylogenies to evaluate the relative degree of BSp or BgR acting on each rust stage. The working hypothesis of this study is that the aecial host—the host on which genetic recombination in the form of fertilization occurs—is under the strongest selective pressure for conserving host associations, and therefore should retain the greatest signal of BSp and coevolution, whereas less selective pressure on the telial stage would result in a better fit to a pattern of BgR.
Materials and methods
DNA extraction, polymerase chain reaction, and sequencing
Fungal DNA was obtained from Pucciniales sori excised from dried host material and isolated using the UltraClean Plant DNA isolation kit (MoBio Laboratories, Solana Beach, CA, USA) following Aime (2006). The majority of material used in this study came from collections housed in the Arthur Herbarium (PUR) at Purdue University (Supplementary Information S1).
Polymerase chain reactions (PCR) used 12.5 μl PCR Master Mix (Promega, Madison, WI, USA), 1.25 μl each 10 μM forward and reverse primers, and 10 μL diluted (10- to 100 fold) DNA template. The nuclear ribosomal small subunit (18S) and the 5′ end of the nuclear ribosomal large subunit (28S) were PCR amplified using primer pairs Rust18SR (Aime 2006) & NS1 (White et al. 1990), and Rust2inv (Aime 2006) & LR6 (Vilgalys and Hester 1990), respectively. PCR product showing strong bands was sequenced directly using the same primers. Otherwise, samples were cleaned using ExoSap (Affymetrix, Santa Clara, CA) and a nested PCR was performed for 18S using rust-specific primers RustNS2-F (GGG AGG TAG TGA CMA TAA ATA ACA ATG) & NS6 (White et al. 1990), and for 28S using primers Rust28SF (TTT TAA GAC CTC AAA TCA GGT G) & LR5 (Vilgalys and Hester 1990). Thermal cycler temperatures and times for both 18S and 28S markers follow those described by Aime (2006). The mitochondrial gene cytochrome oxydase subunit 3 (CO3) was amplified with primer pair CO3F1 & CO3R1 using the primer sequences and protocols described by Beenken et al. (2012). Amplified PCR product was sent to Beckman-Coulter (Danvers, MA) for cycle sequencing using BIG dye v. 3.1 (Perkin Elmer Corp., Waltham, MA), with post reaction dye terminator removal using Agencourt CleanSEQ and sequence delineation performed on an ABI PRISM 3730xl with base calling and data compilation. Sequence fragments were edited and combined to form consensus sequences using Sequencher v. 5.2.3 (GeneCodes, Anne Arbor, MI, USA). The taxonomic and locus identities of the consensus sequences were verified by performing a BLAST search of GenBank.
Data sets
Datasets for individual genes were assembled using MUSCLE (Edgar 2004) with additional manual editing performed in Mesquite v. 2.75 (Maddison & Maddison 2011). Datasets were analyzed using RAxML v. 7.6.3 (Stamatakis 2006) to compare individual gene trees for taxonomic congruence. Maximum likelihood (ML) analysis was performed using the default settings, with 1000 bootstrap replicates, implemented on the CIPRES web portal (Miller et al. 2010; http://www.phylo.org/portal2/).
Four multigene datasets were created in order to estimate divergence times for Pucciniales and to evaluate patterns of codiversification between Pucciniales taxa and their plant hosts. Detailed taxonomic composition and sequence information for the datasets is provided in Supplementary Information S1. These are described more generally in the following paragraphs.
The Dikarya dataset (Supplementary Information S1-Table 1) was created for subsequent calibration of the other datasets in this study. It is comprised of 34 taxa: nine Ascomycota taxa (5 Pezizomycotina and 4 Saccharomycotina) and 25 Basidiomycota taxa (10 Pucciniomycotina, 5 Ustilaginomycotina, and 10 Agaricomycotina). This dataset is represented by 6 genes: 18S, 28S, RNA polymerase II, subunits 1 (RPB1) and 2 (RPB2), elongation factor 1-alpha (ef1α), and mitochondrial ATPase subunit 6 (ATP6). The nuclear ribosomal genes 18S and 28S datasets are represented as nucleotide sequences and the remaining protein coding genes (RPB1, RPB2, ef1α, and ATP6) are represented as amino acid sequences. Sequences used for this dataset, not originally produced for this study, were acquired from the AFTOL molecular database (www.aftol.org), from Genbank, and from the Broad Institute's genome database (www.broadinstitute.org) and the Joint Genome Institute's MycoCosm database (genome.jgi.doe.gov) (Supplementary Information S1-Table 1).
The Pucciniales dataset (Supplementary Information S1-Table 2) represents taxa from across the order including two nested Pucciniales subgroups described below. This dataset consists of 29 taxa, 27 being ingroup taxa selected to represent all known major lineages of rust fungi based on multiple studies (Aime, 2006, Minnis et al., 2012, McTaggart et al., 2016, Aime et al., 2017, Beenken, 2017). The two outgroup taxa represent the sister order to Pucciniales fide Aime et al. (2006). Characters are composed of three genes: 18S, 28S, and CO3. Nucleotide sequences were used for all three genes. For the CO3 alignment the intron regions were removed and the beginning trimmed to start the alignment with the first codon of the reading frame.
Two additional Pucciniales datasets, comprised of two lineages of different ages were constructed to further test for patterns of diversification at different evolutionary time scales. The Melampsorineae dataset consists of an earlier diverging lineage of four rust families. This is composed of 99 taxa representing the Coleosporiaceae, Pucciniastraceae and Melampsoraceae (Supplementary Information S1-Table 3). The 95 taxon ingroup is represented by the genera Coleosporium, Chrysomyxa, Cronartium, Melampsora, Melampsorella, Melampsoridium, Milesina, Naohidemyces, Pucciniastrum, Thekopsora, Urediniopsis, and some asexual morphs currently placed in Caeoma and Uredo; four outgroups were selected based on the results from analyses of the Pucciniales dataset. The Melampsorineae dataset uses 28S and CO3 genes. Both gene datasets are represented as nucleotide sequences with the CO3 alignment modified in the same manner described for the Pucciniales dataset (see above).
The Pucciniaceae dataset (Supplementary Information S1-Table 4) represents a recently diverged lineage and the most specious family of rust fungi. This dataset consists of 62 taxa predominantly from the genera Puccinia and Uromyces; Puccinia malvacearum was chosen as the outgroup based on the study of van der Merwe et al. (2008). Characters are composed of 28S and ef1α nucleotide sequences with ef1α data acquired from the study by van der Merwe et al. (2008).
Phylogenetic analysis and divergence time estimation
Estimations of divergence times was performed using a secondary calibration procedure (Renner 2005). This has been employed for fungi due to limited fossil records from which to establish internal calibration points (Matheny et al., 2009, Floudas et al., 2012, Wilson et al., 2012). Calibration of nodes in the Dikarya dataset used the following fossils: the marasmioid fungi (Marasmius rotula and Mycena amabillissima) based on a 90 million year old (My) fossil Archaeomarasmius leggetti from mid-Cretaceous amber (Hibbett et al. 1997); the Suillineae (Suillus pictus and Gomphidius roseus) was dated at 50 My based on a permineralized suilloid ectomycorrhiza fossil associated with Pinaceae roots (LePage et al. 1997); lastly, the split between the Pezizomycotina and Saccharomycotina in the Ascomycota was calibrated following Floudas et al. (2012), using the early Devonian fossil (452 My) Paleopyrenomycites devonicus (Taylor et al. 2005). The details of the time to most recent common ancestor (tMRCA) prior setup used for analyses of Dikarya, Pucciniales, Melampsorineae, and Pucciniaceae datasets are given in Supplementary Information S2.
Molecular dating analyses were performed using BEAST v. 1.7.5 (Drummond et al., 2006, Drummond and Rambaut, 2007) with XML files created in BEAUTi v. 1.7.5 Site models for nucleotide sequences were analyzed using the GTR+I+G substitution and site heterogeneity model with estimated base frequencies. Nuclear ribosomal markers 18S and 28S were combined under the same site model. Parameter rates were estimated separately for CO3 and ef1α in the Pucciniaceae dataset analyses. Intron regions were removed in order to perform the analysis using a site model using two codon partitions ((1 + 2), 3). Dating of the Dikarya dataset used amino acid sequence characters for protein coding genes, which was analyzed with a WAG+I+G substitution and heterogeneity model. All datasets were analyzed with a clock model set to an uncorrelated relaxed clock and a lognormal rate distribution. Tree priors were set to “Speciation: Birth-Death Process”.
For each analysis, a preliminary BEAST run was performed using a random start tree, to produce a start tree for use in subsequent BEAST analyses. Each BEAST analyses consisted of 10 million generations, and performed up to 10 independent times. The posterior distribution of optimal trees was sampled for each of the 10 BEAST runs. Each analysis was evaluated for an estimated sample size >200 for every prior before being incorporated into a single dataset for molecular dating estimation.
Host phylogeny reconstruction
In order to perform reconciliation analyses, we assembled matching host (consisting of aecial and telial hosts) datasets for each of the three Pucciniales (Pucciniales, Melampsorineae, and Pucciniaceae) datasets. Assembly of these was complicated largely by the availability of sequence data for host taxa. In ca. 20 % of cases, specific species or even genera were not available, and the missing taxon was substituted with one that would approximate its phylogenetic position. For most cases, host associations were assigned at the generic level. This simplified the host datasets, especially where a Pucciniales taxon was known to associate with multiple species within a host genus.
Host data were compiled for each Pucciniales taxon as follows: the host and stage of a specimen were recorded for each collection; the alternate stage was deduced from all available literature; and where more than one species is known to serve as an alternate host we used the highest taxonomic level (genus, family) to represent the host lineage for that Pucciniales taxon (Supplementary Information S4). In cases where hosts were not known, we devised “liberal” and “conservative” models for performing host reconciliation analysis and this is described below. The plastid region ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene was analyzed for host analyses because it is most widely used plant systematic molecular marker for which sequence data are: 1) available on GenBank; 2) easily aligned across the diversity of taxa used in this study; and 3) adequately reconstructs systematic relationships among the Tracheophyta plant hosts of the Pucciniales.
The composition of host datasets includes: 118 taxa for the Pucciniales host dataset, 109 taxa for the Melampsorineae host dataset, and 97 taxa for the Pucciniaceae host dataset. Host taxonomic identity of family, species, and Genbank ID information for each dataset are available in Supplementary Information S7.
Host phylogenetic analyses were done using PAUP* v. 4.0a146 (Swofford 2003). For each data set, phylogenetic model selection was conducted using PAUP* for 64 models of nucleotide substitution. A models fit was evaluated using Akaike Information Criterion (AIC) values. The results of this analysis indicated the GTR+I+G was the most appropriate model of molecular evolution for each of the three data sets. Maximum likelihood searches were conducted using heuristic search methods with tree bisection reconnection (TBgR) branch swapping, collapse of zero-length branches, and all characters weighted equally. The analyses were repeated 100 times with the RANDOM ADDITION option. Sets of equally most parsimonious trees were summarized with a strict consensus tree. Bootstrap tests (Felsenstein 1985) were performed using 300 replicates with heuristic search settings identical to those of the original search.
Coevolution complexity analysis of Pucciniales host associations
Reciprocal pathogen and host datasets were analyzed using reconciliation analysis to compare the coevolutionary signal between the aecial and telial stages of the Pucciniales species separately. Reconciliation analyses were performed using Jane 4 (Conow et al. 2010). Both Pucciniales and host phylogenies were reproduced using Jane's tree editor. Then, for a given host stage, links were drawn from a Pucciniales taxon to each of its hosts in the host phylogeny. In the ca. 20 % of taxa where a specific host taxon was not available in the host phylogeny, the link between the Pucciniales taxon and host that best approximates the missing host's lineage was used instead as previously detailed.
Jane was run using the default costs (in steps) for each of the evolutionary events required for reconciliation: cospeciation = 0; duplication = 1; host switch = 2; loss = 1; failure to diverge = 1. The total number of steps required to reconcile the phylogenies given the aecial and telial host associations were compared and the stage requiring the fewest number of steps was determined to be more parsimonious and thus under greater evolutionary influence by the host phylogeny.
The models created for reconciliation analysis of each dataset include: 1) the liberal model, which uses all available host-pathogen associations possible for analyses of the aecial and telial stages; and 2) the conservative model, which attempts to resolve bias in the parsimony-based reconciliation analysis by removing Pucciniales taxa with only one known host between the two life stages. These taxa would increase the number of steps needed to reconcile a particular life stage, thus their removal would provide less biased analysis from a parsimony standpoint, but it also reduces the representation of Pucciniales diversity and expected coevolutionary complexity that comes with it.
Results
Pucciniales Datasets – The size of the datasets and the size range of sequences generated for this study are available at the end of each table in Supplementary Information S1. The results of maximum likelihood analysis and ML bootstrap results for the multigene Pucciniales, Melampsorineae, and Pucciniaceae datasets are available in Supplementary Information S2-Figs 2–4.
Divergence time estimation
The tMRCA results for nodes in the Pucciniales, Melampsorineae, and the Pucciniaceae are supplied in Fig. 2, Fig. 3, Fig. 4. The tMRCA results for estimated mean and median ages, along with their 95 % highest posterior density (HPD) for each dataset analyzed, are supplied in Supplementary Information S3.
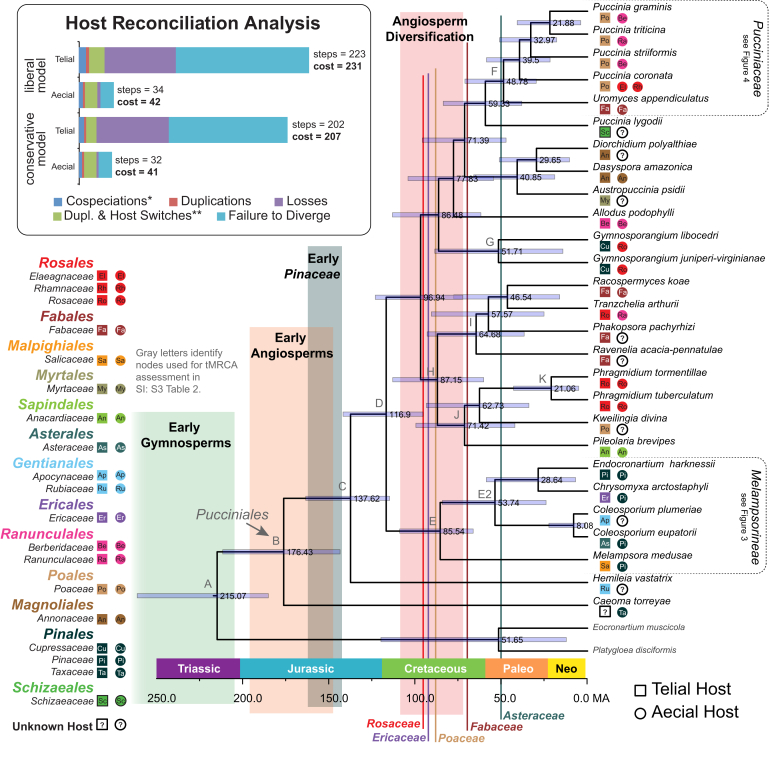
Fig. 2.
The Pucciniales. Letters identify the same nodes found in Fig. 3, Fig. 4. Numbers adjacent to nodes represent median ages in millions of years. Blue bars represent 95 % highest posterior density around the nodes. Taxa are labelled with their corresponding aecial (circles) and telial (squares) host families. The Host Reconciliation Analysis illustrates the steps involved in reconciling host and rust phylogenies. Each step is a different coevolutionary event (e.g. cospeciation, duplications, losses, etc.) that can involve different costs. In host reconciliation analysis, all evolutionary events are given a cost of 1, except: * = cospeciation is given a cost of 0; and ** = duplication and host switching is given a cost of 2. See text and Supplementary Information Table S5 for details.
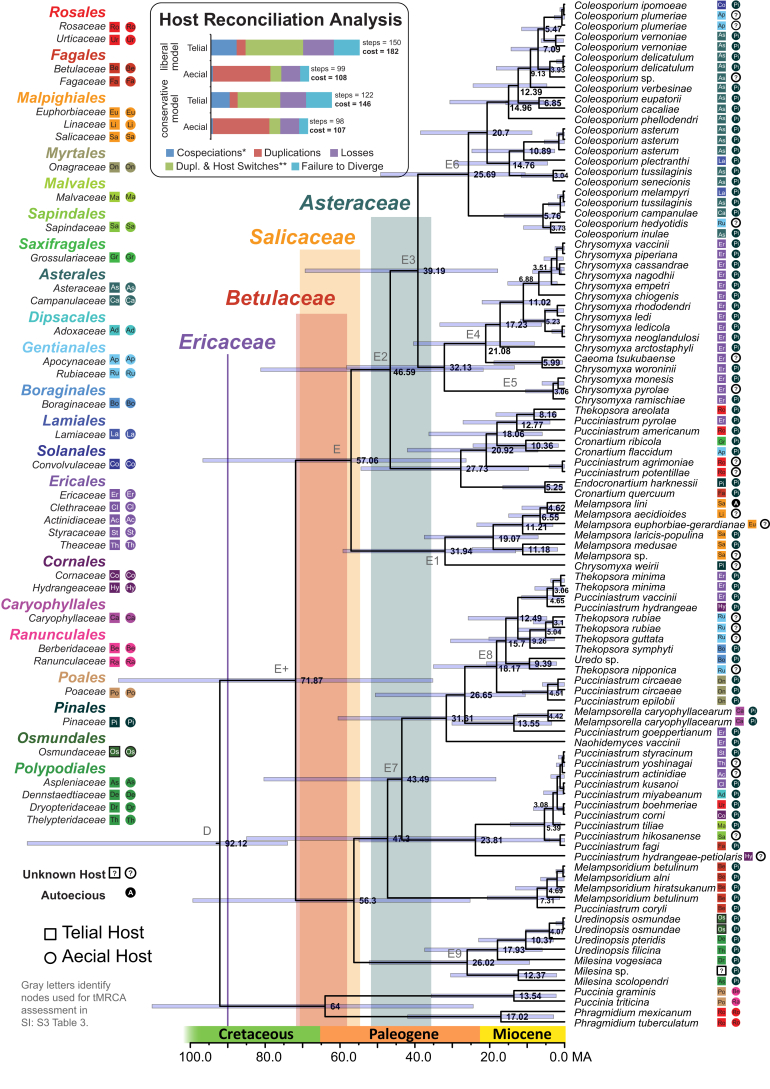
Fig. 3.
The Melampsorineae. Letters identify the same nodes found in Fig. 2, Fig. 4. All other details are the same as those described in the caption for Fig. 2.
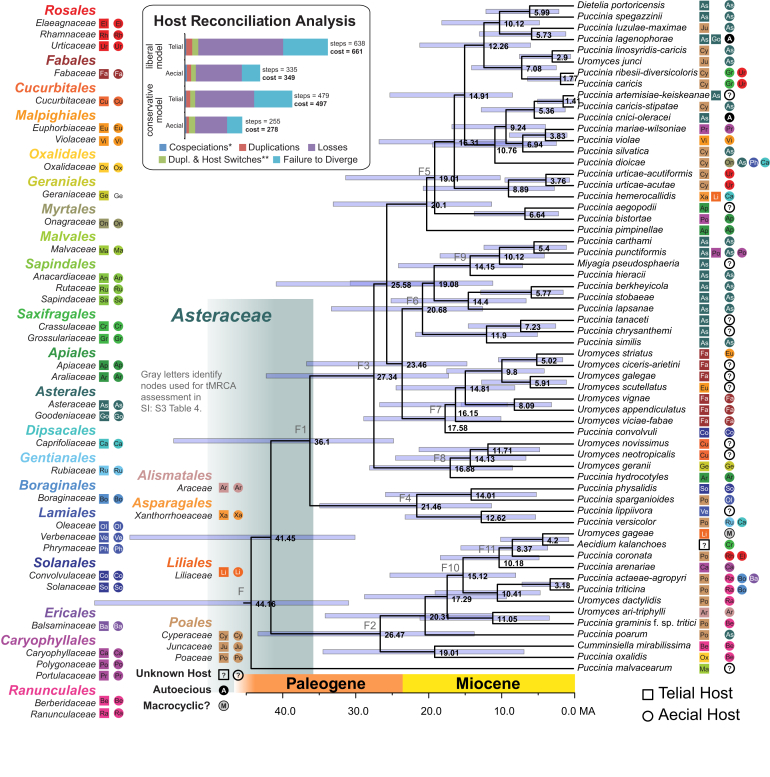
Fig. 4.
The Pucciniaceae. Letters identify the same nodes found in Fig. 2, Fig. 3. All other details are the same as those described in the caption for Fig. 2.
The ages from the Dikarya phylogenetic analysis were estimated from six independent BEAST runs that were combined to produce a consensus tree from 70 000 Bayesian posterior sampled trees (Supplementary Information S2-Fig. 1). Each of the tMRCA results are generally more recent than the results of Floudas et al. (2012), but, the relative median, and mean estimates fall within/overlap the HPD range of that study (Supplementary Information S3-Table 1). This provided confidence in using the estimated ages for nodes A–F (Supplementary Information S2-Fig. 1) for calibration in the Pucciniales, Melampsorineae, and Pucciniaceae tMRCA analyses.
The age estimates for the Pucciniales analysis are the result of three independent BEAST runs, combined to produce a consensus tree from 27 000 trees (Fig. 2). The tMRCA for the Pucciniomycetes (node A) is estimated at 238 million years ago (Ma) (167–316 HPD) in the Dikarya analysis (Supplementary Information S2-Fig. 1) compared to 215 Ma (185–261 HPD) in the Pucciniales analysis (Fig. 2). The Pucciniales tMRCA (node B) is estimated at 175 Ma (122–233 HPD) in the Dikarya analysis and 176 Ma (143–211 HPD) in the Pucciniales analysis. Median estimates for tMRCAs of calibrated nodes tend to be slightly older in the Pucciniales analysis relative to the estimates from the Dikarya analysis. This somewhat offsets the more recent estimated age results for the Dikarya tMRCAs in this study compared to those of Floudas et al. (2012).
The Melampsorineae has a tMRCA median age estimate of 91 Ma (57–131 HPD) in the Dikarya analysis and 85 Ma (66–109 HPD) in the Pucciniales analysis (Supplementary Information S2-Fig. 1; Fig. 2). However, in the analysis of the Melampsorineae dataset the median age for the corresponding node E is 57 Ma (23–97 HPD) (Fig. 3). The discrepancy in the Melampsorineae analyses in tMRCA age estimates for node E potentially stems from the fact that representatives of the Pucciniastraceae were only included in the Melampsorineae (represented as node E+ in Fig. 3) and not the Dikarya or Pucciniales datasets. The large number of taxa that lie between nodes D and E in the Melampsorineae dataset, along with the limited availability of sequence character data used in age estimations, are likely to have skewed this particular result toward a younger estimate than that produced in the Dikarya and Pucciniales analyses.
The Pucciniaceae dataset has a tMRCA median age estimate for node F of 44 Ma (31–66 HPD) (Fig. 4; Supplementary information S3-Table 4). These estimations are consistent with those for node of the Dikarya (45 Ma, 25–73 HPD) and Pucciniales (48 Ma, 30–71 HPD) (Supplementary Information S3-Tables 1, 2; Fig. 2).
Pucciniales Hosts and Host Datasets
A list of hosts for the Pucciniales taxa in the Pucciniales, Melampsorineae, and Pucciniaceae datasets are provided in Supplementary Information S4. The taxa in the Pucciniales dataset associate with plant hosts from 134 different genera representing 31 families/subfamilies. Rust fungi in the Melampsorineae dataset are known to associate with up to 89 host genera from 44 families/subfamilies. Taxa of the Pucciniaceae dataset infect hosts from at least 215 host genera representing 46 families/subfamilies. The sequence data used in the host datasets are provided in Supplementary Information S7.
Coevolutionary complexity analysis of Pucciniales host associations
The size and complexity of Pucciniales and host datasets required that Jane program files for reconciliation analysis had to be done manually. Also, Jane would crash when reading the tree files into the tree editor; as a result, the use of time boundaries was not practical, or potentially possible for analysis. Nonetheless, Jane files for both liberal and conservative models for aecial or telial stage host associations were created for reconciliation analysis (Supplementary Information S6).
Pucciniales reconciliation analysis under the liberal model invoked a cost of 42 under the aecial stage, and 231 under the telial stage (Table 1; inset of Fig. 2). In other words, the stage requiring the fewest number of steps to reconcile with host phylogeny and therefore appearing to be under the greatest evolutionary influence by host is the aecial stage. These results were similar to the conservative model with an aecial stage reconciliation cost of 41, and a telial stage cost of 207. Reconciliation in the Melampsorineae under the liberal model invoked a cost of 108 under the aecial stage, and 182 under the telial stage (Table 1; inset of Fig. 3). Under the conservative model the reconciliation cost in the aecial stage was 107, and the telial stage was 146. Lastly, reconciliation in the Pucciniaceae under the liberal model invoked a cost of 349 for the aecial stage, and a cost of 661 for the telial stage (Table 1; inset of Fig. 4). Under the conservative model reconciliation in the aecial stage had a cost of 278, and the telial stage had a cost of 497. Thus, in all analyses, evidence for codiversification between rust fungi and their aecial hosts was significantly stronger than between the same fungi and their telial hosts, although this pattern was not as strong in the rapidly radiating Pucciniaceae.
Table 1.
Results of reconciliation analysis between Pucciniales datasets and their host phylogenies for aecial and telial host stages.
| Stage | Cost1 |
Total Steps | Cospeciations |
Duplications |
Dupl & host switches |
Losses |
Failure to diverge |
|
|---|---|---|---|---|---|---|---|---|
| =∑ (penalty x steps) | Penalty = 0 | Penalty = 1 (0)1 |
Penalty = 2 | Penalty = 1 | Penalty = 1 | |||
| Pucciniales | Steps | Steps | Steps | Steps | Steps | |||
| Liberal model | Telial | 231 (228) | 223 | 7 | 3 | 15 | 69 | 129 |
| Aecial | 42 (40) | 34 | 4 | 2 | 12 | 3 | 13 | |
| Conservative model | Telial | 207 (205) | 202 | 5 | 2 | 10 | 70 | 115 |
| Aecial | 41 (39) | 32 | 3 | 2 | 12 | 2 | 13 | |
| Melampsorineae | ||||||||
| Liberal model | Telial | 182 (173) | 150 | 26 | 9 | 58 | 31 | 26 |
| Aecial | 108 (50) | 99 | 2 | 58 | 11 | 19 | 9 | |
| Conservative model | Telial | 146 (138) | 122 | 19 | 8 | 43 | 26 | 26 |
| Aecial | 107 (50) | 98 | 2 | 57 | 11 | 19 | 9 | |
| Pucciniaceae | ||||||||
| Liberal model | Telial | 661 (634) | 638 | 4 | 27 | 27 | 380 | 200 |
| Aecial | 349 (330) | 335 | 7 | 19 | 21 | 206 | 82 | |
| Conservative model | Telial | 497 (479) | 479 | 4 | 18 | 22 | 264 | 171 |
| Aecial | 278 (267) | 255 | 5 | 11 | 28 | 143 | 68 | |
Numbers in bold indicate the costs for each model with the penalties for each step as indicated; number in parentheses are results from separate analysis applying a ‘0’ penalty to Duplication events.
Discussion
The expansive phylogenetic sampling of Pucciniales taxa and sequence data presented in this study allow a comprehensive estimate of diversification and timing in the evolution of the rust fungi. The tMRCA estimates for the Pucciniales in Fig. 2 are consistent with those from the Dikarya dataset from this and other studies, but are older than the recent Pucciniales age estimates (McTaggart et al. 2016), which are around 60 Ma younger (ca. 113–115 Ma) than the estimates of this study (see Supplementary Information S3). This is most likely due to differences in how the phylogenies were calibrated. McTaggart et al. (2016) used internal calibrations based on host ages. This study uses external calibration points based on fungal fossils to estimate the age of the Pucciniales (Supplementary Information S2). While further study is needed to understand the effect of calibration choices on estimating ages in Fungi, the results of the current study corroborate well with those of genomic studies in the Basidiomycota (Floudas et al. 2012), as well as the diversification times of major host lineages.
The Pucciniales split from their closest relatives around 215–230 million years ago (Fig. 2, Node A). This is well after the earliest seed plants (313–343 Ma HPD) but before the earliest pines, which arose between 150–160 Ma (Magallón et al. 2013). The MRCA to all extant Pucciniales diversified around 176 Ma (HPD 143.7–211.8 Ma), which comes slightly after the earliest gymnosperms, but coincides with the earliest angiosperms (Fig. 2, Node B).
Several of the oldest diverging lineages are associated with gymnosperms. For example, the earliest diverging extant Pucciniales species to be sequenced is Caeoma torreyae, which forms aecia on Torreya californica. Members of the Melampsorineae form aecia almost exclusively on pinaceous hosts. Another early diverging taxon, Hemileia vastatrix (Mikronegeriaceae), is an economically important pathogen that forms uredinia on Coffea spp. While their aecial host is unknown, a hypothesis that the aecial host for species of Hemileia are from an early diverging gymnosperm is consistent with what is seen in other Mikronegeriaceae (Aime 2006) and C. torreyae.
The results of molecular dating suggest that the diversification of the Pucciniales and the angiosperms is correlated. The core Pucciniales split between 95–140 Ma (Fig. 2, Node D). This spans the late Jurassic and early Cretaceous, just before the estimated diversification of major angiosperm lineages (Magallón & Castillo 2009). Codiversification with angiosperms has been observed many times with insects as pollinators (Grimaldi 1999), herbivores (Farrell, 1998, Currano et al., 2008), or symbiotic generalists (Moreau et al. 2006).
Several groups of fungi are plant-associated. Mycorrhizal fungi are associated with the roots of plants as symbiotic mutualists. However, these fungi consist of various different forms that are either not nearly as diverse as their plant hosts [e.g. the Glomeromycota (Schüβler et al. 2001)], or have evolved multiple independent times producing a diffuse pattern that does not coincide with the diversification of their hosts (Ryberg & Matheny 2012). In contrast, the Pucciniales are exceptionally diverse compared to other groups of plant pathogens, and because individual Pucciniales species can infect more than one host, the diversity of plant species that they infect is even greater.
How the Pucciniales achieved the diversity observed in the order despite having such complex lifecycles is a question that many mycologists have pondered. One part of this question deals with determining which aspects of their life history strategy function as being restrictive, or innovative, in regards to diversification. The alternating stages of this life history (aecial and telial) are believed to be the drivers of either biological specialization (BSp) or biogenic radiation (BgR) in the Pucciniales (Leppik, 1965, Leppik, 1967). This begs the question of whether selective pressures act equally across all stages of the rust life cycle or whether these pressures are in disequilibrium. Our hypothesis for this study is that the aecial host is under the strongest selective pressure for conserving host associations, and should demonstrate the greatest coevolutionary signal, which we interpret as a function of BSp.
Reconciliation analysis shows that reconciling the Pucciniales phylogeny to the telial host phylogeny requires more than five times as many steps as reconciling with the aecial host phylogeny (Table 1; inset for Fig. 2). This demonstrates the importance of the aecial stage in shaping the codiversification between the Pucciniales and their hosts. This pattern is likely the result of specialization of a Pucciniales taxon on a plant host. As a result this shows how the aecial stage follows the BSp model described by Leppik. However, the relative difference in the cost of reconciliation between the aecial and telial stages is not consistent at different time scales (Table 1). The relatively large number of observed host jumps in the telial stage of the Melampsorineae potentially reflects the role of BgR in this lineage (Fig. 3). Host reconciliation in the Pucciniaceae, the youngest lineage tested, shows the differences between aecial and telial hosts could be more strongly dictated by lineage loss or the failure to diversify with its host (Fig. 4).
How natural selection favours shaping the Pucciniales phylogeny to more closely mirror their aecial hosts can be explained in a couple of ways: 1) As illustrated in Fig. 1, the spermogonial stage is that where compatible gametes recombine to initiate the dikaryotic part of the lifecycle. However, spermatia are small and short-lived, constraining the process of fertilization both temporally and spatially; 2) The formation of the dikaryon in the aecial host represents the second process that likely facilitates BgR. Since dikaryotic cells are the result of fertilization, newly combined alleles could theoretically provide an enhanced capacity for fungal dikaryons to infect new telial hosts directly via the aeciospores or during the repetitive cycling of urediniospores (green arrows in Fig. 1). Over greater evolutionary time, these two factors play a more significant role in etching out the pattern of codiversification in the pathogen-host phylogenies. However, in more recent lineages, the relative differences between aecial and telial host reconciliation is less significant suggesting that the natural selective forces of BSp and BgR have not had enough time to influence rust systematic diversity in these.
The hologenetic ladder (HL) was hypothesized by Leppik (1953) as a mechanism for the Pucciniales to use its alternating life stages as rungs on a ladder in order to codiversify with plant hosts. However, this is not clearly evident from our results. In the Melampsorineae, we can see Leppik's argument reflected in the phylogeny with taxa forming telia on ferns (Fig. 3, Node E0), to aecia on Pinaceae (Node E+), to telia on Salicaceae (Node E1). However, following telia on Salicaceae, there is a problem of a “missing step” because the HL predicts the next step as aecia on angiosperms but there is no evidence of this in the Melampsorineae (excepting autoecious species; Fig. 3). In addition, it is difficult to perceive how the “steps” of the HL would work by alternating aecial and telial hosts given the current understanding of the aecial stage's role in BSp and telial stage's role in BgR on the Pucciniales as a whole.
In the reconciliation analysis, more recent Pucciniales lineages (inset Fig. 3, Fig. 4) have a reduced relative difference in cost between the aecial and telial stages relative to the broader Pucciniales (inset Fig. 2). This illustrates a narrowing of the relative impact between BgR and BSp in younger or rapidly diversifying lineages of rust fungi. Here the HL could be involved in shaping the Pucciniaceae as the aecial and telial stages are more similar in respect to their ability to radiate to new hosts. As new rust lineages mature over time, the difference between BgR and BSp will grow resulting in clearer differences between aecial and telial reconciliation patters, as seen in the Pucciniales. Whether this current hypothesis—the influence of the HL in shaping younger Pucciniales lineages but having a lesser impact on the order as a whole—accurately describes what is being observed in the evolution of the Pucciniaceae requires more detailed investigation.
Conclusions
The Pucciniales represents the most diverse, environmentally and economically important group of plant pathogenic fungi. The fact that they also have the most complex life histories in fungi that include an alternation of generations with concomitant host specificity for each, has made interpretation of their evolution difficult. The evolution of the Pucciniales reflects that of their hosts in a number of facets. The age of early diversifying lineages coincides with the ages of coniferous hosts, while the majority of extant Pucciniales diversity co-diversified with that of their angiosperm hosts during the Cretaceous period. Reconciliation analyses demonstrate support for the hypothesis that the aecial stage is under more selective pressure than the telial stage, and shows stronger evidence of codiversification with hosts than the latter. This corresponds to the relative importance of BSp acting on this stage of the life cycle and suggests co-evolution could be at responsible for the patterns seen between rust fungi and their aecial hosts. In contrast, host reconciliation analyses suggest markedly less constraints acting on the telial host stage, where BgR is likely to play a greater role in speciation.
Acknowledgements
The authors are grateful to Dr. Merje Toome-Heller for permission to adapt one of her illustrations for Fig. 1. We are also grateful to numerous graduate students, post docs, and technicians who have assisted with the generation of sequence data and accessioning of rust collections in the Aime Lab through the years. Funding for this research was awarded to MCA from the following sources: NSF DEB-0732968 to D. Hibbett and MCA; USDA DEB-1458290 to MCA; and USDA Hatch 1010662.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.simyco.2018.02.002.
Contributor Information
M.C. Aime, Email: maime@purdue.edu.
A.W. Wilson, Email: andrew.wilson@botanicgardens.org.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Aime M.C. Toward resolving family-level relationships in rust fungi (Uredinales) Mycoscience. 2006;47:112–122. [Google Scholar]
- Aime M.C., Matheny P.B., Henk D.A. An overview of the higher-level classification of Pucciniomycotina based on analyses of nuclear large and small subunit rDNA sequences. Mycologia. 2006;98:896–905. doi: 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- Aime M.C., McTaggart A.R., Mondo S.J. Phylogenetics and phylogenomics of rust fungi. Advances in Genetics. 2017;100:267–307. doi: 10.1016/bs.adgen.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Bauer R., Bergerow D., Sampaio J.P. The simple-septate basidiomycetes: a synopsis. Mycological Progress. 2006;5:41–66. [Google Scholar]
- Beenken L. Austropuccinia: a new genus name for the myrtle rust Puccinia psidii placed within the redefined family Sphaerophragmiaceae (Pucciniales) Phytotaxa. 2017;297:53–61. [Google Scholar]
- Beenken L., Zoller S., Berndt R. Rust fungi on Annonaceae II: the genus Dasyspora Berk. & M.A. Curtis. Mycologia. 2012;104:659–681. doi: 10.3852/11-068. [DOI] [PubMed] [Google Scholar]
- Conow C., Fielder D., Ovadia Y. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms for Molecular Biology. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currano E.D., Wilf P., Wing S.L. Sharply increased insect herbivory during the Paleocene–Eocene thermal maximum. Proceedings of National Academy of Sciences. 2008;105:1960–1964. doi: 10.1073/pnas.0708646105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.J., Castlebury L.A., Aime M.C. Phylogenetic relationships of sugarcane rust fungi. Mycological Progress. 2010;9:459–468. [Google Scholar]
- Drummond A.J., Ho S., Phillips M. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell B.D. “Inordinate Fondness” explained: why are there so many beetles? Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- Feau N., Vialle A., Mathieu A. DNA barcoding in the rust genus Chrysomyxa and its implications for the phylogeny of the genus. Mycologia. 2011;103:1250–1266. doi: 10.3852/10-426. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cofidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Flor H.H. The complementary genetic systems in flax rust. Advances in Genetics. 1956;8:29–59. [Google Scholar]
- Floudas D., Binder M., Riley R. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- Grimaldi D. The co-radiations of pollinating insects and angiosperms in the Cretaceous. Annals of the Missouri Botanical Garden. 1999;86:373–406. [Google Scholar]
- Hibbett D.S., Grimaldi D., Donoghue M.J. Fossil mushrooms from Cretaceous and Miocene ambers and the evolution of homobasidiomycetes. American Journal of Botany. 1997;84:981–991. [PubMed] [Google Scholar]
- LePage B.A., Currah R.S., Stockey R.A. Fossil ectomycorrhizae from the Middle Eocene. American Journal of Botany. 1997;84 470–412. [PubMed] [Google Scholar]
- Leppik E.E. Some viewpoints on the phylogeny of the rust fungi. I. Conifer rusts. Mycologia. 1953;45:46–74. [Google Scholar]
- Leppik E.E. Some viewpoints on the phylogeny of rust fungi. V. Evolution of biological specialization. Mycologia. 1965;57:6–22. [Google Scholar]
- Leppik E.E. Some viewpoints on the phylogeny of rust fungi. VI. Biogenic radiation. Mycologia. 1967;54:568–579. [Google Scholar]
- Maddison W.P., Maddison D.R. 2011. Mesquite: a modular system for evolutionary analysis, Version 2.75.http://mesquiteproject.org [Google Scholar]
- Magallón S., Castillo A. Angiosperm diversification through time. American Journal of Botany. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Magallón S., Hilu K.W., Quandt D. Land plant evolutionary timeline: gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. American Journal of Botany. 2013;100:556–573. doi: 10.3732/ajb.1200416. [DOI] [PubMed] [Google Scholar]
- Maier W., Begerow D., Weiß M. Phylogeny of the rust fungi: an approach using nuclear large subunit ribosomal DNA sequences. Canadian Journal of Botany. 2003;81:12–23. [Google Scholar]
- Matheny B.P., Aime M.C., Bougher N. Out of the Paleotropics? Historical biogeography and diversification of the cosmopolitan ectomycorrhizal mushroom family Inocybaceae. Journal of Biogeography. 2009;36:569–799. [Google Scholar]
- McTaggart A.R., Shivas R.G., Nest M.A. Host jumps shaped the diversity of extant rust fungi (Pucciniales) New Phytologist. 2016;209:1149–1158. doi: 10.1111/nph.13686. [DOI] [PubMed] [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA: 1–8. [Google Scholar]
- Minnis A.M., McTaggart A., Rossman A. Taxonomy of mayapple rust: the genus Allodus resurrected. Mycologia. 2012;104:942–950. doi: 10.3852/11-350. [DOI] [PubMed] [Google Scholar]
- Moreau C.S., Bell C.D., Vila R. Phylogeny of the ants: diversification in the age of Angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- Renner S.S. Relaxed molecular clocks for dating historical plant dispersal events. Trends in Plant Science. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Ryberg M., Matheny P.B. Asynchronous origins of ectomycorrhizal clades of Agaricales. Proceedings of the Royal Society B. 2012;279:2003–2011. doi: 10.1098/rspb.2011.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüβler A., Schwarzott D., Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution** Dedicated to Manfred Kluge (Technische Universität Darmstadt) on the occasion of his retirement. Mycological Research. 2001;105:1413–1421. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, Massachusetts: 2003. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods), version 4 edn. [Google Scholar]
- Taylor T.N., Hass H., Kerp H. Perithecial ascomycetes from the 400 million year old Rhynie chert: an example of ancestral polymorphism. Mycologia. 2005;97:269–285. [PubMed] [Google Scholar]
- Toome M. Estoniana University of Life Sciences; 2010. Leaf rust (Melampsora) on willows: ecological and plant response studies. Ph.D. thesis. 123 pp. [Google Scholar]
- van der Merwe M.M., Walker J., Ericson L. Coevolution with higher taxonomic host groups within the Puccinia/Uromyces rust lineage obscured by host jumps. Mycological Research. 2008;112:1387–1408. doi: 10.1016/j.mycres.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T.D., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phyogenies. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- Wilson A.W., Binder M., Hibbett D.S. Diversity and evolution of ectomycorrhizal host associations in the Sclerodermatineae (Boletales, Basidiomycota) New Phytologist. 2012;194:1079–1095. doi: 10.1111/j.1469-8137.2012.04109.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.