Abstract
Accumulating evidence indicates that Checkpoint kinase 1 (CHEK1) plays an essential role in tumor cells and that it could induce cell proliferation and could be related to prognosis in multiple types of cancer. However, the biological role and molecular mechanism of CHEK1 in GBM still remain unclear. In this study, we identified that CHEK1 expression was enriched in glioblastoma (GBM) tumors and was functionally required for tumor proliferation and that its expression was associated to poor prognosis in GBM patients. Mechanically, CHEK1 induced radio resistance in GBM cells, and CHEK1 knockdown increased cell apoptosis when combined with radiotherapy via regulation of the DNA repair/recombination protein 54L (RAD54L) expression. Therapeutically, we found that CHEK1 inhibitor attenuated tumor growth both in vitro and in vivo. Collectively, CHEK1 promotes proliferation, induces radio resistance in GBM, and could become a potential therapeutic target for GBM.
Introduction
Glioblastoma (GBM) is one of the most frequently diagnosed and lethal of primary brain tumors [1]. Despite the maximal therapies including surgical resection followed by radiation and chemotherapy, the median survival of GBM patients is still only 14.6 months [2]. Recent studies indicate that therapeutic benefit of irradiation and TMZ treatments is only transient due in most part to the resistance mechanisms elicited by GBM [2], [3]. A large range of kinases has been proved to promote radio resistance and proliferation, thus inducing recurrence of GBM cells [4], [5], [6]. Novel therapeutic approaches that could target core oncogenic pathways and/or pathways that confer treatment resistance to tumor cells are urgently needed.
DNA double-strand breaks (DSB) are the major mechanisms which induce cell apoptosis and inhibit cell proliferation in GBM after irradiation [7]. However, accumulating data suggested that multiple pathways which induce DNA damage repair could be activated after tumor cells received irradiation and protect cells from apoptosis and cell arrest [8]. The DNA repair/recombination protein (RAD54) family of DNA translocase proteins functions in concert with RAD51 to promote recombinational [9]. RAD54 translocation has also been proposed to act following homology recognition as a “heteroduplex pump” to incorporate the invading ssDNA into the D-loop while simultaneously removing RAD51 during the generation of the heteroduplex product [10]. Mason et al. [11] proved that RAD54L and RAD54B deficiency or expression of a dominant-negative RAD54 allele displays elevated levels of RAD51 foci on DNA repair. Thus, RAD54 appears to contribute to DNA repair by stabilizing association of RAD51 with ssDNA prior to RAD51-mediated strand exchange and then disassembling RAD51 from the dsDNA exchange product [12], [13]. However, the mechanism of transcriptional regulation of RAD54L still remains unclear.
Checkpoint kinase 1 (CHEK1) is an serine/threonine-specific protein kinase that coordinates the DNA damage response and cell cycle checkpoint response [14]. Activation of CHEK1 results in the initiation of cell cycle checkpoints, cell cycle arrest, DNA repair, and cell death to prevent damaged cells from progressing through the cell cycle [15]. Activated CHEK1 in turn phosphorylates a number of downstream effectors to trigger a pleiotropic cellular response including transcription regulation, energy consumption alteration, cell-cycle arrest or delay, DNA repair, or cell death if the damage is too severe to repair [15]. Yang et al. reported that inhibition of CHEK1 reduced cell proliferation of GBM cells. Despite these findings, more detailed studies need to be done to clarify the functional role and downstream targets of CHEK1 in GBM cells.
In this study, we identified CHEK1 as the most upregulated kinase encoding genes in GBM and that it is functionally required for tumor growth, tumorigenesis, and infiltration of GBM. Moreover, CHEK1-dependent transcriptional regulation of RAD54L is critical in DNA damage repair after radiotherapy, thus promoting radioresistance and tumor recurrence. Furthermore, we found that inhibition of CHEK1 via shRNA or inhibitor could suppress tumor growth and enhance radiosensitivity of GBM cells both in vitro and in vivo. Altogether, CHEK1 is a key regulator of tumor growth and radio resistance and could be a potential therapeutic target for GBM.
Materials and Methods
Ethics
Usage of experimental animals and cell lines (nude mice) in this study was approved by the Scientific Ethics Committee of Xi'an Jiaotong University, Xi'an, China.
Reagents and Antibodies
The following reagents and primary antibodies are used in this study: DMEM-F12 (Gibco, 10565-018), fetal bovine serum (Gibco, 10082-147), Accutase solution (Sigma, A6964-100), Alamar Blue (Invitrogen, DAL1100), RIPA buffer (Sigma, R0278), phosphatase inhibitor cocktail (Sigma, P0044), protease inhibitor cocktail (P8340), Bradford (BIORAD, 500-0006), BSA used in Bradford assay (BioLabs, B9001S), PageRuler plus prestained protein (Thermo scientific, 26619), iScript Reverse Transcription supermix for qRT-PCR (Bio-rad, 170-8841), shCHEK1 lentivirus particles (pGFP-C-shLenti, Origene, TR320302), CHIR-124 (Selleckchem, S2683), anti-CHEK1 (Novus Biologicals, NB100-464, rabbit, used for WB and IHC), anti-RAD54L (Novus Biologicals, NBP2-33916, rabbit, used for WB), and β-Actin (Sigma, A5316, mouse, used for WB).
In Vitro Cell Cultures
Glioblastoma cell lines U87, U373, U251, and U138 and normal human astrocytes (NHA) are provided by Xi'an Jiaotong University. Cell lines are cultured in DMEM/F12 medium containing 10% FBS supplement (vol%). The culture medium is replaced every 5 to 10 days.
RNA Isolation and Quantitative Real-Time PCR
RNA is isolated by using RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. RNA concentration is determined using a Nanodrop 2000 (Thermo Scientific). cDNA is synthesized by using iScript reverse transcription supermix for qRT-PCR (Bio-rad) according to the manufacturer's protocol. The reverse-transcribed cDNA is analyzed by quantitative RT-PCR (qRT-PCR), and 18S is used as an internal control. Each qRT-PCR includes a 10-μl reaction mixture per well that includes 2.5 μl cDNA, 0.5 μl forward primer (0.5 μM), 0.5 μl reverse primer (0.5 μM), 1.5 μl of DNase/RNase-free distilled water, and 5 μl SYBR green reagent (QIAGEN). The following cycles are performed during DNA amplification: 94°C for 2 minutes, 40 cycles of 94°C (30 seconds), 60°C (30 seconds), and 72°C (40 seconds). 18S is used as an internal control. The primer sequences are showed below: CHEK1 forward: TCATCCATTTCTAACAAATTCACTT, CHEK1 reverse: TGGGCTATCAATGGAAGAAAA, RAD54L forward: GAGCCCAGAGGACCTTGATA, RAD54L reverse: AACCACCTTGTCTGGACAGC, 18S forward: GGCCCTGTAATTGGAATGAGTC and 18S reverse: CCAAGATCCAACTACGAGCTT.
Western Blotting
The cell lysates are prepared in RIPA buffer containing 1% protease and 1% phosphatase inhibitor cocktail (Sigma Aldrich) on ice. The sample protein concentrations are determined by the Bradford method. Equal amounts of protein lysates (10 μg/lane) are fractionated on NuPAGE Novex 4%-12% Bis-Tris Protein gel (Invitrogen) and transferred to a PVDF membrane (Invitrogen). Subsequently, the membranes are blocked with 5% skimmed milk for 1 hours and then treated with the relevant antibody at 4°C overnight. Protein expression is visualized with Amersham ECL Western Blot System (GE Healthcare Life Sciences). β-Actin serves as a loading control.
In Vivo Intracranial Xenograft Tumor Models
Six-week-old nude mice are used for GBM intracranial implantation. All animal experiments are carried out in Xi'an Jiaotong University. The GBM suspension (1 × 105 cells in 5 μl of PBS) transduced with nontarget or shCHEK1 lentivirus is injected into the brains of nude mice after anesthesia. At least six mice are used for each group. Drug treatment is done through tail vein injection and starts from 5 days after tumor cells are implanted. Mice were monitored once a day for symptoms related to tumor growth including an arched back, unsteady gait, paralysis of legs, and body weight loss. Mice were euthanized by overdose anesthesia of ketamine and xylazine after a total body weight loss of 40% or severe symptoms were observed.
Statistical Analysis
All the data are presented as mean ± SD. The number of replicates for each experiment is stated in the figure legend. Statistical differences between the two groups are evaluated by two-tailed t test. The comparison among multiple groups is performed by one-way analysis of variance followed by Dunnett's posttest. The statistical significance of Kaplan-Meier survival plot is determined by log-rank analysis. Statistical analysis is performed by Prism 6 (GraphPad Prism) unless mentioned otherwise in the figure legend. P < .05 is considered as statistically significant.
Results
CHEK1 Expression Was Highly Enriched in GBM Cells
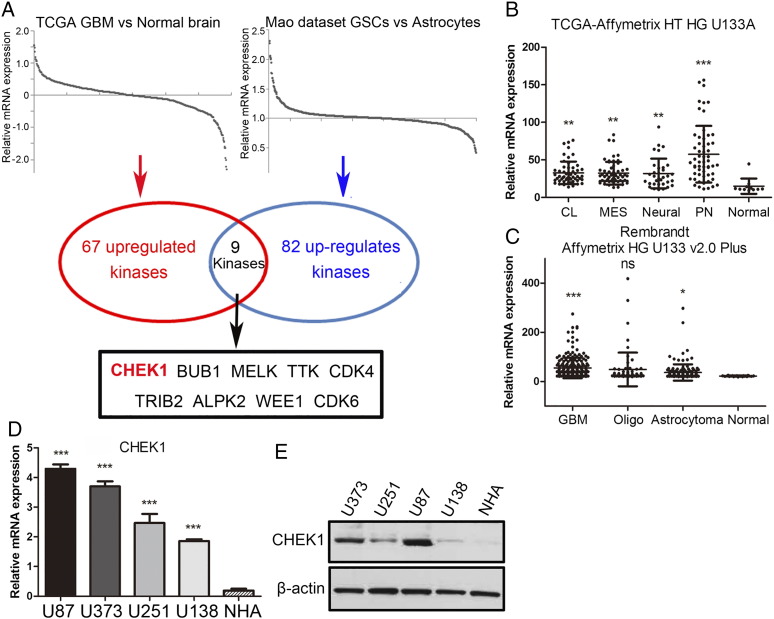
In this study, we first sought to characterize the role of CHEK1 in GBM tumors. To this end, we analyzed the data from microarray database published in 2013 [16] and TCGA dataset. We find that CHEK1 was one of the most upregulated kinase-encoding genes in both datasets when comparing GBM samples to normal human astrocyte cells (Figure 1A). Additionally, we found that CHEK1 expression was significantly enriched in all the four subtypes of GBM according to the TCGA database (classical, mesenchymal, neural, and proneural) compared with nontumor tissue (Figure 1B). Moreover, we analyzed Rembrandt dataset and identified that CHEK1 expression was significantly increased in GBM when compared to low-grade glioma and normal brain tissue (Figure 1C). To confirm this, qRT-PCR using four GBM cell lines (U87, U373, U251 and U138) and NHA was performed, and the result exhibited higher expression of CHEK1 mRNA in GBM (Figure 1D). Similar to the mRNA expression, Western blotting data demonstrated the same result (Figure 1E). Taken together, these data indicated that CHEK1 was preferentially expressed in GBM.
Figure 1.
CHEK1 expression was highly enriched in GBM cells.
(A) Genome-wide transcriptome microarray analysis from TCGA dataset and Mao's dataset shows that CHEK1 is one of the most upregulated kinase encoding genes in GBM samples compared to normal tissue or astrocytes. (B) Analysis of TCGA database indicated that CHEK1 was highly expressed in all the four subgroups of GBM (classical, mesenchymal, neural, and proneural) compared with nontumor tissue. (C) Analysis of Rembrandt database indicated that CHEK1 was highly expressed in GBM when compared to oligodendroglioma, astrotoma, and nontumor tissue. (D) qRT-PCR analysis showed CHEK1 mRNA expression was elevated in four GBM cell lines (U87, U373, U251, and U138) compared with normal human astrocytes (P < .01). (E) Western blotting analysis indicated that CHEK1 protein expression was enriched in GBM tissue compared with normal brain from the paired patient samples. β-Actin served as a control.
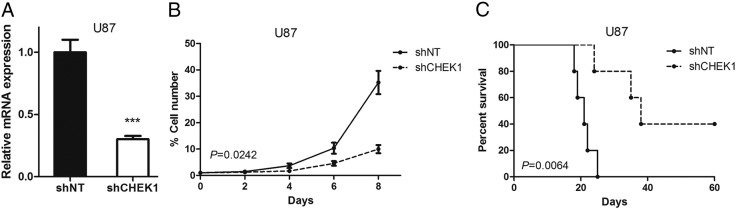
CHEK1 Was Functionally Required for GBM Proliferation Both In Vitro and In Vivo
To examine the biological role of CHEK1 in GBM, we picked one in vitro GBM cell line (U87) which had the highest CHEK1 expression and transduced with either lentiviral shRNA clone for CHEK1 (sh CHEK1) or a nontargeting shRNA (shNT, negative control). The efficiency for lentivirus infection was confirmed by qRT-PCR analysis. The result indicated that CHEK1 mRNA expression was significantly reduced in shCHEK1 U87 cells (Figure 2A). In vitro cell growth kinetics of shSHEK1 transfected U87 cells were diminished proportionally to the reduction levels of CHEK1 (Figure 2B). Next, we investigated the effect of CHEK1 knockdown on in vivo tumor formation. To this end, we used U87 cell–derived mouse intracranial tumor models. The results showed that mice xenografted with shNT-transfected U87 rapidly formed lethal hypervascular GBM-like tumors within 60 days, while a longer survival could be observed in mice xenografted shCHEK1 group (Figure 2C). These findings indicated that CHEK1 was required for GBM proliferation both in vitro and in vivo.
Figure 2.
CHEK1 was functionally required for GBM proliferation both in vitro and in vivo.
(A) qRT-PCR analysis of U87 cells transduced with shRNA against CHEK1 (sh CHEK1) or nontargeting control (shNT) (P < .01, n = 3, with t test). (B) In vitro cell growth assay showed that shRNA against CHEK1 (shCHEK1) inhibited cell proliferation of U87 cells (P < .05, n = 6, with t test). (C) Kaplan-Meier analysis of nude mice harboring intracranial tumor derived from U87 cells transduced with shNT (n = 5) or shCHEK1 (n = 5) (P = .0064, with log-rank test).
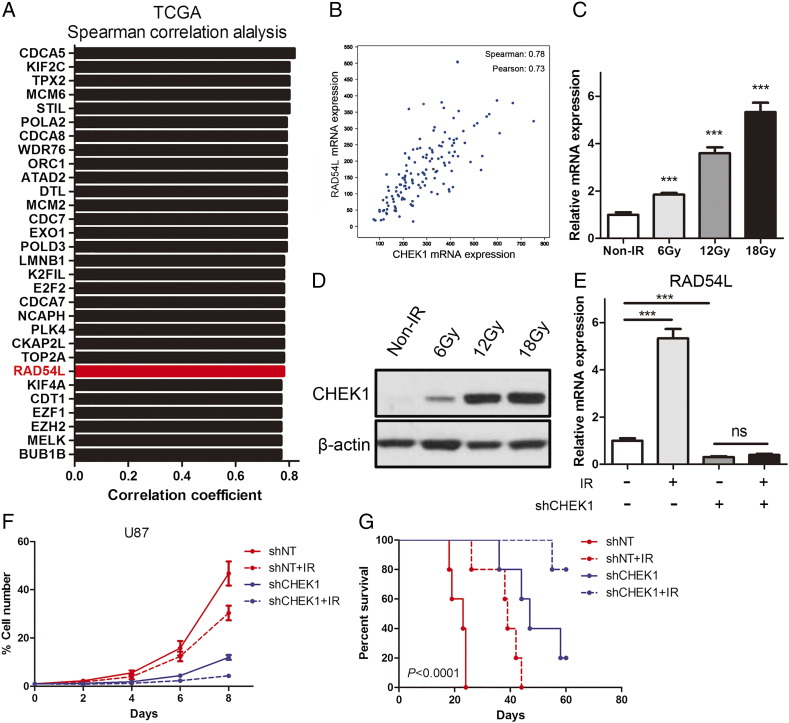
CHEK1-Dependent RAD54L Regulation is Essential for Proliferation and Radioresistance of GBM Cells
To further assess the downstream target of CHEK1 in GBM, we performed correlation analysis by using the data from the TCGA dataset, and the result indicated that RAD54L was one of the top genes which could be correlated to CHEK1 mRNA (Figure 3, A and B). Furthermore, irradiation resulted in a marked increase in CHEK1 expression at both mRNA level (Figure 3C) and protein level (Figure 3D). Additionally, we observed that RAD54L expression was elevated after U87 cells were exposed to radiotherapy, while this effect could be demolished by CHEK1 knockdown (Figure 3E). Similar to qRT-PCR data, in vitro cell growth assay indicated that artificial knockdown of CHEK1 could enhance the radiosensitivity of U87 GBM cells (Figure 3F). Moreover, by using mice xenograft model, we found that inhibition of CHEK1 by shRNA significantly reduced tumorigenesis and radioresistance of U87 cells (Figure 3G). These data suggest that CHEK1 was critical for RAD54L regulation and promotes cell growth and radioresistance of GBM cells.
Figure 3.
CHEK1-dependent RAD54L regulation is essential for proliferation and radioresistance of GBM cells.
(A) Top 30 genes correlated to CHEK1 mRNA expression in TCGA dataset. (B) Pearson or Spearman correlation analysis was performed to evaluate correlation between CHEK1 and RAD54L in TCGA dataset. (C) qRT-PCR analysis for CHEK1 mRNA expression in U87 cells treated with different dose of irradiation (P < .01, n = 3 with t test). (D) Western blotting analysis of U87 cells treated with different dose of irradiation. β-Actin served as a control. (E) qRT-PCR analysis indicated that RAD54L expression was elevated after U87 cells were exposed to radiotherapy, while these effects could be demolished by CHEK1 knockdown (P < .05, n = 3 with t test). (F) In vitro cell growth assay indicated that artificial knockdown of CHEK1 could enhance the radiosensitivity of U87 GBM cells (P < .05, n = 3 with t test). (G) Kaplan-Meier analysis was performed for the comparison of survival in U87-implanted mice treated with shNT or shCHEK1 and then compared with irradiation (P < .0001, with log-rank test).
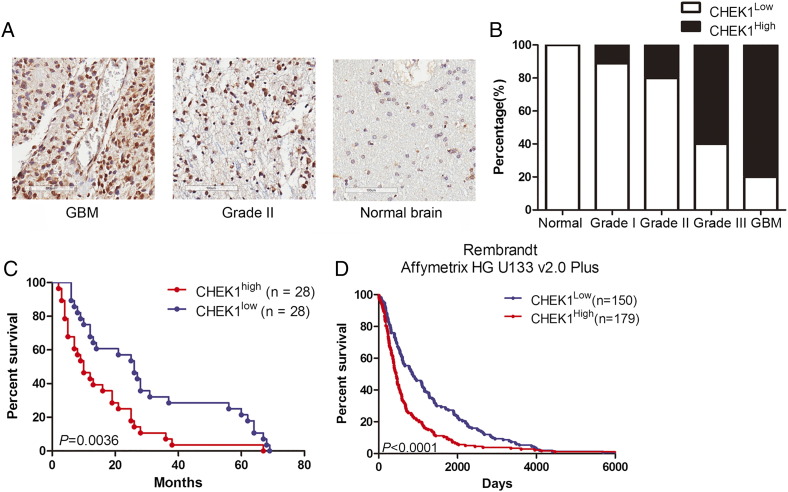
CHEK1 Expression Was Associated with Poor Prognosis in GBM Patients
To clarify the function of CHEK1 in GBM, we collected 56 GBM samples from patients who underwent surgery in our institution from 2009 to 2016 and immunohistostaining to evaluate the expression of CHEK1. The results indicated that CHEK1 was highly expressed in GBM samples (Figure 4, A and B), and high CHEK1 expression could be related to poor prognosis in GBM patients when compared to patients with low CHEK1 expression (Figure 4C). Similar results could be observed in Rembrandt dataset (Figure 4D), which indicated that CHEK1 expression was highly enriched in GBM and could be associated with poor prognosis in GBM patients.
Figure 4.
CHEK1 expression was associated with poor prognosis in GBM patients.
(A) Representative images of IHC-stained patient samples. (B) CHEK1 was highly expressed in GBM when compared to low-grade glioma or normal human brain. (C) Kaplan-Meier analysis of patient samples from Xi'an Jiaotong University indicated the inverted correlation between CHEK1 expression and postsurgical survival of GBM patients (P = .0036, with log-rank test). (D) Kaplan-Meier analysis of patient samples from the Rembrandt dataset indicated the inverted correlation between CHEK1 expression and postsurgical survival of GBM patients (P < .0001, with log-rank test).
CHEK1 Inhibitor Reduced Tumor Growth and Radioresistance of GBM Cells
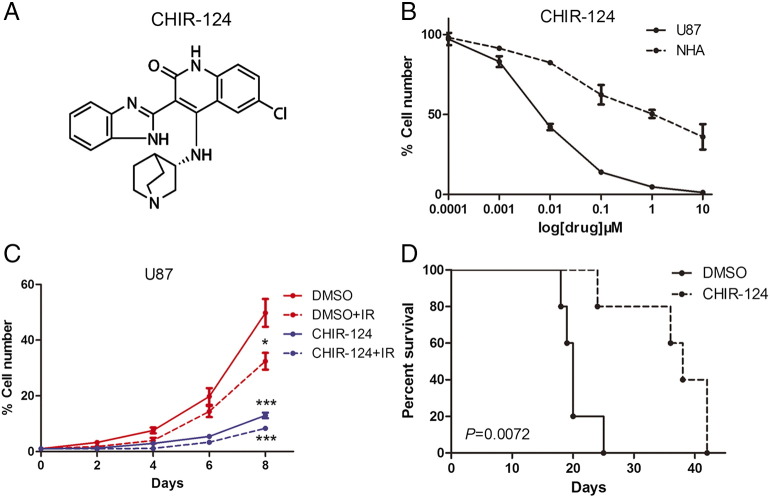
Based on the inhibitory effects of CHEK1 silencing on the GBM cell proliferation and tumorigenicity, we sought to find a potential target for clinical treatment for GBM. To this end, CHEK1 inhibitor CHIR-124 was used to clarify the effects of CHEK1 inhibition on GBM (Figure 5A). In vitro IC50 assay indicated that cell viability of U87 cells could be blocked by CHIR-124 treatment, and the U87 showed higher sensitivity to CHIR-124 compared to NHA cells (Figure 5B). Additionally, in vitro cell proliferation was significantly eliminated by CHIR-124 when combined with radiation compared with radiotherapy alone (Figure 5C). More importantly, systemic treatment of U87-derived mouse brain tumors with CHIR-124 significantly attenuated the tumor growth and radiosensitivity, thereby extending the survival of tumor-bearing mice compared to the vehicle-treated counterparts (Figure 5D). Altogether, CHIR-124 inhibitor reduced tumor growth and radio resistance of GBM cells both in vitro and in vivo.
Figure 5.
The CHEK1 inhibitor reduced tumorigenicity and radioresistance of GBM.
(A) Chemical structure of CHEK1 inhibitor CHIR-241. (B) In vitro cell viability assay for CHIR-241 with U87 GBM cells and NHA. (C) In vitro cell growth assay showed that CHIR-241 decreased cell proliferation of U87 when combined with radiation (P < .01, with t tests). (D) Kaplan-Meier analysis was performed for the comparison of survival in U87-implanted mice treated with or without CHIR-241 (P = .0072, with log-rank test).
Discussion
In this study, we identified CHEK1-RAD54L pathway as an activated molecular mechanism after radiotherapy and that it was functionally required for tumorigenesis and radioresistance in GBM. Mechanically, we found that CHEK1 regulates RAD54L expression and then induces radioresistance in GBM. Therapeutically, we confirmed that inhibition of CHEK1 via either shRNAs or small molecule inhibitor could reduce tumor growth and enhance radio sensitivity in GBM. Altogether, CHEK1 is a regulator for RAD54L which is essential for tumor growth and radioresistance.
Radiotherapy is effective for tumor cells mainly through targeting DNA double strand and inducing DNA damage; the DNA repair pathways of tumor cells can influence the efficacy of radiotherapy and play an important functional role in therapy resistance and recurrence in tumor [17]. Recently, evidence showed that various DNA repair pathways can be differentially activated after radiation [18], [19], [20]. RAD54L has been identified as one of the key proteins necessary for homologous recombination and DNA repair in many organisms [13], [21]. Leone et al. [21] reported that RAD54L is a risk factor and/or a genetic marker in human meningiomas and could be correlated to poor prognosis. Li et al. [12] found that RecQ1, RAD54L, and ATM genes are associated with reduced survival and that deletion of RAD54L could enhance the therapy sensitivity of pancreatic cancer. These data are similar with our findings that RAD54L is essential for tumor growth and radioresistance in GBM, which implied the same functional role of RAD54L in human cancer cells.
CHEK1 is an serine/threonine-specific protein kinase that coordinates the DNA damage response and cell cycle checkpoint response [14]. Activation of CHEK1 results in the initiation of cell cycle checkpoints, cell cycle arrest, DNA repair, and cell death to prevent damaged cells from progressing through the cell cycle [15]. However, the downstream pathway of CHEK1 in DNA repair response still remains unclear. To this end, by using DNA microarray and correlation analysis, we identified RAD54L as the potential downstream molecule which functions in activation of DNA damage repair in GBM cell after receiving radiotherapy. Moreover, we found that CHEK1 inhibition affects transcription of RAD54L, and this mechanism was confirmed in this study. However, the transcription factor which could directly bind to RAD54L promoter area still remains unclear. Further experiments including chromatin immunoprecipitation sequencing for RAD54L promoter and co-immunoprecipitation for CHEK1 protein need to be done to clarify the complete pathway between CHEK1 and RAD54L.
Acknowledgements
We thank all the members of the Department of Neurosurgery, The First Affiliated Hospital of Xi'an Jiaotong University.
References
- 1.Linz U. Commentary on Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial (Lancet Oncol. 2009;10:459-466) Cancer. 2010;116(8):1844–1846. doi: 10.1002/cncr.24950. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, Shin K, Seo YJ, Choi YS, Lee JI. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLoS One. 2015;10(10):e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Araujo PR, Gorthi A, da Silva AE, Tonapi SS, Vo DT, Burns SC, Qiao M, Uren PJ, Yuan ZM, Bishop AJ. Musashi1 Impacts Radio-Resistance in Glioblastoma by Controlling DNA-Protein Kinase Catalytic Subunit. Am J Pathol. 2016;186(9):2271–2278. doi: 10.1016/j.ajpath.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Yang T, Xu G, Liu H, Ren C, Xie W, Wang M. Cyclin-Dependent Kinase 2 Promotes Tumor Proliferation and Induces Radio Resistance in Glioblastoma. Transl Oncol. 2016;9(6):548–556. doi: 10.1016/j.tranon.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones NM, Rowe MR, Shepherd PR, McConnell MJ. Targeted inhibition of dominant PI3-kinase catalytic isoforms increase expression of stem cell genes in glioblastoma cancer stem cell models. Int J Oncol. 2016;49(1):207–216. doi: 10.3892/ijo.2016.3510. [DOI] [PubMed] [Google Scholar]
- 6.Park SY, Piao Y, Thomas C, Fuller GN, de Groot JF. Cdc2-like kinase 2 is a key regulator of the cell cycle via FOXO3a/p27 in glioblastoma. Oncotarget. 2016;7(18):26793–26805. doi: 10.18632/oncotarget.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varghese RT, Liang Y, Guan T, Franck CT, Kelly DF, Sheng Z. Survival kinase genes present prognostic significance in glioblastoma. Oncotarget. 2016;7(15):20140–20151. doi: 10.18632/oncotarget.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274(5):2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 9.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34(15):4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghamrasni SE, Cardoso R, Li L, Guturi KK, Bjerregaard VA, Liu Y, Venkatesan S, Hande MP, Henderson JT, Sanchez O. Rad54 and Mus81 cooperation promotes DNA damage repair and restrains chromosome missegregation. Oncogene. 2016;35(37):4836–4845. doi: 10.1038/onc.2016.16. [DOI] [PubMed] [Google Scholar]
- 11.Mason JM, Dusad K, Wright WD, Grubb J, Budke B, Heyer WD, Connell PP, Weichselbaum RR, Bishop DK. RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells. Nucleic Acids Res. 2015;43(6):3180–3196. doi: 10.1093/nar/gkv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24(11):1720–1728. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong Y, Merino D, Nimmervoll B, Gupta K, Wang YD, Finkelstein D, Dalton J, Ellison DW, Ma X, Zhang J. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer Cell. 2015;27(5):712–727. doi: 10.1016/j.ccell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142(1):1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134(5):1013–1023. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JK, Jeon HY, Kim H. The molecular mechanisms underlying the therapeutic resistance of cancer stem cells. Arch Pharm Res. 2015;38(3):389–401. doi: 10.1007/s12272-014-0531-1. [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, Zhang Q, Atsaves V, Yang H, Claret FX. Suppression of Jab1/CSN5 induces radio- and chemo-sensitivity in nasopharyngeal carcinoma through changes to the DNA damage and repair pathways. Oncogene. 2013;32(22):2756–2766. doi: 10.1038/onc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusin P, Walczak A, Zwierzchlejska A, Olszewski J, Morawiec-Bajda A, Kaczmarczyk D, Kusmierczyk K, Garncarek P, Pytel D, Sliwinnski T. DNA damage and repair of head and neck cancer cells after radio- and chemotherapy. Z Naturforsch C. 2009;64(7-8):601–610. doi: 10.1515/znc-2009-7-821. [DOI] [PubMed] [Google Scholar]
- 20.Zhestianikov VD, Akimov AA, Savel'eva GE, Girshovich MZ, Ivin BA. The suppression of the repair of radio-induced DNA damage in Escherichia coli cells by indazolin derivatives and proksifein. Tsitologiia. 1996;38(11):1203–1210. [PubMed] [Google Scholar]
- 21.Leone PE, Mendiola M, Alonso J, Paz-y-Mino C, Pestana A. Implications of a RAD54L polymorphism (2290C/T) in human meningiomas as a risk factor and/or a genetic marker. BMC Cancer. 2003;3:6. doi: 10.1186/1471-2407-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]