Abstract
Liver is a major contributor of protein production physiologically. The aberrant state of protein synthesis leads to tumor progression. Eukaryotic elongation factor 1 alpha 1 (eEF1A1) is a major member of the eukaryotic elongation factor family that regulates protein synthesis. Although eEF1A1 plays an essential role in controlling the cell fate, its clinical significance in tumor development and progression has not been reported. Here, we aimed to uncover the expression and prognostic significance of eEF1A1 in hepatocellular carcinoma (HCC). Our data indicated that eEF1A1 expression was elevated in HCC cell lines and clinical samples at both the mRNA and protein levels. Immunohistochemistry revealed that eEF1A1 expression was upregulated in HCC samples compared with corresponding non-tumorous tissues. In 50 HCC cases with portal vein embolus, higher eEF1A1 immunoreactivity was detected in tumor metastases compared with the primary lesions. Kaplan–Meier analysis indicated that increased eEF1A1 expression was closely associated with unfavorable post-surgical overall and disease-free survival in 453 HCC patients. Moreover, multivariate analysis indicated eEF1A1 as an independent predictor for overall and disease-free survival. Collectively, our study suggests eEF1A1 as a novel prognostic biomarker and potential therapeutic target for HCC patients.
Introduction
Hepatocellular carcinoma (HCC), a predominant form of primary liver cancer, is the third most frequent cause of cancer death globally with high morbidity and mortality rates [1], [2]. Despite recent reports revealing many causative cancer-related factors, which could improve our understanding of hepatocarcinogenesis and lead to the development of novel treatment approaches [3], the current treatment options for HCC patients are still limited to surgical resection and the anti-angiogenic multikinase inhibitor sorafenib [4], [5]. The five-year survival rate of HCC patients has not considerably improved over the past few years because of post-surgical recurrence and metastasis [6], [7], [8]. Currently, tumor prognostic evaluations are useful for the surveillance of the natural disease progression [9]. However, there is limited evidence for a promising biomarker for HCC prognostic prediction [10], [11]. Therefore, it is of great significance to identify more reliable diagnostic and prognostic biomarkers for HCC that might offer better treatment options for HCC patients.
Eukaryotic protein synthesis is a dynamic process that traditionally involves three essential steps: initiation, elongation, and termination [12]. Each step is highly and restrictedly regulated by a series of soluble factors. The sequential action of these factors plays essential roles in controlling the survival of cells. Among these processes, the eukaryotic elongation factor 1 (eEF1) complex plays an indispensable function in eukaryotic protein peptide chain elongation in all eukaryotic cells [13], [14]. eEF1A1, eukaryotic elongation factor 1 alpha 1 (eEF1A2) and eEF1Bαβγ subcomplex [15]. Human eEF1A1 is a core subunit of the eEF1 complex that shows ability for GTP binding as well as amino acid-tRNA delivery [16].
Aside from its canonical function in translation elongation by ribosomes, eEF1A1 has been implicated in a wide variety of cellular processes. Previous studies indicated that eEF1A1 can shuttle between the nucleus and cytoplasm, and is involved in nuclear tRNAs exportation [17], signaling transduction [18], cellular apoptosis [19], heat shock response [20], cytoskeleton regulation [21], and RNA virus replication [13], making its bio-function far more complex than originally thought. Recent studies demonstrated that eEF1A1 also participates in tumor progression. Li et al. [22] revealed that the co-localization and binding of eEF1A1 and PAK4 promoted the metastasis and progression of gastric cancer cells. The ubiquitin-like protein FAT10 competes with ubiquitin for eEF1A1 binding to stabilize eEF1A1 expression and promote tumor proliferation [23]. Blanch et al. [24] suggested that eEF1A1 was a negative regulator of p53 and p73, since silencing of eEF1A1 partially rescued the anti-apoptosis and chemoresistance observed in response to p53 or p73 knockdown. Farra et al. [15] reported that eEF1A1 depletion in HCC downregulated the transcription factor E2F1 to induce the G1/G0 cell cycle block. However, the clinical significance of eEF1A1 in tumor development and progression has not yet been reported.
With the aim to identify a novel prognostic biomarker to fight HCC, we sought to provide the first demonstration of the expression and clinical significance of eEF1A1 in HCC. In the present study, we elucidate the expression character of eEF1A1 in HCC cell lines and a large cohort of clinical samples. The association between eEF1A1 expression and clinical pathological parameters were further analyzed. Our data identified eEF1A1 as a novel prognostic biomarker and potential therapeutic target for HCC patients.
Materials and Methods
Patients, Tissue Specimens, and Follow-up
Archived paraffin-embedded pathological specimens (collected between January 2000 and December 2010) from 453 primary HCC patients at Sun Yat-sen University Cancer Center (SYSUCC), Guangzhou, China, were analyzed along with clinical and pathological information obtained from patient records. The mean follow-up time was 40.5 months, and the sample included 408 (90.1%) males and 45 (9.9%) females. The mean age was 49 years, ranging from 14 to 77 years. Paired freshly resected HCC tissues and corresponding adjacent non-tumorous samples were collected for quantitative real-time polymerase chain reaction (qRT-PCR) and western blot. The fresh tissues samples were resected from patients and cut into bean size into EP tube to avoid free/thaw cycle. Liquid nitrogen was used to protect the tissues from degradation during the transportation. Samples were stored at −80 °C for long term. None of the patients had received radiotherapy or chemotherapy before surgery. All samples were anonymously labeled. This study was approved by the Institute Research Medical Ethics Committee.
Tissue Microarray (TMA) Construction and Immunohistochemistry (IHC)
HCC and adjacent non-tumorous liver tissues specimens were re-embedded into new paraffin blocks for TMA. The TMA blocks were cut into 4-μm sections and subjected to IHC staining. The slides were dewaxed in xylene and a graded alcohol series, and then treated with 3% hydrogen peroxide in methanol. After avidin-biotin blocking overnight at 4°C, the slides were incubated with EF1A1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing in phosphate buffered saline three times, the slides were incubated with biotinylated goat anti-mouse antibodies. The slides were then stained with DAKO liquid 3,′3-diaminobenzidine tetrahydrochloride (DAB) and finally counterstained with Mayer's hematoxylin. Protein expression levels of EF1A1-stained TMA slides were observed under a microscope and assessed by two independent pathologists (Shi-Xun Lu and Li-Li Liu). The positively stained samples were scored as follows: 0, less than 5% positively stained cells; 1, 6–24% positively stained cells; 2, 25–49% positively stained cells; 3, 50–74% positively stained cells; 4, 75–100% positively stained cells. The intensity was scored as follows: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The final score was calculated by multiplying the percentage score by the staining intensity score. The median IHC score of 8 was chosen as the cut-off value for defining high and low EF1A1 expression.
Statistical Analysis
Statistical analyses were performed using the SPSS 19.0 software (SPSS, Chicago, IL, USA). The Student's t-test was used to determine the significance of differences in EF1A1 expression levels. Pearson's χ2 test or Fisher's exact test was performed to analyze the correlations between EF1A1 expression level and the clinicopathological parameters. The Kaplan–Meier method and the multivariate Cox proportional hazards regression model were conducted for survival analysis and univariate analysis. Data reported in bar or column charts are depicted as mean ± SEM. P-values are indicated directly in graphs and were considered significant at P < .05 and were considered very significant at P < .01.
Cell lines and Cell Culture
HCC cells were obtained from the Type Culture Collection Cell Bank, Chinese Academy of Science Committee (Shanghai, China) and routinely cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco). All cells were maintained in a humidified incubator at 37°C and 5% CO2.
Western Blot and Antibodies
Total proteins from clinical samples and cultured cells were extracted and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then electrophoretically transferred onto a polyvinylidene fluoride membrane (Millipore, Bedford, MA). After blocking in 5% non-fat milk for 1 h at room temperature(20–25 °C), the membranes were incubated with appropriately diluted primary antibodies overnight at 4°C. After washing with Tris-buffered saline with Tween 20, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody at 1:20,000 dilutions for 1 h at room temperature. The membranes were visualized by the enhanced Phototope TM-HRP Detection Kit and exposed to the Bio-Rad processor. Antibodies used in this study were eEF1A1 (1:1000, Santa Cruz, CA, USA) and β-actin (1:1000, Santa Cruz, CA, USA).
qRT-PCR
Total RNA from clinical samples and cultured cells was extracted by Trizol reagent (BIOO Scientific Co., USA). Reverse transcription and SYBR Green-based real-time PCR were carried out subsequently. The primers were designed as follows: eEF1A1 forward: 5′-TGTCGTCATTGGACACGTAGA-3′ and reverse: 5′-ACGCTCAGCTTTCAGTTTATCC-3′; 18S forward: 5′-TGAGAAACGGCTACCACATCC-3′ and reverse: 5′-ACCAGACTTGCCCTCCAATG-3′.
Results
eEF1A1 Expression was Elevated in HCC Cell Lines and Fresh Tissue Samples
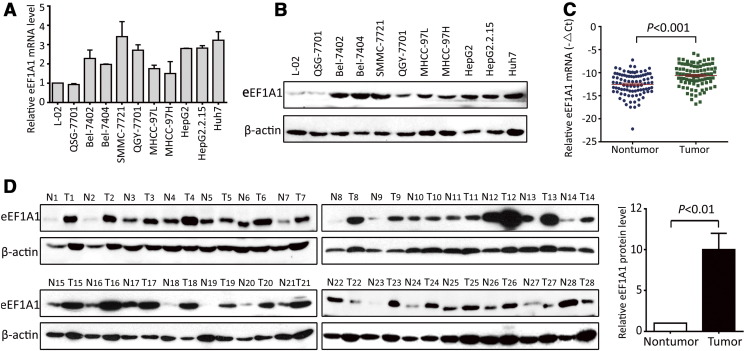
To identify the expression of eEF1A1 in HCC, cell lines and fresh tissue samples were detected by qRT-PCR and western blot. The results showed that eEF1A1 mRNA expression was up-regulated in the HCC cell lines compared with that in the immortalized hepatic cell lines L-02 and QSG-7701, in agreement with a previous work [25] (Figure 1A). Similarly, eEF1A1 showed the same elevated protein expression levels in all HCC cell lines (Figure 1B). To further evaluate the expression of eEF1A1, 90 pairs of HCC fresh tissues resected from patients with primary HCC were collected. eEF1A1 mRNA expression was markedly up-regulated in the HCC samples compared with that in the matched non-tumor tissues (Figure 1C). Consistently, the protein level of eEF1A1 was markedly increased by 10.2-fold in 82.1% (23/28) of the HCC fresh tissues compared with the corresponding adjacent non-tumorous paired samples (Figure 1D).
Figure 1.
eEF1A1 expression was elevated in HCC cell lines and fresh tissue samples A. The mRNA expression levels of eEF1A1 in HCC and immortalized liver cell lines examined by qRT-PCR. The assay was conducted three times independently. B. The protein expression levels of eEF1A1 in HCC and immortalized liver cell lines detected by western blot. C. Expression profile of eEF1A1 mRNA in 90 pairs of HCC and corresponding adjacent liver tissues; 18S RNA was used as a reference to normalize the fold change in expression levels. The eEF1A1 mRNA value was subtracted from 18S RNA value and indicated as “-△Ct”. D. eEF1A1 protein expression in 28 paired HCC and adjacent non-tumorous tissues. The average expression levels of eEF1A1 in tumor and non-tumor tissues were calculated by the fold change accordingly. The protein expression of eEF1A1 was normalized by calculating the ratio of eEF1A1/β-actin of the gray values. Quantitative data are presented as mean ± SD.
Overexpression of eEF1A1 in HCC Detected by IHC
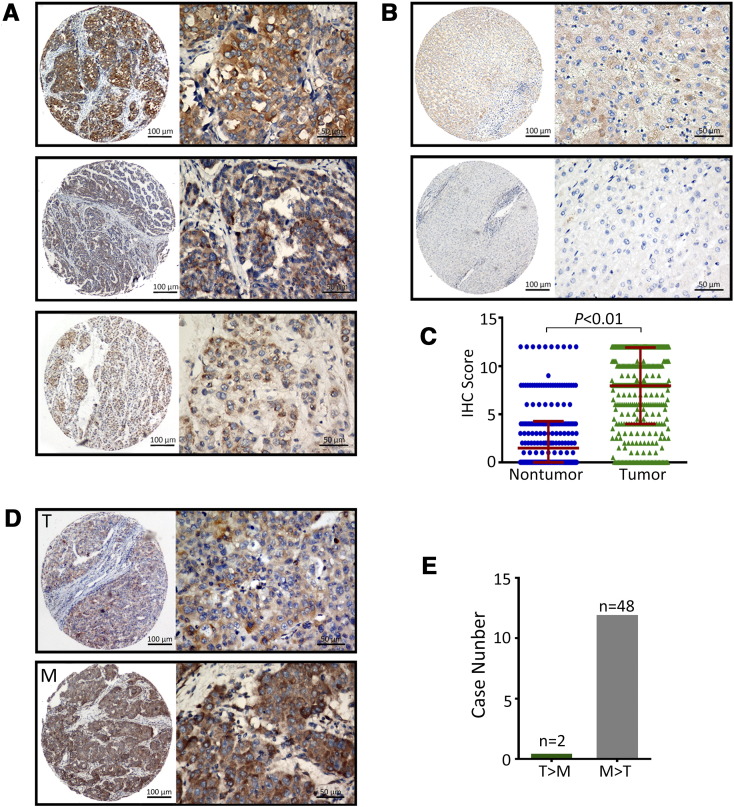
To further confirm the expression profile of eEF1A1 in HCC, 453 archived paraffin-embedded HCC samples were collected and constructed into a TMA cohort along with clinical and pathological information. According to the IHC data, the positive staining of eEF1A1 in HCC, cirrhotic and normal tissues were 90.3%,29.3% and 26.7%, respectively. The data suggest eEF1A1 overexpression may contribute to the tumor initiation and progression. The representative IHC images of eEF1A1 expression in the tumor were observed with high, medium, and low staining (Figure 2A). eEF1A1 was primarily located in the cytoplasm. Positive staining of eEF1A1 expression was detected in 91.2% (413/453) of HCC tissues, in ..% of cirrhotic tissues and ..% of normal liver tissues. eEF1A1 expression in HCC was remarkably higher than that in non-tumorous tissues (Figure 2B and C).
Figure 2.
Overexpression of eEF1A1 in HCC detected by IHC A. Representative images of IHC staining for eEF1A1 expression in a TMA cohort. Representative images of strong (top), moderate (middle), and weak (bottom) intensity staining for tumor tissues are shown. B. Representative IHC images of positive (top) and negative (bottom) non-tumor tissues are presented. C. The IHC scores of the TMA cohort, including 453 HCC patients. D. eEF1A1 expression in 50 HCC metastasis cases analyzed by IHC. Representative photomicrographs are shown for the primary tumor (T) and metastatic (M) lesions. E. Comparison of eEF1A1 levels between the primary tumor and metastatic lesions. Quantitative data are presented as mean ± SD.
In another TMA cohort consisting of 50 HCC cases with portal vein embolus, higher eEF1A1 immunoreactivity was detected in tumor embolus metastases compared with the corresponding primary lesions. As shown by the IHC data, the scores of 48 tumor embolus samples were higher in the metastases than in the primary lesions, with only two cases showing the opposite results (Figure 2D). Taken together, our data indicated that eEF1A1 is overexpressed in HCC compared with non-tumor tissues and is even higher in tumor metastasis lesions.
Association of eEF1A1 Expression and HCC Patient Survival
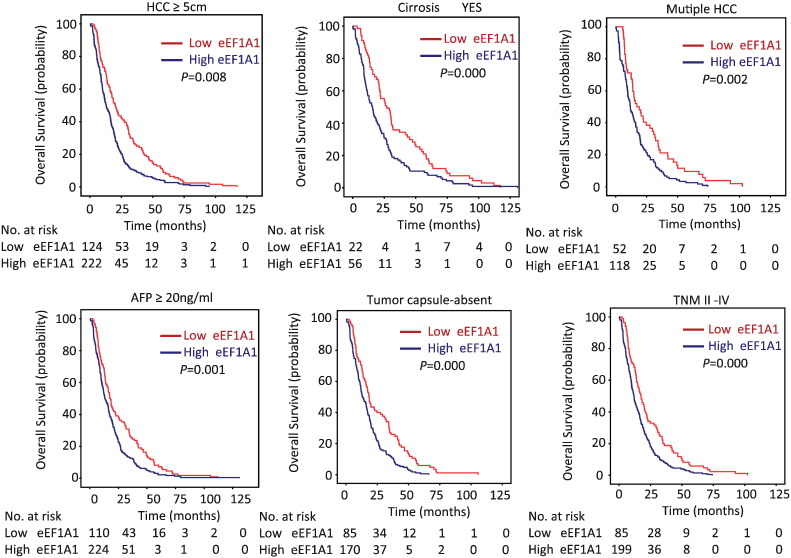
To determine the clinical significance of eEF1A1 in HCC, the correlation between eEF1A1 expression and the clinicopathological variables of HCC patients was analyzed. According to the median IHC staining score (8) of eEF1A1, the HCC cases were divided into two groups: high eEF1A1 expression and low eEF1A1 expression. High expression of eEF1A1 was observed in 64.7% (293/453) of the cases. Statistical analysis indicated that high eEF1A1 expression was associated with poor tumor differentiation but not any of the other parameters (Table 1). Next, we determined the prognostic implication of eEF1A1 in HCC patients. Kaplan–Meier analysis showed that patients with high eEF1A1 expression had shorter overall survival compared with the low eEF1A1 group (median, 22.49 months vs. 13.53 months, respectively, P < .01; Figure 3A). High eEF1A1 expression was also associated with poor disease-free survival (median, 13.48 months vs. 7.52 months, respectively, P < .01; Figure 3B) and the tendency for relapse in HCC patients (Figure 3C). Stratified survival analysis further confirmed the prognostic value of EF1A1. Statistical analyses further demonstrated that eEF1A1 expression was associated with a series of pathological parameters related to overall survival (Figure 4 and Supplementary Figure S1). Therefore, our data demonstrated that eEF1A1 can predict the clinical outcome of patients with HCC.
Table 1.
Correlation of Clinicopathological Parameters and eEF1A1 Expression.
| Variable | eEF1A1 Expression |
|||
|---|---|---|---|---|
| All Cases | Low Expression | High Expression | P Valuea | |
| Age (years)b | 0.220 | |||
| <49 | 213 | 69 (32.4%) | 144 (67.6%) | |
| ≥49 | 240 | 91 (37.9%) | 149 (62.1%) | |
| Gender | 0.108 | |||
| Male | 408 | 149(36.5%) | 259 (63.5%) | |
| Female | 45 | 11 (24.4%) | 34 (75.6%) | |
| HBV | 0.473 | |||
| Positive | 379 | 136(35.9%) | 243(64.1%) | |
| Negative | 73 | 23 (31.5%) | 50 (68.5%) | |
| AFP (ng/ml) | 0.232 | |||
| <20 | 106 | 44(41.5%) | 62 (58.5%) | |
| ≥20 | 346 | 115 (33.3%) | 231 (66.7%) | |
| Cirrhosis | 0.285 | |||
| Yes | 372 | 135 (36.3%) | 237 (63.7%) | |
| No | 80 | 24(30.0%) | 56 (70.0%) | |
| Tumor size (cm) | 0.347 | |||
| <5 | 96 | 30 (31.3%) | 66 (68.7%) | |
| ≥5 | 357 | 130 (36.4%) | 227 (63.6%) | |
| Tumor multiplicity | 0.363 | |||
| Single | 276 | 102 (37.0%) | 174 (63.0%) | |
| Multiple | 177 | 58 (32.8%) | 119 (67.2%) | |
| Differentiation | 0.026 | |||
| Well-Moderate | 234 | 94(63.3%) | 140 (36.7%) | |
| Poor-undifferentiated | 219 | 66 (30.1%) | 153 (69.9%) | |
| Stage | 0.948 | |||
| I | 164 | 110 (67.1%) | 54 (32.9%) | |
| II-IV | 307 | 205 (66.8%) | 102 (33.2%) | |
| Vascular invasion | 0.089 | |||
| Yes | 100 | 28(28.0%) | 72(72.0%) | |
| No | 352 | 131 (37.2%) | 221(62.8%) | |
| Tumor Capsule | 0.338 | |||
| Complete | 189 | 71 (37.6%) | 118 (62.4%) | |
| Incomplete | 262 | 87 (33.2%) | 175 (66.8%) | |
| TNM | 0.847 | |||
| No | 426 | 150 (35.2%) | 276 (64.8%) | |
| Yes | 27 | 10 (37.0%) | 17 (63.0%) | |
Chi-square test;
Median age; AFP, alpha-fetoprotein; HBV, hepatitis B virus infection.
Figure 3.
Association of eEF1A1 expression and HCC patient survival A. The correlation of eEF1A1 expression and overall survival was determined in a TMA cohort including 453 patients by Kaplan–Meier analysis. B. Disease-free survival of the same eEF1A1 TMA cohort. C. The curve for relapse evaluated according to eEF1A1 expression level. The life table is shown in each graph below.
Figure 4.
Stratified analysis of eEF1A1 expression related to overall survival . The correlation of eEF1A1 expression and overall survival in the indicated groups.
Univariate and Multivariate Analyses of Prognostic Variables in HCC
To further evaluate the representativeness of our samples in HCC prognostic prediction, Cox regression analysis was carried out. eEF1A1 expression was identified as a prognostic factor, along with tumor size, tumor multiplicity, tumor capsule, liver cirrhosis, serum level of alpha fetoprotein (AFP), tumor differentiation, vascular invasion, and TNM stage (Table 2). After adjusting for the prognostic factors established in the univariate analysis, multiple Cox regression analysis was carried out, which revealed eEF1A1 as an independent factor for worse overall survival (hazard ratio = 1.849, 95% confidence interval: 1.501–2.278, P < .01) and poor disease-free survival (hazard ratio = 1.354, 95% confidence interval: 1.027–1.786, P = .032) (Table 2, Table 3). In conclusion, our data indicated eEF1A1 as an independent prognostic marker for HCC patients.
Table 2.
Univariate and Multivariate Analyses of Clinicopathological and eEF1A1 Expression for Overall Survival (n = 453).
| Variables | Univariate Analysis |
Multivariate Analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Overall survival | ||||
| Age (<49 vs. ≥49 years) | 0.884 (0.732–1.067) | 0.199 | ||
| Gender (female vs. male) | 1.052 (0.767–1.444) | 0.753 | ||
| HBV (positive vs. negative) | 1.245(0.961–1.612) | 0.089 | ||
| Tumor size (<5 vs. ≥5 cm) | 1.544 (1.225–1.947) | 0.000 | 1.031 (1.007–1.055) | 0.012 |
| Tumor multiplicity (single vs. multiple) | 1.331 (1.097–1.615) | 0.004 | 0.843 (0.671–1.086) | 0.196 |
| Tumor capsule (absent vs. present) | 0.697 (0.573–0.848) | 0.000 | 0.871(0.708–1.072) | 0.192 |
| Liver cirrhosis (yes vs. no) | 0.673 (0.525–0.861) | 0.002 | 0.110 (0.626–1.049) | 0.811 |
| AFP (<20 vs. ≥20 ng/mL) | 1.532 (1.228–1.912) | 0.000 | 1.255 (0.998–1.578) | 0.052 |
| Vascular invasion (yes vs. no) | 1.906 (1.515–2.398) | 0.000 | 1.269(0.987–1.631) | 0.063 |
| Tumor differentiation | 1.494 (1.236–1.807) | 0.000 | 1.201 (0.968–1.489) | 0.096 |
| TNM (I vs. II-IV) | 1.938 (1.583–2.374) | 0.000 | 1.805 (1.384–2.354) | 0.000 |
| eEF1A1 expression (low vs. high) | 1.638 (1.342–2.001) | 0.000 | 1.849 (1.501–2.278) | 0.000 |
AFP, a-fetoprotein; HBV, hepatitis B virus infection; HR, hazard ratio; CI, confidence interval.
Table 3.
Univariate and Multivariate Analyses of Clinicopathological and eEF1A1 Expression for Disease-Free Survival (n = 453).
| Variables | Univariate Analysis |
Multivariate Analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Overall survival | ||||
| Age (<49 vs. ≥49 years) | 0.892 (0.686–1.159) | 0.391 | ||
| Gender (female vs. male) | 1.190 (0.778–1.819) | 0.422 | ||
| HBV (positive vs. negative) | 1.245(0.961–1.612) | 0.089 | ||
| Tumor size (<5 vs. ≥5 cm) | 1.011 (0.745–1.372) | 0.943 | ||
| Tumor multiplicity (single vs. multiple) | 0.880 (0.667–1.161) | 0.366 | ||
| Tumor capsule (absent vs. present) | 1.045 (0.802–1.361) | 0.744 | ||
| Liver cirrhosis (yes vs. no) | 0.823 (0.586–1.157) | 0.263 | ||
| AFP (<20 vs. ≥20 ng/mL) | 1.214 (0.900–1.637) | 0.204 | ||
| Vascular invasion (yes vs. no) | 1.127 (0.807–1.574) | 0.483 | ||
| Tumor differentiation | 0.929 (0.684–1.261) | 0.637 | ||
| TNM (I vs. II-IV) | 1.864 (0.663–1.126) | 0.279 | ||
| eEF1A1 expression (low vs. high) | 1.354 (1.027–1.786) | 0.032 | ||
AFP, a-fetoprotein; HBV, hepatitis B virus infection; HR, hazard ratio; CI, confidence interval.
Discussion
HCC is a lethal disease representing a global challenge because of its high morbidity and mortality rate. The five-year survival rate of this neoplasm remains dismal, and there are limited biomarkers available for HCC prognostic prediction. To improve the outcome of HCC patients, the potential underlying molecular mechanism remains to be elucidated to aid in the discovery of more reliable diagnostic and prognostic biomarkers. Since liver is a major contributor of whole body protein production physiologically, we tend to identify whether protein synthesis leads to HCC progression. In this study, we demonstrated that eEF1A1 was overexpressed in HCC and was strongly correlated with unfavorable overall and disease-free survival. Moreover, multivariate analysis indicated eEF1A1 as an independent prognostic predictor. Thus, this is the first study to unveil the clinical significance of eEF1A1.
Dysregulation of the eEF1 complex has been reported in human cancers, and the role of eEF1A2, the homolog of eEF1A1, is particularly well documented in this respect [26], [27]. eEF1A2 was shown to be overexpressed in human tumors, demonstrating oncogenic properties [28]. Moreover, overexpression of eEF1A2 was found to be associated with poorer prognosis in gastric cancer patients [29]. However, the expression profile and clinical significance of eEF1A1 has been rarely demonstrated in tumors overall. In this study, eEF1A1 expression was found to be increased in a cohort of 453 patients with HCC. HCC patients with high eEF1A1 expression had a shorter life, both in overall survival and disease-free survival, compared with the low expression group. The fact that eEF1A1 expression was increased in the portal vein embolus in 48 HCC samples strongly indicated that eEF1A1 is involved in tumor progression. These data suggest that eEF1A1 might be a novel prognostic biomarker for HCC patients. Interestingly, more cytosolic eEF1A protein was found in well-differentiated HCC cell lines. Using a specific antibody for eEF1A1, we showed that eEF1A1 mainly localized in the cytoplasm of HCC cells. We have detected elevated eEF1A1 protein expression in 219 poor-undifferentiated samples compared with 234 well-moderate samples (please refer to Table 1. Therefore, it should be of interest to determine whether eEF1A2 is involved in the HCC cell differentiation.
Tumor inhibitors related to the EF1A complex have been previously reported. Using biochemical approaches, Losada et al. [28] indicated that the eEF1A1 homolog eEF1A2 has a high-affinity interaction with plitidepsin, an antitumor agent of marine origin that exerts an anti-proliferative effect in cancer cells. Scaggiante et al. [30] tested the anti-tumor potential of a nucleotide aptamer containing a GT repetition (GT75) that specially interferes with eEF1A activity in three HCC cell lines. They indicated that GT75 impairs the viability of hepatocarcinoma cells and potentiates the effects of bortezomib and idarubicin. To improve the therapeutic effect of HCC, great efforts are still required in the advent of systemic treatment. We hope that targeted therapies related to eEF1A1 might come to the spotlight in the future.
In summary, our data demonstrated increased eEF1A1 expression in HCC cell lines and clinical samples. An elevated eEF1A1 expression level predicted unfavorable overall and disease-free survival. Collectively, our study suggests eEF1A1 as a new promising biomarker for the prognosis of patients with HCC.
The RDD nubmer of this paper is RDDB2017000217.
The following is the supplementary data related to this article.
Acknowledgments
Acknowledgements
We thank pathology department of Sun Yat-sen university cancer center for collating patient information in this study. National Key R&D Program of China (2017YFC1309000) and The National Natural Science Foundation of China (No. 81372572, 81572405, 81572406, 81602135) supports to our findings.
Conflict of Interests
The authors declare no conflict of interest.
Author Contributions
Conception and design of the study: Shi-Lu Chen, Jing-Ping Yun; Generation, collection, assembly, analysis of data: Shi-Lu Chen, Shi-Xun Lu, Li-Li Liu, Chun-Hua Wang; Scoring and evaluation of IHC stained slides: Shi-Xun Lu, Li-Li Liu, Xia Yang; Drafting and revision of the manuscript: Shi-Lu Chen, Chris Zhiyi Zhang, Jing-Ping Yun; Approval of the final version of the manuscript: all authors.
Contributor Information
Hui-Zhong Zhang, Email: zhanghuizh@sysucc.org.cn.
Jing-ping Yun, Email: yunjp@sysucc.org.cn.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 3.Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64:842–848. doi: 10.1136/gutjnl-2014-307990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol. 2015;62:1420–1429. doi: 10.1016/j.jhep.2015.02.038. [DOI] [PubMed] [Google Scholar]
- 5.Worns MA, Galle PR. HCC therapies--lessons learned. Nat Rev Gastroenterol Hepatol. 2014;11:447–452. doi: 10.1038/nrgastro.2014.10. [DOI] [PubMed] [Google Scholar]
- 6.Giordano S, Columbano A. Met as a therapeutic target in HCC: facts and hopes. J Hepatol. 2014;60:442–452. doi: 10.1016/j.jhep.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Hou G, Chen L, Liu G, Li L, Yang Y, Yan HX, Zhang HL, Tang J, Yang YC, Lin X. Aldehyde dehydrogenase-2 (ALDH2) opposes hepatocellular carcinoma progression by regulating AMP-activated protein kinase signaling in mice. Hepatology. 2017;65:1628–1644. doi: 10.1002/hep.29006. [DOI] [PubMed] [Google Scholar]
- 8.Dong W, Zhang T, Wang ZG, Liu H. Clinical outcome of small hepatocellular carcinoma after different treatments: a meta-analysis. World J Gastroenterol. 2014;20:10174–10182. doi: 10.3748/wjg.v20.i29.10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Gramont A, Watson S, Ellis LM, Rodon J, Tabernero J, de Gramont A, Hamilton SR. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12:197–212. doi: 10.1038/nrclinonc.2014.202. [DOI] [PubMed] [Google Scholar]
- 10.Giannini EG, Farinati F, Ciccarese F, Pecorelli A, Rapaccini GL, Di Marco M, Benvegnu L, Caturelli E, Zoli M, Borzio F. Prognosis of untreated hepatocellular carcinoma. Hepatology. 2015;61:184–190. doi: 10.1002/hep.27443. [DOI] [PubMed] [Google Scholar]
- 11.Li KW, Li X, Wen TF, Lu WS. The effect of postoperative TACE on prognosis of HCC: an update. Hepatogastroenterology. 2013;60:248–251. doi: 10.5754/hge12665. [DOI] [PubMed] [Google Scholar]
- 12.Sasikumar AN, Perez WB, Kinzy TG. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA. 2012;3:543–555. doi: 10.1002/wrna.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Wei T, Abbott CM, Harrich D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol Mol Biol Rev. 2013;77:253–266. doi: 10.1128/MMBR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novosylna O, Jurewicz E, Pydiura N, Goral A, Filipek A, Negrutskii B, El'skaya A. Translation elongation factor eEF1A1 is a novel partner of a multifunctional protein Sgt1. Biochimie. 2015;119:137–145. doi: 10.1016/j.biochi.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Farra R, Scaggiante B, Guerra C, Pozzato G, Grassi M, Zanconati F, Perrone F, Ferrari C, Trotta F, Grassi G. Dissecting the role of the elongation factor 1A isoforms in hepatocellular carcinoma cells by liposome-mediated delivery of siRNAs. Int J Pharm. 2017;525:367–376. doi: 10.1016/j.ijpharm.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 16.Greganova E, Altmann M, Butikofer P. Unique modifications of translation elongation factors. FEBS J. 2011;278:2613–2624. doi: 10.1111/j.1742-4658.2011.08199.x. [DOI] [PubMed] [Google Scholar]
- 17.Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci Biotechnol Biochem. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- 18.Negrutskii B, Vlasenko D, El'skaya A. From global phosphoproteomics to individual proteins: the case of translation elongation factor eEF1A. Expert Rev Proteomics. 2012;9:71–83. doi: 10.1586/epr.11.71. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi Y, Yonehara S. Novel cell death by downregulation of eEF1A1 expression in tetraploids. Cell Death Differ. 2009;16:139–150. doi: 10.1038/cdd.2008.136. [DOI] [PubMed] [Google Scholar]
- 20.Vera M, Pani B, Griffiths LA, Muchardt C, Abbott CM, Singer RH, Nudler E. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife. 2014;3:e03164. doi: 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Guo H, Mi Z, Gao C, Bhattacharya S, Li J, Kuo PC. EF1A1-actin interactions alter mRNA stability to determine differential osteopontin expression in HepG2 and Hep3B cells. Exp Cell Res. 2009;315:304–312. doi: 10.1016/j.yexcr.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Li J, Li F. P21 activated kinase 4 binds translation elongation factor eEF1A1 to promote gastric cancer cell migration and invasion. Oncol Rep. 2017;37:2857–2864. doi: 10.3892/or.2017.5543. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Chen L, Ge J, Yan C, Huang Z, Hu J, Wen C, Li M, Huang D, Qiu Y. The Ubiquitin-like Protein FAT10 Stabilizes eEF1A1 Expression to Promote Tumor Proliferation in a Complex Manner. Cancer Res. 2016;76:4897–4907. doi: 10.1158/0008-5472.CAN-15-3118. [DOI] [PubMed] [Google Scholar]
- 24.Blanch A, Robinson F, Watson IR, Cheng LS, Irwin MS. Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS One. 2013;8:e66436. doi: 10.1371/journal.pone.0066436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grassi G, Scaggiante B, Farra R, Dapas B, Agostini F, Baiz D, Rosso N, Tiribelli C. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Biochimie. 2007;89:1544–1552. doi: 10.1016/j.biochi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Veremieva M, Khoruzhenko A, Zaicev S, Negrutskii B, El'skaya A. Unbalanced expression of the translation complex eEF1 subunits in human cardioesophageal carcinoma. Eur J Clin Invest. 2011;41:269–276. doi: 10.1111/j.1365-2362.2010.02404.x. [DOI] [PubMed] [Google Scholar]
- 27.Veremieva M, Kapustian L, Khoruzhenko A, Zakharychev V, Negrutskii B, El'skaya A. Independent overexpression of the subunits of translation elongation factor complex eEF1H in human lung cancer. BMC Cancer. 2014;14:913. doi: 10.1186/1471-2407-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losada A, Munoz-Alonso MJ, Garcia C, Sanchez-Murcia PA, Martinez-Leal JF, Dominguez JM, Lillo MP, Gago F, Galmarini CM. Translation Elongation Factor eEF1A2 is a Novel Anticancer Target for the Marine Natural Product Plitidepsin. Sci Rep. 2016;6:35100. doi: 10.1038/srep35100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Lu M, Chen Y, Meng D, Sun R, Yun D, Zhao Z, Lu D, Li Y. Overexpression of eukaryotic elongation factor 1 alpha-2 is associated with poorer prognosis in patients with gastric cancer. J Cancer Res Clin Oncol. 2015;141:1265–1275. doi: 10.1007/s00432-014-1897-7. [DOI] [PubMed] [Google Scholar]
- 30.Scaggiante B, Farra R, Dapas B, Baj G, Pozzato G, Grassi M, Zanconati F, Grassi G. Aptamer targeting of the elongation factor 1A impairs hepatocarcinoma cells viability and potentiates bortezomib and idarubicin effects. Int J Pharm. 2016;506:268–279. doi: 10.1016/j.ijpharm.2016.04.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.