Acute Myeloid Leukemia (AML) is an aggressive hematological malignancy caused by somatically acquired changes affecting a well-defined set of genes1. While rare high-risk variants affecting specific transcription factors account for a proportion of myelodysplastic syndrome (MDS) and AML associated with a family history, the contribution of other germline variants conferring low-intermediate risk has not yet been determined, partly because these are more difficult to identify from pedigree analysis. Here we use an Australian AML patient cohort to analyze rare, deleterious variants affecting genes involved in the rare recessive bone marrow failure syndrome Fanconi Anemia (FA). FA is caused by bi-allelic germline mutations in any of the 22 FANC genes (except for FANCB and FANCR which are X-linked and autosomal dominant), and is associated with profoundly increased risk of AML2. The proteins encoded by the FANC genes participate in the removal of interstrand crosslinks (ICL) and the protection and resolution of stalled replication forks, an essential step for faithful DNA replication2,3. Deficiency for these genes, combined with other mutations, results in pre-leukemia or leukemia in mouse models4.
Novel, rare, somatic and germline, coding and splicing FANC gene variants (Tier 1 mutations1, MAF < 0.001) were identified using whole exome sequencing (WES) of 131 samples from adult Caucasian AML patients (cohort characteristics shown in Supplementary Table S1). Variants were also identified in WES data from an ethnically matched, all-female, healthy control cohort (n = 323)5. Following pathogenicity filtering, we identified a total of 53 heterozygous FANC variants in 45 patients (Supplementary Table S2). In contrast to what has been observed for FA AML, for which over-representation of +1q, del7/7q and +3q has been reported6, we observed significant under-representation of monosomy 7/del(7q) (P = 0.028; Fig. 1a and Supplementary Table S1).
Fig. 1. FANC variants in the Australian AML cohort.
a Association of FANC mutations with recurrent somatic AML mutations and cytogenetics in the Australian AML Cohort. The columns in the figure represent each patient in the Australian AML cohort (n = 131). The first row in the figure represents the 22 FANC genes as a group. The subsequent rows represent individual FANC genes, AML recurrently mutated genes, karyotype and cytogenetic characteristics. Patients carrying one or more FANC mutations (n = 44) are represented with red boxes. Black boxes indicate patients carrying a D-C mutation for the specific FANC gene. Patients with specific AML mutations are represented with blue boxes. Patients with normal, abnormal (1 or 2 abnormalities) and complex (3 or more abnormalities) karyotypes are represented with green boxes. Patients with specific karyotypic abnormalities are represented by orange boxes. Del7/7q is under-represented (blue highlight) in the FANC-mutant AML patient group (P = 0.028). b D-C mutations present in the Australian AML cohort. Frequency of each D-C mutation is presented for the Australian AML and control cohorts, and for the ExAc (non-Finish European) database
Sanger sequencing of selected FANC tumor variants, for which matched non-tumor material was available, showed that the majority were present in the non-tumor material (11 out of 12, Supplementary Fig. S1), indicating that AML FANC variants are predominantly germline. Further sequencing of cDNA from selected AML samples revealed expression of both wild type (WT) and mutant sequences for the majority of variants (Supplementary Fig. S2). A number of FANC variants of significant interest were also identified based on known disease associations, protein structure, or clinical characteristics (summarized in Supplementary Table S3).
For a stringent analysis of pathogenic FANC variants, we next identified known disease-causing (D-C) variants in the AML and healthy cohorts by cross-referencing to pathogenic variants in the FA (FAMutdb: http://www2.rockefeller.edu/fanconi/) and breast cancer (kConFab: http://www.kconfab.org/Index.shtml and BIC: https://research.nhgri.nih.gov/bic/) databases (Fig. 1b). For case-control comparison with our healthy all-female cohort, we limited analysis to female AML patients. While the overall frequencies of female patients and controls with FANC variants were similar (35% vs. 32%, respectively), we observed an elevated frequency of female patients carrying D-C variants [13.7% vs. 4.5% controls; Odds Ratio (OR) = 3.3, 95% CI: 1.3–8.6; P = 0.018]. We also observed increased frequency of D-C FANC variants in the Australian and TCGA AML cohorts compared to that reported for healthy male and female European-Americans in the ESP database (n = 4298 individuals)7 (Australian cohort: 6.9% vs. 2.1% controls, OR = 3.4, 95% CI: 1.7–7.0, P = 0.0003; TCGA cohort: 4.9% vs. 2.1% controls, P = 0.054). The frequency of damaging variants for several FANC genes (FANCC, FANCL, and FANCM, Supplementary Fig. S3) was significantly increased in the Australian AML cohort relative to that in the non-Finnish European population in the Exome Aggregation Consortium (ExAC) database (Supplementary Table S4). Finally, comparison of all rare damaging variants in both the Australian and TCGA AML cohorts to the healthy Australian control cohort, using burden analysis, showed an elevated frequency of FANCL variants in the Australian AML cohort (P = 0.01, Supplementary Fig. S4), and of FANCC and FANCO variants in the TCGA cohort (P = 0.037 and P = 0.01, respectively), consistent with a previous report8. Taken together, the elevated frequencies of D-C and damaging FANC variants suggest that subtle changes to FA pathway function in HSC may play a role in AML predisposition and pathogenesis. There are a number of important differences that need to be considered when relating our study to previous studies of FA families which have not found increased incidence of AML in FA carriers9,10. Firstly, such familial studies are biased towards analysis of only 3 FANC genes (FANCA, FANCC, and FANCG), which represent the vast majority of FA cases. It is possible that bi-allelic loss of function for some FANC genes may not be tolerated at the germline level (i.e., those genes rarely found mutated in FA) while rare heterozygous mutations may still confer a cancer risk for carriers. Secondly, while familial studies address the question of high-risk D-C FANC variants in carriers, modest-risk or low-risk variants in these genes would be predicted to induce AML with low penetrance, masking the familial pre-disposition. Thus, further analysis focusing on assessing familial cancer and AML incidence for families across all FA complementation groups, and additional prospective sequencing studies in larger multi-center AML and control cohorts, will help to clarify this issue. Such studies are also important to establish the implications for FA families, particularly for selection of sibling donors for stem cell transplantation of FA patients, and for identification of carriers who may have modest-intermediate risk of AML or other cancers. Other studies have reported damaging germline FANC variants in MDS/sAML, therapy related AML, and in cohorts of patients with a range of other familial cancers (Supplementary Table S5).
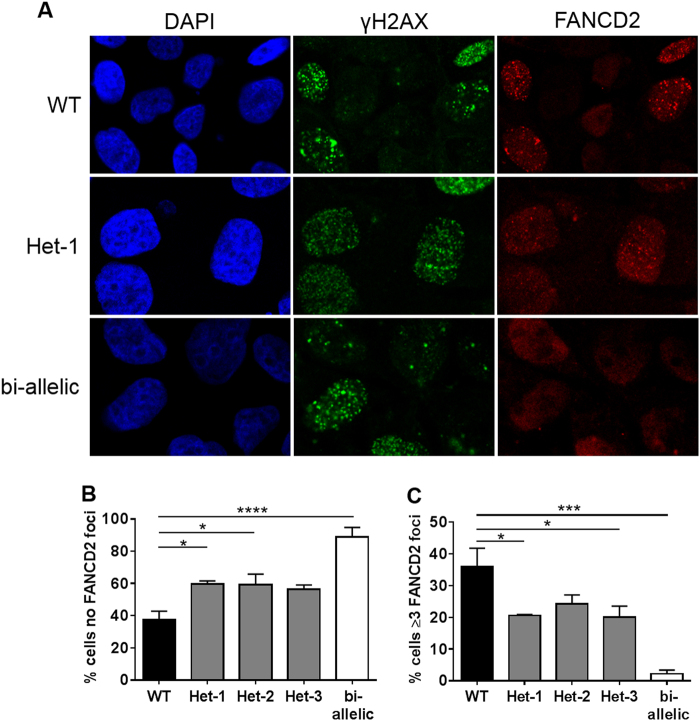
To investigate whether heterozygous FANC mutations can indeed impair the cellular response to DNA damaging events, we generated MCF10A clones with heterozygous FANCL deficiency using CRISPR-Cas9. We compared responses to treatment with an ICL agent (mitomycin C, MMC) for a single WT clone, 3 independent heterozygous FANCL-mutant clones, and a bi-allelic FANCL-mutant clone carrying a nonsense mutation on one allele and a single amino acid deletion on the other (Supplementary Fig. S5). FANCL protein level showed a 75% reduction compared to the WT control for the bi-allelic FANCL clone, and 35–45% reduction for the heterozygous clones (Supplementary Fig. S6). Treatment with MMC did not alter the cell cycle distribution of the heterozygous clones compared to the WT clone; however, the bi-allelic clone displayed accumulation of cells in G2/M phase (Supplementary Fig. S7), consistent with delayed repair of ICLs. Moreover, while the WT clone displayed robust FANCD2-foci formation following MMC treatment, this was not observed for the bi-allelic FANCL-mutant clone (Fig. 2a–c), consistent with loss of FANCL activity11. Importantly, for the three heterozygous clones, the frequencies of cells lacking FANCD2 foci (Fig. 2b) and with ≥ 3 FANCD2 foci (Fig. 2c) were intermediate compared to the WT and bi-allelic clones indicating reduced FANCL activity in the heterozygous clones compared to WT, but more activity compared to the bi-allelic mutant clone. Overall, these data are consistent with a reduction in, but not abrogation of, DNA repair activity in cells carrying heterozygous damaging mutations in FANCL. While in this study we focused on FA DNA repair pathway activation in response to ICL-induced DNA damage, recent reports indicate a role for some FANC genes in replication fork protection3; therefore, it will also be of interest to measure the response of FANC heterozygous cells under conditions of replicative stress. It is well established that BRCA1/2 haploinsufficiency is associated with an impaired DNA damage response, telomere erosion, genomic instability, and premature senescence12, and further detailed functional studies of other FANC heterozygous models are now important to link the epidemiological and functional data.
Fig. 2. FANCD2 foci formation in a FANCL heterozygous cell line model.
a Immunofluorescent images captured at 63 × magnification for three representative MCF10A CRISPR-FANCL clones; WT, heterozygous (Het-1), and bi-allelic. Cells were treated with mitomycin C (MMC) and probed with DAPI (blue) or antibodies for γH2AX (green), and FANCD2 (red). b Percentage of γH2AX positive cells with no detectable FANCD2 foci in MCF10A CRISPR-FANCL clones. Black: WT clone, gray: heterozygous clones (Het-1, Het-2, Het-3); white: bi-allelic clone. For statistical comparison, One-way ANOVA with Tukey multiple comparison was performed. *P < 0.05, ***P < 0.001, ****P < 0.0001. c Percentage of γH2AX positive cells with FANCD2 foci ≥ 3 in the 5 MCF10A CRISPR-FANCL clones. Black: WT clone, gray: heterozygous clones (Het-1, Het-2, Het-3); white: bi-allelic clone. For statistical comparison, One-way ANOVA with Tukey multiple comparison was performed. *P < 0.05, *** P < 0.001, **** P < 0.0001
In summary, our findings suggest that decreased function of the FA DNA repair pathway in HSC is due to deleterious germline heterozygous FANC variants which may result in a reduced capacity to maintain genome integrity, which may in turn contribute to increased risk of AML. This may be particularly significant under conditions associated with increased replicative stress or DNA damage. Such a deficiency would be predicted to have important consequences during emergency hematopoiesis13 or when HSC are confronted with endogenous or exogenous cross-linking toxins14. The effect of gene variants affecting the FA pathway will also be influenced by other factors, such as environmental exposure, infectious agents and/or, other genetic variants that affect metabolism of aldehydes, or individual DNA damage response. Over time, a subtle change to genomic stability in HSC may contribute to acquisition of somatic driver mutations explaining enrichment of pathogenic FA variants in AML cases. Finally, our findings may have therapeutic implications. Recent studies have demonstrated that the FA pathway, together with Poly-ADP ribose polymerase 1 (PARP1), play a key role protecting replication forks and preventing genomic instability during replication3. Thus, AML cells with partial FA pathway deficiency may display enhanced sensitivity to agents that boost replicative stress and DNA damage, particularly when combined with PARP1 inhibitor; this combination has been proposed as a potential therapeutic approach in AML15.
Electronic supplementary material
Acknowledgements
We would like to acknowledge the Leukemia Foundation of Australia for their support of the Australian Leukemia and Lymphoma Group (ALLG) tissue bank. The South Australian Cancer Research Biobank (SACRB) is supported by the Cancer Council SA Beat Cancer Project, Medvet Laboratories Pty Ltd and the Government of South Australia. We would also like to acknowledge the assistance from the Detmold Family Cytometry Facility, Hanson Institute, SA Pathology. The authors thank Brooke Gardiner and colleagues at UQ Centre for Clinical Genomics (UQ Diamantina Institute) for carrying out exome capture and sequencing. We also thank Megan Ellis (ALLG tissue bank) and Carolyn Butcher (SACRB) for invaluable assistance in identifying and preparing samples, Diana Iarossi, Michelle Perugini, Silke Danner and Sarah Moore for assistance with molecular and clinical annotation of AML samples, Crystal McGirr for general technical assistance and Charlotte Hodson for help with analysis of mutation effect on function. We would like to gratefully acknowledge the funding support from the Queen Elizabeth Hospital Hematology-Oncology Private Practice Fund; the Queensland Health Study, Educational and Research Trust Fund; the Royal Adelaide Hospital Contributing Hematologists’ Fund, and the National Health and Medical Research Council (Australia). K.Z.Y.M. is supported by a Queen Elizabeth Hospital Hematology-Oncology Research Award, J.X.G. was supported by a Fellowship from the Princess Alexandra Hospital and Queensland Health, and A.J.D. is supported by the Victorian Cancer Agency fellowship.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Kyaw Ze Ya Maung, Paul J. Leo.
These authors jointly supervised this work: Richard J D’Andrea, Thomas J Gonda.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41408-018-0090-7).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Richard J D’Andrea, Email: richard.dandrea@unisa.edu.au.
Thomas J Gonda, Email: Thomas.Gonda@unisa.edu.au.
References
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat. Rev. Cancer. 2018;18:168–185. doi: 10.1038/nrc.2017.116. [DOI] [PubMed] [Google Scholar]
- 3.Schlacher K. PARPi focus the spotlight on replication fork protection in cancer. Nat. Cell Biol. 2017;19:1309–1310. doi: 10.1038/ncb3638. [DOI] [PubMed] [Google Scholar]
- 4.Du W, Amarachintha S, Wilson AF, Pang Q. Hyper-active non-homologous end joining selects for synthetic lethality resistant and pathological Fanconi anemia hematopoietic stem and progenitor cells. Sci. Rep. 2016;6:22167. doi: 10.1038/srep22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan EL, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochowski A, et al. Patients with Fanconi anemia and AML have different cytogenetic clones than de novo cases of AML. Pediatr. Blood Cancer. 2012;59:922–924. doi: 10.1002/pbc.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers KJ, Fu W, Akey JM, Monnat RJ., Jr. Global and disease-associated genetic variation in the human Fanconi anemia gene family. Hum. Mol. Genet. 2014;23:6815–6825. doi: 10.1093/hmg/ddu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berwick M, et al. Genetic heterogeneity among Fanconi anemia heterozygotes and risk of cancer. Cancer Res. 2007;67:9591–9596. doi: 10.1158/0008-5472.CAN-07-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tischkowitz M, Easton DF, Ball J, Hodgson SV, Mathew CG. Cancer incidence in relatives of British Fanconi Anaemia patients. BMC Cancer. 2008;8:257. doi: 10.1186/1471-2407-8-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 12.Sedic M, Kuperwasser C. BRCA1-hapoinsufficiency: unraveling the molecular and cellular basis for tissue-specific cancer. Cell Cycle. 2016;15:621–627. doi: 10.1080/15384101.2016.1141841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 14.Pontel LB, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falzacappa MV, et al. The combination of the PARP inhibitor rucaparib and 5FU Is an effective strategy for treating acute leukemias. Mol. Cancer Ther. 2015;14:889–898. doi: 10.1158/1535-7163.MCT-14-0276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.