Abstract
Bupi Yishen Formula (BYF), a Chinese medicine preparation, has been clinically applied for the recovery of chronic kidney disease and for delaying its progress. Nevertheless, the chemical components in BYF have yet to be fully clarified. Ultra-high performance liquid chromatography with linear ion trap-Orbitrap mass spectrometry (UHPLC-LTQ-Orbitrap-MSn) and triple-quadrupole tandem mass spectrometry (UHPLC-TQ-MS/MS) methods were developed for qualitative chemical profiling and multi-components quantitative analysis in BYF. The chromatographic separation was performed on a Phenomenex Kinetex C18 column (2.1 × 100 mm i.d., 1.7 μm) using gradient elution of water (A) and acetonitrile (B) both containing 0.1% formic acid. Eighty-six compounds, including flavones, saponins, phenolic acids, and other compounds were authenticated or temporarily deduced according to their retention behaviors, mass mensuration, and characteristic fragment ions with those elucidated reference substances or literatures. Among the herbal medicinal materials of the formula, Astragali Radix, Codonopsis Radix, Salviae Miltiorrhizae Radix Rhizoma, and Polygoni Multiflori Radix Praeparata contributed to the bulk of the dissolved metabolites of the formula extraction. In addition, seven analytes were simultaneously determined by UHPLC-TQ-MS/MS, which was validated and has managed to determine major components in BYF. The study indicated that the established qualitative and quantitative methods would be potent and dependable analytical tools for characterizing multi-constituent in complex prescriptions decoction and provided a basis for the evaluation of bioactive components in BYF.
Keywords: Chinese medicine preparation, chemical analysis, multi-component determination, linear ion trap-orbitrap, quality control
Introduction
Chinese herbal medicine is the main form of clinical prevention and treatment of Traditional Chinese Medicine (TCM), the composition of which is composed of many different ingredients, and the organic combination of these different ingredients is different from adding individual ingredients simply. The material basis of Chinese herbal medicine is to coordinate and interact with each other so as to achieve the integrate function. Different from western medicine research, studies on Chinese herbal compound emphasize the integrity of the complex prescription, which should not split off from intrinsic characteristics of TCM and pursuit monomer compound (Wang et al., 2005). The material base of single herb or prescription is active substance groups. These groups of active substances are compatibly combined according to certain requirements, which act on multiple targets and thus has pleiotropic effects by multiple pathways (Xiong et al., 2015). Therefore, it is imperative to use modern advanced techniques to intrinsically explain the material basis of Chinese herbal medicine and to elaborate the connotation of compatibility and its curative effect.
Bupi Yishen Formula (BYF) is a non-herbal combination preparation of TCM which possesses the basic characterization of formula compatibility of TCM. BYF is prepared from the extract mixture of nine herbs, namely Astragali Radix, Codonopsis Radix, Atractylodis Macrocephalae Rhizoma, Poria, Dioscoreae Rhizoma, Polygoni Multiflori Radix Praeparata, Cuscutae Semen, Coicis Semen, and Salviae Miltiorrhizae Radix Rhizoma (Liu et al., 2012). The clinical application of BYF is treating and delaying the progression of chronic kidney disease, including postponing chronic renal failure symptoms, defering early and mid-renal dysfunction, delaying entering the dialysis time, and protection of residual renal function (Mao et al., 2015). Modern pharmacological studies revealed that the decoction could effectively delay glomerular filtration rate (GFR) of patients on the fourth stage of chronic kidney disease. Unambiguously, detecting and identifying the major components in BYF is a prerequisite and the hinge to disclose the active constituents and how they produce the effectiveness.
In recent years, reports on global characterizations of complicated ingredients in TCM prescriptions continues to grow steadily due to the recently rapid development of multifarious hyphenated and hybrid mass spectrometry (MS). Analytical methods have exhibited good performance in analysis of unknown targets from TCM prescriptions, containing LC-ESI/MS (Dou et al., 2009; Shaw et al., 2012), LC-TOF/MS (Sun et al., 2013), LC/MS-IT-TOF, etc. (Hao et al., 2008; Liu et al., 2016).
Ultra-high performance liquid chromatography (UHPLC) has been utilized in many bioanalytical fields in recent years due to its rapid analysis and excellent separation (Simons et al., 2009; Ha et al., 2013). Equipped with a relatively short column with a low flow rate, UHPLC usually cost a remarkably shorter analysis time to achieve the same separation efficiency as HPLC. The hybrid LTQ-Orbitrap analytical platform, being composed of an ion trap coupled with an Orbitrap mass analyzer, enables two scan types obtained at the same time. The Orbitrap provides relatively higher mass accuracy (< 3 ppm) and mass resolution than a number of other mass spectrometers, which is available for determining exact molecular formulas (Dunn et al., 2008; Tchoumtchoua et al., 2013). Moreover, multi-stage MSn mass spectra can be detected using ion trap by data-dependent scan and also minimize total analysis time, owing to its trigger for fragment spectra of target ions, and avoiding duplication by dynamic exclusion settings (Qiu et al., 2013). Thus, the LTQ-Orbitrap platform provides elemental compositions as well as multiple-stage mass data, which allow fast, sensitive, and reliable detecting, thus facilitating the identification of unknown compounds. Constituents of BYF could be structurally classified based on similar carbon skeletons, which should share a similar fragmentation pathway of each type and hence generate common characteristic product ions. Thus, mass spectra analysis for structural identifications could be facilitated by proposed strategies. In our previous study, the combination of UHPLC and LTQ-Orbitrap-MSn has been successfully used in analyzing multiple components in single herbal extracts (Xu et al., 2014; Wang et al., 2015; Zhang et al., 2015). In this study, we attempt to exploit it to detect and identify the TCM prescription, which contain hundreds of different chemical constituents.
The present work attempted to establish an expeditious UHPLC-LTQ-Orbitrap-MSn applicable approach for rapid separation and reliable identification of major constituents in BYF extract. Several strategies were used during the process, such as diagnostic fragment ions screening and fragment monitoring. In the decoction, eighty-six components altogether were identified or tentatively identified according to retention time and MS spectra data. Besides, a quantitative analysis approach has been constructed by Ultra-high performance liquid chromatography with triple quadrupole mass spectrometry (UHPLC-TQ-MS/MS). Seven representative compounds of relatively high contents unequivocally identified, were selected as marker components to evaluate the quality of BYF. The UHPLC-LTQ-Orbitrap MSn, and UHPLC-TQ-MS/MS platforms were proved as potent tools for both rapid qualitative and quantitative detection and analysis of complicated constituents from natural resources and the study facilitated the comprehensive quality control of BYF.
Experimental
Chemicals, reagents, and materials
Chemical references including calycosin-7-O-β-D-glucopyranoside, calycosin, formononetin, astragulin, salvianolic acid B, (E)-2,3,5,4′-Tetrahydroxystilbene-2-O-glucopyranoside ((E)-THSG) Astragaloside I, Astragaloside II, Astragaloside III, Astragaloside IV, soyasaponin I, lobetyolin, emodin were bought from Must (Chengdu, China). Rosmarinic acid, lithospermic acid, formononetin-7-O-glucopyranoside were from Yuanye (Shanghai, China). Salvianolic acid A was purchased from Feiyu (Jiangsu, China). Isomucronulatol-7-O-glucoside, 9, 10-di-methoxypterocarpan-3-O-β-D-glucopyranoside were isolated from Astragalus membranaceus and provided by Prof. Zhu Dayuan from Shanghai Institute of Materia Medica. (S)-THSG, emodin-8-O-β-D-glucoside and physcion-8-O-β-D-glucoside were isolated from Polygonum multiflorum in our lab. The purity of each standard was determined by HPLC (≥95%) and their structures were confirmed by MS, 1H-NMR, and 13C-NMR. All references were deliquated with methanol for at a concentration of 50.0 μg/mL.
HPLC-grade Acetonitrile, methanol, and formic acid were from Sigma Aldrich (MO, USA). Ultra-pure water was prepared by a Milli-Q water system (Millipore, MA, USA). Other reagents and chemicals were of analytical grade.
Astragali Radix (No. 11050419, Neimenggu), Codonopsis Radix (No. 110653271, Gansu), Atractylodis Macrocephalae Rhizoma (No. 110600711, zhejiang), Poria (No. 110506341, Hunan), Dioscoreae Rhizoma (No. 121001014, Henan), Polygoni Multiflori Radix Praeparata (No. 110400831, Henan), Cuscutae Semen (No. 110502581, Shandong), Coicis Semen (No. 110600371, Guizhou), and Salviae Miltiorrhizae Radix Rhizoma (No. 110601741, Anhui) were from Kangmei(Guangdong, China). They were authenticated by Dr. Huang Zhihai and the specimens were preserved in Guangdong Provincial Hospital of TCM. Two batches of BYF concentrated granule was produced in the pilot-scale by Peili Pharmaceutical Co., Ltd. (NanNing, China).
Preparation of calibration standard solutions
Standard of seven compounds was accurately weighed and dissolved in methanol separately to prepare the stock solution of each. A mixed stock solution was obtained, containing seven stock solutions, giving a concentration of 15.30 μg/mL for calycosin-7-O-Glc, 6.45 μg/mL for calycosin, 644.80 μg/mL for (E)-THSG, 6.03 μg/mL for astragulin, 15.60 μg/mL for rosmarinic acid, 8.10 μg/mL for salvianolic acid A, 0.812 μg/mL for salvianolic acid B, respectively. Daidzein (50.0 μg/mL) was also prepared with methanol to obtain the internal standard (IS) stock solution. To construct calibration curves, the mixed stock solution was continuously diluted for series concentrations at 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, and 1/128 of the original one. In a 2 mL volumetric flask, 0.2 mL of each concentration solution above, as well as 100 μL IS solutions were added, and all concentrations were finally diluted to 2 mL with 18% aqueous methanol. The acquired solutions were conserved at 4°C in refrigerator until use. All the solutions were filtered through 0.22 μm membranes before analysis.
Sample preparation
(i) Extraction of crude drugs: A total 125 g dry pieces of nine medicinal materials were mixed by prescription ratio and extracted with boiling water (1:10) for three times (45,30, and 30 min, respectively), filtered through gauze. Then three filtrates were combined and vacuum evaporated to recover the solvent at 56°C, and then BYF extract could be obtained. Extract was transferred into 250 mL volumetric flask, then adjusted to desired level with 10% methanol solution (final crude drug concentration was 0.5 g/mL). Solid-phase extraction (SPE) with C-18 column (ProElut, 200 mg, 3 mL column volume) was used for the pretreatment procedure, which had been conditioned with methanol (2 mL) and water (2 mL). After 1.0 mL of BYF extract was loaded, the column was washed by 10% methanol (2 mL), and eluted with 1.0 mL 100% methanol slowly. The dry pieces of each herb were disposed through the same procedure, thus individual decoction was obtained. All the sample solutions were passed through 0.22 μm membranes prior to analysis.
(ii) Pretreatment of decoction for quantitative study: The lyophilized powder of BYF decoction was produced by freeze-drying. 11.20 g lyophilized powder was acquired from 100 mL of BYF decoction. Five hundred and sixty milligrams of BYF was filtered, a 0.1 mL portion of which was added with 10 μL of the IS solution, and then was diluted with methanol to 5 mL. The sample solutions were passed through 0.22 μm membranes.
(iii) Pretreatment of BYF concentrated granule: Concentrated granule (2.0 g) was accurately weighed and precisely dissolved in 100 mL of 80% aqueous methanol, and then refluxed for 1 h. The extract was cooled down to room temperature, weighed and made up a deficiency by 80% methanol, which was then treated in the same way as (ii).
UHPLC-LTQ-orbitrap-MSn conditions
Chromatographic separation was conducted by a Thermo Accela UHPLC system (San Joes, USA) comprising an autosampler, a quaternary pump, a diode-array detector (DAD), and a column compartment settled to room temperature. A Phenomenex Kinetex C18 column (2.1 × 100 mm i.d., 1.7 μm) was utilized for sample separating. The mobile phase was mixture of water (A) and acetonitrile (B), both containing 0.1% formic acid. The elution gradient was set as follows: 0–12 min (10–25% B), 12–25 min (25–32% B), 15–42 min (32–56% B), 42–51 min (56–95% B). The injection volume of samples was 2 μL with a flow rate of mobile phase at 200 μL/min.
For qualitative experiments, a Thermo Fisher Scientific LTQ-Orbitrap XL hybrid mass spectrometer (Bremen, Germany) was hyphenated to the LC instrument via an electron spray ionization (ESI) interface. The samples were determined in negative mode. The ESI parameters were set (spray voltage was −3.5 KV; capillary temperature was 325°C; tube lens voltage was −76 V; Sheath gas and auxiliary gases were 45 and 6 units, respectively). The Orbitrap mass analyzer was set up the full scan mass range at m/z 120–1,200 of 30,000 resolution in centroided-type mass mode. In data-dependent MSn acquisition, the most intense ions were always selected for online MS2-MS3 analysis by FT and MS4-MS5 analysis by LTQ, and dynamic exclusion detection was also conducted during the process for repetition prevention. Dynamic exclusion parameters was set as follows: Repeat count, 2; Repeat duration, 0.35 min; Exclusion duration, 1.0 min; Exclusion mass width, 3 amu. The collision energy for collision-induced dissociation (CID) was set as 30 % of maximum.
The number and types of expected atoms were fixed as follows for possible elemental composition of components: carbons ≤ 50, hydrogens ≤ 80, oxygens ≤ 30, nitrogens ≤ 2. The accuracy error threshold was set at 3 ppm. The software of Thermo Fisher Scientific Xcalibur 2.1 was applied for data analysis.
UHPLC-TQ-MS/MS analysis
An AccelaTM UPLC system and a Thermo Scientific TSQ Quantum Ultra triple-quadrupole spectrometer (San Jose, USA) fitted with an ESI probe were employed for quantitative analysis. The separation column, column temperature, and the mobile phase were identical with those of qualitative conditions, with a gradient elution of 18–39% B at 0–5 min, 39–65% B at 5–7 min, 65–95% B at 7–9 min at 10–12 min with a flow rate at 250 μL/min. The injection volume was set at 5 μL.
Multiple reaction monitoring (MRM) was used for MS data acquisition and the conditions were designed as below: capillary temperature was 400°C; capillary voltage was 2.5 kV for negative mode and 3.0 kV for positive mode; sheath gas (N2) was pressure 40 psi; auxiliary gas was 8 psi; the dwell time was 100 ms. The detection parameters of target compounds were summarized in Table 1. Peak areas of each analyte and IS acquired in MRM mode were employed for calibration curve establishing. Data were collected and analyzed by Thermo Xcalibur 2.1.0 Software.
Table 1.
Chromatographic retention time, MRM parameters, and collision energy for the seven investigated compounds.
| Analytes | tR (min) | Ionization mode | Precursor ion (m/z) | Product ion (m/z) | Collision energy (eV) |
|---|---|---|---|---|---|
| Calycosin-7-O-Glc | 2.33 | ESI(+) | 447.13 | 285.10 | 27 |
| (E)-THSG | 2.65 | ESI(–) | 405.02 | 243.04 | 21 |
| Astragulin | 3.18 | ESI(–) | 447.02 | 255.04 | 35 |
| Rosmarinic acid | 3.71 | ESI(–) | 359.99 | 161.03 | 20 |
| Salvianolic acid A | 4.73 | ESI(–) | 492.99 | 295.03 | 18 |
| Salvianolic acid B | 4.21 | ESI(–) | 717.00 | 321.03 | 33 |
| Calycosin | 4.88 | ESI(–) | 283.36 | 268.04 | 24 |
| IS daidzein | 4.47 | ESI(+) | 254.90 | 199.01 | 23 |
Validation of quantitative method
The linear calibration curves were established by the analyte/IS ratio of each analyte (peak area ratio between each analyte and IS). Diluted standard solutions were successively analyzed until a signal-to-noise ratio (S/N) 3:1 and 10:1 were reached, respectively, to measure the limit of detection (LOD) and limit of quantification (LOQ) of each target compound. The intra-day precision was evaluated by detecting six times during 1 day, while the inter-day precision was assessed for 3 days in a row. Repeatability was obtained by six independent sample solutions using identical procedure in section Sample Preparation and variations were displayed by the relative standard deviation (RSD). One sample solution was tested at room temperature at different times within 24 h for stability evaluation. The recovery test was validated by adding known amounts of mixed reference solution to sample solutions at three concentration levels.
Results and discussion
Optimization of extraction procedure and analysis conditions
Variable factors during extraction procedures of BYF granule, including extraction solvent (water, 50, 80, and 100% methanol), method (reflux and sonication), solvent volume (30, 60, and 100 mL), and time (15, 30, 45, and 60 min) were optimized so as to extract the compounds efficiently. The optimized method was finally determined to extract the BYF granule with 100 ml of 80% methanol by refluxed for 1 h.
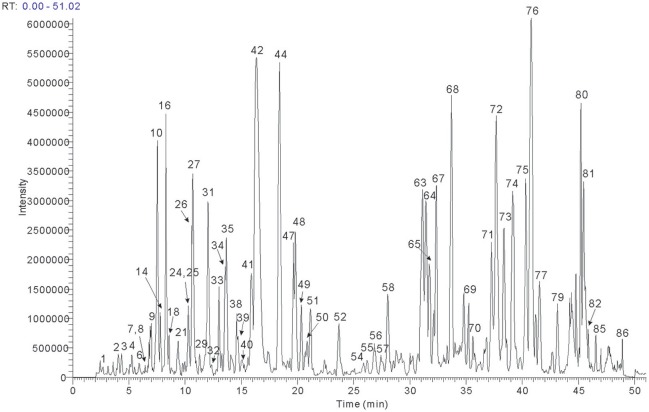
The UHPLC conditions were optimized, containing type of column, column temperature, mobile phase system, and flow rate. The Phenomenex Kinetex C18 column was selected based upon our previous multi-constituents analysis. Besides, different kinds of mobile phases were tested (acetonitrile and methanol with added modifiers, including formic acid, acetic acid, and ammonium acetate). A combination of acetonitrile and water both containing 0.1% (v/v) formic acid was found not only compatible to MS analysis, but also suitable for compounds separation for qualitative analysis. Comparing the TIC of the negative and positive modes, signal response was found more sensitive to the majority of components in negative mode, thus the MSn data were detected in negative mode. The total ion chromatogram of BYF was acquired for structure confirmation (shown in Figure 1).
Figure 1.
The total ion chromatogram in negtive mode of the BYF decoction.
Characterization of constituents in BYF extract
BYF extract was analyzed using the optimized UHPLC-LTQ-Orbitrap-MSn method. To elucidate the chemical components, known compounds were identified by comparing with the data of reference standards. Based on the MSn analysis of the authentic compounds, the characteristic fragmentation behaviors of each type with the same carbon skeleton were conducted, and thus applied the obtained rules to structure characterization of their derivatives. For other unknown compounds, the structures were tentatively identified according to MSn spectra and previous data in literatures. Eighty-six compounds in all were identified or tentatively identified (Table 2), including 15 flavones, 10 saponins, 12 phenolic acids, and other compounds. Nine herbs made markedly different chemical contributions to BYF. Specifically, the major constituents in BYF extract came from Astragali Radix (25 compounds), Codonopsis Radix (18 compounds), Salviae Miltiorrhizae (11 compounds), Cuscutae Semen (16 compounds), and Polygoni Multiflori Radix Praeparata (6 compounds).
Table 2.
Compounds detected and identified in BYF decoction.
| No. | tR/min | [M–H]− (Mass error, ppm) | Characteristic fragment ions | Molecular formula | Compounds | Resources |
|---|---|---|---|---|---|---|
| 1 | 2.67 | 353.086 82 (−0.44) | MS2 [353]: 191.1, 179.0 | C16H18O9 | 3-Caffeoyl-quinic acid | Cs |
| 2 | 4.01 | 353.086 76 (−0.49) | MS2 [353]: 191.1, 179.0 | C16H18O9 | 5-Caffeoyl-quinic acid | Cs |
| MS2 [191]: 173.0, 127.0 | ||||||
| 3 | 4.35 | 353.086 98 (−0.28) | MS2 [353]: 191.1, 179.0, 173.0 | C16H18O9 | 4-Caffeoyl-quinic acid | Cs |
| MS2 [191]: 173.0, 127.0 | ||||||
| 4 | 5.30 | 469.191 35a (−0.22) | MS2 [469]: 423.2 | C18H32O11 | (S)-3-Hexenyl-β-D-sophoriside | Cr |
| 423.185 76 (−0.32) | MS3 [423]: 261.1, 221.1, 179.1, 161.1 | |||||
| 5 | 5.48 | 350.19608b (−0.12) | MS3 [350]: 250.1, 205.1, 161.1 | C19H28NO | Codonopyrrolidium Ac | Cr |
| 6 | 5.84 | 469.19125a (−0.32) | MS2 [469]: 423.2 | C18H32O11 | (E)-2-Hexenyl-β-D-sophoriside | Cr |
| 423.185 91 (−0.60) | MS3 [423]: 261.1, 221.1, 179.1, 161.0 | |||||
| 7 | 6.44 | 405.118 07 (0.06) | MS2 [405]: 243.1 | C20H22O9 | (Z)-THSG | Pm |
| MS2[243]: 225.1, 215.1, 201.0, 173.0, 149.0, 137.0 | ||||||
| 8 | 6.52 | 677.228 09 (−0.65) | MS2[677]: 497.2, 453.2, 261.1 | C29H42O18 | Tangshenoside I | Cr |
| MS3[261]: 99.1, 161.1 | ||||||
| 9 | 6.96 | 471.207 40a (0.18) | MS2[471]: 425.2 | C18H34O11 | Hexyl β-sophoroside | Cr |
| 425.201 57 (−0.17) | MS3[425]: 263.1, 161.0 | |||||
| 10 | 7.50 | 491.118 65a (0.25) | MS2[417]: 283.1 | C22H22O10 | Calycosin-7-O-β-D-glup | Ar |
| MS3[283]: 268.0 | ||||||
| MS4[268]: 240.0, 224.0, 211.0, 184.0 | ||||||
| 11 | 7.45 | 595.129 33 (−0.03) | MS2[595]: 463.1, 301.0, 300.0 | C26H28O16 | Quercetin 3-O-(2-O-apisyl)-galactoside | Cs |
| MS3[300]: 271.0, 255.0 | ||||||
| MS4[271]: 243.0, 227.01, 199.0 | ||||||
| 12 | 7.70 | 469.133 51 (−0.54) | MS2 [469]: 325.1, 265.1, 235.1 | C21H26O12 | Tangshenoside IV isomer | Cr |
| MS3 [325]: 265.1, 235.1 | ||||||
| 13 | 7.74 | 441.196 66a (0.01) | MS2 [395]: 263.1, 161.0 | C17H32O10 | Hexyl-(pen)-glup | Cr |
| 395.191 10 (−0.21) | ||||||
| 14 | 7.79 | 633.180 91 (−0.49) | MS2 [633]: 471.1, 307.1 | C30H34O15 | Unknown | Cs |
| MS3[307]: 247.1, 205, 187, 163.1, 145.0 | ||||||
| 15 | 7.97 | 521.128 72a (0.25) | MS2[521]: 475.1, 359.1, 313.1, 298.1, 207.0 | C23H24O11 | Odoratin-glup | Ar |
| MS3[313]: 298.0, 283.0, 270.0 | ||||||
| 16 | 8.32 | 405.118 47 (−0.09) | MS2 [405]: 243.1 | C20H22O9 | (E)-THSG | Pm |
| MS2[243]: 225.1, 215.1, 201.0, 173.0, 149.0, 137.0 | ||||||
| 17 | 8.40 | 309.154 85a (0.46) | MS2 [309]: 263.1 | C12H24O6 | Hexyl β-D-glup | Cr |
| MS3 [263]: 161.1 | ||||||
| 18 | 8.54 | 463.087 52 (0.42) | MS2[595]: 463.1, 301.0, 300.0 | C21H20O12 | Quercetin 3-O-galactoside (hyperoside) | Cs |
| MS3[301]: 271.0, 255.0, 179.0, 151.0 | ||||||
| 19 | 8.88 | 603.227 84a (−0.51) | MS2 [603]: 557.2, 467.2, 341.1, 323.1 | C26H38O13 | Lobetyolinin | Cr |
| 20 | 9.10 | 557.128 11 (−0.86) | MS2[557]: 405.1, 313.1, 243.1 | C27H25O13 | 2,3,5,4′-Tetrahydroxystilbene-2-(galloyl)-O-glc | Pm |
| MS3[313]: 169.0, 295.0 | ||||||
| 21 | 9.32 | 261.133 79 (0.53) | MS2[261]: 187.1, 125.1 | C12H22O6 | Hexenyl-β-D-glup | Cr |
| 22 | 9.40 | 565.191 10a (−1.01) | MS2[519]: 357.1 | C26H32O11 | Unknown | |
| 519.185 85 (−0.79) | MS2[357]: 151.0, 136.0 | |||||
| 23 | 9.72 | 469.133 82 (−0.23) | MS2[469]: 407.1, 367.1, 325.1 | C21H26O12 | Tangshenoside V | Cr |
| MS3[325]: 163.0, 119.1 | ||||||
| 24 | 10.21 | 417.081 63[M-H]−(0.01) | MS2[417]: 373.1, 175.0 | C20H17O10 | Salvianolic acid D | Sm |
| MS2[307]: 175.0 | ||||||
| 25 | 10.30 | 537.102 29 (−1.9) | MS2[537]: 493.1, 339.0, 295.1 | C27H22O12 | Salvianolic acid H or I | Sm |
| MS3[339]: 321.0, 295.1, 185.0 | ||||||
| 26 | 10.60 | 187.097 35 (0.89) | MS2[187]: 125.1, 97.1 | C9H16O4 | Azelaic acid | Co / Cr |
| 27 | 10.69 | 447.092 41 (−0.33) | MS2[447]: 327.0, 285.0, 284.0 | C21H20O11 | Astragulin | Cs |
| MS3[284]: 255.0, 227.0 | ||||||
| 28 | 10.83 | 507.113 01a (−0.31) | MS2[507]: 461.1, 299.1 | C22H22O11 | Pratensein-7-O-Glu | Ar |
| MS3[299]: 284.0 | ||||||
| MS4[284]: 256.0, 227.0, 212.0 | ||||||
| 29 | 11.30 | 477.102 60 (−1.5) | MS2[477]: 314.0 | C22H22O12 | Unknown | / |
| MS3[314]: 285.0, 271.0, 243.0 | ||||||
| 30 | 11.86 | 441.175 29a (−0.23) | MS2 [441]: 395.1, 305.1, 215.1, 185.1, 179.1 | C20H28O8 | Lobetyolin | Cr |
| 395.169 65 (−0.40) | ||||||
| 31 | 12.03 | 359.076 51 (0.36) | MS2 [359]: 223.0, 197.1, 179.0, 161.0 | C18H6O8 | Rosmarimic acid | Sm |
| 32 | 12.33 | 537.102 48 (−0.27) | MS2[537]: 493.1, 339.0, 295.1 | C27H22O12 | Salvianolic acid H or I | Sm |
| MS2[493]: 321.0, 295.1 | ||||||
| 33 | 13.03 | 723.499 08a (0.78) | MS2[677]: 451.3 | C48H68O5 | Unknown | Cr/Am/Dr/ Ar |
| 677.493 90 (1.08) | MS2[677]: 433.4, 225.1 | |||||
| 34 | 13.56 | 537.102 97 (0.22) | MS2[537]: 493.1 | C27H22O12 | Lithospermic acid | Sm |
| MS3[493]: 295.1 | ||||||
| MS4[295]: 277.1, 159.0, 109.0 | ||||||
| 35 | 13.69 | 475.123 66a (0.17) | MS2[475]: 429.1, 267.1 | C22H22O9 | Formononetin-7-O-Glc | Ar |
| MS3[267]: 252.0, 22.9, 208.1 | ||||||
| 36 | 14.43 | 505.133 64a (−0.42) | MS2[475]: 297.1 | C23H24O10 | 6,4′-Dimethoxyisoflavone-7-O-Glc | Ar |
| MS3[297]: 282.1 | ||||||
| MS4[282]: 267.1, 254.1, 239.1 | ||||||
| 37 | 14.48 | 823.265 87 (0.35) | MS2 [823]: 497.2, 453.2, 261.1 | C38H48O20 | 6‴-Trans-p-coumaroyl-tangshenoside I | Cr |
| 38 | 14.60 | 471.128 48 (−1.4) | MS2 [471]: 307.1 | C24H24O10 | Unknown | Cs |
| MS3[307]: 247.1, 187.0, 163.1, 145.0 | ||||||
| 39 | 14.74 | 533.128 66a (−1.6) | MS2[533]: 487.1, 445.1, 283.1 | C24H24O11 | Calycosin-7-O-glc-6″-O-acetate | Ar |
| MS3[283]: 268.0 | ||||||
| MS4[268]: 240.0, 224.0, 211.0 | ||||||
| 40 | 15.13 | 823.265 81 (0.34) | MS2 [823]: 497.2, 453.2, 261.1 | C38H48O20 | 6‴-Cis-p-coumaroyl-tangshenoside I | Cr |
| 41 | 15.92 | 269.092 59 (−0.03) | MS2[269]: 225.1 | C15H14O3N2 | Cuscutamine | Cs |
| MS3[225]: 207.1, 183.1, 166.1, 156.1 | ||||||
| 42 | 16.38 | 717.144 65 (-0.36) | MS2[717]: 519.1, 321.0 | C36H30O16 | Salvianolic acid B | Sm |
| MS3[519]: 339.0, 321.0 | ||||||
| MS4[321]: 293.0, 279.1, 277.1 | ||||||
| MS5[279]: 251.1 | ||||||
| 43 | 16.43 | 507.150 36a (0.66) | MS2[507]: 461.1, 299.1, 284.1 | C23H26O10 | 9,10-Di-methoxypterocarpan-3-O-β-D-glucopyranoside | Ar |
| MS3[299]: 284.1, 269.0, 241.0 | ||||||
| 44 | 18.40 | 283.060 73 (0.63) | MS2[283]: 268.0, 240.0, 224.0, 211.0 | C16H12O5 | Calycosin | Ar |
| 45 | 18.54 | 509.165 44a (0.08) | MS2[509]: 463.1, 445.1, 346.1, 301.1, 286.0 | C23H28O10 | Isomucronulatol-7-O-β-D-glucopyranoside | Ar |
| MS3[301]: 286.1, 135.0, 109.0 | ||||||
| 46 | 19.37 | 523.123 23 (–0.27) | MS2[523]: 491.1, 343.1, 325.1, 293.0 | – | – | – |
| 47 | 19.63 | 493.113 16 (−0.6) | MS2[493]: 295.1 | C26H22O10 | Salvianolic acid A | Sm |
| MS3[295]: 277.1, 267, 185, 159.0, 109.0 | ||||||
| 48 | 19.81 | 717.143 92 | MS2[717]: 519.1 | C36H30O16 | Salvianolic acid E | Sm |
| (−1.09) | MS3[519]: 339.0, 321,.0 | |||||
| MS4[321]: 279.1, 251.1 | ||||||
| 49 | 20.34 | 551.118 35 (−1.0) | MS2[551]: 519.1 | C28H24O12 | Monomethyl lithospermater | Sm |
| MS3[519]: 353.1, 321.0 | ||||||
| 50 | 20.81 | 431.097 08(−0.19) | MS2[431]: 269.0 | C21H20O10 | Emodin-8-O-glc | Pm |
| MS3[269]: 225.0, 241.0 | ||||||
| MS4[225]: 181.0, 210.0 | ||||||
| 51 | 21.14 | 493.112 70 (−0.22) | MS2[493]: 295.1 | C26H22O10 | Isomer of Salvianolic acid A | Sm |
| MS3[295]: 277.1, 267.1, 185.1 | C36H30O15 | |||||
| 52 | 23.68 | 491.097 23 (−0.04) | MS2[491]: 293.0, 311.1 | C26H20O10 | Salvianolic acid C | Sm |
| MS3[293]: 276.0, 265.0, 249.1 | ||||||
| 53 | 24.23 | 577.154 30 (−0.88) | – | C27H30O14 | Unknown | |
| 54 | 25.32 | 297.076 02 (0.27) | MS2[297]: 282.1, 267.1, 254.1, 239.1 | C17H14O5 | 7-Hydroxy-6,4′-dimethoxyisoflavone | Ar |
| 55 | 26.18 | 517.133 67a (−0.38) | MS2[517]: 429.1, 267.1 | C24H24O10 | Formononetin-7-O-β-D-glycoside-6″-O-acetate | Ar |
| MS3[267]: 252.0 | ||||||
| MS4[252]: 223.1, 208.1 | ||||||
| 56 | 26.85 | 285.039 64 (−0.27) | MS2[285]: 257.0, 229.1, 213.1, 169.1, 199.1, 151.0 | C15H10O6 | Kaempferol or luteolin | Cs |
| 57 | 27.86 | 299.055 33 (0.32) | MS2[299]: 284.1, 271.1, 255.1, 240.1, 227.0 | C16H12O6 | Rhamnocitrin | Ar |
| 58 | 28.03 | 327.217 10 (0.50) | MS2 [327]: 291.2, 229.1, 211.1, 171.1 | C18H32O5 | 9,12,13-Trihydroxy-octadec-10,15-dienoic acid | Sm / Cr |
| MS3 [229]: 211.1 | ||||||
| 59 | 28.28 | 549.159 85a (−0.42) | MS2[549]: 485.1, 459.1, 415.1, 299.1 | C25H28O11 | 9,10-Di-methoxypterocarpan-3-O-β-D-glucopyranoside-acetate | Ar |
| MS3[299]: 284.1, 269.0, 241.0 | ||||||
| 60 | 29.56 | 991.509 34a (−1.50) | MS2[991]: 783.5, 765.5, 489.4 | C47H78O19 | Astragaloside VI or VII | Ar |
| 61 | 29.99 | 671.138 73 (−0.80) | MS2[671]: 473.1 | C35H29O14 | Unknown | - |
| MS3[473]: 339.1, 321.0 | ||||||
| 62 | 30.51 | 491.118 59a (0.19) | MS2[445]: 283.1 | C22H22O10 | Physcion-8-O-glc | Pm |
| 445.112 92 (−0.01) | MS2[283]: 268.0, 240.0 | |||||
| MS2[240]: 212.0, 184.1 | ||||||
| 63 | 31.13 | 267.065 92 (0.7) | MS2[267]: 252.0 | C16H12O4 | Formononetin | Ar |
| MS3[252]: 223.0, 208.0, 132.0 | ||||||
| 64 | 31.42 | 329.232 70 (0.45) | MS2 [329]: 311.2, 293.2, 229.1, 211.1 | C18H34O5 | 9,12,13-Trihydroxy-octadec-10-enoic acid | Cr |
| MS3 [171]: 153.0, 127.1, 125.1 | ||||||
| 65 | 31.72 | 329.232 64 (−0.16) | MS2 [329]: 311.2, 293.2, 229.1, 211.1 | C18H34O5 | 5,6,9-Trihydroxy-octadec-7-enoic acid | Cr / Co |
| MS3 [229]: 211.1, 209.1 | ||||||
| 66 | 33.03 | 991.508 61a (−2.22) | MS2[991]: 783.5, 765.5, 621.4, 489.4 | C47H78O19 | Astragaloside V | Ar |
| 67 | 32.35 | 843.421 45 (−0.57) | MS2[843]: 797.4, 779.4, 633.5 | C38H68O20 | Cuscutic acid C/Isomer | Cs |
| 68 | 33.70 | 843.421 08 (−0.94) | MS2[843]: 797.4, 779.4, 633.5 | C38H68O20 | Cuscutic acid C/Isomer | Cs |
| 69 | 35.23 | 829.456 18a (−1.83) | MS2[783]: 651.4, 621.4, 489.4 | C41H68O14 | Astragaloside IV | Ar |
| 783.450 20 (−2.33) | ||||||
| 70 | 35.62 | 829.455 26a(−2.75) | MS2[783]: 621.4, 489.4 | C41H68O14 | Astragaloside III | Ar |
| 783.450 26 (−2.27) | ||||||
| 71 | 37.26 | 885.431 03 (−2.10) | MS2[885]: 839.4, 821.4, 633.3 | C40H70O21 | Unknown | Cs |
| 72 | 37.70 | 885.431 27 (−1.31) | MS2[885]: 839.4, 821.4, 633.3 | C40H70O21 | Unknown | Cs |
| 73 | 38.35 | 885.431 09 (−1.49) | MS2[885]: 839.4, 821.4, 633.3 | C40H70O21 | Unknown | Cs |
| 74 | 39.13 | 885.431 27 (−1.31) | MS2[885]: 839.4, 821.4, 635.3 | C40H70O21 | Unknown | Cs |
| 75 | 40.30 | 871.467 29a (−1.29) | MS2[825]: 783.5, 765.4, 489.4 | C43H70O15 | Astragaloside II | Ar |
| 825.460 75 (−2.35) | ||||||
| 76 | 40.78 | 885.431 40 (−1.18) | MS2[885]: 839.4, 821.4, 633.5 | C40H70O21 | Acely-cuscutic acid C | Cs |
| 77 | 41.54 | 681.367 55 (−1.6) | – | C32H58O15 | Unknown | |
| 78 | 42.40 | 987.513 61a (−2.31) | MS2[941]: 923.5, 795.5, 615.4, 457.4, 437.4 | C48H78O18 | Soyasaponin I | Ar |
| 941.508 18 (−2.26) | ||||||
| 79 | 43.14 | 871.466 67a (−1.91) | MS2[871]:783.5, 765.5, 717.4, 633.4, 603.4, 489.4 | C43H70O15 | Isoastragaloside II | Ar |
| 825.46057 (−2.52) | ||||||
| 80 | 45.19 | 913.477 66a (-1.48) | MS2[867]: 825.5, 807.5, 765.5, 747.5, 729.5, 717.4, 705.5, 699.4, 633.4, 567.4, 489.4 | C45H72O16 | Astragaloside I | Ar |
| 867.471 07 (−2.59) | ||||||
| 81 | 45.45 | 295.227 36 (0.59) | MS2[295]: 277.2, 195.1, 171.1 | C18H32O3 | Coronaric acid | Co |
| MS3[277]: 233.2 | ||||||
| 82 | 45.49 | 913.476 5a (−2.58) | MS3[867]: 825.5, 807.5, 765.5, 747.5, 729.5, 717.4, 705.4, 699.4, 633.4, 567.4, 489.4 | C45H72O16 | Isoastragaloside I | Ar |
| 83 | 45.80 | 269.045 04 (0.59) | MS2[269]: 225.0 | C15H10O5 | Emodin | Pm |
| MS3[225]: 181.0, 210.0 | ||||||
| 84 | 45.86 | 915.493 23a (−1.56) | – | C45H72O16 | Dioscin | Dr |
| 85 | 46.55 | 955.487 79 (−1.92) | MS2[955]: 891.5, 763.5, 701.4, 613.4, 523.4 | C47H74O17 | Acetylastragaloside I | Ar |
| 86 | 48.91 | 339.231 75 (−0.3) | MS2[339]: 163.1 | C23H32O2 | Unknown | – |
[M+HCOO]−
[M+H]+
only detected in positive mode.
Cs, Cuscutae Semen; Cr, Codonopsis Radix; Pm, Polygoni Multiflori Radix Praeparata; Ar, Astragali Radix; Sm, Salviae Miltiorrhizae Radix Rhizoma; Co, Coicis Semen; Am, Atractylodis Macrocephalae Rhizoma; Dr, Dioscoreae Rhizoma; P, Poria; pen, pentoside; glup, glucopyranoside.
Compounds from astragali radix
Isoflavones and saponins, the major bioactive compounds in Astragali Radix, have various effects such as tonic, immunostimulant, cardioprotective diuretic, and hepatoprotective properties (Xu et al., 2006; Auyeung et al., 2009). In our work, 20 compounds from RA were totally characterized in BYF, including 12 isoflavones and 8 saponins. By comparing with information of reference standards, calycosin-7-O-β-D-glycoside, ononin, calycosin, formononetin, isomucronulatol-7-O-β-D-glucoside, 9,10-diMP-3-O-glucoside, 9,10-di-methoxypterocarpan-3-O-β-D-glucopyranoside, Astragaloside I, Astragaloside II, Astragaloside III, Astragaloside IV, and soyasaponin I were identified. Based on the MSn analysis of these authentic compounds, the characteristic fragmentation behaviors of isoflavones and saponins were proposed in our previous study (Zhang et al., 2015), which were applied for the structure elucidation of their derivatives.
The MS2 spectra of Compound 39 and Compound 55 exhibited characteristic product ions [M-C2H2O]− (m/z 475.1 and 429.1) and [M-glu-C2H2O]− (m/z 283.1 and 267.1), and their characteristic product ions yielded from the aglycone ion coincided with those of calycosin and formononetin. Based on the cleavage rules of loss of acetyl (42 Da) and acetylglucosyl (204 Da) groups, the two compounds were deduced as acetyl-glucoside of calycosin and formononetin.
Astragalosides from BYF decoction were mainly constituted by cycloastragenol aglycone, while aglycone ions or ions originated from the neutral loss of different glycosyl moiety in their MS2 spectra. Take Compound 69 as an example, the [M-H]− ion was m/z 783.450 20 (C41H67O), which easily lose the sugar units in its MS2 spectra and gained typical product ions at m/z 651, 621, 489 from the loss of one xylose ([M-132]−), one glucose ([M-164 (glu)]−), one xylose and glucose ([M-132 (xyl)-164 (glu)]−), respectively. In addition, one soyasaponin (Compound 78) of lower content from Astragali Radix was found in BYF decoction.
Compounds from codonopsis radix
The identified compounds of Codonopsis Radix in BYF can be classified into four main classes, namely, phenylpropanoid glycosides, acetylene glycosides, hexyl (hexenol) glycosides (Lin et al., 2013). In (-)ESI-MS spectra of BYF decoction, apart from phenylpropanoid glycosides (including compounds 8, 12, 23, 37, and 40) existing in [M-H]− ion forms, others displayed as both [M+HCOO]− and [M-H]− ions.
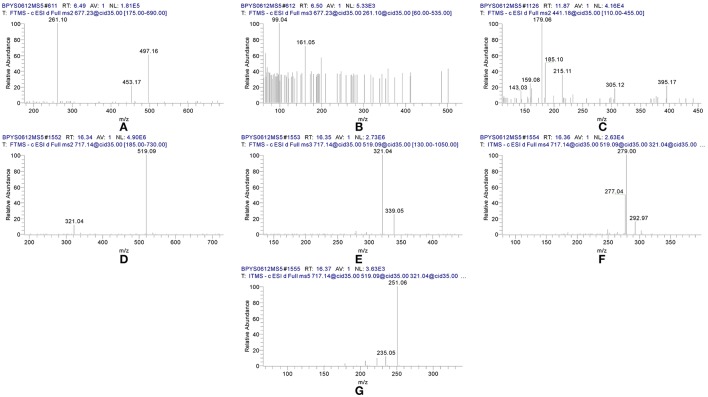
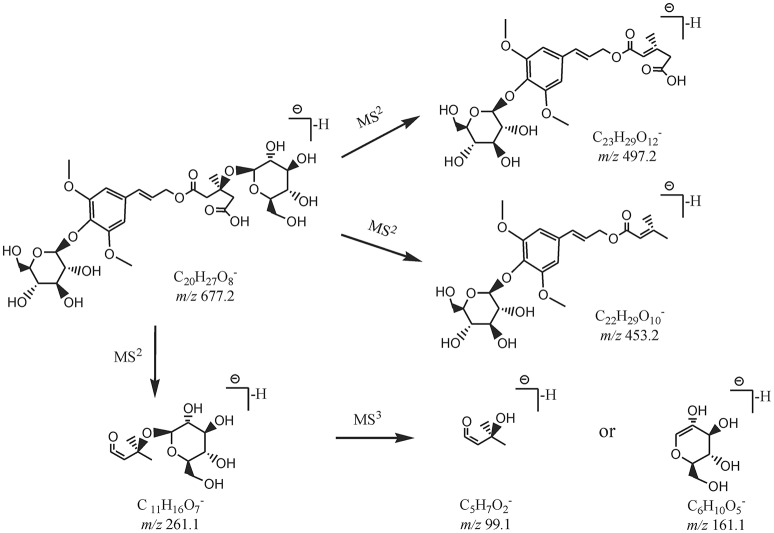
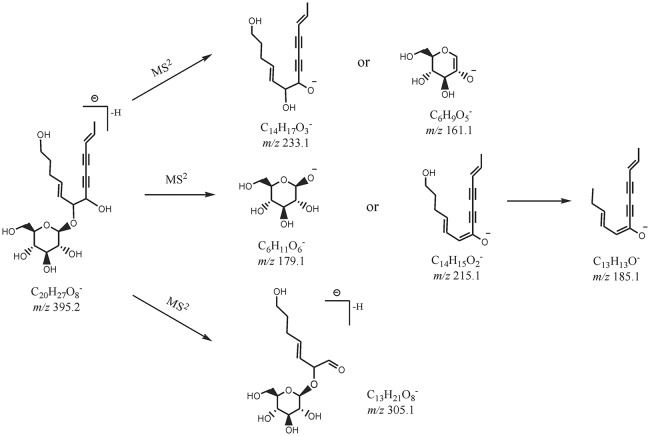
By comparing the retention time values and mass data with those of the references, Compound 8 and 30 were unambiguously identified as tangshenoside I and lobetyolin, which are the representative compounds of phenylpropanoid glycosides and acetylene glycosides in Radix Codonopsis. Their MSn spectra and proposed fragmentation pathways were summarized in Figures 2A–C, 3, 4, respectively. The [M-H]− ion and the typical ions in the MS2 spectra of Compound 19 (C26H37O), namely, m/z 557.2 [M-H]−, 467.2 [M-C7H6]−, 341.1 [M-C14H17O2]−, were all 162 Da less compared to those of Compound 30, demonstrating that they have identical site cleavage. Compound 19 was therefore characterized as lobetyolinin by comparison with the literature (Kanji et al., 2003).
Figure 2.
MSn spectra of tangshenoside I, lobetyolin, and salvianolic acid B. (A) MS2 of 667; (B) MS3 of 261; (C) MS2 of 441; (D) MS2 of 717; (E) MS3 of 519; (F) MS4 of 321; (G) MS5 of 279.
Figure 3.
The proposed MSn fragmentation pathways of tangshenoside I.
Figure 4.
The proposed MSn fragmentation pathways of lobetyolin.
Compounds 37 and 40 were identified as diastereomers by the same deprotonated ions at m/z 823.265 8 (C38H47O) and the same productions at m/z 497.2, m/z 453.2, and m/z 261.1, and they could be differentiated by their elution order. Their log P calculated by Discovery Studio were 0.47 and 0.59. As cis-isomers with lower polarity could by eluted relatively later than trans-isomers, compounds 37 and 40 were identified as 6′′′-trans- and 6′′′-cis-p-coumaroyl-tangshenoside I.
Compounds 9, 13, and 17 exhibited the [M+HCOO−]− precursor ion at m/z 471.207 40, 441.196 66, and 309.154 85, and their MS2 and MS3 spectra all yielded ions at m/z 263.1 (C12H23O) and 161.1 (C6H9O) as the base peak, respectively. It is inferred that Compounds 9 and 13 were, respectively, substituted by an additional glucopyranoside and pentoside compared to Compound 17. Compounds 9, 13, and 17 were tentatively characterized as hexyl β-sophoroside, hexyl-(pen)-glucopyranoside, and hexyl β-D-glucopyranoside.
Isomers (Compounds 4 and 6) were obtained by the EIC of m/z 423. Both of Them displayed [M-H]− ion at m/z 423.185 76 (C18H31O) and their MS2 spectra all exhibited ion at m/z 261.1 [M-H-C6H10O5]− and 161.1 (C6H9O). Based on the fragmentation information and related literature (Tsai and Lin, 2010), they were primarily identified as (S)-3-hexenyl-β-D-sophoriside and (E)-2-hexenyl-β-D-sophoriside. Their log P calculated by Discovery Studio were −1.9 and −2.0 and cis-isomers was eluted relatively later, therefore, compounds 4 and 6 were assigned as (S)-3-hexenyl-β-D-sophoriside and (E)-2-hexenyl-β-D-sophoriside.
Compounds from salviae miltiorrhizae radix rhizoma
Salviae Miltiorrhizae Radix Rhizoma was mainly composed of hydrophilic salvianolic acids and lipophilic diterpenoid quinines (Wu et al., 2007). This research adopted the water extraction method so that the major ingredients of Salviae Miltiorrhizae Radix Rhizoma in the BYF are primarily salvianolic acids. This type of compounds has high molecular weight and a lot of homologs, which display similar ESI-MSn behaviors for their differentiations.
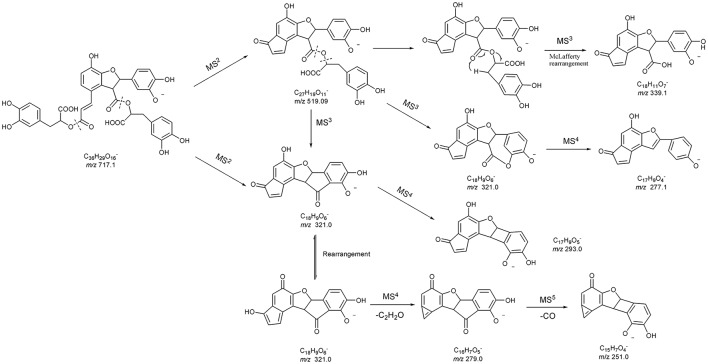
Compound 42 displayed the [M-H]− ions at m/z 717.144 65 with the elemental composition of C36H30O16. Its MS2 spectrum gave diagnostic fragment ions at m/z 519.1 and 321.0, caused by the loss of one and two molecular unit of Danshensu, respectively. In the MS3 spectrum, two distinctive ions at m/z 339.1 and 321.0, resulted from neutral loss of one danshensu and the McLafferty rearrangement, respectively, were observed. The CID of ion m/z 321.0 could furtherly produced MS4 and MS5 spectra. Its MSn spectra, as well as fragmentation pattern were shown in Figures 2D–G, 5. As its retention time and fragmentation ions were identical with those of the reference compound, Compound 42 was identified as salvianolic acid B.
Figure 5.
The proposed MSn fragmentation pathways of salvianolic acid B.
As for the fragmentation pathway of Salvianolic acids, with Danshensu as their parent nucleus, their loss of H2O and CO, as well as successive losses of Danshensu, occurred based on their MS spectra. Based on molecular weight and multi-stage information provided by MSn, combining with literatures (Hu et al., 2005; Liu et al., 2007; Zhu et al., 2010), 11 phenolic acids in the prescription were identified accurately.
Compounds from other component herbs
Phenolic constituents, including stilbenes and anthraquinones, were regarded as the main active components in Polygoni Multiflori Radix Praeparata (Chen et al., 2012). Most of these stilbenes previously reported were mainly 2,3,5,4′-tetrahydroxy substituted type (Liu et al., 2011). Compounds 7, 16, 50, 62, and 82 were indubitably distinguished as (E)-THSG, (S)-THSG, emodin-8-O-β-D-glucoside, physcion-8-O-β-D-glucoside, and emodin, by comparing their tR-values and mass information with those of the standards. The MS and MS2 spectra of Compounds 7, 16, and 20 showed characteristic ions at m/z 405.118 29 and 243.065 55, representing the corresponding elemental composition of C20H21O and C14H11O, which was consistent with our previous studies (Qiu et al., 2013). Fragmentation behaviors of anthraquinones were also accordance with previous results.
The components in Cuscutae Semen are mainly phenolic acids and flavonoids. For example, The MS2 and MS3 spectra of Compounds 1, 2, and 3 were in accordance with those of 3-CQA (caffeoyl-quinic acid), 5-CQA, and 4-CQA available from the literature (Zhang et al., 2007). Although the fragmentation data of 5-CQA and 4-CQA were the same, retention time of 5-CQA was shorter in reversed-phase chromatography. Elution orders of the three isomers were consistent with the reported.
Compounds 67 and 68 both exhibited [M-H]− ion at m/z 843.421 75 (C38H67O) and their MS2 spectra all identical ions, they were tentatively identified as cuscutic acid C and its isomers. Five isomers of acely-Cuscutic acid C (Compounds 71, 72, 73, 74, and 76) were presented by ion extraction at m/z 885 from TIC. They showed identical precursor ion and MS2 spectrum, while substituted position of their acetyl group was remained to be further studied.
Validation of quantitative method
Seven compounds, unequivocally identified with relatively high content in both the decoction and the granule, were selected as marker components to evaluate the quality of BYF. As the extraction process that we applied was through traditional method, which is extracted by water, the major components of high content were mainly water-soluble and highly polar compounds. These compounds have been observed at the early 25 min of the UHPLC-LTQ-Orbitrap-MS spectra. Meanwhile, those less polar compounds of much lower content emerged between 25 and 50 min, However, the relatively high peak area of such compounds in mass spectra has no direct relationship to their actual content in samples. Seven compounds for quantitative analysis were mainly phenolic and flavonoid compounds. Thus daidzein, a flavonoid, was chosen as the IS due to its structural and polar similarity with the analytes, and no daidzein exist nor be detected in BYF.
Nice linearity with coefficients of determination (R2 > 0.9994) were gained for the seven compounds. LOD and LOQ tests were carried out and listed in Table 3. The intra- or inter-day variations (RSD) were within the range of 0.24–2.99, 0.64–3.04, and 0.53–2.32%, 2.16–3.72% for mixed standard solution and sample solution, respectively (Table 4). Analytes in the sample solution were found stable for 24 h with a RSD < 3.38%. Recoveries of the fourteen compounds ranged from 95.86 to 104.04% with RSD from 1.12 to 4.02% (shown in Table 5). As a result, the developed UHPLC-TQ-MS/MS method was considered as a sensitive, repeatable and accurate tool for the quantitative analysis of main compounds in BYF.
Table 3.
Regression equations, linearity ranges, correlation coefficients, LOD, and LOQ data of the seven analytes.
| Analytes | Regression equations | Linear range (ng/mL) | R2 | LODa (ng/mL) | LOQb (ng/mL) |
|---|---|---|---|---|---|
| Calycosin-7-O-Glc | Y = 0.00591514X + 0.00256763 | 11.48~765.0 | 0.9999 | 0.03 | 0.08 |
| (E)-THSG | Y = 0.000746494X − 0.0040087 | 48.36~3,224.0 | 1.0000 | 0.02 | 0.04 |
| Astragulin | Y = 0.000933611X − 0.00599332 | 4.523~301.5 | 0.9996 | 0.02 | 0.75 |
| Rosmarinic acid | Y = 0.000437025X − 0.00906707 | 11.70~780.0 | 0.9994 | 0.98 | 3.91 |
| Salvianolic acid A | Y = 0.000390993X − 0.0022037 | 6.075~405.0 | 0.9995 | 0.51 | 2.53 |
| Salvianolic acid B | Y = 0.000286894X + 0.004386 | 609.0~40,600.0 | 0.9999 | 1.27 | 5.08 |
| Calycosin | Y = 0.00265233X + 0.00139572 | 5.038~322.4 | 0.9995 | 0.04 | 0.15 |
LOD, limit of detection.
LOQ, limit of quantification.
Table 4.
Precision, repeatability, and stability of the seven investigated compounds.
| Analytes | Precision (RSD %) | Repeatability (RSD%; n = 6) | Stability (RSD%; n = 6) | |||
|---|---|---|---|---|---|---|
| Standard solution | BYF extract | |||||
| Intra-day (n = 6) | Inter-day (n = 3) | Intra-day (n = 6) | Inter-day (n = 3) | |||
| Calycosin-7-O-Glc | 2.99 | 2.02 | 0.84 | 2.16 | 1.06 | 1.95 |
| (E)-THSG | 1.48 | 0.64 | 0.77 | 2.03 | 0.45 | 0.36 |
| Astragulin | 2.27 | 2.80 | 0.53 | 3.72 | 1.18 | 2.38 |
| Rosmarinic acid | 1.07 | 2.74 | 0.73 | 2.90 | 1.24 | 1.11 |
| Salvianolic acid A | 0.97 | 2.15 | 2.32 | 4.51 | 3.00 | 1.70 |
| Salvianolic acid B | 0.24 | 2.74 | 1.02 | 2.83 | 0.74 | 1.42 |
| Calycosin | 2.09 | 3.04 | 0.77 | 3.04 | 0.78 | 3.38 |
Table 5.
Recovery of the analytes.
| Analytes | Initial (mg) | Added (mg) | Detected (mg) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| Calycosin-7-O-Glc | 36.452 | 36.117 | 73.102 | 100.69 | 2.36 |
| 45.465 | 45.147 | 90.417 | 100.76 | ||
| 54.716 | 54.176 | 110.166 | 103.44 | ||
| (E)-THSG | 130.131 | 129.649 | 257.833 | 98.50 | 4.02 |
| 161.804 | 161.901 | 328.248 | 102.80 | ||
| 194.186 | 194.367 | 386.662 | 99.03 | ||
| Astragulin | 12.832 | 12.884 | 25.643 | 99.43 | 3.96 |
| 16.183 | 16.100 | 32.122 | 99.00 | ||
| 19.397 | 19.316 | 37.913 | 95.86 | ||
| Rosmarinic acid | 35.905 | 35.932 | 71.485 | 99.02 | 1.24 |
| 45.219 | 44.928 | 89.772 | 99.16 | ||
| 53.911 | 53.924 | 107.457 | 99.30 | ||
| Salvianolic acid A | 13.590 | 13.694 | 27.325 | 100.75 | 1.12 |
| 17.258 | 17.107 | 34.256 | 100.74 | ||
| 20.036 | 20.542 | 40.565 | 101.07 | ||
| Salvianolic acid B | 930.601 | 928.765 | 1, 869.590 | 101.10 | 2.11 |
| 1, 162.726 | 1, 160.186 | 2, 309.808 | 98.87 | ||
| 1, 395.901 | 1, 393.148 | 2, 806.526 | 101.25 | ||
| Calycosin | 15.531 | 15.505 | 31.663 | 104.04 | 2.58 |
| 19.362 | 19.374 | 39.527 | 104.08 | ||
| 23.197 | 23.243 | 46.378 | 99.73 |
Application to analysis of BYF samples
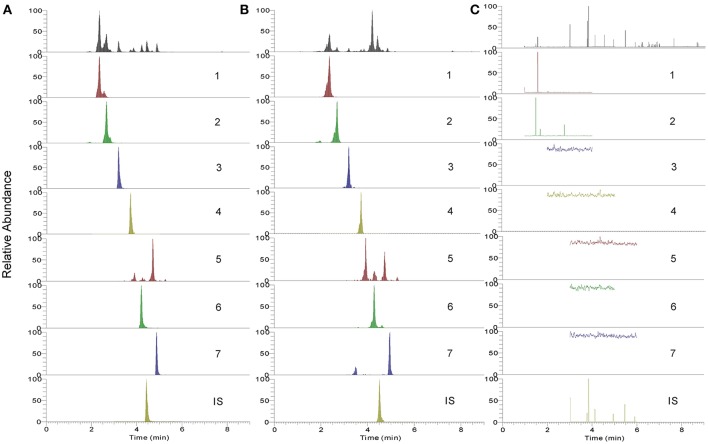
The established UHPLC-TQ-MS/MS method was subsequently applied for quantitative analysis of both BYF decoction and its preparations. Two different batches of BYF extract powder and three different batches of BYF granule were detected using the developed method. MRM chromatograms of seven main compounds in BYF were displayed in Figure 6. The contents of the investigated compounds were determined and the outcomes were shown in Table 6.
Figure 6.
The UHPLC-TQ-MS/MS analysis MRM chromatogram of 7 analytes. (A) mixed standard references solution; (B) sample solution of BYF; (C) negative samples; 1, Calycosin-7-O-Glc; 2, (E)-THSG; 3, astragulin; 4, rosmarinic acid; 5, salvianolic acid A; 6, salvianolic acid B; 7, Calycosin; IS, Daidzein.
Table 6.
Contents (μg/g, n = 3) of the seven investigated compounds in the samples of BYF extract powder and BYF granule.
| Analytes | Contents of BYF extract powder | Contents of BYF granule | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | ||
| Calycosin-7-O-Glc | 244.124 | 246.061 | 251.827 | 122.741 | 137.846 | |
| (E)-THSG | 382.673 | 466.849 | 438.201 | 270.693 | 305.022 | |
| Astragulin | 52.899 | 61.430 | 57.947 | 27.192 | 39.693 | |
| Rosmarinic acid | 10.447 | 29.943 | 23.690 | 63.700 | 103.399 | |
| Salvianolic acid A | 11.303 | 18.126 | 19.853 | 97.021 | 108.978 | |
| Salvianolic acid B | 5, 395.621 | 5, 769.462 | 5, 507.373 | 2, 059.143 | 2, 671.567 | |
| Calycosin | 38.498 | 43.058 | 40.046 | 16.167 | 11.441 | |
As shown in Table 6, salvianolic acid B was found as the most abundant compound, and compared with BYF decoction, the contents of most investigated compounds are relative low in concentrated granule. This difference might result from manufacturing procedures, namely, concentration, mixing, granulation, and drying processes. Meanwhile, the contents of rosmarinic acid and salvianolic acid A were much higher in concentrated granule than in BYF decoction. The variability could be explained because salvianolic acid B could be degradated and oxidized in the manufacturing procedures thus transform to other phenolic acids, such as rosmarinic acid, salvianolic acid A, lithospermic acid, etc. (Zheng and Qu, 2012).
Conclusion
In this paper, chemical constituents of BYF were systematically investigated by UHPLC-LTQ-Orbitrap-MSn and UHPLC-TQ-MS/MS methods, which provided comprehensively both qualitative and quantitative information for analysis of major components in BYF. Eighty-six compounds including flavones, saponins, phenolic acids, and other compounds were identified. The quantitative method was proved to have nice linearity, good accuracy, sensitivity, and repeatability. Although the bioactive components have not be determined, the present method will be helpful for providing the chemical basis for the further pharmacokinetic studies and effective quality evaluation of BYF, which would be of great importance for its safety use and mechanisms of action.
Author contributions
JZ and WX performed the experiments, analyzed the data, and wrote the paper. XL, ZH, and XQ conceived and designed the experiment, contributed reagent, materials, analysis tools, and revised the manuscript. PW, JH, and JB provided constructive suggestions for this research. All authors gave approval to the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Science and Technology Planning Project of Guangdong Province, China (2016A020226045, 2017A030313709, 2014A020221101, 2016A020226037, 2017B030314166), Pearl River S&T Nova Program of Guangzhou, China (201806010048), Special Subject of TCM Science and Technology Research of Guangdong Provincial Hospital of TCM, China (YN2016QJ07, YN2016QJ01) and Construction Project of TCM Hospital Preparation by Special Fund of Strong Province Construction in TCM, Guangdong, China (No. 6).
References
- Auyeung K. K., Cho C. H., Ko J. K. (2009). A novel anticancer effect of Astragalus saponins: transcriptional activation of NSAID-activated gene. Int. J. Cancer. 125, 1082–1091. 10.1002/ijc.24397 [DOI] [PubMed] [Google Scholar]
- Chen Q., Zhang S. Z., Ying H. Z., Dai X. Y., Li X. X., Yu C. H., et al. (2012). Chemical characterization and immunostimulatory effects of a polysaccharide from Polygoni Multiflori Radix Praeparata, in cyclophosphamide-induced anemic mice. Carbohyd. Polym. 88, 1476–1482. 10.1016/j.carbpol.2012.02.055 [DOI] [Google Scholar]
- Dou S. S., Liu L., Jiang P., Zhang W. D., Liu R. H. (2009). LC-DAD and LC-ESI-MS chromatographic fingerprinting and quantitative analysis for evaluation of the quality of Huang-Lian-Jie-Du-Tang. Chromatographia 69, 659–664. 10.1365/s10337-008-0945-3 [DOI] [Google Scholar]
- Dunn W. B., Broadhurst D., Brown M., Baker P. N., Redmand C. W., Kenny L. C., et al. (2008). Metabolic profiling of serum using Ultra Performance Liquid Chromatography and the LTQ-Orbitrap mass spectrometry system. J. Chromatogr. B 871, 288–298. 10.1016/j.jchromb.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Ha J., Shim Y. S., Seo D., Kim K., Ito M., Nakagawa H. (2013). Determination of 22 ginsenosides in ginseng products using ultra-high-performance liquid chromatography. J. Chromatogr. Sci. 51, 355–360. 10.1093/chromsci/bms148 [DOI] [PubMed] [Google Scholar]
- Hao H. P., Cui N., Wang G. J., Xiang B. R., Liang Y., Xu X. Y., et al. (2008). Global detection and identification of nontarget components from herbal preparations by liquid chromatography hybrid ion trap time-of-flight mass spectrometry and a strategy. Anal. Chem. 80, 8187–8194. 10.1021/ac801356s [DOI] [PubMed] [Google Scholar]
- Hu P., Liang Q. L., Luo G. A., Zhao Z. Z., Jiang Z. H. (2005). Multi-component HPLC fingerprinting of radix Salviae miltiorrhizae and its LC-MS-MS identification. Chem. Pharm. Bull. 53, 677–683. 10.1248/cpb.53.677 [DOI] [PubMed] [Google Scholar]
- Kanji I., Maiko O., Li Y., Toshihiro F., Kunihide M., Norie T. (2003). Polyacetylene glycosides from Pratia nummularia cultures. Phytochemistry 62, 643–646. 10.1016/S0031-9422(02)00669-6 [DOI] [PubMed] [Google Scholar]
- Lin L. C., Tsai T. H., Kuo C. L. (2013). Chemical constituents comparison of Codonopsis tangshen, Codonopsis pilosula var. modesta and Codonopsis pilosula. Nat. Prod. Res. 27:1812. 10.1080/14786419.2013.778849 [DOI] [PubMed] [Google Scholar]
- Liu A. H., Lin Y. H., Yang M., Guo H., Guan S. H., Sun J. H., et al. (2007). Development of the fingerprints for the quality of the roots of Salvia miltiorrhiza and its related preparations by HPLC-DAD and LC-MSn. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 846, 32–41. 10.1016/j.jchromb.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Liu C. H., Ju A. C., Zhou D. Z., Li D., Kou J. P., Yu B. Y., et al. (2016). Simultaneous qualitative and quantitative analysis of multiple chemical constituents in YiQiFuMai injection by ultra-fast liquid chromatography coupled with ion trap time-of-flight mass spectrometry. Molecules 21:640. 10.3390/molecules21050640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. S., Mao W., Yang N. Z., Zhou C., Li C., Zhang L., et al. (2012). Traditional Chinese Medicine Preparation for Treating Chronic Kidney Disease and its Preparation Method. P.R.China, Patent No 201210029862.8. Beijing: State Intellectual Property Office of the P.R.C. [Google Scholar]
- Liu Z., Liu Y. Y., Wang C., Guo N., Song Z. Q., Wang C., et al. (2011). Comparative analyses of chromatographic fingerprints of the roots of Polygonum multiflorum Thunb. and their processed products using RRLC/DAD/ESI-MSn. Planta Med. 77, 1855–1860. 10.1055/s-0030-1271200 [DOI] [PubMed] [Google Scholar]
- Mao W., Zhang L., Zou C., Li C., Wu Y. F., Su G. B., et al. (2015). Rationale and design of the Helping Ease Renal failure with Bupi Yishen compared with the Angiotensin II Antagonist Losartan (HERBAAL) trial: a randomized controlled trial in non-diabetes stage 4 chronic kidney disease. BMC Complement. Altern. Med. 15:316. 10.1186/s12906-015-0830-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. H., Zhang J., Huang Z. H., Zhu D. Y., Xu W. (2013). Profiling of phenolic constituents in Polygonum multiflorum Thunb. by combination of ultra-high-pressure liquid chromatography with linear ion trap-Orbitrap mass spectrometry. J. Chromatogr. A 1292, 121–131. 10.1016/j.chroma.2012.11.051 [DOI] [PubMed] [Google Scholar]
- Shaw L. H., Lin L. C., Tsai T. H. (2012). HPLC-MS/MS analysis of a traditional Chinese medical formulation of Bu-Yang-Huan-Wu-Tang and its pharmacokinetics after oral administration to rats (pharmacokinetics of herbal formulation). PLoS ONE 7:e43848 10.1371/journal.pone.0043848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R., Vincken J. P., Bakx E. J., Verbruggen M. A., Gruppen H. (2009). A rapid screening method for prenylated flavonoids with ultra-high-performance liquid chromatography/electrospray ionisation mass spectrometry in licorice root extracts. Rapid Commun. Mass Spectrom. 23, 3083–3093. 10.1002/rcm.4215 [DOI] [PubMed] [Google Scholar]
- Sun L., Wei H., Zhang F., Gao S. H., Zeng Q. H., Lu W. Q., et al. (2013). Qualitative analysis and quality control of traditional chinese medicine preparation tanreqing injection by LC-TOF/MS and HPLC-DAD-ELSD. Anal. Methods 5, 6431–6440. 10.1039/C3AY40681D [DOI] [Google Scholar]
- Tchoumtchoua J., Njamen D., Mbanya J. C., Skaltsounis A., Halabalaki M. (2013). Structure-oriented UHPLC-LTQ Orbitrap-based approach as a dereplication strategy for the identification of isoflavonoids from Amphimas pterocarpoides crude extract. J.Mass Spectrom. 48, 561–575. 10.1002/jms.3167 [DOI] [PubMed] [Google Scholar]
- Tsai T. H., Lin L. C. (2010). Phenolic glycosides and pyrrolidine alkaloids from Codonopsis tangshen. Chem. Pharm. Bull. 40, 1546–1550. 10.1248/cpb.56.1546 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y. Y., Yang G. (2005). Methods and modes about the theory of traditional Chinese prescription composition. China J. Chin. Mater. Med. 30, 6–8. 10.3321/j.issn:1001-5302.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Wang T. H., Zhang J., Qiu X. H., Bai J. Q., Gao Y. H., Xu W. (2015). Application of ultra-high-performance liquid chromatography coupled with LTQ-orbitrap mass spectrometry for the qualitative and quantitative analysis of Polygonum multiflorum thumb. and its processed products. Molecules 21:E40. 10.3390/molecules21010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. L., Yee L. P., Jiang Z. H., Cai Z. (2007). One single LC-MS/MS analysis for both phenolic components and tanshinones in Radix Salviae miltiorrihizae and its medicinal products. Talanta 73, 656–661. 10.1016/j.talanta.2007.04.038 [DOI] [PubMed] [Google Scholar]
- Xiong X., Wang P., Li X., Zhang Y. (2015). Shenqi pill, a traditional Chinese herbal formula, for the treatment of hypertension: a systematic review. Complement. Ther. Med. 23, 484–493. 10.1016/j.ctim.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Xu F., Zhang Y., Xiao S., Lu X., Yang D., Yang X., et al. (2006). Absorption and metabolism of astragali radix decoction: in silico, in vitro, and a case study in vivo. Drug Metab. Dispos. 34, 913–924. 10.1124/dmd.105.008300 [DOI] [PubMed] [Google Scholar]
- Xu W., Zhang J., Zhu D. Y., Huang J., Huang Z. H., Bai J. Q., et al. (2014). Rapid separation and characterization of diterpenoid alkaloids in processed roots of Aconitum carmichaeli using ultra high performance liquid chromatography coupled with hybrid linear ion trap-Orbitrap tandem mass spectrometry. J. Sep. Sci. 37, 2864–2873. 10.1002/jssc.201400365 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu X. J., Xu W., Huang J., Zhu D. Y., Qiu X. H. (2015). Rapid characterization and identification of flavonoids in Radix Astragali by ultra-high-pressure liquid chromatography coupled with linear ion trap-orbitrap mass spectrometry. J. Chromatogr. Sci. 53, 945–952. 10.1093/chromsci/bmu155 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Shi P., Qu H., Cheng Y. (2007). Characterization of phenolic compounds in Erigeron breviscapus by liquid chromatography coupled to electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2971–2984. 10.1002/rcm.3166 [DOI] [PubMed] [Google Scholar]
- Zheng X. T., Qu H. B. (2012). Characterisation of the degradation of salvianolic acid B using an on-line spectroscopic analysis system and multivariate curve resolution. Phytochem. Anal. 23, 103–109. 10.1002/pca.1330 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zhang H., Zhao L., Dong X., Li X., Chai Y. F., et al. (2010). Rapid separation and identification of phenolic and diterpenoid constituents from Radix Salvia miltiorrhizae by high-performance liquid chromatography diode-array detection, electrospray ionization time-of-flight mass spectrometry and electrospray. Rapid Commun. Mass Spectrom. 21, 1855–1865. 10.1002/rcm.3023 [DOI] [PubMed] [Google Scholar]