Abstract
The complement system plays a critical role in immune responses against pathogens. However, its identity and regulation in the lung are not fully understood. This study aimed to explore the role of tripartite motif protein (TRIM) 72 in regulating complement receptor (CR) of the Ig superfamily (CRIg) in alveolar macrophage (AM) and innate immunity of the lung. Imaging, absorbance quantification, and flow cytometry were used to evaluate in vitro and in vivo AM phagocytosis with normal, or altered, TRIM72 expression. Pulldown, coimmunoprecipitation, and gradient binding assays were applied to examine TRIM72 and CRIg interaction. A pneumonia model was established by intratracheal injection of Pseudomonas aeruginosa. Mortality, lung bacterial burden, and cytokine levels in BAL fluid and lung tissues were examined. Our data show that TRIM72 inhibited CR-mediated phagocytosis, and release of TRIM72 inhibition led to increased AM phagocytosis. Biochemical assays identified CRIg as a binding partner of TRIM72, and TRIM72 inhibited formation of the CRIg-phagosome. Genetic ablation of TRIM72 led to improved pathogen clearance, reduced cytokine storm, and improved survival in murine models of severe pneumonia, specificity of which was confirmed by adoptive transfer of wild-type or TRIM72KO AMs to AM-depleted TRIM72KO mice. TRIM72 overexpression promoted bacteria-induced NF-κB activation in murine alveolar macrophage cells. Our data revealed a quiescent, noninflammatory bacterial clearance mechanism in the lung via AM CRIg, which is suppressed by TRIM72. In vivo data suggest that targeted suppression of TRIM72 in AM may be an effective measure to treat fatal pulmonary bacterial infections.
Keywords: complement receptor of the Ig family, tripartite motif-containing protein 72, phagocytosis, NF-κB signaling, bacterial pneumonia
Pulmonary infections are the most common cause of hospitalization, and frequently lead to acute respiratory distress syndrome (1). Treatment of pulmonary infections has become increasingly challenging due to a burgeoning proportion of immunocompromised patients and the emergence of antibiotic-resistant strains of bacteria (2), many of which are common lung pathogens, such as Pseudomonas aeruginosa (P.a.). In addition to efforts to develop new antibiotics, elucidating the unrevealed mechanisms of lung host defense is crucial for preventing organ injury and morbidity after severe pulmonary infections.
Alveolar macrophages (AMs) are the major sentinel cells of the lung. An initial step of the lung’s innate immune response is phagocytosis of the invading pathogens by AMs. This may be mediated by pathogen recognition receptors binding to pathogen-associated molecular patterns, or by recognition of opsonized pathogens by the complement receptors (CR) or Fcγ receptors (FcγR) (3, 4), triggering downstream signals to mediate a wide range of responses, including proinflammatory, noninflammatory, and antiinflammatory (5). Previous studies suggest that a dynamic cross-talk exists among the various signaling pathways activated by the same pathogen, and balance of this cross-talk determines the efficiency of pathogen clearance and the severity of inflammation during the innate immune response (6, 7).
In the alveoli, abundant surfactant proteins, members of the collagen-like lectins (collectins), opsonize airborne pathogens and facilitate activation of the complement system via the lectin pathway (8). Studies found that complement proteins are present in the alveoli, whereas C3b levels were elevated in alveolar lining fluid, from aging mice and humans (9). The complement pathway has also been shown to be involved in host defense, allergy, and tissue injury of the lung (10–12). Receptors for complement fragment-opsonized pathogens (i.e., CRs) mediate the immune effector function of the activated complement system, which is grouped into three families: the short consensus repeat family CR1 and CR2, the β2-integrin family CR3 and CR4, and the CR of the Ig superfamily (CRIg) (4). Of these CRs, CR2 mainly functions in enhancing B cell immunity (13), whereas CR3 and CR4 play wide roles in leukocyte trafficking and migration, synapse formation, and costimulation (14) due to their ability to adhere cells to the extracellular matrix. CR-mediated phagocytosis was generally regarded as less robust than FcγR-mediated phagocytosis (15) in removing invading pathogens. However, CR1 was shown to play a role in phagocytosis by neutrophils, but not by tissue macrophages (16). In addition, the CR identified in 2006 (17, 18), CRIg, was shown to selectively express in tissue macrophages and function to remove C3b or iC3b-opsonized pathogens (17). Nevertheless, the contribution of CRIg in phagocytosis of AMs and innate immunity of the lung has not been reported.
Our previous studies found that tripartite motif family protein (TRIM) 72 is a critical membrane repair gene for alveolar epithelial cells (19, 20), and other studies demonstrated that the E3 ligase activity of TRIM72 is involved in tissue development (21), glucose metabolism (22), and focal adhesion (23). Despite a great number of TRIM family members being shown to participate in innate host defense (24, 25), the biological function of TRIM72 in innate immune response has never been reported before. In this study, we revealed that CRIg is expressed in AMs, and contributes to a noninflammatory type of bacterial phagocytosis. Pulldown and protein–protein binding assays identified an interaction between CRIg and TRIM72. Biochemical and imaging analysis showed that CR phagocytosis, and CRIg-mediated phagosome formation, were inhibited by the presence of TRIM72. Removing TRIM72 inhibition of CRIg through genetic ablation increased resilience to severe pulmonary infections in mice, supporting the substantial potential of the normally quiescent CRIg-mediated bacterial clearance and its overall impact in innate immune defense of the lung.
Methods
Beads and Zymosan Phagocytosis
Primary AMs were incubated with medium containing FITC beads in noninactivated serum (Φ = 2 μm, 50 beads/cell) or Alexa Fluor 488-Zymosan-A bioParticles (100 particles/cell). After 1 hour, cells were extensively washed and fixed with 4% paraformaldehyde. AMs containing fluorescent beads/particles were counted to determine % phagocytic cells. FITC+ AMs were also quantified by flow cytometry and counted for phagocytic indexes (number of ingested beads/macrophages) (26).
FcγR- and CR-mediated Phagocytosis
Lentiviral L309C- or L309C-TRIM72–transduced murine alveolar macrophage cells (MH-S) were seeded in 96-well plates at 10,000 cells/well. After attachment, 100 μl/well 1 × 107/ml IgG- or IgM-opsonized sheep red blood cells (sRBCs) were added to the cell monolayer for 1 hour. Unbound sRBCs were removed with ammonium-chloride-potassium lysis buffer. MH-S cells were then lysed and treated with 50 μl 2,7-diaminofluorene containing 3% hydrogen peroxide and 6 M urea to facilitate hemoglobin-catalyzed fluorene blue formation. Absorbance was measured at 620 nm, and compared with standards. For the phagosome imaging study, human embryonic kidney (HEK) 293 cells were cotransfected with human CRIg (hCRIg)-mCherry plus GFP–human TRIM72 (hTRIM72) or controls, and bare sRBCs or IgM-opsonized sRBCs were incubated with the cells for 10 minutes before imaging.
In Vivo Bacterial Phagocytosis
50 μl of 5 × 106 cfu/ml P.a. GFP was intratracheally administered into lungs of anesthetized mice. After 1 hour, BAL fluid (BALF) was pelleted to prepare cytospin slides, which were stained with Kwik diff-staining. GFP+ AMs were counted using a fluorescent microscope (% phagocytic cells). Differential cell counting was performed for macrophages and neutrophils based on cell morphology (27).
Pulldown Assay and Gradient Protein–Protein Interaction
His-tagged TRIM72 recombinant protein (bait) was immobilized on HisPur Cobalt resin in a Pierce Pull-Down PolyHis Protein:Protein Interaction Kit, and incubated with 800 μl of deidentified human lung samples (prey) overnight at 4°C. The bait–prey complex was eluted, separated on SDS-PAGE, and visualized with silver staining. Positive bands were excised for mass spectrometry protein identification. For gradient protein–protein interaction (28), equal molar concentrations of maltose binding protein (MBP) or MBP-TRIM72 immobilized on amylose resin was incubated with 0, 1, 2.5, 7.5, or 10 μg His-tagged hCRIg recombinant protein overnight at 4°C. After washing, the protein complex was eluted with buffer containing 10 mM maltose. Eluent was separated on SDS-PAGE, transferred to 0.2-μm nitrocellulose membrane, and blotted with anti-CRIg antibody.
Bacterial Pneumonia Models
First, sublethal (2.5 × 105 cfu/ml) or lethal (3 × 107 cfu/ml) doses of P.a. were injected into wild-type (WT) and TRIM72KO mice intratracheally (29). In a separate experiment, 50 μl of clodronate liposomes were injected intratracheally to TRIM72KO mice to deplete AMs, and adoptive transfer of 1 × 106 WT or TRIM72KO AMs was conducted via endotracheal instillation after 72 hours (30, 31); 48 hours later, the mice were challenged with a lethal dose of P.a. Body weight was measured daily. Mortality, lung bacterial burden, and cytokines in BALF or lung tissue were determined at Day 2 after infection.
Statistical Analysis
All data were analyzed using Origin 6.0 and IBM SPSS Statistics version 25, and presented as mean (±SE). Statistical significance was assumed at P less than 0.05 or P less than 0.005. See the data supplement for details.
Results
CRIg Is a Novel Interacting Partner of TRIM72 in the Lung
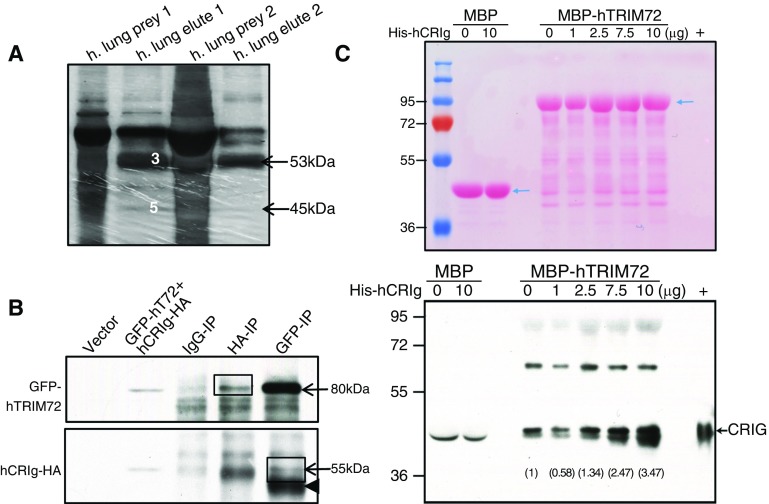
To identify novel interacting partners of TRIM72 in the lung, we performed pulldown experiments using His-tagged hTRIM72 recombinant protein as bait, and cryopreserved normal human lung tissues as prey. Pierce cobalt chelate resin His protein pulldown kit was used for on-column bait–prey interaction with reduced nonspecific binding. Pulldown bands from two separate experiments were sent for protein identification (Figure 1A). A novel tissue macrophage–specific phagocytic receptor (i.e., CRIg [17, 18]) was identified in both experiments (along with complement proteins). The interaction between TRIM72 and CRIg was further confirmed by coimmunoprecipitation of GFP-tagged hTRIM72 and human influenza hemagglutinin-tagged hCRIg (hCRIg-HA) in HEK293 cells (Figure 1B). Furthermore, we conducted gradient binding assays, using set amounts of MBP-tagged recombinant hTRIM72 or MBP as a control, on an amylose column to capture increasing amounts of hCRIg recombinant protein input (Figure 1C). Our results suggest specific binding between the two proteins, as CRIg protein was captured by TRIM72 proportionally to the amount of input (Figure 1C).
Figure 1.
Complement receptor (CR) of the Ig superfamily (CRIg) is a novel interacting partner of tripartite motif protein (TRIM) 72 in the lung. (A) Silver staining of prey (human lung lysates) and elutes in two in-column poly-his–tagged protein interaction assays. Target proteins were identified by mass spectrometry, including human TRIM (hTRIM) 72 (band 3 in prey1 and arrow in prey2, ∼53 kD) and human CRIg (hCRIg; short form, band 5 in prey1 and arrow in prey2, ∼45 kD). (B) Coimmunoprecipitation of GFP-tagged hTRIM72 (GFP-hT72) and hemagglutinin-tagged hCRIg (hCRIg-HA) in human embryonic kidney (HEK) 293 cells; arrows, GFP-T72 at roughly 80 kD, hCRIg-HA at roughly 55 kD (long form); arrowhead, IgG heavy chain; IP = immunoprecipitation; box, specific interaction bands. (C) Ponceau S staining (upper panel) and Western blot detection of CRIg (lower panel) in the gradient binding elutes using approximately 0.4 μM recombinant maltose binding protein (MBP) or MBP-hTRIM72 with a designated amount of His-hCRIg protein (short form, ∼45 kD); arrows, MBP or MBP-hTRIM72 (upper panel), CRIg (lower panel); +, His-hCRIg. Note: a doublet was detected by anti-CRIg close to the “+” control, but only the upper band is a specific CRIg band. Gray values (numbers) of the CRIg band were quantified and labeled on the Western image. Experiments were repeated three times.
CRIg Expresses in the Lung
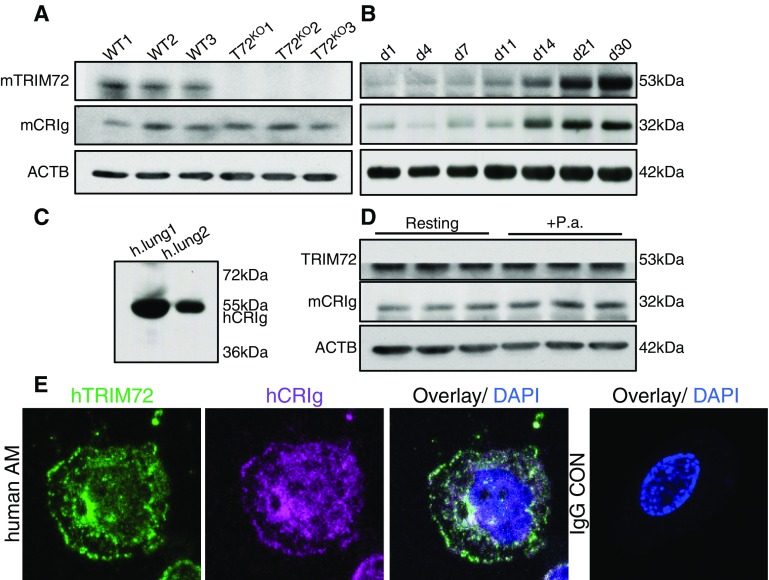
It was reported previously that CRIg expresses in tissue macrophages in the liver (17, 18). To further characterize the expression of CRIg in the lung, we first detected CRIg protein in primary AMs by Western blot. As shown in Figure 2A, a roughly 32-kD CRIg band was detected in AMs from WT lungs, and CRIg expression was not altered by genetic TRIM72 ablation. In addition, Figure 2B showed that expression of TRIM72 and CRIg in neonatal lungs during the first 30 postnatal days increase in parallel, indicating an important role of these two proteins in mature lungs. A roughly 55-kD CRIg band was detected in human lung homogenates (Figure 2C). This species difference in CRIg molecular size is likely due to the presence of both a long and a short form of CRIg variant in human tissues, but only a short-form variant in mouse tissues, as reported previously (17). Furthermore, exposure to P.a. bacteria caused a slight trend of increased CRIg expression (not statistically significant), but no change in TRIM72 expression was observed (Figure 2D). The subcellular localization of TRIM72 and CRIg in resting primary AMs was examined by immunostaining. As shown in Figure 2E, TRIM72 and CRIg are present on the plasma membrane and paranuclear regions in primary AMs isolated from human lungs, where partial colocalization of TRIM72 and CRIg was observed.
Figure 2.
CRIg is expressed in the lung. (A) Western blot detection of mouse TRIM72 (mTRIM72) and mouse CRIg (mCRIg; ∼32 kD) in primary alveolar macrophages (AMs) from the wild-type (WT) and TRIM72KO (T72KO) mice. (B) CRIg and TRIM72 were detected in developing mouse lungs from Postnatal Day 1 to Postnatal Day 30 (d1–d30) by Western blot; levels of β-actin (ACTB) were used as loading controls. (C) Detection of endogenous CRIg in human (h) lungs (long form, ∼55 kD). (D) TRIM72 and CRIg were detected in WT AMs at resting stage, or exposed to Pseudomonas aeruginosa (P.a.) for 60 minutes. (E) immunostaining detection of TRIM72 (green) and CRIg (magenta) in primary human AMs. Cell was costained with DAPI (blue), and negative control using IgG only was included. Experiments were repeated three times.
TRIM72 Regulates CR-mediated Phagocytosis in AMs
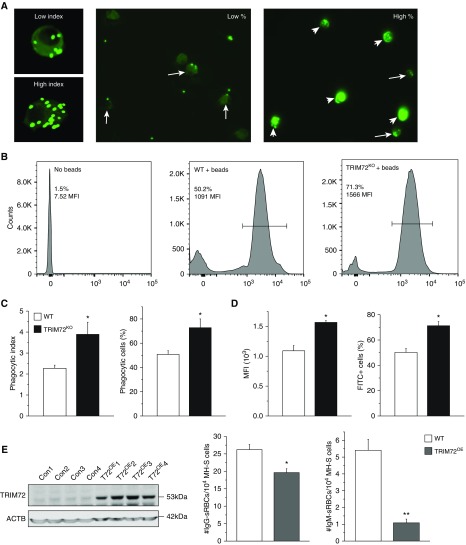
To dissect the physiological role of the interaction between TRIM72 and CRIg, we first conducted experiments to examine AM phagocytosis. Indiscriminate opsonized phagocytosis was examined using non–heat-inactivated FBS-coated FITC beads (26). Our results show that, in the absence of TRIM72, both the phagocytic index and percentage of phagocytic cells were significantly increased as compared with the WT. This was confirmed by both imaging analysis and flow cytometry analysis of FITC+ AMs (Figures 3A–3D). This suggests that TRIM72 plays an inhibitory role in AM phagocytosis. As opsonized phagocytosis may be mediated by either CR or FcγR, we next tested the specificity of TRIM72 inhibition on these two types of receptor-mediated phagocytosis. Murine AM cell line, MH-S, cells were infected with lentivirus-containing full-length TRIM72 (TRIM72OE) or control viral particles (Figure 3E). FcγR- and CR-mediated phagocytosis was examined by quantification of AM-internalized sRBCs that were coated with either anti-sRBC IgG or anti-sRBC IgM, respectively (32). Our results showed that TRIM72OE led to a slight reduction in FcγR-mediated phagocytosis, but a more than fivefold decrease in CR-mediated phagocytosis (Figure 3E), suggesting that TRIM72 is a preferential inhibitor of CR-mediated phagocytosis. The status of phagocytic pathogen recognition receptor–mediated phagocytosis was examined by ingestion of Alexa Fluor 488–conjugated Zymosan particles by WT and TRIM72KO AMs. Our data show that this particular phagocytic mechanism was highly active in AMs (78–80% phagocytic cells), which was not affected by TRIM72 ablation (see Figure E1 in the data supplement).
Figure 3.
TRIM72 regulates CR-mediated phagocytosis in AMs. (A) Representative images of low versus high phagocytic indexes showing primary AMs containing green fluorescent beads (left); representative images showing low and high % phagocytic AMs. Arrows, low phagocytic index AMs; arrowheads, high phagocytic index AMs. (B) Representative flow cytometry detection of phagocytizing cells in no beads control, WT + beads, and TRIM72KO + beads AMs. Bar defines bead-containing cell population (%) and mean fluorescence intensity (MFI). (C) Statistics of average phagocytic index and % phagocytic AMs in WT and TRIM72KO AMs; n = 5 for both groups, *P < 0.05. (D) Statistics of flow cytometry MFI and % FITC+ cells in WT and TRIM72KO AMs; n = 3 for both groups, *P < 0.05. (E) Western blot detection of TRIM72 overexpression (T72OE) after lentivirus L309C infection of murine alveolar macrophage cells (MH-S) with control and TRIM72 containing vectors (left); ACTB was detected as a loading control. Quantification of opsonized sheep red blood cell (sRBC) phagocytosis by MH-S cells (right) in the presence of IgG (Fcγ receptor [FcγR]–mediated phagocytosis) or IgM (CR-mediated phagocytosis); n = 6 for each group, *P < 0.05 and **P < 0.005 compared with WT control. Data are presented as mean (±SE). Con = control; TRIM72KO = TRIM72 knockout; TRIM72OE = TRIM72 overexpression.
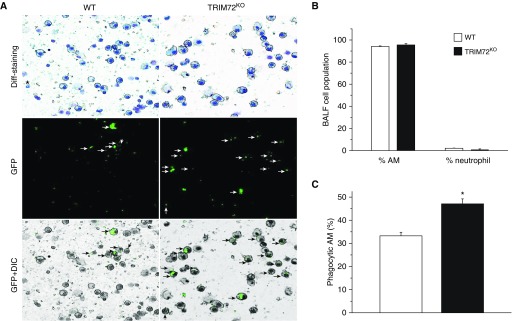
Next, we evaluated the effect of TRIM72KO on phagocytosis of lung pathogens in vivo after intratracheal instillation of P.a. GFP. The percentage of AMs and neutrophils in BALF at 1 hour after P.a. injection was quantified. As shown in Figures 4A and 4B, a majority of BALF cells were AMs, with some lymphocytes and sparse neutrophils. This was consistent with the timeline of neutrophil recruitment into the airspace after P.a. infection (33, 34). Nevertheless, we observed an increase in the percentage of phagocytic AMs in TRIM72KO mice as compared with the WT mice (Figure 4C), similar to the in vitro results presented previously here. Because neutrophils are reported as one of the major cell types mediating host immune responses to P.a. infection (35), we sorted neutrophils from the WT lungs using Ly6G as a cell surface marker for neutrophils (36), and examined TRIM72 expression in these cells (Figure E2). LPS was administered to the lung for 24 hours to induce substantial neutrophil infiltration into the lung (34). As seen in Figure E2, we did not detect any TRIM72 protein in Ly6G+ cell populations, even in the LPS-treated lungs, so we conclude that TRIM72 does not express in neutrophils, and thus TRIM72 knockout is not likely to affect neutrophil phagocytosis of the injected P.a. bacteria.
Figure 4.
TRIM72 ablation enhances in vivo AM phagocytosis of P.a. (A) Representative images of BAL fluid (BALF) cell cytospin slides from WT and TRIM72KO mice 1 hour after injection of P.a. GFP. Kwik-diff staining identifies AMs (large, round cells) and neutrophils (star); GFP identifies phagocytic cells (white arrows) and GFP+ differential interference contrast (DIC) identifies internalized GFP bacteria (black arrows) in AMs. (B) Quantification of percentage of AMs and neutrophils in BALF of WT and TRIM72KO mice 1 hour after P.a. injection. (C) Quantification of percentage of phagocytic AM in WT and TRIM72KO mice; n = 3 for each group, *P < 0.05. Data are presented as mean (±SE).
TRIM72 Directly Inhibits CRIg-mediated Phagosome Formation
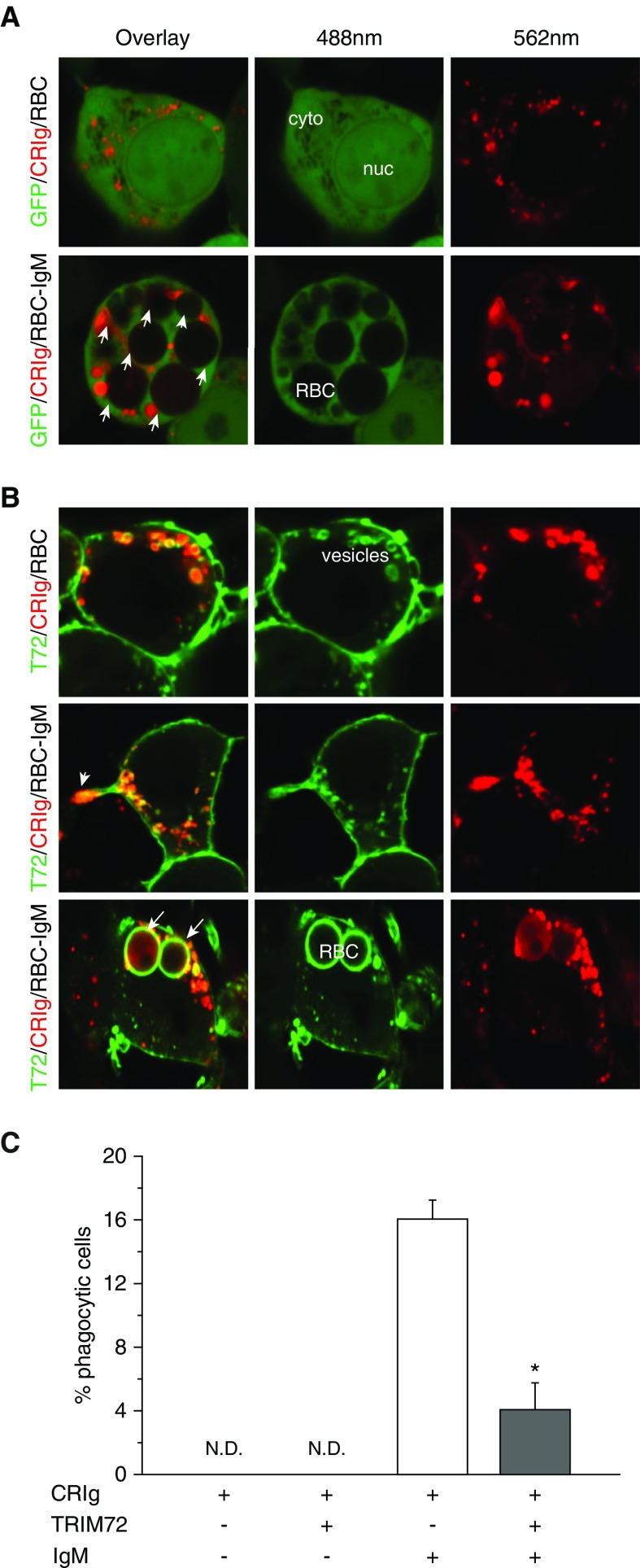
It was reported that CRIg phagosome endocytosis, and recycling from and to the plasma membrane, were constitutively active via receptor endocytosis (17, 37). Because we showed that TRIM72 modulates caveolar endocytosis (20), we therefore examined whether TRIM72 might inhibit the function of CRIg by interfering with its membrane recycling process. Clathrin-mediated endocytosis, the reported endocytic pathway for CRIg recycling, was stimulated by incubating the cells at 37°C in primary human AMs, and probed with FITC-transferrin, a specific cargo for the clathrin pathway (20), followed by coimmunostaining of CRIg and TRIM72. Our results showed that CRIg distribution became highly variable with the stimulation of receptor endocytosis, but no significant amount of CRIg/TRIM72/FITC-transferrin colocalization was observed (except for where transferrin is still bound to the plasma membrane; Figure E3), suggesting that TRIM72 might not play a major role in altering CRIg phagosome recycling. However, this still needs to be further confirmed by quantitative measurements (17). To examine the role of TRIM72 in the effector process of CRIg-mediated phagocytosis, we compared CR-mediated phagocytosis in nonphagocytic HEK293 cells transfected with hCRIg-mCherry with or without cotransfection of GFP-hTRIM72. Figure 5A and Figure 5C demonstrated active phagocytosis of IgM-opsonized sRBCs by hCRIg-mCherry–expressing cells, and coating of CRIg on the surface of ingested sRBCs. In contrast, in cells cotransfected with GFP-hTRIM72 and hCRIg-mCherry (Figures 5B and 5C), the number of cells that ingested sRBCs was greatly reduced compared with cells with only ectopic CRIg expression. In addition, TRIM72 was seen to localize on CRIg-containing phagocytic processes and around the intracellular phagosomes containing internalized IgM-opsonized sRBC, which were decorated with CRIg (Figure 5B).
Figure 5.
TRIM72 directly inhibits CRIg-mediated phagosome formation. (A) Representative images of HEK293 cells cotransfected with mCherry-tagged CRIg (CRIg-mCherry) and empty GFP vector, and incubated with bare sRBCs (upper) or IgM-opsonized sRBCs (lower). (B) Representative images of HEK293 cells cotransfected with CRIg-mCherry and GFP-tagged TRIM72 (GFP-TRIM72), and incubated with bare sRBCs (upper) or IgM-opsonized sRBCs (lower). Arrowhead, phagocytic process; arrows, ingested sRBCs. cyto = cytosol; nuc = nucleus. (C) Quantification of phagocytic cells containing intracellular sRBCs that are positive for both GFP and mCherry fluorescence (%); n = 3 for both bare sRBC controls, n = 10 for the two IgM-opsonized groups, *P < 0.05. N.D. = not determined. Data are presented as mean (±SE).
TRIM72 Ablation Improves Bacterial Clearance and Survival in P.a. Pneumonia
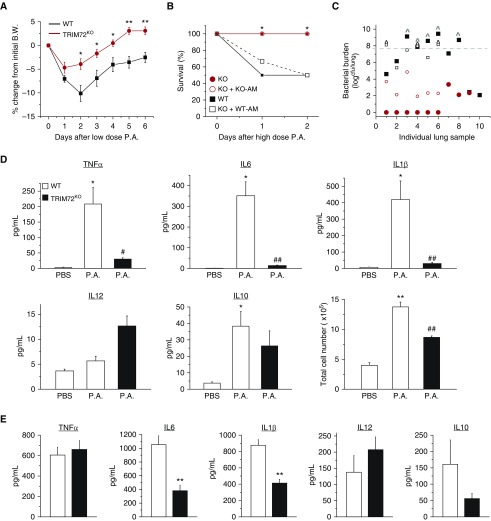
To test the in vivo effects of altered CRIg phagocytosis as a result of TRIM72 ablation, we used an acute P.a. pneumonia model (29). When naive WT and TRIM72KO mice receiving a sublethal dose (2.5 × 105 cfu/ml) of PAO1 (a clinical isolate of P.a.), body weight loss of mice started immediately (Figure 6A). TRIM72KO mice showed less weight loss than the WT mice throughout the first 8 days after P.a. injection. Interestingly, when a second dose of PAO1 was administered to these mice, no difference in body weight loss was seen between the WT and TRIM72KO mice (Figure E4), suggesting that lack of TRIM72 does not affect adaptive immune responses. Mortality of the WT and TRIM72KO mice was compared using a higher dose of P.a. (1.7–4.2 × 107 cfu/ml). Strikingly, at Day 2 after PAO1 injection, all 10 TRIM72KO mice survived, whereas 5 (50%) of the WT mice died (Figure 6B). Quantification of the lung bacterial burden in the five deceased WT mice indicated bacterial dissemination (i.e., higher bacterial loads than the injected dose) in all cases (Figure 6C), whereas TRIM72KO mice had significantly lower lung bacterial burden, showing a good correlation between mortality and bacterial load. This suggests that effective bacterial clearance is responsible for the improved mortality in TRIM72KO mice. We also tested the levels of cytokines in BALF of the severe P.a. pneumonia mice. In WT mice, P.a. pneumonia induced significant increases in major proinflammatory cytokines, including TNF-α, IL-6, IL-1β, and in IL-10, a feedback antiinflammatory cytokine (38) (Figure 6D). Total cell numbers in BALF were also increased (Figure 6D) in P.a.-injected WT mice compared with noninfected controls, indicating an influx of immune cells into the lung. In contrast, P.a.-injected TRIM72KO mice had significantly less TNF-α, IL-6, IL-1β, as well as fewer cells in BALF than the P.a.-injected WT mice, whereas the levels of IL-12 and IL-10 were not significantly different.
Figure 6.
TRIM72 ablation improves bacterial clearance and survival in P.a. pneumonia. (A) Percent body weight (B.W.) loss of naive WT and TRIM72KO mice after the first intraperitoneal injection of roughly 2.5 × 105 cfu/ml PAO1 (a clinical isolate of P.a.); n = 13 for WT (black squares), n = 8 for TRIM72KO (red circles), *P < 0.05, **P < 0.005 compared with WT. (B) Percentage of survival at Day 2 after roughly 3 × 107 cfu/ml P.a. intraperitoneal injection; n = 10 for WT (solid black squares) and TRIM72KO (solid red circles), n = 6 for adoptive transfer of WT AM to TRIM72KO (open black squares) and for KO AM to TRIM72KO (open red circles); *P < 0.05 for WT versus TRIM72KO groups, and for WT AM to TRIM72KO versus KO AM to TRIM72KO groups. (C) Scatter plot of whole-lung bacterial burden at Day 2 P.a. injection in WT, TRIM72KO, WT AM to TRIM72KO, and KO AM to TRIM72KO groups. Gray dashed line designates injected bacterial dose; ^ designates mice that have died. P < 0.05 for WT versus TRIM72KO groups, and for WT AM to TRIM72KO versus KO AM to TRIM72KO, n = 6–10 (as described in B). (D) At Day 2 after P.a. injection, ELISA detection of cytokine levels of TNF-α, IL-6, IL-1β, IL-12, and IL-10 in BALF; total cell number in BALF was measured by hemocytometer counting, n = 5 for WT PBS (open bar), n = 10 for WT P.a. (open bar), and n = 9 for TRIM72KO P.a. (solid bar), *P < 0.05, **P < 0.005 compared with WT PBS; #P < 0.05, ##P < 0.005 compared with WT P.a. (E) Lung tissue levels of TNF-α, IL-6, IL-1β, IL-12, and IL-10 in WT AM to TRIM72KO and KO AM to TRIM72KO groups at Day 2 after P.a. injection; n = 6, **P < 0.005. Data are presented as mean (±SE).
To examine if AM-specific TRIM72 loss is responsible for the resistance to P.a. pneumonia seen in the global TRIM72KO mice, we further characterized the above parameters in AM-depleted (39) TRIM72KO mice that received adoptive transfer of WT or TRIM72KO AMs (40). In Figure 6B, all TRIM72KO mice that received KO AMs survived at Day 2 after a high-dose P.a. injection (open circle), overlapping with the mortality trend of the TRIM72KO mice without adoptive transfer of AMs. In contrast, TRIM72KO mice receiving WT AMs had a survival rate of 66.7% at Day 1 and 50% at Day 2 after P.a. injection, highly similar to that of the WT mice without adoptive transfer of AMs (open square, Figure 6B). Consistently, lung bacterial burdens of the TRIM72KO mice receiving KO AMs (open circle, Figure 6C) were significantly lower than those that received WT AMs (open square, Figure 6C). We also measured lung tissue cytokine levels in these two groups of mice and found that TRIM72KO mice receiving KO AMs had significantly lower levels of IL-6 and IL-1β than the TRIM72KO mice receiving WT AMs (Figure 6E). Collectively, these data show that AM-specific TRIM72 ablation is responsible for the enhanced P.a. clearance from the lung and improved mouse survival in the P.a. pneumonia model, without substantially increasing proinflammatory cytokine production.
TRIM72 Overexpression Enhances NF-κB Activation
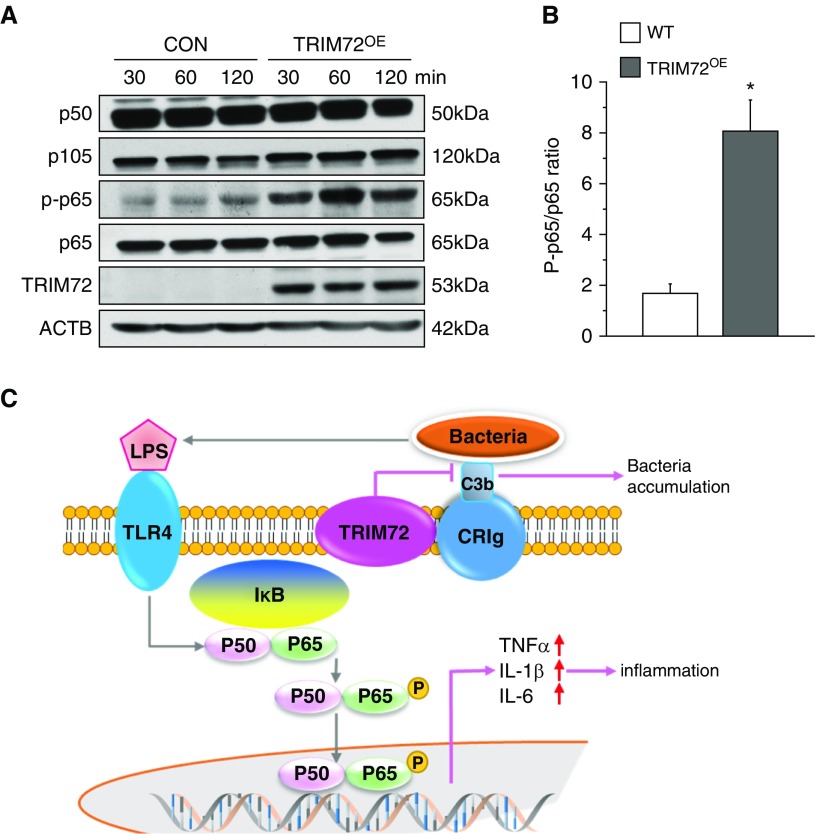
NF-κB acts as a master switch for the effector mechanisms of innate immune responses (41), and phosphorylation and nuclear translocation of the canonical p50/p65 heterodimer of NF-κB are associated with increased transcription of proinflammatory cytokine genes and cytokine secretion (42). To test if altered TRIM72 expression changes NF-κB activation, we first conducted lentivirus-mediated TRIM72 overexpression in MH-S cells, and then measured the ratio of p50 to its inactive precursor, p105, as well as the ratio of phosphorylated p65 to total cellular p65 upon exposure of the cells to P.a. (Figure 7A). A more than fourfold increase in p65 phosphorylation was observed in TRIM72OE MH-S cells as compared with controls (Figures 7A and 7B), echoing the reduced cytokine production seen in TRIM72KO mice. The essential roles of TRIM72 in mediating effector functions of the innate immunity were summarized in Figure 7C. This shows that the presence of TRIM72 on AMs keeps macrophage-specific phagocytic receptor CRIg quiescent, and that the relatively higher bacterial loads as a result of TRIM72-mediated CRIg phagocytosis suppression leads to enhanced activation of the inflammatory switch, NF-κB (Figure 7C).
Figure 7.
TRIM72 overexpression enhances NF-κB p65 phosphorylation. (A) Immunoblot analysis of p50, p50 precursor p105, phosphorylated (p) and total p65 in MH-S cells with TRIM72 overexpression (TRIM72OE), and controls (CON), stimulated with PAO1 for designated time. TRIM72 immunoblot was included to show proper overexpression, and ACTB was included as loading control. (B) Ratio of p-p65 to total p65; white bar, WT; gray bar, TRIM72OE; n = 3, *P < 0.05 compared with WT. (C) Working model for the role of TRIM72 in regulating bacterial phagocytosis and inflammation. Upon bacteria invasion, complement in the lung opsonizes bacteria, which binds to CRIg on the surface of AMs. This initiates phagocytosis and pathogen clearance, whereas bacterial components (e.g., LPS) activate Toll-like receptor (TLR) 4 and NF-κB heterodimer p50/p65. TRIM72 is a binding partner of CRIg on AMs that inhibits this process, which prefers bacterial accumulation and production of inflammatory cytokines. Data are presented as mean (±SE).
Discussion
In this study, we describe a novel mechanism through which CRIg in AMs is able to mediate a noninflammatory type of phagocytosis, and potentially contribute to innate immune responses against bacterial pneumonia. We found that the expression of TRIM72 on AMs, a previously identified plasma membrane repair molecule for alveolar epithelial cells, represents a newly discovered regulatory mechanism of this normally quiescent, but effective, complement system–mediated pathogen removal mechanism in the lung.
Complement proteins abundantly present in the bloodstream, and their classic roles in removing circulating pathogens, are well known. The complement system may be activated via the alternative pathway through spontaneous C3 hydrolysis to quickly opsonize small amounts of circulating pathogens and possibly contribute to tissue injuries (43), or via the antibody-mediated classical pathway, to further amplify the adaptive immune responses (44). In addition, the complement system may also be activated via the lectin pathway, but the physiological significance of this complement activation pathway is less clear. The distinct tissue distribution pattern of many lectins (45), for example, its abundant presence in the lung, suggests a possible role of the complement in the maintenance of tissue homeostasis. Paradoxically, both deficiency of complement components and loss of function mutations in inhibitors of the complement system are associated with tissue injury and autoimmune diseases in human and murine models (43, 44). Therefore, specific regulatory mechanisms of the complement pathway in tissues need further in-depth investigation.
In the lungs, of the five CRs expressed on innate immune cells, AMs express CR4 (CD18/CD11c) (46), but no appreciable amounts of CR1–3 (47). Helmy and colleagues (17) state that CRIg is expressed in human AMs, although they did not show their data. Our results in this study confirm CRIg expression in both human and mouse AMs (Figure 2). Previous studies suggest that CRIg plays a critical role in macrophage phagocytosis in the liver and intestine (17, 48). The ability of CRIg to directly recognize bacterial pathogen–associated molecular patterns, such as lipoteichoic acid (49), may enhance its capturing of circulating bacteria. In addition, studies show that CRIg elicits an inhibitory role on adaptive immune responses (4, 18, 48, 50), but the intracellular killing of pathogens after CRIg-mediated phagocytosis is not reduced (51). In fact, an engineered soluble form of CRIg can inhibit the alternative pathway, and ameliorate inflammation and tissue destruction due to excessive inflammation in animal models of arthritis, lupus nephritis, and cutaneous disease (52, 53), suggesting that CRIg largely mediates noninflammatory immune responses.
Despite the fact that nonspecific complement-mediated phagocytosis was generally regarded as less efficient than antibody-specific FcγR-mediated phagocytosis (15), we highlighted the importance of CR in pathogen clearance and lung host defense in the present study. As a potential inhibitor of CRIg, TRIM72 physically interacts with CRIg (Figure 1), resulting in suppression of CRIg-mediated phagocytosis (Figures 3–5). Most importantly, using a P.a. pneumonia mortality model, we observed a low inflammatory phenotype, yet dramatic improvement in bacteria clearance and mortality when TRIM72 inhibition of CRIg was removed (Figure 6). This indicates a significant contribution of the CRIg and TRIM72 interaction in determining the outcome of severe pulmonary infections through a balance between pathogen clearance and proinflammatory tissue injury. However, we didn’t characterize if TRIM72 interacts with CR4, nor do we have access to CRIg−/− mice, so the possible contribution of another CRIg-independent pathways for TRIM72 regulation of CR phagocytosis is yet to be confirmed. In addition, although a previous report suggests that neutrophils are a major innate immune cell type fighting P.a. infection (35), we believe that TRIM72 does not regulate neutrophil function, because it is not expressed in Ly6G+ cells. As AM phagocytosis of invading pathogens occurred earlier than any significant amount of neutrophil recruitment to the airspace (Figure 4), up-regulated AM phagocytosis after TRIM72 ablation and reduced cytokine production should have reduced the need for a substantial amount of neutrophil infiltration into the lung (Figure 6D), which was known to cause subsequent tissue injury (54).
Our data also show that, when TRIM72 is overexpressed, receptor-medicated phagocytosis is suppressed (Figure 3E), and bacterial exposure leads to increased activation of the p65 subunit of NF-κB. The overall role of CRIg/TRIM72 interaction in AMs is illustrated in Figure 7C, highlighting a novel immunomodulatory role of TRIM72 in shifting the innate immunity equilibrium toward the proinflammatory direction by inhibiting CRIg-mediated bacterial clearance, and maintaining activation of the proinflammatory pathway upon pathogen invasion.
TRIM family proteins contain a conserved RING, B-box, and coiled-coil motif at the N terminus, with a variable SPRY domain at the C terminus to distinguish specific isoforms (24). Functions of many TRIM proteins are dictated by the intrinsic E3 ligase activity on their RING domains. However, a few important functions of some TRIM proteins are not related to their E3 ligase activity, such as membrane repair (55) and acid secretion (56). Here, we did not find evidence of CRIg E3 ligase degradation by TRIM72, because a parallel increase in the two proteins’ expression was seen in developing lungs, and yet no CRIg up-regulation was seen in TRIM72KO AMs (Figure 2). Interestingly, previous studies have reported the formation of an IgG binding pocket in the PRY-SPRY domain of TRIM proteins (57) that are involved in immune responses against viruses (58), including TRIM21 (Ro52), TRIM5α, and TRIM20 (pyrin). Park and colleagues (59) revealed a great similarity in the crystalline structure of the PRY-SPRY domains of TRIM72 and TRIM21. Collectively, this may provide a structural basis for direct TRIM72 and CRIg interaction, as the latter is known to contain Ig-like domains (17).
The evolutionary rationale for the immune cell to possess such an inhibitory mechanism for CRIg is an interesting aspect to consider. We previously showed that TRIM72 expression in alveolar epithelial cells is essential for successful repair of plasma membrane injury (19, 20). One possibility is that CRIg activation may be enhanced in TRIM72-defective tissues that are prone to cellular damage. This would allow debris to be quickly removed via CRIg activity without excessive inflammatory responses when tissue damage occurs. Our data in this study show that TRIM72 may be able to play a central role in mediating these processes.
In summary, we reveal an important role of CRIg in host defense of the lung against pulmonary infections and a novel regulatory mechanism for CRIg. We show that TRIM72 is a direct interacting partner of CRIg and an inhibitor of its function. This interaction determines the status of cytokine storm, pathogen clearance and mortality in vivo. Our study suggests that targeted suppression of TRIM72 or stimulation of CRIg in AMs may be valuable new measures for the treatment of severe pulmonary infections.
Acknowledgments
Acknowledgment
The authors thank Dr. Rolf D. Hubmayr for critical review of the manuscript content.
Footnotes
This work was supported by National Institutes of Health (NIH) R01HL116826, R21AI133465, and CRCF MF17-039-LS (X.Z.), NIH R01HL075557 (J.W.C.), and by Department of Defense (DOD) W81XWH-14-2-0168 and G2015-115 Global Health Innovative Technology Fund (A.R.S.).
Author Contributions: Conception and design—X.Z. and A.R.S.; performing experiments—N.N., X.C., C.T., I.P., J.M.S., and X.Z.; analysis and interpretation—H.F. (statistics), X.Z., J.M.S., J.W.C., and A.R.S.; drafting the manuscript—X.Z., J.W.C., and A.R.S.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2017-0236OC on December 21, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Kojicic M, Li G, Hanson AC, Lee KM, Thakur L, Vedre J, et al. Risk factors for the development of acute lung injury in patients with infectious pneumonia. Crit Care. 2012;16:R46. doi: 10.1186/cc11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31:S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 3.Groves E, Dart AE, Covarelli V, Caron E. Molecular mechanisms of phagocytic uptake in mammalian cells. Cell Mol Life Sci. 2008;65:1957–1976. doi: 10.1007/s00018-008-7578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He JQ, Wiesmann C, van Lookeren Campagne M. A role of macrophage complement receptor CRIg in immune clearance and inflammation. Mol Immunol. 2008;45:4041–4047. doi: 10.1016/j.molimm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187:S340–S345. doi: 10.1086/374747. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song WC. Crosstalk between complement and Toll-like receptors. Toxicol Pathol. 2012;40:174–182. doi: 10.1177/0192623311428478. [DOI] [PubMed] [Google Scholar]
- 8.Matalon S, Wright JR. Surfactant proteins and inflammation: the yin and the yang. Am J Respir Cell Mol Biol. 2004;31:585–586. doi: 10.1165/rcmb.F286. [DOI] [PubMed] [Google Scholar]
- 9.Moliva JI, Rajaram MV, Sidiki S, Sasindran SJ, Guirado E, Pan XJ, et al. Molecular composition of the alveolar lining fluid in the aging lung. Age (Dordr) 2014;36:9633. doi: 10.1007/s11357-014-9633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watford WT, Ghio AJ, Wright JR. Complement-mediated host defense in the lung. Am J Physiol Lung Cell Mol Physiol. 2000;279:L790–L798. doi: 10.1152/ajplung.2000.279.5.L790. [DOI] [PubMed] [Google Scholar]
- 11.Pandya PH, Wilkes DS. Complement system in lung disease. Am J Respir Cell Mol Biol. 2014;51:467–473. doi: 10.1165/rcmb.2013-0485TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russkamp NF, Ruemmler R, Roewe J, Moore BB, Ward PA, Bosmann M. Experimental design of complement component 5a–induced acute lung injury (C5a-ALI): a role of CC-chemokine receptor type 5 during immune activation by anaphylatoxin. FASEB J. 2015;29:3762–3772. doi: 10.1096/fj.15-271635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weis JJ, Tedder TF, Fearon DT. Identification of a 145,000 Mr membrane protein as the C3d receptor (CR2) of human B lymphocytes. Proc Natl Acad Sci USA. 1984;81:881–885. doi: 10.1073/pnas.81.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross GD. Regulation of the adhesion versus cytotoxic functions of the Mac-1/CR3/αMβ2-integrin glycoprotein. Crit Rev Immunol. 2000;20:197–222. [PubMed] [Google Scholar]
- 15.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 16.Sengeløv H, Kjeldsen L, Kroeze W, Berger M, Borregaard N. Secretory vesicles are the intracellular reservoir of complement receptor 1 in human neutrophils. J Immunol. 1994;153:804–810. [PubMed] [Google Scholar]
- 17.Helmy KY, Katschke KJ, Jr, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 18.Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SC, Kellett T, Wang S, Nishi M, Nagre N, Zhou B, et al. TRIM72 is required for effective repair of alveolar epithelial cell wounding. Am J Physiol Lung Cell Mol Physiol. 2014;307:L449–L459. doi: 10.1152/ajplung.00172.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagre N, Wang S, Kellett T, Kanagasabai R, Deng J, Nishi M, et al. TRIM72 modulates caveolar endocytosis in repair of lung cells. Am J Physiol Lung Cell Mol Physiol. 2016;310:L452–L464. doi: 10.1152/ajplung.00089.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrera F, Meroni G. TRIM proteins in development. Adv Exp Med Biol. 2012;770:131–141. doi: 10.1007/978-1-4614-5398-7_10. [DOI] [PubMed] [Google Scholar]
- 22.Song R, Peng W, Zhang Y, Lv F, Wu HK, Guo J, et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature. 2013;494:375–379. doi: 10.1038/nature11834. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen N, Yi JS, Park H, Lee JS, Ko YG. Mitsugumin 53 (MG53) ligase ubiquitinates focal adhesion kinase during skeletal myogenesis. J Biol Chem. 2014;289:3209–3216. doi: 10.1074/jbc.M113.525154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozato K, Shin DM, Chang TH, Morse HC., III TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomar D, Singh R. TRIM family proteins: emerging class of RING E3 ligases as regulator of NF-κB pathway. Biol Cell. 2015;107:22–40. doi: 10.1111/boc.201400046. [DOI] [PubMed] [Google Scholar]
- 26.Lasbury ME, Tang X, Durant PJ, Lee CH. Effect of transcription factor GATA-2 on phagocytic activity of alveolar macrophages from Pneumocystis carinii–infected hosts. Infect Immun. 2003;71:4943–4952. doi: 10.1128/IAI.71.9.4943-4952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannoni E, Sawa T, Allen L, Wiener-Kronish J, Hawgood S. Surfactant proteins A and D enhance pulmonary clearance of Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2006;34:704–710. doi: 10.1165/rcmb.2005-0461OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapetina SG-HH. A guide to simple, direct, and quantitative in vitro binding assays. J Biol Methods. 2017;4:e62. doi: 10.14440/jbm.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ojielo CI, Cooke K, Mancuso P, Standiford TJ, Olkiewicz KM, Clouthier S, et al. Defective phagocytosis and clearance of Pseudomonas aeruginosa in the lung following bone marrow transplantation. J Immunol. 2003;171:4416–4424. doi: 10.4049/jimmunol.171.8.4416. [DOI] [PubMed] [Google Scholar]
- 30.van Rooijen N, van Kesteren-Hendrikx E. “In vivo” depletion of macrophages by liposome-mediated “suicide”. Methods Enzymol. 2003;373:3–16. doi: 10.1016/s0076-6879(03)73001-8. [DOI] [PubMed] [Google Scholar]
- 31.Walters DM, Breysse PN, Wills-Karp M. Ambient urban Baltimore particulate-induced airway hyperresponsiveness and inflammation in mice. Am J Respir Crit Care Med. 2001;164:1438–1443. doi: 10.1164/ajrccm.164.8.2007121. [DOI] [PubMed] [Google Scholar]
- 32.Mosser DM, Zhang X. Measuring opsonic phagocytosis via Fcγ receptors and complement receptors on macrophages. Curr Protoc Immunol. 2011 doi: 10.1002/0471142735.im1427s95. Chapter 14:Unit 14.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, Guo RF, Newstead MW, Standiford TJ, Macariola DR, Shanley TP. Effect of IL-10 on neutrophil recruitment and survival after Pseudomonas aeruginosa challenge. Am J Respir Cell Mol Biol. 2009;41:76–84. doi: 10.1165/rcmb.2008-0202OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reutershan J, Basit A, Galkina EV, Ley K. Sequential recruitment of neutrophils into lung and bronchoalveolar lavage fluid in LPS-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L807–L815. doi: 10.1152/ajplung.00477.2004. [DOI] [PubMed] [Google Scholar]
- 35.Koh AY, Priebe GP, Ray C, Van Rooijen N, Pier GB. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun. 2009;77:5300–5310. doi: 10.1128/IAI.00501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Lookeren Campagne M, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cell Microbiol. 2007;9:2095–2102. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 38.King A, Balaji S, Le LD, Crombleholme TM, Keswani SG. Regenerative wound healing: the role of interleukin-10. Adv Wound Care (New Rochelle) 2014;3:315–323. doi: 10.1089/wound.2013.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leemans JC, Juffermans NP, Florquin S, van Rooijen N, Vervoordeldonk MJ, Verbon A, et al. Depletion of alveolar macrophages exerts protective effects in pulmonary tuberculosis in mice. J Immunol. 2001;166:4604–4611. doi: 10.4049/jimmunol.166.7.4604. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, et al. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 42.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 43.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 44.Ballanti E, Perricone C, Greco E, Ballanti M, Di Muzio G, Chimenti MS, et al. Complement and autoimmunity. Immunol Res. 2013;56:477–491. doi: 10.1007/s12026-013-8422-y. [DOI] [PubMed] [Google Scholar]
- 45.Garred P, Genster N, Pilely K, Bayarri-Olmos R, Rosbjerg A, Ma YJ, et al. A journey through the lectin pathway of complement—MBL and beyond. Immunol Rev. 2016;274:74–97. doi: 10.1111/imr.12468. [DOI] [PubMed] [Google Scholar]
- 46.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 47.Berger M, Norvell TM, Tosi MF, Emancipator SN, Konstan MW, Schreiber JR. Tissue-specific Fcγ and complement receptor expression by alveolar macrophages determines relative importance of IgG and complement in promoting phagocytosis of Pseudomonas aeruginosa. Pediatr Res. 1994;35:68–77. doi: 10.1203/00006450-199401000-00015. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka M, Nagai T, Usami M, Hasui K, Takao S, Matsuyama T. Phenotypic and functional profiles of CRIg (Z39Ig)-expressing macrophages in the large intestine. Innate Immun. 2012;18:258–267. doi: 10.1177/1753425911400641. [DOI] [PubMed] [Google Scholar]
- 49.Zeng Z, Surewaard BG, Wong CH, Geoghegan JA, Jenne CN, Kubes P. CRIg functions as a macrophage pattern recognition receptor to directly bind and capture blood-borne gram-positive bacteria. Cell Host Microbe. 2016;20:99–106. doi: 10.1016/j.chom.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Jung K, Seo SK, Choi I. Endogenous VSIG4 negatively regulates the helper T cell–mediated antibody response. Immunol Lett. 2015;165:78–83. doi: 10.1016/j.imlet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Kim KH, Choi BK, Kim YH, Han C, Oh HS, Lee DG, et al. Extracellular stimulation of VSIG4/complement receptor Ig suppresses intracellular bacterial infection by inducing autophagy. Autophagy. 2016;12:1647–1659. doi: 10.1080/15548627.2016.1196314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katschke KJ, Jr, Helmy KY, Steffek M, Xi H, Yin J, Lee WP, et al. A novel inhibitor of the alternative pathway of complement reverses inflammation and bone destruction in experimental arthritis. J Exp Med. 2007;204:1319–1325. doi: 10.1084/jem.20070432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lieberman LA, Mizui M, Nalbandian A, Bossé R, Crispín JC, Tsokos GC. Complement receptor of the immunoglobulin superfamily reduces murine lupus nephritis and cutaneous disease. Clin Immunol. 2015;160:286–291. doi: 10.1016/j.clim.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Anderson BO, Brown JM, Harken AH. Mechanisms of neutrophil-mediated tissue injury. J Surg Res. 1991;51:170–179. doi: 10.1016/0022-4804(91)90090-9. [DOI] [PubMed] [Google Scholar]
- 55.Cai C, Masumiya H, Weisleder N, Matsuda N, Nishi M, Hwang M, et al. MG53 nucleates assembly of cell membrane repair machinery. Nat Cell Biol. 2009;11:56–64. doi: 10.1038/ncb1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishi M, Aoyama F, Kisa F, Zhu H, Sun M, Lin P, et al. TRIM50 protein regulates vesicular trafficking for acid secretion in gastric parietal cells. J Biol Chem. 2012;287:33523–33532. doi: 10.1074/jbc.M112.370551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yap MW, Stoye JP. TRIM proteins and the innate immune response to viruses. Adv Exp Med Biol. 2012;770:93–104. doi: 10.1007/978-1-4614-5398-7_7. [DOI] [PubMed] [Google Scholar]
- 59.Park EY, Kwon OB, Jeong BC, Yi JS, Lee CS, Ko YG, et al. Crystal structure of PRY-SPRY domain of human TRIM72. Proteins. 2010;78:790–795. doi: 10.1002/prot.22647. [DOI] [PubMed] [Google Scholar]