ABSTRACT
A balance between protein synthesis and degradation is necessary to maintain cellular homeostasis. Failure to triage aberrant proteins may result in their accumulation and aggregation in the cytosol. The valosin-containing protein (VCP)–BCL2-associated athanogene 6 (BAG6) complex facilitates a wide variety of ubiquitin-mediated quality control events at the endoplasmic reticulum (ER), both prior to ER translocation and during ER-associated degradation (ERAD). However, how ubiquitylated clients associated with BAG6 are recognized by VCP for proteasomal degradation is presently unknown. We have identified UBXN1 as the VCP adaptor in BAG6-dependent processes occurring prior to ER insertion but not during ERAD. The loss of VCP-UBXN1 results in the inappropriate stabilization of ubiquitylated BAG6 clients and their accumulation in insoluble aggregates and sensitizes cells to proteotoxic stress. Our results identify how VCP is specifically targeted to ubiquitylated substrates in the BAG6 triage pathway and suggest that the degradation of ubiquitylated clients by the proteasome is reliant on the association of UBXN1 with ubiquitylated substrates and the catalytic activity of VCP.
KEYWORDS: VCP, proteasome, protein quality control, ubiquitin
INTRODUCTION
Protein quality control (PQC) is of critical importance in maintaining cellular and organismal homeostasis. A multitude of pathways within the cell monitor protein structure and function from translation to degradation to ensure optimal function. Failure to maintain these processes can result in the aberrant accumulation of damaged proteins that have a propensity to aggregate; such aggregates can be detrimental to cell viability, as they can hinder vital cellular processes, such as cell division, trafficking, and organelle function (1–3). Indeed, a variety of neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS), Huntington's disease, and Alzheimer's disease, arise due to defects in protein homeostasis and subsequent protein aggregation (4).
Valosin-containing protein (VCP) (also known as p97 or Cdc48p in yeast) is an abundant, evolutionarily conserved AAA-ATPase that participates in a number of ubiquitin-dependent PQC pathways. VCP utilizes the energy harnessed from ATP hydrolysis to unfold or extract ubiquitylated client proteins from membranes, DNA, or multisubunit complexes (5). While VCP has been reported to act upon soluble substrates, it can also act upon substrates in topologically constrained environments that are otherwise inaccessible to the 26S proteasome. VCP assembles with many dedicated adaptor proteins that enable its integration into a host of cellular pathways, including endoplasmic reticulum (ER)-associated degradation (ERAD), the DNA damage response, vesicle trafficking, cell cycle control, ciliogenesis, and autophagy (6, 7). Mutations in VCP cause a number of neurodegenerative diseases, including a multisystem disorder known as inclusion body myopathy, Paget's disease of the bone with frontotemporal dementia (IBMPFD), and ALS. Many of the documented mutations reside at the N terminus of VCP in a region used for associations with numerous adaptors, and some studies suggest that these mutations alter the association of adaptors with VCP, thereby skewing the repertoire of VCP complexes within the cell and resulting in the stabilization of specific substrates. Over 40 VCP adaptors that either serve to recruit substrates to VCP or further modify substrates already associated with VCP have been identified. However, with the exception of the ubiquitin fusion degradation 1 (UFD1)-nuclear protein localization 4 (NLP4) dimeric adaptor complex, many VCP adaptors remain poorly characterized in terms of the pathways that they participate in or the substrates that they recognize. Our recent investigation of the VCP-adaptor interaction network in mammalian cells identified a novel interaction between a poorly understood VCP adaptor, UBXN1 (also known as stress-activated protein kinase substrate 1 [SAKS1]), and the BCL2-associated athanogene 6 (BAG6) chaperone complex (6).
BAG6 is a multifunctional chaperone that oversees multiple protein quality control events at the ER, including (i) the degradation of aberrant newly synthesized proteins, (ii) the posttranslational targeting of proteins to the ER, (iii) the degradation of ER-destined proteins if targeting fails, and (iv) ER-associated degradation (8–10). BAG6 forms a stoichiometric complex with the transmembrane domain recognition complex of 35 kDa (TRC35) and ubiquitin-like protein 4A (UBL4A) that function together in known BAG6-dependent processes. This complex interfaces with proteins specific for each of the above-mentioned pathways to mediate distinct outcomes. One of the best-understood roles for BAG6 is in the targeting or triage of proteins destined for the ER. Through a series of elegant biochemical studies using ER-targeted transmembrane domain (TMD)-containing clients, Shao and colleagues demonstrated previously that the TMD is first captured by small glutamine-rich tetratricopeptide alpha (SGTA) at the ribosome and is subsequently transferred via the BAG6-TRC35-UBL4A complex to the transmembrane domain recognition complex of 40 kDa (TRC40) for insertion into the ER (Fig. 1A) (11). The failure to appropriately transfer the client to TRC40 (for example, when the ER-resident translocon is saturated) commits the client to ubiquitin-mediated proteolysis, whereby SGTA transfers the client directly to BAG6, followed by the recruitment of the E3 ligase RNF126 to rapidly ubiquitylate client proteins (Fig. 1A) (11, 12).
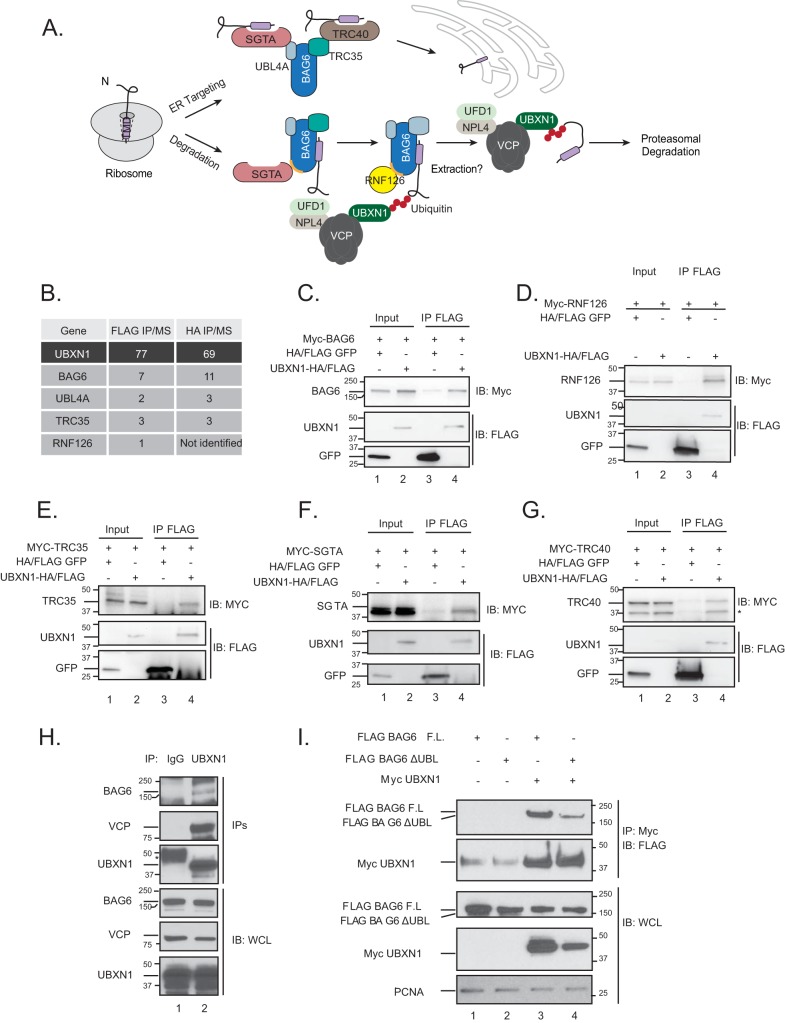
FIG 1.
VCP-UBXN1 associates with the BAG6 chaperone complex. (A) Model for substrate triage by the BAG6 complex. (B) Summary of interactions identified in UBXN1 affinity-purified complexes analyzed by mass spectrometry (IP/MS). Numbers indicate average numbers of peptides identified in either FLAG or HA-UBXN1 purifications (for more details, see reference 6). (C to G) UBXN1-HA/FLAG or FLAG-GFP (negative control) was transiently expressed in HEK-293T cells along with the indicated Myc-tagged constructs. FLAG purifications were probed for the Myc-tagged interactors. UBXN1 associates with Myc-BAG6 (C), Myc-RNF126 (D), Myc-TRC35 (E), Myc-SGTA (F), and Myc-TRC40 (G). IB, immunoblotting. (H) Endogenous UBXN1 was purified from HEK-293T lysates and probed for the indicated proteins. Rabbit IgG was used as a negative control. WCL, whole cell lysate. (I) Wild-type FLAG-BAG6 or the ΔUBL mutant was transiently transfected into HEK-293T cells along with Myc-UBXN1. Myc-UBXN1 IPs were probed for associated FLAG-BAG6 (n = 3). F.L., full length.
VCP has been reported to associate with BAG6 in specific contexts. For instance, BAG6 prevents retrotranslocated VCP substrates in the ERAD pathway from aggregation in the cytosol (9). More recently, BAG6 and VCP have been reported to participate in the attenuation of ER translocation during the ER stress response (13). How VCP is specifically recruited to these BAG6-containing complexes is not known but is likely dependent on dedicated VCP adaptors.
Here, we provide evidence of a specific role for the VCP-UBXN1 complex in the degradation of select ubiquitylated BAG6 clients. Our studies suggest that BAG6 clients, once ubiquitylated, are not directly degraded by the proteasome; rather, they must first be recognized by the VCP-UBXN1 complex prior to degradation. The failure of ubiquitylated substrates to engage VCP-UBXN1 leads to their accumulation as insoluble aggregates, and the loss of UBXN1 sensitizes cells to agents that induce proteotoxic stress. The rapid and timely degradation of these substrates is essential, as their accumulation in cells will overwhelm chaperone systems, cause inappropriate interactions with cytosolic components, and ultimately lead to aggregation. These aggregates can further seed the formation of insoluble deposits that are detrimental to cellular and, ultimately, organismal viability. Thus, both proper targeting and the degradation of newly synthesized proteins that fail to appropriately localize are controlled by a dynamic multiprotein network that constantly surveys clients and the surrounding cellular environment.
RESULTS
VCP-UBXN1 associates with the BAG6 complex.
We recently interrogated protein complexes that associate with VCP adaptors belonging to the ubiquitin X domain (UBXD) family in HEK-293T cells and identified a novel interaction between UBXN1 and members of the BAG6 chaperone complex, including BAG6, UBL4A, and TRC35 (Fig. 1B) (6). Notably, we also identified RNF126, the E3 ligase that ubiquitylates substrates bound to BAG6 (Fig. 1B). We verified these interactions using transient-transfection studies with epitope-tagged constructs in HEK-293T cells. Tagged UBXN1 associated with BAG6, TRC35, SGTA, TRC40, and RNF126, suggesting that the BAG6 complex is stably associated in cells (Fig. 1C to F). We confirmed the interaction at the endogenous level and demonstrate that UBXN1 associates with BAG6 and VCP in cells (Fig. 1H). These findings support previously reported observations of copurification of VCP with BAG6 (9, 13).
The N-terminal UBL domain of BAG6 associates with RNF126 and, to a lesser extent, SGTA and is critical for the role of BAG6 in protein triage. BAG6 lacking the UBL domain (ΔUBL-BAG6) is incapable of mediating client ubiquitylation, which we presumed would be the signal for VCP-UBXN1 recruitment (11). We therefore asked if the UBL domain of BAG6 was necessary for the UBXN1 interaction in cells. While full-length BAG6 efficiently interacted with UBXN1, we observed a deficit in binding to ΔUBL-BAG6 (Fig. 1I). These studies indicate that VCP-UBXN1 associates with the BAG6 complex in a BAG6 UBL domain-dependent manner. In an effort to identify the components of the VCP-UBXN1-BAG6 complex that directly interact, we purified individual proteins for in vitro binding assays. However, we were unable to detect direct interactions between BAG6, RNF126, SGTA, and the UBL4A-TRC35 dimer with VCP or UBXN1 in these studies (data not shown). Thus, the mechanism by which this multisubunit complex is assembled remains to be elucidated (see Discussion).
UBXN1 is dispensable for ERAD.
VCP has a well-defined role in ERAD, where it mediates the retrotranslocation of ubiquitylated substrates from within the ER. Previous studies indicate that BAG6 also contributes to ERAD, where it functions as a chaperone “holdase” downstream of VCP-mediated extraction to shield hydrophobic segments of unfolded polypeptides (9). The VCP adaptors UBXD8 and UBXD2 along with UFD1-NPL4 participate in ERAD (14–16). In contrast, UBXN1 was reported in two previous studies to be a negative regulator of ERAD (17, 18). Nevertheless, we asked if UBXN1 is required for ERAD using well-established fluorescence reporter cell lines expressing membrane and luminal ER proteins: (i) green fluorescent protein-tagged T cell receptor α (GFP-TCRα), (ii) an alpha 1 antitrypsin null Hong Kong variant (A1AT NHK-GFP), and (iii) transthyretin D18G (TTR D18G-GFP). These reporters are normally cleared by ERAD in a VCP-dependent manner; however, the inhibition of ERAD (for example, by VCP depletion) results in an increased GFP signal that can be quantified by flow cytometry. As expected, the depletion of VCP as well as UBXD8 caused an increase in the GFP signal, consistent with stabilization (Fig. 2A to C and E). However, we observed negligible stabilization of the ERAD substrates in UBXN1-depleted cells (Fig. 2A to C and E). To further verify this observation, we performed translation shutoff studies with GFP-TCRα cells depleted of either VCP or UBXN1. Treatment of cells with cycloheximide resulted in decreases in GFP-TCRα protein levels within 2 h, whereas the depletion of VCP caused a stabilization of GFP-TCRα (Fig. 2F). However, we observed no stabilization of the substrate in UBXN1-depleted cells (Fig. 2G). In contrast, the depletion of either UBXN1 or VCP caused the stabilization of a soluble proteasomal substrate, GFP-ubiquitin G76V (GFPu) (Fig. 2D), suggesting that while UBXN1 may be required for the degradation of soluble ubiquitylated substrates (independent of BAG6), it is dispensable for ERAD. Notably, Xu and colleagues reported previously that BAG6 and UBXD8 associate with the E3 ligase GP78 at the ER membrane to facilitate ERAD. In that study, GP78 did not associate with other VCP adaptors, including UBXN1 (10).
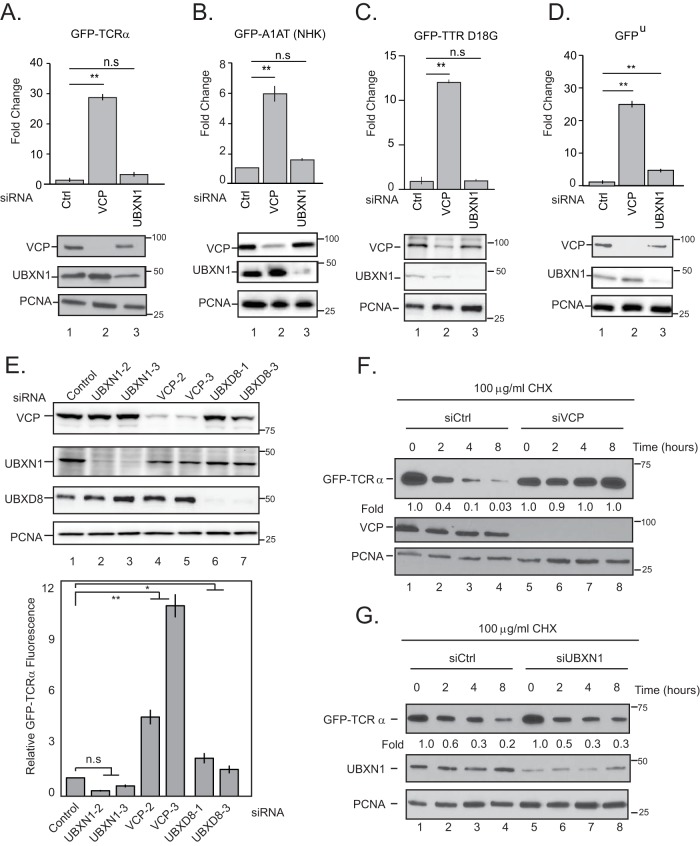
FIG 2.
UBXN1 is not required for ER-associated degradation. (A to D) HEK-293T cells stably expressing the ERAD reporters GFP-TCRα (A), A1AT NHK-GFP (B), and GFP-TTR D18G (C) or the soluble proteasomal substrate GFP-ubiquitin G76V (GFPu) (D) were transiently transfected with siRNAs targeting VCP or UBXN1. The level of stabilized GFP in each sample was measured by flow cytometry. Immunoblots of the relative knockdown of VCP and UBXN1 in each cell line are shown. (E) GFP-TCRα cells were transfected with the indicated siRNAs (including UBXD8), and GFP levels were measured by flow cytometry. (F and G) GFP-TCRα cells were transfected with control, VCP (F), or UBXN1 (G) siRNAs and treated with 100 μg/ml cycloheximide (CHX) for the indicated times. Cell lysates were probed for GFP levels. Each study was performed in triplicate three independent times. Error bars represent standard deviations. *, P ≤ 0.05; **, P ≤ 0.01; n.s, not significant (as determined by two-tailed Student's t test) (n = 3).
VCP-UBXN1 is required for the degradation of BAG6 substrates that fail to target the ER.
Having ruled out a role for UBXN1 in ERAD, we next asked if VCP-UBXN1 is required for BAG6 processes prior to ER insertion. For these studies, we utilized two distinct substrates that have been extensively validated in the BAG6 pathway in previous studies. The first substrate, prion protein (PrP), contains mutations in its signal sequence to impede proper targeting to the ER (19). To facilitate detection, we introduced an internal hemagglutinin (HA) epitope that was demonstrated previously to not interfere with PrP function and designate this substrate HA-PrPmut (Fig. 3A) (19). The second substrate, OPG-TASK, contains an N-terminal opsin glycosylation sequence from rhodopsin fused to a transmembrane domain-containing fragment of TASK1 (also known as KCNK3) (Fig. 3B) (20). As reported previously, these two substrates are almost exclusively mislocalized to the cytosol and are rapidly degraded, with a half-life (t1/2) of less than 1 h (Fig. 3). In contrast, the inhibition of the proteasome with bortezomib resulted in a significant stabilization of both substrates (Fig. 3). We asked if the depletion of VCP or UBXN1 impacted steady-state levels of these substrates. Cells were transiently transfected with small interfering RNAs (siRNAs) targeting VCP or UBXN1 as well as OPG-TASK or HA-PrPmut, and lysates were probed for the relevant substrate. As a positive control, we depleted BAG6, which resulted in increased substrate levels, as expected (Fig. 4A). The depletion of VCP or UBXN1 with multiple siRNAs caused substantial increases in steady-state levels of OPG-TASK (Fig. 4A) and HA-PrPmut (Fig. 4B and C). To confirm that the increased HA-PrPmut levels observed in cells lacking VCP are due to an inhibition of degradation, we treated cells with cycloheximide in combination with an ATP-competitive VCP inhibitor (CB-5083) and observed the stabilization of HA-PrPmut (Fig. 4D). Our inability to completely deplete UBXN1 protein levels with the available siRNAs prompted us to generate a UBXN1 knockout (KO) cell line using clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 gene editing. Treatment of wild-type or UBXN1 KO cells with cycloheximide resulted in the stabilization of HA-PrPmut and OPG-TASK only in KO cells (Fig. 4E and F). Importantly, the codepletion of VCP or UBXN1 with BAG6 did not lead to a greater stabilization of OPG-TASK than that observed by the depletion of these proteins alone, suggesting that they are part of the same pathway (Fig. 4G). In order to determine the specificity of UBXN1 in the degradation of BAG6 substrates, we depleted five other VCP adaptors and asked how they impacted substrate degradation. The depletion of p47 (also known as NSFL1C), UBXD8, and UBXD7 did not impact HA-PrPmut levels (Fig. 4H and I). In contrast, siRNAs targeting the UFD1-NPL4 dimer implicated in many VCP-dependent pathways resulted in increases in steady-state levels of HA-PrPmut, suggesting their involvement in substrate degradation (Fig. 4I).
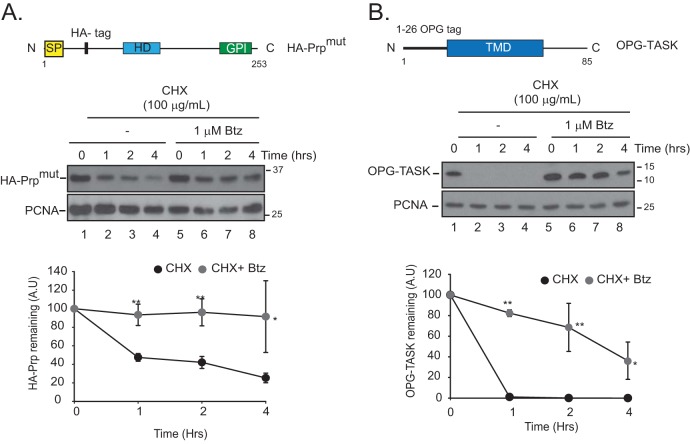
FIG 3.
BAG6 clients that fail to target to the ER and are inherently unstable and degraded by the proteasome. HEK-293T cells were transfected with HA-PrPmut (A) or OPG-TASK (B) and then treated with cycloheximide and bortezomib (Btz) for the indicated times. Bortezomib was added 1 h prior to cycloheximide treatment. Cells were lysed in 1% SDS, lysates were probed for the indicated proteins, and levels of each substrate were quantified by densitometry and normalized to PCNA levels. The top panels show illustrations of BAG6 substrates used in this study. SP, signal peptide; HD, hydrophobic domain; GPI, glycosylphosphatidylinositol anchor; TMD, transmembrane domain. *, P ≤ 0.05; **, P ≤ 0.01 (as determined by two-tailed Student's t test) (n = 3).
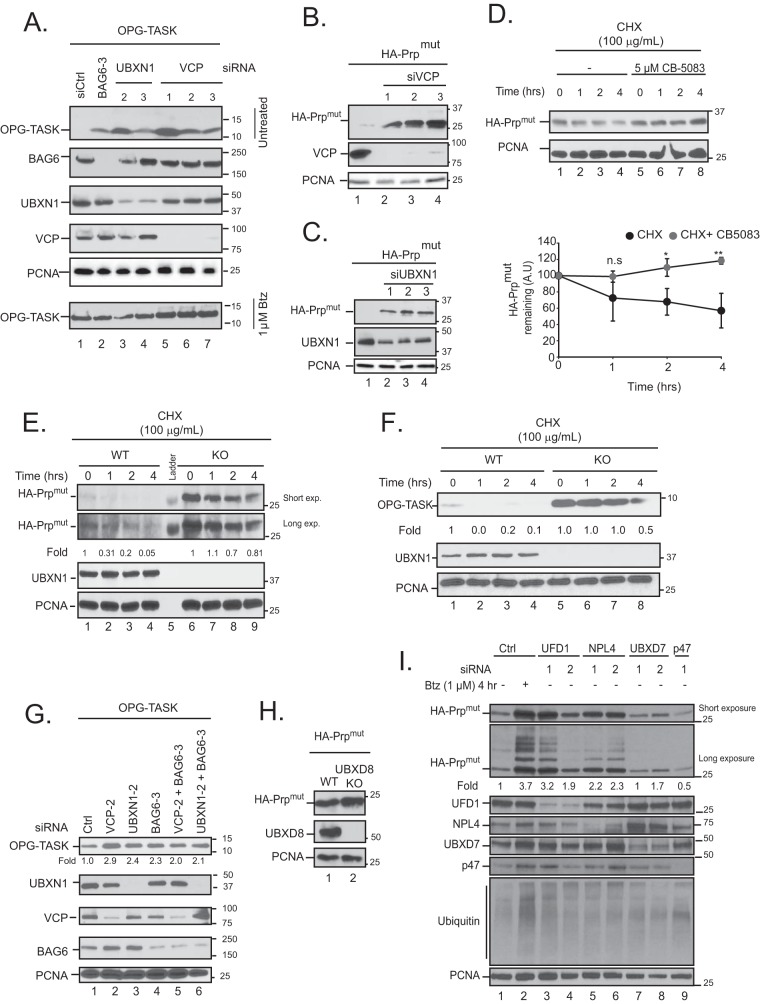
FIG 4.
VCP-UBXN1 is required for the turnover of BAG6 clients. (A) HEK-293T cells were transiently transfected with the indicated siRNAs and OPG-TASK. A duplicate set of samples derived from the same set of transfections was treated with bortezomib to demonstrate equal levels of transfection (bottom). Cell lysates were probed for OPG-TASK levels. (B and C) HEK-293T cells were transiently transfected with control, VCP (B), or UBXN1 (C) siRNAs and HA-PrPmut. Levels of HA-PrPmut were determined. (D) HEK-293T cells were transfected with HA-PrPmut and then treated with cycloheximide in the presence or absence of the VCP inhibitor CB-5083 for the indicated times. Cells were lysed in 1% SDS, lysates were probed for the indicated proteins, and levels of the substrate were quantified by densitometry and normalized to PCNA levels. A.U, arbitrary units. (E and F) Wild-type (WT) and UBXN1 knockout (KO) HeLa Flp-in T-REX cells were transfected with HA-PrPmut (E) or OPG-TASK (F) and treated with cycloheximide for the indicated times. Cells were lysed in 1% SDS, lysates were probed for the indicated proteins, and the levels of each substrate were quantified by densitometry and normalized to PCNA levels. (G) HEK-293T cells were transiently transfected with control, VCP, UBXN1, and BAG6 siRNAs, either alone or in combination, along with OPG-TASK. The levels of OPG-TASK were determined and quantified relative to control values. (H) HeLa Flp-in T-REX (wild-type and UBXD8 KO) cells were transfected with HA-PrPmut, and the levels of PrP in lysates were determined. (I) HEK-293T cells were transfected with the indicated siRNAs and HA-PrPmut. Cell lysates were probed with the indicated proteins. The levels of HA-PrPmut were calculated by densitometry and normalized to PCNA levels (n = 3). n.s, not significant; *, P ≤ 0.05; **, P ≤ 0.01 (as determined by two-tailed Student's t test) (n = 3).
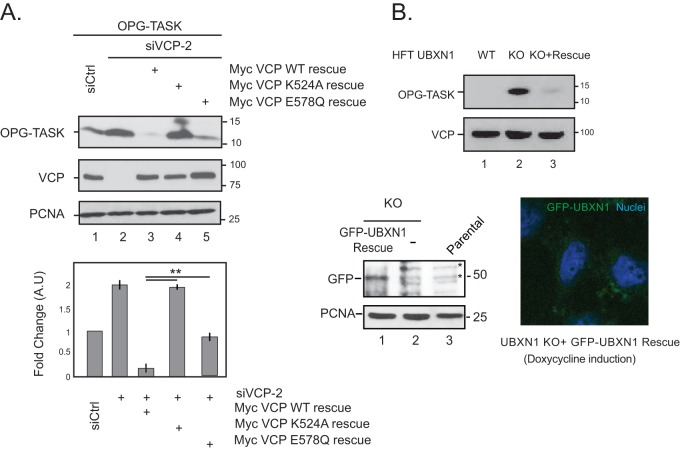
To verify that the stabilization of BAG6 clients is indeed due to the specific depletion of VCP or UBXN1, we attempted to rescue the defect by reexpressing VCP or UBXN1 constructs in cells lacking these two proteins. The expression of wild-type, siRNA-resistant VCP in cells depleted of endogenous VCP reinstated the degradation of OPG-TASK (Fig. 5A, compare lanes 2 and 3). Importantly, the ATPase activity of VCP is required for substrate degradation, as the expression of the siRNA-resistant VCP K524A mutant and, to a lesser extent, the VCP E578Q mutant (both of which are within the D2 ATPase domain and required for ATP binding and hydrolysis, respectively) was unable to rescue substrate degradation compared to wild-type VCP (Fig. 5A, lanes 3 to 5). We were also able to reinstate OPG-TASK degradation in UBXN1 KO cells by the reexpression of GFP-UBXN1 (Fig. 5B). Together, these findings implicate the VCP catalytic activity in association with UBXN1 and UFD1-NPL4 in the degradation of BAG6 clients.
FIG 5.
VCP catalytic activity is required for substrate degradation. (A) HEK-293T cells were depleted of VCP by siRNA-mediated knockdown and transiently transfected with OPG-TASK and VCP siRNA rescue plasmids expressing wild-type VCP or two separate ATPase-defective mutants in the D2 domain (K524A and E578Q). The level of OPG-TASK in each sample was determined. The bottom panel shows data for densitometry analysis from 3 independent experiments. (B) Rescue of OPG-TASK degradation in UBXN1 KO cells. UBXN1 KO cells inducibly expressing GFP-UBXN1 were transfected with OPG-TASK. The expression of GFP-UBXN1 by the addition of 1 μg/ml doxycycline for 72 h reinstated OPG-TASK degradation. *, nonspecific bands. The micrograph shows GFP-UBXN1 induction by doxycycline. **, P ≤ 0.01 (as determined by two-tailed Student's t test) (n = 3).
UBXN1 recognizes ubiquitylated BAG6 clients.
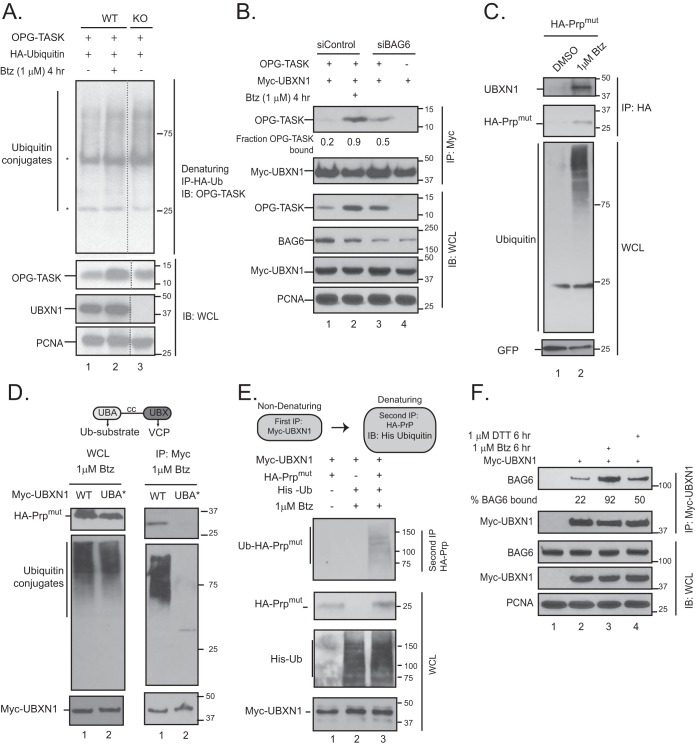
VCP-adaptor complexes primarily recognize ubiquitylated substrates; therefore, we asked if UBXN1 functioned downstream of substrate ubiquitylation and associated with ubiquitylated substrates. We expressed HA-ubiquitin and OPG-TASK in wild-type and UBXN1 KO cells and performed denaturing immunoprecipitations (IPs) with HA-ubiquitin. Wild-type cells were treated with bortezomib as a positive control. In comparison to wild-type cells, OPG-TASK stabilized in UBXN1 KO cells was ubiquitylated based on the appearance of higher-molecular-weight bands in the HA-ubiquitin immunoprecipitates probed for OPG-TASK in a manner equivalent to that observed for wild-type cells treated with bortezomib (Fig. 6A, compare lane 1 with lanes 2 and 3). PrPmut stabilized in UBXN1 KO or VCP-inhibited cells was also similarly modified with ubiquitin (Fig. 7E). We were able to readily detect an interaction between UBXN1 and OPG-TASK or HA-PrPmut in cells treated with bortezomib (Fig. 6B and C). Importantly, the association between UBXN1 and OPG-TASK was dependent on BAG6, as the siRNA-mediated depletion of BAG6 caused a reduction in the level of OPG-TASK associated with UBXN1 despite nearly equivalent levels of stabilization (Fig. 6B, compare lanes 2 and 3). To determine if the interaction between UBXN1 and HA-PrPmut was mediated by the ubiquitin-associated (UBA) domain on UBXN1, we mutated the key residues Met13 and Phe15 to alanine within the UBA domain (denoted UBA*). We observed diminished binding of total cellular ubiquitin conjugates with the UBA mutant, as expected (Fig. 6D). We also observed a significant deficit in the ability of HA-PrPmut to interact with the UBXN1 UBA* mutant in cells, consistent with the importance of the UBA domain for substrate recognition (Fig. 6D). However, in these studies, UBXN1 appeared to be associated with unmodified HA-PrPmut. We reasoned that this may be due to relatively low levels of the ubiquitin-modified substrate as well as the deubiquitination of the substrate upon cell lysis. To confirm that UBXN1 associated with the ubiquitylated substrate, we transfected cells with Myc-UBXN1, HA-PrPmut, and His-ubiquitin and lysed cells in the presence of iodoacetamide and PR-619 to broadly inhibit deubiquitinase activity. Myc-UBXN1 was first purified under nondenaturing conditions to preserve the interaction with HA-PrPmut. The immune complexes were denatured with 1% SDS and diluted, and a second purification for HA-PrP was performed. As shown in Fig. 6E, HA-PrP bound to UBXN1 was indeed ubiquitylated. Finally, we asked if the association between UBXN1 and BAG6 was sensitive to agents that would cause the accumulation of ubiquitylated BAG6 substrates. To this end, we transfected cells with Myc-UBXN1 and treated cells with bortezomib to globally accumulate ubiquitin conjugates or dithiothreitol (DTT) to induce ER stress. Previous studies established that one aspect of the unfolded protein response is the degradation of translocating polypeptides at the ER translocon, a process that is dependent on BAG6 and VCP (13, 21, 22). We observe an increased association between Myc-UBXN1 and endogenous BAG6 in cells treated with bortezomib and, to a slightly lesser extent, upon DTT treatment, suggesting that the accumulation of ubiquitylated proteins enhances the association between UBXN1 and BAG6 (Fig. 6F, compare lane 2 with lanes 3 and 4). Taken together, the data from these studies suggest that the VCP-UBXN1 complex is part of the BAG6 triage pathway at a step downstream of substrate ubiquitylation.
FIG 6.
UBXN1 recognizes ubiquitylated substrates in a BAG6-dependent manner. (A) Wild-type and UBXN1 KO cells were transfected with OPG-TASK and HA-ubiquitin (Ub). Cells were denatured in 2% SDS lysis buffer, and HA-ubiquitin was affinity purified and probed for ubiquitylated OPG-TASK (*, heavy and light chains). (B) HEK-293T cells were transfected with BAG6 siRNA, OPG-TASK, and Myc-UBXN1. Myc-UBXN1 immunoprecipitates were probed for OPG-TASK. The fraction of OPG-TASK bound to UBXN1 was calculated by densitometry and normalized to the levels of OPG-TASK in the cell lysate. (C) HA-PrPmut was transiently expressed in HEK-293T cells that were treated with 1 μM bortezomib for 2 h. Stabilized HA-PrPmut was immunoprecipitated via the HA tag and probed for UBXN1. GFP was used as a transfection control. (D) Domains within UBXN1. UBA, ubiquitin associated; cc, coiled coil; UBX, ubiquitin X. HEK-293T cells were transfected with HA-PrPmut and wild-type Myc-UBXN1 or the UBA M13A F15A mutant (UBA*). Cells were treated with bortezomib for 2 h, and Myc-UBXN1 was immunopurified. (E) HeLa Flp-in T-REX cells were transfected with HA-PrPmut, Myc-UBXN1, and His-ubiquitin and treated with 1 μM bortezomib, as indicated. Consecutive nondenaturing (for Myc-UBXN1) and denaturing (for HA-PrPmut) immunoprecipitations were performed, and immunoprecipitates were probed for His-ubiquitin to demonstrate that PrP bound to UBXN1 is ubiquitylated. (F) HEK-293T cells were transfected with Myc-UBXN1 and treated with 1 μM bortezomib or 1.5 μM DTT for 6 h. Myc-UBXN1 was purified from cell lysates and probed for BAG6. The levels of BAG6 bound to UBXN1 were determined by densitometry and normalized to BAG6 levels in the whole-cell lysate (n = 3).
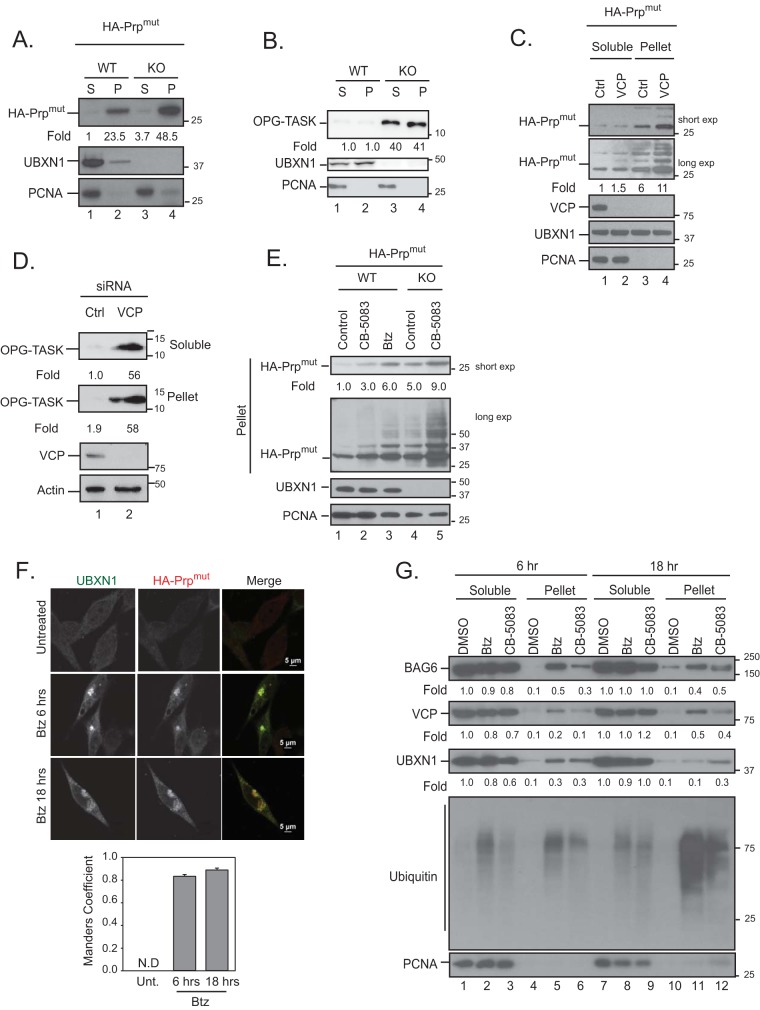
FIG 7.
Loss of VCP-UBXN1 leads to the accumulation of HA-PrPmut in insoluble compartments. (A and B) Wild-type or UBXN1 KO cells were transiently transfected with HA-PrPmut (A) or OPG-TASK (B) and fractionated into soluble (S) and insoluble pellet (P) fractions. Levels of the substrate in each compartment were probed. (C and D) HeLa Flp-in T-REX cells were transfected with control or VCP siRNA and HA-PrPmut (C) or OPG-TASK (D) and fractionated as described above for panel A. (E) Wild-type and UBXN1 KO cells were transfected with HA-PrPmut and treated with 5 μM CB-5083 or 1 μM bortezomib (Btz) for 4 h. The levels of PrP were determined. (F) HA-PrPmut was transfected into HeLa Flp-in T-REX cells, and cells were treated with 1 μM bortezomib for 6 or 18 h. Cells were fixed, stained for endogenous UBXN1 and HA, and imaged. Colocalization was determined by the Manders overlap coefficient for 25 cells in 3 replicate experiments. N.D, not determined. (G) HeLa Flp-in T-REX cells were treated with 5 μM CB-5083 (VCP inhibitor) or 1 μM bortezomib for 6 or 18 h. Cells were fractionated into soluble and pellet fractions, and the levels of VCP, UBXN1, and BAG6 in each fraction were determined. Fold changes for all panels were determined by densitometry and normalized to PCNA levels (n = 3).
Depletion of VCP-UBXN1 leads to the accumulation of insoluble BAG6 clients.
Ubiquitylated substrate triage by BAG6 is essential, as clients that contain TMDs would otherwise aggregate in the cytosol. We asked if the substrates stabilized in VCP- or UBXN1-depleted cells accumulated as insoluble aggregates. Cells were lysed to obtain the soluble (S) component, and the detergent-resistant pellet was designated the insoluble compartment (P) and was resuspended in SDS-containing buffer. The soluble and insoluble fractions were isolated from VCP-depleted or UBXN1 KO cells expressing HA-PrPmut or OPG-TASK. Other substrates accumulated in the soluble fraction, as observed previously; however, we observed an increase in the level of each substrate in the insoluble fraction of cells lacking VCP or UBXN1 compared to control cells (Fig. 7A to D). The stabilization of HA-PrPmut in the insoluble fraction in UBXN1 KO cells was further enhanced upon treatment with CB-5083 (Fig. 7E, compare lanes 4 and 5). This was expected given our observed requirement for UFD1-NPL4 for substrate degradation. Our observation that UBXN1 associated with stabilized OPG-TASK and HA-PrPmut prompted us to determine whether UBXN1 is also present in the insoluble compartment. We imaged cells expressing HA-PrPmut and observed a significant colocalization (Manders overlap coefficient of ∼0.84) of endogenous UBXN1 and HA-PrPmut in aggregate-like structures in cells treated with bortezomib at both early and late time points (Fig. 7F). Furthermore, we observed the recruitment of UBXN1 and VCP, as well as BAG6, to the insoluble fraction in cells treated with bortezomib or CB-5083 at both 6 and 18 h posttreatment (Fig. 7G). Thus, our findings identify an important role of the VCP-UBXN1 complex in the elimination of substrates by the proteasome in order to prevent their inappropriate accumulation as insoluble and potentially toxic aggregates in the cytosol.
Loss of UBXN1 renders cells sensitive to agents that induce proteotoxic stress.
We next asked what was the physiological consequence of UBXN1 deletion. We reasoned that an inability to clear BAG6 clients in the absence of VCP or UBXN1 would be detrimental to cell viability. We took two approaches to explore this idea: (i) observing the clearance of newly synthesized misfolded proteins generated by the translation poisons in wild-type and KO cells and (ii) assessing the viability of wild-type and KO cells with agents that inhibit protein translocation into the ER.
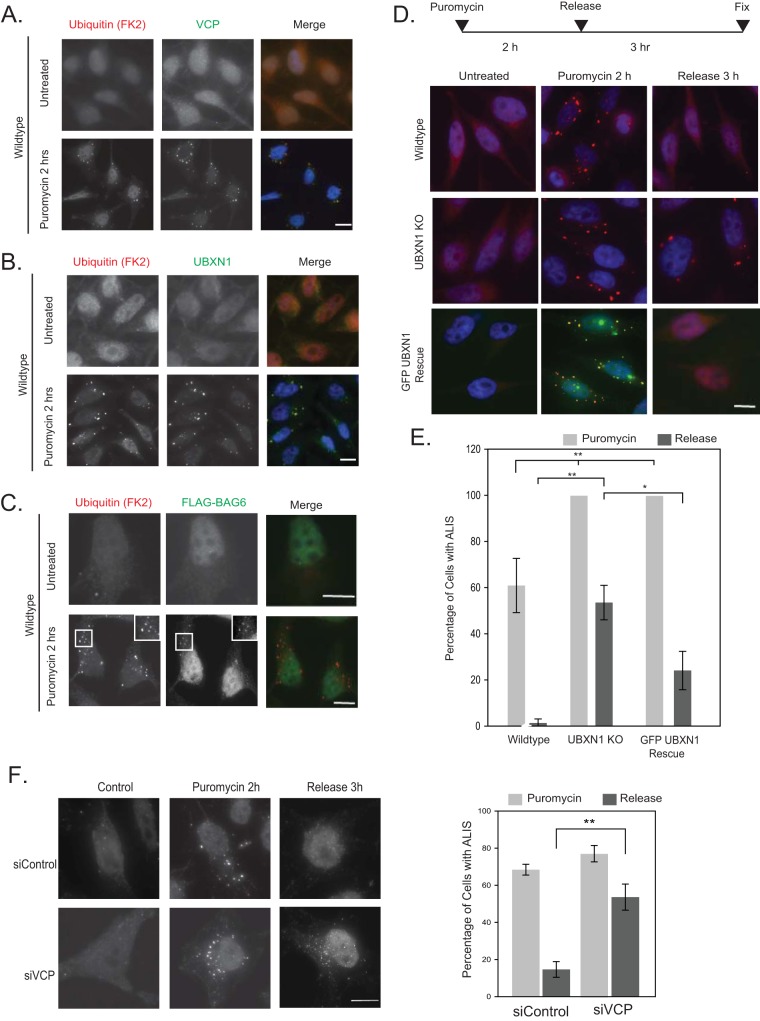
For the first approach, cells were treated with the protein synthesis inhibitor puromycin to cause premature chain termination during translation and the accumulation of defective ribosomal products (DRiPs). DRiPs are frequently sequestered into punctate, ubiquitin-positive structures in the cytosol, referred to as aggresome-like induced structures (ALISs). Endogenous VCP, UBXN1, and FLAG-BAG6 redistributed to ubiquitin-positive ALISs after puromycin treatment (Fig. 8A and C). We also asked if endogenous UFD1-NPL4 was present at ALISs after puromycin treatment and observed a limited recruitment of NPL4 but not UFD1 (data not shown). We next set out to determine the involvement of VCP and UBXN1 in the clearance of ALISs by the proteasome. We note that both VCP and BAG6 have been reported to be involved in DRiP clearance (23, 24). UBXN1 KO cells contained more ALISs (100% of cells imaged for KO cells versus 60% for wild-type cells) after puromycin treatment (Fig. 8D and E). To induce clearance, cells were treated with puromycin for 2 h, followed by the release of the cells into drug-free medium for a further 3 h. While the number of cells with ALISs decreased after 3 h of release in wild-type cells, these structures persisted in UBXN1 KO as well as VCP-depleted cells (Fig. 8D to F). GFP-UBXN1 expressed in KO cells colocalized with ALISs and rescued the phenotype by reinstating clearance (Fig. 8D and E).
FIG 8.
VCP-UBXN1 is required for the clearance of defective ribosomal products. (A to C) HeLa Flp-in T-REX cells were treated with 5 μg/ml puromycin for 1 h and stained for ubiquitin (FK2) and VCP (A), UBXN1 (B), or FLAG-BAG6 (transiently transfected) (C). (D) Wild-type, UBXN1 KO, or UBXN1 KOGFP-UBXN1 Rescue cells were treated with puromycin for 1 h and released into drug-free medium for 3 h to enable the clearance of ALISs. Cells were stained for ubiquitin (FK2) (red) and nuclei (Hoechst dye). The number of cells containing ubiquitin-positive ALIS puncta was quantified by using ImageJ. (E) Quantification of data in panel D. Data for approximately 220 cells were quantified in 3 independent experiments. (F) HeLa Flp-in T-REX cells were transfected with control or VCP siRNAs, treated with puromycin for 2 h, and released into drug-free medium for 3 h to enable the clearance of ALISs. Cells were stained for ubiquitin (FK2) and imaged. The number of cells containing ubiquitin-positive ALIS puncta was quantified by using ImageJ. We note that the puncta in VCP-depleted cells were smaller than those observed in UBXN1 KO cells. Nevertheless, clearance of ALISs is defective in cells lacking both UBXN1 and VCP. Data for 75 cells were quantified in 3 independent experiments. *, P ≤ 0.05; **, P ≤ 0.01 (as determined by Student's t test). Bars, 10 μm.
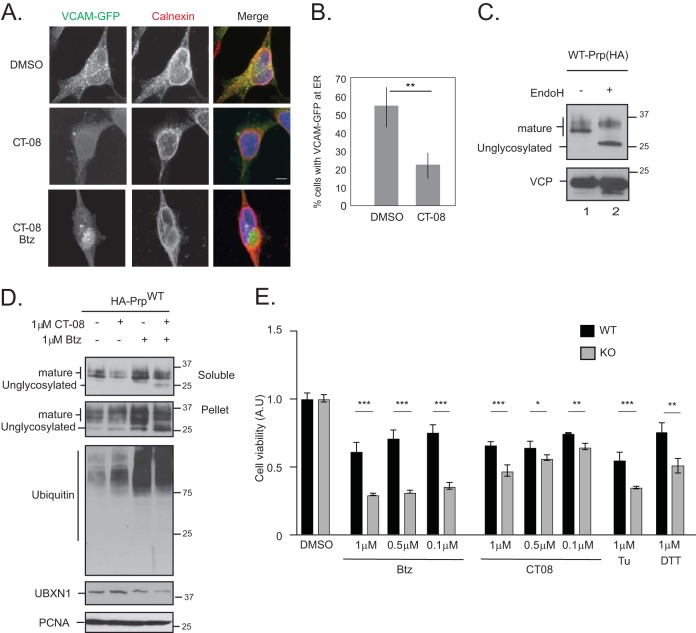
In the second approach, we treated cells with CT08, a specific and potent variant of cotransin, an inhibitor of the Sec61 translocon, as a mechanism to generate ubiquitylated BAG6 substrates (21, 25, 26). CT08 inhibits the translocation of vascular cell adhesion molecule 1 (VCAM1) and tumor necrosis factor alpha (TNF-α) into the ER at nanomolar concentrations. However, at higher concentrations, it broadly inhibits the translocation of secreted and transmembrane ER proteins (25). The inhibitory effect of CT08 on ER translocation was verified by monitoring the ER translocation of a VCAM-GFP construct by microscopy (Fig. 9A and B) (25). Furthermore, CT08 treatment inhibited the translocation of wild-type PrP into the ER, as observed by the decrease in the level of mature glycosylated PrP (verified by endoglycosidase H [EndoH] treatment of lysates) (Fig. 9C and D). Nontranslocated PrP is degraded by the proteasome, as observed by the accumulation of nonglycosylated species in cells treated with both bortezomib and CT08 (Fig. 9D, lane 4) (27). The accumulation of unglycosylated PrP was more pronounced in the insoluble fraction, as reported previously (27). We reasoned that in cells lacking UBXN1, the generation of ubiquitylated substrates by bortezomib or CT08 may be detrimental to cell survival due to the inability of these cells to mediate substrate degradation, leading their subsequent aggregation. Indeed, treatment of wild-type cells with CT08 resulted in a decline in cell viability across a range of drug concentrations; however, UBXN1 KO cells were more sensitive to both bortezomib and CT08 treatment than their wild-type counterparts (Fig. 9E). A similar phenotype was observed with DTT and tunicamycin treatment as well (Fig. 9E). Cotreatment with CT08 and the VCP inhibitor CB-5083 was extremely cytotoxic, with 100% cell death (data not shown). These findings strongly suggest that the VCP-UBXN1 complex is required for the timely degradation of ubiquitylated BAG6 clients and that the loss of UBXN1 renders cells sensitive to proteotoxic (in particular ER) stress.
FIG 9.
Loss of UBXN1 sensitizes cells to proteotoxic stress. (A) HeLa Flp-in T-REX cells were transfected with VCAM1-GFP and treated with 100 nM CT08 for 24 h. A total of 1 μM bortezomib was added during the final 8 h of CT08 treatment. Cells were fixed, stained for calnexin, and imaged. In CT08-treated cells, VCAM-GFP no longer localized to the ER. Cotreatment with bortezomib and CT08 was cytotoxic, and few cells remained to be quantified; however, in surviving cells, VCAM-GFP was localized to an aggresome-like structure next to the nucleus. Data for 50 cells were quantified in each of 2 independent experiments. (B) Quantification of data in panel A. (C) To determine the migration of mature and unglycosylated wild-type PrP, cell lysates expressing HA-PrP were treated with EndoH and resolved on SDS-PAGE gels. (D) Wild-type HA-PrP-expressing HeLa Flp-in T-REX cells were treated with 1 μM CT08 or 1 μM bortezomib for 6 h, as indicated. Cells were fractionated into soluble and pellet fractions. Mature and unglycosylated PrPs are indicated. (E) Wild-type and UBXN1 KO cells were plated in triplicate into 96-well plates and treated with the indicated concentrations of bortezomib, CT08, DTT, or tunicamycin (Tu) for 24 h. Cell viability was measured and normalized to the value for DMSO-treated controls for each cell line. **, P ≤ 0.01 (as determined by one-way ANOVA) (n = 2).
DISCUSSION
In this study, we identify the VCP-UBXN1 complex as a novel and important regulator of BAG6 client triage. UBXN1 is an abundant but poorly studied VCP adaptor that was reported previously to function in immune signaling pathways. Upon viral infection, UBXN1 binds to MAVS and interferes with the formation of the MAVS/TRAF3/TRAF6 inflammasome (28). Whether this occurs in the context of VCP associations is unknown. UBXN1 is also a negative regulator of NF-κB signaling in a VCP-independent manner (29). Our study identifies a novel VCP-dependent function of UBXN1 in protein quality control. Based on an unbiased proteomic screen for complexes associated with VCP adaptors in mammalian cells, we identified BAG6 and its adaptors TRC35 and UBL4A as interactors with UBXN1 (6). The BAG6 complex has multiple distinct roles in protein quality control: it mediates the degradation of aberrant newly synthesized proteins, facilitates the targeting of tail-anchored proteins to the ER, recruits the ubiquitylation machinery for the degradation of proteins that fail to be inserted, and participates in the ER-associated degradation of misfolded proteins. Thus, the nature of the BAG6-bound client and the formation of distinct protein complexes likely dictate the participation of BAG6 in a given pathway. We provide evidence that VCP and UBXN1 associate with the BAG6 complex to specifically regulate substrate degradation prior to ER translocation. A variety of model substrates that mislocalize to the cytosol due to the introduction of targeted mutations or truncations demonstrate increased steady-state levels upon the depletion of VCP or UBXN1. While VCP has multiple roles in ER homeostasis, for example, in the retrotranslocation of misfolded proteins from the ER lumen, UBXN1 does not appear to be required for ERAD based on our studies using multiple, distinct ERAD reporter cell lines but instead appears to function in regulating protein quality control prior to ER insertion. We show that VCP-UBXN1 recognizes ubiquitylated BAG6 clients and mediates their proteasomal degradation. The loss of UBXN1 results in the aggregation of substrates in insoluble inclusions. We have been unable to identify the direct interaction partners in this complex; however, it is possible that VCP-UBXN1 recognizes the BAG6 module when it is occupied with a client that is ubiquitylated, a scenario that was not reflected by our in vitro binding studies. Indeed, data from our binding studies in cells demonstrating that UBXN1 associates with ubiquitin-modified substrates and that the association between BAG6 and UBXN1 is stimulated by proteotoxic stress support this idea. Alternatively, it may also be possible that VCP-UBXN1 recognizes a component of the BAG6 complex that remains to be identified. The ATPase activity of VCP and its homohexameric structure allow it to physically segregate ubiquitylated substrates that are in an environment that the proteasome cannot easily access. This functionality is attributed to its dislocation role in ERAD, mitochondrial outer membrane protein degradation, as well as the extraction of ubiquitylated proteins on DNA. Our results with VCP catalytic mutants and the ATP competitive inhibitor CB-5083 demonstrate the requirement of VCP ATPase activity for substrate degradation. Clients once transferred to BAG6 by SGTA are committed for degradation but dissociate slowly from BAG6, suggesting that they may need to be extracted prior to degradation (11). Thus, ATP hydrolysis by VCP may enable the extraction and local unfolding of ubiquitylated substrates bound to BAG6 and their subsequent delivery to the proteasome. Physiologically, the loss of UBXN1 negatively impacts the ability of cells to respond to agents that induce proteotoxic stress. We note that while VCP-UBXN1 is critical for substrate triage, there are likely other (VCP-dependent and -independent) pathways to degrade substrates. We observe that the VCP adaptor UFD1-NPL4 that docks with many distinct VCP adaptor complexes also appears to regulate BAG6 substrate triage. Future work will determine whether this reflects two distinct VCP complexes or a larger complex consisting of all three adaptors (as the UFD1-NPL4 dimer docks onto a single monomer within the VCP hexamer).
It is likely that there are multiple overlapping quality control systems in the cytosol to deal with errors that arise during the targeting of newly synthesized proteins to specific organelle compartments. Each pathway therefore has a distinct repertoire of clients that it handles, but it is likely that there is a degree of redundancy that enables substitution if specific components become saturated. The extent to which VCP-UBXN1 participates in these parallel pathways remains to be determined. Given the abundance of VCP and the diverse repertoire of adaptors that it can access, we speculate that VCP can regulate the triage of other organelle-specific substrates in the context of specific adaptors and/or clients. The availability of in vitro systems that recapitulate key steps of substrate triage will enable the reconstitution of the VCP-UBXN1-mediated degradation of ubiquitylated substrates. Certainly, the identification of ubiquitylated substrates that are dependent on VCP-UBXN1 for degradation will illuminate the dependency of different quality control pathways on this complex.
MATERIALS AND METHODS
Cell culture, transfections, and immunoprecipitation.
HEK-293T and HeLa Flp-in T-REX cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin. Cells were maintained in a humidified, 5% CO2 atmosphere at 37°C. ERAD reporter cell lines were a generous gift from Ron Kopito (Stanford). For siRNA transfections, cells were trypsinized and reverse transfected with 30 nM siRNAs by using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested at 60 h posttransfection. For DNA transfections, cells in 6-well plates were transfected with 0.5 μg OPG-TASK, 2 μg of HA-PrP, 1 μg of HA-ubiquitin, or 1 μg of VCAM1-GFP using Lipofectamine 3000 and typically harvested at 24 to 36 h posttransfection. Cells were lysed in mammalian cell lysis buffer (50 mM Tris-Cl [pH 6.8], 150 mM NaCl, 0.5% Nonidet P-40, Halt protease inhibitors [Pierce], and 1 mM DTT) or in 1% SDS plus 50 mM Tris-Cl (pH 6.8). See the figure legends for details. Cells were incubated at 4°C for 10 min and then centrifuged at 14,000 rpm for 15 min. The supernatant was removed, and the protein concentration was estimated by using the Bradford method (Bio-Rad). Protein G-Sepharose and the indicated antibodies were used for immunoprecipitation at 4°C for 3 to 5 h (Fig. 1) or overnight (Fig. 5). Beads were washed 3 to 5 times in 1 ml mammalian cell lysis buffer and resuspended in 2× SDS sample buffer. Iodoacetamide and PR-619 were used at a final concentration of 50 μM. For denaturing immunoprecipitation studies, immune complexes were denatured in a solution containing 1% SDS and 50 mM Tris-Cl (pH 6.8) and then diluted 10-fold in 50 mM Tris-Cl (pH 6.8). Diluted samples were used for subsequent purifications. Cells were treated with 0.5 to 1 μM bortezomib, 5 μM CB-5083, 100 μg/ml cycloheximide, 5 μg/ml of puromycin, or 0.1 to 1 μM CT08 for the indicated times (see the figure legends for details).
Immunoblot quantification was performed by densitometry using ImageJ. Protein levels were normalized to the value for a loading control (typically PCNA). Quantification of immunoblots is available in Table S1 in the supplemental material.
Generation of CRISPR cell lines.
The CRISPR-Cas9 gene-editing system was used to generate UBXN1 knockout cell lines in HeLa Flp-in T-REX cells. The guide sequence 5′-GCCGTCCCAGGATATGTCCAA-3′ was cloned into the pX459 vector carrying hSpCas9 and transiently transfected into HeLa cells by using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. At 36 h posttransfection, the cells were pulsed briefly with 1 μg/ml puromycin for a further 24 to 36 h. The surviving cells were then serially diluted in 96-well plates for clonal selection, and expression levels were monitored by immunoblotting. The rescue cell line was generated by transfecting pcDNA FRT/TO-GFP-UBXN1 and pOG-44 into UBXN1 KO cells by using Lipofectamine 3000 for integration into the single FLP recombination target (FRT) site. Cells were treated with 200 μg/ml hygromycin to select for stably integrated cells. GFP-UBXN1 expression was induced with 1 μg/ml doxycycline for 72 h.
Antibodies, siRNAs, and reagents.
The rabbit VCP, UBXN1, BAG6, UFD1, NPL4, UBXD8, and calnexin antibodies were obtained from Proteintech; mouse opsin antibody was a generous gift from Stephen High (University of Manchester); mouse ubiquitin antibody (clone P4D1 used for immunoblotting) and mouse FLAG were obtained from Cell Signaling Technology; mouse PCNA and mouse Myc (clone 9E10) antibodies were obtained from Santa Cruz; mouse HA was obtained from Covance; mouse FK2 used for immunofluorescence was obtained from EMD Millipore; and His antibody was obtained from Clontech. CB-5083 was a gift from Cleave Biosciences, and bortezomib was obtained from Selleckchem. CT08, wild-type PrP (HA), and VCAM1-GFP were generous gifts from Ramanujan Hegde (MRC, UK). siRNAs against BAG6 (catalog number s15467), UBXN1-2 (catalog number s27292), UBXN1-3 (catalog number s27293) UFD1-1 (catalog number s14635), UFD1-2 (catalog number s228150), NPL4 (catalog number s31209), NPL4-2 (catalog number s31210), UBXD7-1 (catalog number s24996), and UBXD7-2 (catalog number 24997) were purchased from Ambion (Thermo Fisher Scientific). VCP siRNAs (siRNA 1 [catalog number HSS11262], siRNA 2 [catalog number HSS111263], and siRNA 3 [catalog number HSS111264]) were obtained from Invitrogen (Thermo Fisher Scientific). VCP rescue constructs were described previously (30) and were resistant to siRNA 2.
Immunofluorescence and microscopy.
HeLa Flp-in T-REX cells were grown on coverslips (no. 1.5) in a 12-well plate and transfected with HA-PrPmut by using Lipofectamine 3000. At 24 h posttransfection, cells were untreated or treated with 1 μM bortezomib for 6 or 18 h. Cells were washed briefly in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde at room temperature for 15 min. Cells were washed in PBS, permeabilized in 0.2% Triton X-100 for 5 min, washed again, and then blocked in 1% bovine serum albumin (BSA) in PBS for 1 h. For DRiP clearance studies, cells were blocked in 3% chicken serum plus 0.1% Triton X-100 in PBS for 1 h. The coverslips were incubated with the indicated antibodies overnight at 4°C in a humidified chamber, washed, and incubated for a further hour with the appropriate Alexa Fluor-conjugated secondary antibodies (Molecular Probes) for 1 h in the dark at room temperature. Cells were washed with PBS, and nuclei were stained with Hoechst dye and mounted onto slides. All images were collected by using a Nikon A1R scan head with a spectral detector and resonant scanners on a Ti-E motorized inverted microscope equipped with a 60× Plan Apo 1.4-numerical-aperture (NA) objective lens. The indicated fluorophores were excited with either a 405-nm, 488-nm, or 594-nm laser line. Images were analyzed by using FIJI (https://imagej.net/Fiji). Colocalization analysis was performed by using the Coloc2 plug-in in FIJI.
Flow cytometry.
HEK-293 cells stably expressing ERAD reporter cell lines were transfected with the indicated siRNAs in triplicate by using Lipofectamine 3000. Due to incomplete UBXN1 depletion, we transfected cells with UBXN1 siRNAs on two consecutive days to obtain greater knockdown. Sixty hours after the first transfection, cells were harvested and collected into DMEM–10% FBS, and the GFP signal was measured by using the FACSCalibur system (BD Biosciences). A total of 10,000 live, gated cells were counted per sample, and the number of GFP-positive cells was determined based on the fluorescence levels observed in untreated versus bortezomib-treated cells. Means and standard deviations for triplicate measurements were calculated, and statistical significance was calculated by using the Student t test.
Cell fractionation.
HEK-293T or HeLa Flp-in T-REX cells were transfected with the indicated siRNAs and relevant substrates by using Lipofectamine 3000. At 60 h posttransfection, cells were harvested and fractionated to generate soluble and insoluble fractions. Cells were lysed in mammalian cell lysis buffer, as described above. Cells were lysed at 4°C for 10 min and then centrifuged at 14,000 rpm for 10 min. The supernatant (soluble fraction) was removed, and the pellet was lysed in buffer B (50 mM Tris-Cl [pH 6.8], 150 mM NaCl, 2% SDS), vortexed, and boiled briefly. The resulting lysate was designated the insoluble pellet fraction. Protein concentrations for both soluble and pellet fractions were determined by a bicinchoninic acid (BCA) assay (Pierce). Where indicated, the cells were treated with 0.5 to 1 μM bortezomib or 1 μM CB-5083 for either 4, 6, or 18 h (see the figure legends) prior to fractionation.
DRiP clearance studies.
HeLa Flp-in T-REX cells were grown on coverslips and treated with 5 μg/ml of puromycin for 2 h at 37°C. For release studies, cells were washed three times in DMEM–10% FBS and released into drug-free medium for 3 h at 37°C. For rescue experiments, GFP-UBXN1 expression was induced with 1 μg/ml doxycycline for 18 h prior to puromycin treatment. Doxycycline was maintained throughout the experiment. Coverslips were fixed and processed for microscopy, as outlined above. The particle analysis plug-in in ImageJ was used to quantify ALIS formation. At least 150 cells per sample were analyzed in three biological replicate studies. Means and standard deviations were calculated, and statistical significance was calculated by using the Student t test.
Cell viability studies.
Wild-type and UBXN1 KO cells were plated in triplicate into 96-well plates (4,000 cells per well). The next day, cells were treated with either dimethyl sulfoxide (DMSO), 1 μM bortezomib, 1 μM CT08, or 1.5 μM DTT for 24 h. Cell viability was measured by using the Cell Titer-Glo system (Promega) according to the manufacturer's instructions. Values were normalized to the value for DMSO-treated samples for each cell line. Means and standard deviations were calculated, and statistical significance was calculated by one-way analysis of variance (ANOVA).
Supplementary Material
ACKNOWLEDGMENTS
R.G. and M.R. conceived and designed the study. R.G., S.M., F.S., and M.R. performed the experiments, and M.R. wrote the manuscript.
This work was supported by NIH grant K22-CA175142 and an Earl P. Charlton award to M.R.
We thank Alenka Lovy at the Imaging and Cell Analysis Core and Stephen Kwok and Allen Parmelee at the Flow Cytometry Core at Tufts University School of Medicine for their help. We are extremely grateful to Ramanujan Hegde (MRC), Stephen High (University of Manchester), Ron Kopito (Stanford), and Sichen Shao (Harvard Medical School) for generously sharing reagents.
We have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00154-18.
REFERENCES
- 1.Shao S, Hegde RS. 2016. Target selection during protein quality control. Trends Biochem Sci 41:124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Bennett EJ. 2016. Proteome complexity and the forces that drive proteome imbalance. Nature 537:328–338. doi: 10.1038/nature19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolff S, Weissman JS, Dillin A. 2014. Differential scales of protein quality control. Cell 157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW. 2015. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discov 14:759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 6.Raman M, Sergeev M, Garnaas M, Lydeard JR, Huttlin EL, Goessling W, Shah JV, Harper JW. 2015. Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat Cell Biol 17:1356–1369. doi: 10.1038/ncb3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. 2008. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. 2011. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. 2011. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell 42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Liu Y, Lee JG, Ye Y. 2013. A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation. J Biol Chem 288:18068–18076. doi: 10.1074/jbc.M112.449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS. 2017. Mechanistic basis for a molecular triage reaction. Science 355:298–302. doi: 10.1126/science.aah6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. 2014. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell 55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadowaki H, Nagai A, Maruyama T, Takami Y, Satrimafitrah P, Kato H, Honda A, Hatta T, Natsume T, Sato T, Kai H, Ichijo H, Nishitoh H. 2015. Pre-emptive quality control protects the ER from protein overload via the proximity of ERAD components and SRP. Cell Rep 13:944–956. doi: 10.1016/j.celrep.2015.09.047. [DOI] [PubMed] [Google Scholar]
- 14.Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ. 2006. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci 119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- 15.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci U S A 105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki M, Otsuka T, Ohsaki Y, Cheng J, Taniguchi T, Hashimoto H, Taniguchi H, Fujimoto T. 2012. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell 23:800–810. doi: 10.1091/mbc.E11-11-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaLonde DP, Bretscher A. 2011. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J Biol Chem 286:4892–4901. doi: 10.1074/jbc.M110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park ES, Yoo YJ, Elangovan M. 2017. The opposite role of two UBA-UBX containing proteins, p47 and SAKS1 in the degradation of a single ERAD substrate, alpha-TCR. Mol Cell Biochem 425:37–45. doi: 10.1007/s11010-016-2860-5. [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Rahbar R, Hegde RS. 2001. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem 276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- 20.Wunderley L, Leznicki P, Payapilly A, High S. 2014. SGTA regulates the cytosolic quality control of hydrophobic substrates. J Cell Sci 127:4728–4739. doi: 10.1242/jcs.155648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, Taunton J, Katze MG, Ron D. 2006. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 23.Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. 2010. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol 190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. 2013. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maifeld SV, MacKinnon AL, Garrison JL, Sharma A, Kunkel EJ, Hegde RS, Taunton J. 2011. Secretory protein profiling reveals TNF-alpha inactivation by selective and promiscuous Sec61 modulators. Chem Biol 18:1082–1088. doi: 10.1016/j.chembiol.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrison JL, Kunkel EJ, Hegde RS, Taunton J. 2005. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 27.Rane NS, Yonkovich JL, Hegde RS. 2004. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J 23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P, Yang L, Cheng G, Yang G, Xu Z, You F, Sun Q, Lin R, Fikrig E, Sutton RE. 2013. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep 3:1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang YB, Tan B, Mu R, Chang Y, Wu M, Tu HQ, Zhang YC, Guo SS, Qin XH, Li T, Li WH, Li AL, Zhang XM, Li HY. 2015. Ubiquitin-associated domain-containing ubiquitin regulatory X (UBX) protein UBXN1 is a negative regulator of nuclear factor kappaB (NF-kappaB) signaling. J Biol Chem 290:10395–10405. doi: 10.1074/jbc.M114.631689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman M, Havens CG, Walter JC, Harper JW. 2011. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol Cell 44:72–84. doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.