ABSTRACT
Orthologs of the herpes simplex virus (HSV) UL16 gene are conserved throughout the Herpesviridae. Because of this conservation, one might expect that the proteins perform similar functions for all herpesviruses. Previous studies on a UL16-null mutant derived from HSV-2 strain 186 revealed a roughly 100-fold replication defect and a critical role for UL16 in the nuclear egress of capsids. These findings were in stark contrast to what has been observed with UL16 mutants of HSV-1 and pseudorabies virus, where roughly 10-fold replication deficiencies that were accompanied by defects in the secondary envelopment of cytoplasmic capsids were reported. One possible explanation for this discrepancy is that HSV-2 strain 186 is not representative of the HSV-2 species. To address this possibility, multiple UL16-null mutants were constructed in multiple HSV-2 and HSV-1 strains by CRISPR/Cas9 mutagenesis, and their phenotypes were characterized side by side. This analysis showed that all the HSV-2 UL16 mutants had 50- to 100-fold replication deficiencies that were accompanied by defects in the nuclear egress of capsids, as well as defects in the secondary envelopment of cytoplasmic capsids. By contrast, most HSV-1 UL16 mutants had 10-fold replication deficiencies that were accompanied by defects in secondary envelopment of cytoplasmic capsids. These findings indicated that UL16 has HSV species-specific functions. Interestingly, HSV-1 UL16 could promote the nuclear egress of HSV-2 UL16-null strains, suggesting that, unlike HSV-1, HSV-2 lacks an activity that can promote nuclear egress in the absence of UL16.
IMPORTANCE HSV-2 and HSV-1 are important human pathogens that cause distinct diseases in their hosts. A complete understanding of the morphogenesis of these viruses is expected to reveal vulnerabilities that can be exploited in the treatment of HSV disease. UL16 is a virion structural component that is conserved throughout the Herpesviridae and functions in virus morphogenesis; however, previous studies have suggested different roles for UL16 in the morphogenesis of HSV-2 and HSV-1. This study sought to resolve this apparent discrepancy by analyzing multiple UL16 mutant viruses derived from multiple strains of HSV-2 and HSV-1. The data indicate that UL16 has HSV species-specific functions, as HSV-2 has a requirement for UL16 in the escape of capsids from the nucleus whereas both HSV-2 and HSV-1 require UL16 for final envelopment of capsids at cytoplasmic membranes.
KEYWORDS: herpes simplex virus, tegument, UL16, nuclear egress, secondary envelopment
INTRODUCTION
While the early stages of herpesvirus assembly take place in the nucleus, the final stages of virion assembly occur in the cytoplasm of infected cells. Viral DNA is packaged into preformed procapsids in the infected-cell nucleus, resulting in the formation of C capsids that are competent for subsequent stages in virion maturation. To reach the cytoplasm, genome-containing C capsids must transit across the inner and outer nuclear membranes, utilizing a process referred to as nuclear egress—a subject of recent and intense investigation by numerous laboratories (1, 2). Nuclear egress of C capsids occurs through primary envelopment of capsids at the inner nuclear membrane, followed by de-envelopment and release of capsids into the cytoplasm through fusion of the perinuclear virion envelope with the outer nuclear membrane. Once in the cytoplasm, the C capsid acquires its final envelope by budding into membrane vesicles derived from the trans-Golgi network, or an endocytic compartment, in a process referred to as secondary envelopment (3–5). Finally, enveloped virions contained within vesicles are transported to the cell surface, where they fuse with the plasma membrane, releasing the mature virion into the extracellular space.
This study concerns the functions of the herpes simplex virus (HSV) UL16 protein in virion assembly. Orthologs of the HSV UL16 protein are conserved throughout the Herpesviridae; however, its specific roles in the virus replicative cycle are poorly understood. Contributing to this lack of clarity are seemingly conflicting reports on the functions of the HSV-2 and HSV-1 UL16 orthologs. Our laboratory recently reported that deletion of UL16 from HSV-2 strain 186 resulted in a roughly 100-fold reduction in virus replication and a failure of C capsids to undergo efficient nuclear egress (6). In contrast to these findings, several groups have reported that HSV-1 UL16 mutants have more modest (roughly 10-fold) replication deficiencies and are defective in secondary envelopment rather than nuclear egress (7, 8). It is noteworthy that studies on the UL16 ortholog from pseudorabies virus (PRV), a virus distantly related to HSV, closely resembled the findings seen with HSV-1, where roughly 10-fold replication deficiencies associated with defective secondary envelopment were reported (9). What could explain these conflicting reports? One possibility was that the single strain of HSV-2 studied by Gao and colleagues (6), strain 186, was an outlier and the results obtained with this strain were not representative of the HSV-2 species as a whole. Another possibility was that the HSV-1 and PRV strains analyzed previously were constructed in a way that promoted the selection of suppressor mutations that might overcome the replication deficiencies and nuclear egress phenotypes exhibited by our original HSV-2 186 strain UL16-null mutant [HSV-2 (186) Δ16], which, in contrast, was isolated on complementing cells to mitigate the selection of suppressor mutations. A third possibility was that UL16 has species-specific functions during the morphogenesis of HSV-2 and HSV-1.
The goal of this study was to resolve this controversy by performing a side-by-side analysis of a panel of newly constructed UL16 mutants derived from multiple strains of HSV-2 and HSV-1. All the strains were constructed using the same procedures. CRISPR/Cas9-based mutagenesis was used to create the mutant virus genomes, and all the UL16 mutant viruses were propagated on UL16-expressing cells to avoid the enrichment of suppressor mutants during strain isolation. To extend the previous analysis of HSV-2 strain 186, we chose to build UL16 mutants into strains HG52 and SD90e. Strain HG52 is a well-studied HSV-2 reference strain that was the first to be completely sequenced (10), whereas strain SD90e is a low-passage-number clinical isolate that has been proposed to serve as a new HSV-2 reference strain (11, 12). For the construction of new HSV-1 UL16 mutants, we chose to utilize strains F and KOS, two well-studied laboratory strains that have been used by others to study the function of UL16 (7, 8). Our analysis indicates that UL16 plays a critical role in both the nuclear egress and secondary envelopment of HSV-2 strains, whereas HSV-1 UL16 functions primarily in the secondary envelopment of cytoplasmic capsids. Interestingly, trans-complementation experiments revealed that HSV-2 and HSV-1 UL16 proteins can substitute for each other, suggesting that the genetic basis for the species-specific requirements of UL16 reside outside the UL16 locus.
(This article was submitted to the bioRxiv online preprint archive [https://doi.org/10.1101/274548].)
RESULTS
Construction of HSV UL16 mutant viruses.
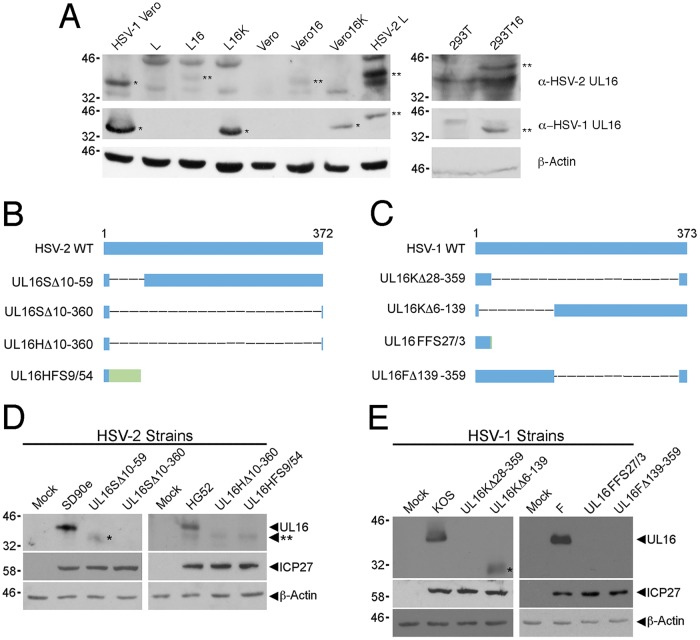
We constructed UL16 deletions in multiple HSV-2 and HSV-1 strains to enable a comparative analysis of the viruses. To avoid any selective pressure during the isolation of the strains, UL16-expressing cell lines were used (Fig. 1A). UL16 deletions were constructed in HSV-2 strains SD90e and HG52 and HSV-1 strains KOS and F by CRISPR/Cas9-based mutagenesis as described in Materials and Methods. Two mutants derived independently from each strain were selected for further study, and all the mutants studied were used at low passage numbers. DNA sequencing of HSV-2 and HSV-1 deletion mutants revealed the exact nature of the UL16 mutants isolated (Fig. 1B and C). To verify that the UL16 mutants did not express UL16, Vero cells were infected with the different HSV-2 and HSV-1 UL16 mutants at a multiplicity of infection (MOI) of 0.5, and cell lysates were prepared 24 h postinfection (hpi). Western blots of the cell lysates were probed for HSV-2 and HSV-1 UL16 proteins, the HSV immediate-early protein ICP27 (infection control), and β-actin (loading control) (Fig. 1D and E). Full-length UL16 was observed in all wild-type-virus-infected cell lysates, while no full-length UL16 protein was expressed in lysates from any UL16 mutant-infected cells. Notably, HSV-2 strain UL16SΔ10-59- and HSV-1 strain UL16KΔ6-139-infected cell lysates contained truncated UL16 proteins (Fig. 1D and E, single asterisks). These data confirmed that all the UL16 mutants failed to produce full-length UL16 protein.
FIG 1.
Construction and verification of HSV UL16 mutants. (A) Western blot analysis of UL16-expressing cell lines used for the construction and analysis of UL16 mutants. Cell lysates were harvested and analyzed by Western blotting for HSV-2 UL16 and HSV-1 UL16. Lysates from Vero cells infected with HSV-1 and L cells infected with HSV-2 were used as positive controls. The single asterisks indicate HSV-1 UL16 protein (∼41 kDa), and the double asterisks indicate HSV-2 UL16 protein (∼42 kDa). β-Actin was used as a loading control. (B and C) Diagrams of full-length (wild-type [WT]) and mutant UL16 proteins from HSV-2 (B) and HSV-1 (C). Four UL16 deletion mutants of each HSV species were selected for further analysis. The blue bars represent UL16 protein sequence, the dashed lines represent deleted sequences, and the green bar represents non-UL16 amino acids that arise due to frameshift. In the nomenclature used, the first letter after UL16 indicates the parental strain (S, SD90e; H, HG52; K, KOS; F, F); Δ refers to an in-frame deletion, and the numbers following Δ indicate the positions of the codons that were deleted from the UL16 gene; FS refers to a frameshift, the first number following FS refers to the position of the codon where the frameshift occurred, and the last number refers to the number of non-UL16 amino acids. (D and E) Western blots of cell lysates from Vero cells infected with the strains shown in panels B and C were probed using antiserum against HSV-2 UL16 (D) or HSV-1 UL16 (E). ICP27 antiserum was used as a positive control for viral infection, while β-actin antiserum was used as a loading control. The single asterisks indicate truncated forms of UL16. The double asterisk indicates the position of a nonspecific band detected in HG52-infected cell lysates in panel D.
UL16 is required for efficient cell-to-cell spread of both HSV-2 and HSV-1 strains.
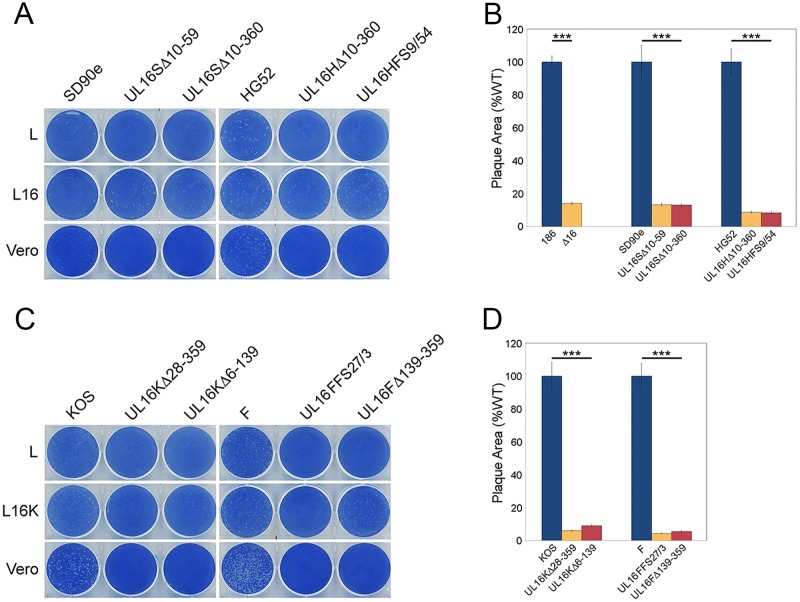
To determine if the cell-to-cell spread properties of the new UL16 deletion mutants were consistent with our results with HSV-2 strain 186 (6) and those reported by others for HSV-1 strains KOS and F (8), monolayers of L, L16 (expressing HSV-2 UL16), L16K (expressing HSV-1 UL16), or Vero cells were infected with UL16 mutants and their parental viruses (Fig. 2). At 72 hpi, the cells were fixed and stained with methylene blue. All the HSV-2 UL16-null mutants formed visible plaques on complementing L16 cells but did not form visible plaques on noncomplementing L cells (Fig. 2A). Similarly, all the HSV-1 UL16 mutants formed visible plaques on complementing L16K cells but not on L cells (Fig. 2C). Importantly, all the UL16-null strains formed visible plaques on noncomplementing Vero cells, albeit much smaller than those formed by their parental strains (Fig. 2A and C), indicating some capacity for spread between Vero cells that was not seen on L cell monolayers. The absence of macroscopically visible UL16-null mutant plaques on L cell monolayers may reflect poorer replication of these strains on L cells than on Vero cells, as was observed for our HSV-2 (186) Δ16 strain (6). These data suggested that HSV-1 and HSV-2 UL16 mutants have similar deficiencies in cell-to-cell spread.
FIG 2.
Cell-to-cell spread capabilities of HSV UL16 mutants. (A and C) Identical dilutions of each HSV strain were used to infect the noncomplementing and complementing cell monolayers indicated. The cells were fixed and stained with 0.5% methylene blue in 70% methanol at 72 hpi. (B and D) Vero cells were infected with the indicated viruses, the cells were fixed, and plaques were stained using antiserum against HSV Us3 by indirect immunofluorescence microscopy at 24 hpi. Plaque sizes were determined as described in Materials and Methods (n = 40 plaques per strain). The error bars represent standard errors of the mean. HSV wild-type strains 186, SD90e, HG52, KOS, and F were normalized to 100%. ***, P < 0.0001.
To quantify the abilities of UL16 deletion viruses to spread, we measured the areas of the plaques produced by the UL16 deletion mutants on noncomplementing Vero cells. At 24 hpi, the plaques were fixed and stained using antisera against HSV Us3, and the areas of the plaques were measured using ImagePro 6.3. The two HSV-2 (SD90e) UL16 mutants formed plaques approximately 13% of the size of those of their parental strain, while HSV-2 (HG52) UL16 mutant plaques were around 8% the size of WT HG52 plaques (Fig. 2B). In addition, the plaque size of our original HSV-2 186 strain UL16-null mutant, Δ16, was 14% of that of the WT 186 strain (Fig. 2B). Surprisingly, all the HSV-1 UL16-null mutants formed plaques roughly 95% smaller than those of their parental strains, similar to what was observed with HSV-2 UL16-null strains (Fig. 2D). Collectively, these findings suggested that UL16 is critical for virus spread on noncomplementing cells, and no obvious differences between HSV-2 and HSV-1 spread were observed in the absence of UL16.
Replication kinetics of HSV-2 and HSV-1 UL16-null strains.
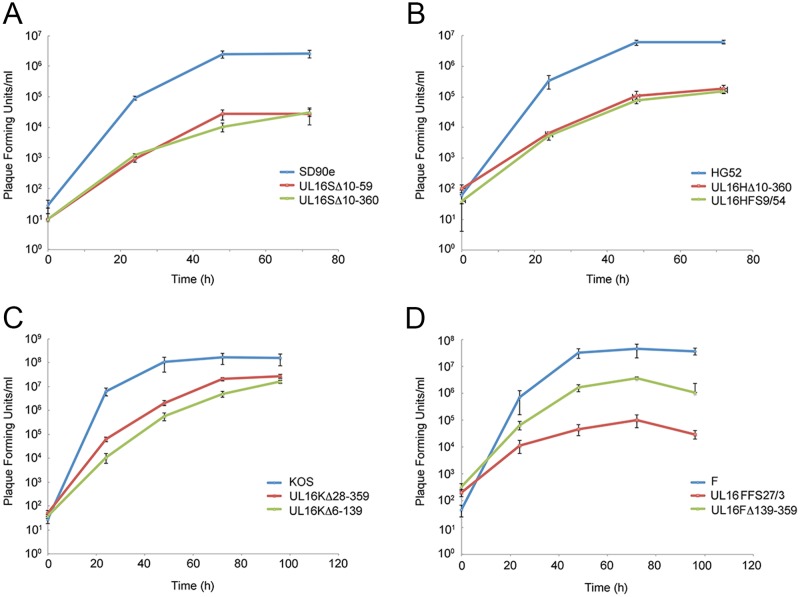
To provide a more comprehensive view of the replication defects of HSV-2 and HSV-1 UL16 deletion mutants, we performed multistep growth analysis. Monolayers of Vero cells were infected with HSV-2 and HSV-1 UL16 mutants and their corresponding parental strains at an MOI of 0.01. Cells and medium were harvested together at the indicated time points after infection and titrated on monolayers of complementing L16 cells. The results showed that the HSV-2 (SD90e and HG52) UL16 deletions had approximately 100-fold and 50-fold reductions in endpoint titers, respectively, compared to their parental strains (Fig. 3). By contrast, with one exception, our KOS and F UL16 mutants had roughly 10-fold reductions in virus replication compared to their parental strains (Fig. 3). UL16FFS27/3 was an outlier, as it replicated much more poorly (400-fold lower than WT F) than the other HSV-1 strains analyzed. With the exception of the UL16FFS27/3 strain, these data are consistent with previous findings (6–8) indicating that HSV-2 UL16 mutants replicate less efficiently than HSV-1 UL16 mutants.
FIG 3.
Replication kinetics of HSV UL16 deletion mutants. Monolayers of Vero cells were infected with parental HSV strains SD90e (A), HG52 (B), KOS (C), and F (D) and their corresponding UL16 deletion mutants at an MOI of 0.01. Cells and medium were harvested together at the indicated times postinfection and titrated on monolayers of L16 cells. Each data point represents the average data from two biological replicates, each of which was titrated in triplicate. The error bars are standard errors of the mean.
Reciprocal complementation between HSV-2 and HSV-1 UL16 proteins.
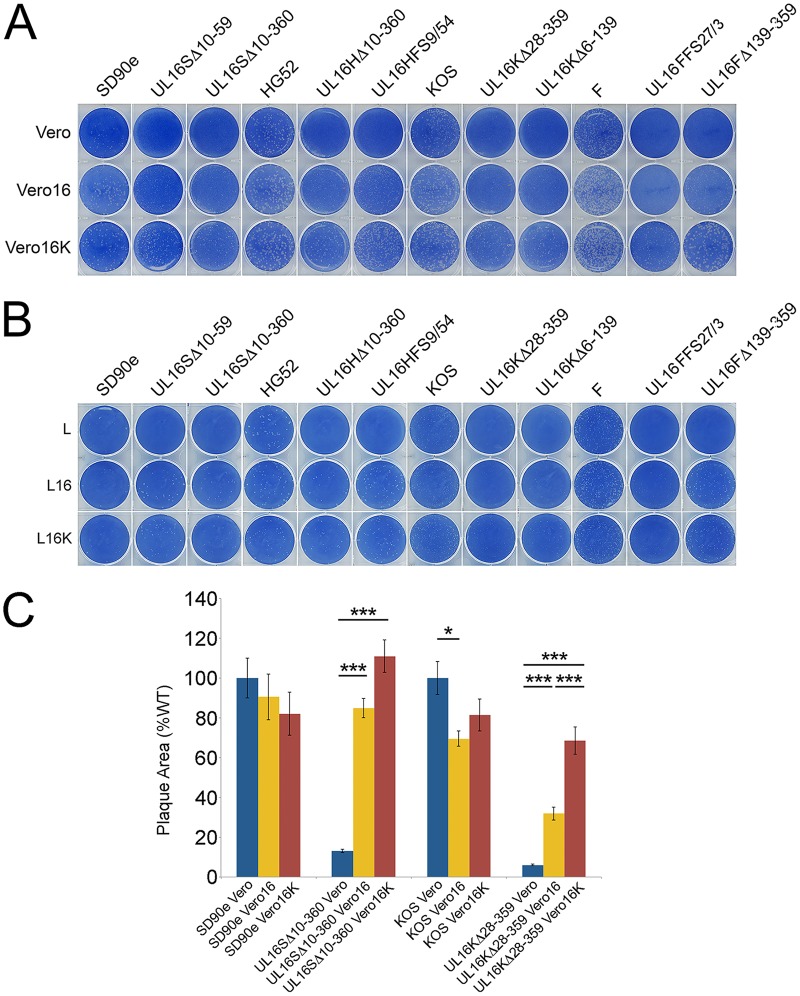
To examine whether HSV-2 and HSV-1 UL16 proteins could functionally compensate for each other, reciprocal-complementation assays were performed. Monolayers of Vero, Vero16 (expressing HSV-2 UL16), and Vero16K (expressing HSV-1 UL16) cells were infected with the same dilutions of HSV-2 and HSV-1 UL16 deletion viruses and their parental strains. At 72 hpi, the cells were fixed and stained with methylene blue. Interestingly, all the HSV-2 and HSV-1 UL16 mutants formed large plaques on Vero16 and Vero16K monolayers (Fig. 4A). A similar experiment was performed on L, L16, and L16K cells that also demonstrated reciprocal complementation of the strains (Fig. 4B). Notably, UL16 mutants formed very small plaques on Vero cells compared to their parental strains, consistent with the data shown in Fig. 2. Parental strains of HSV-1 and HSV-2 had indistinguishable plating efficiencies on Vero, Vero16, and Vero16K cells. By contrast, HSV-2 UL16 mutants had 2.9 (±1.4)-fold higher plating efficiency on Vero16 cells and 3.2 (±1.8)-fold higher plating efficiency on Vero16K cells than on Vero cells. HSV-1 UL16 mutants had 2.9 (±1.0)-fold higher plating efficiency on Vero16 cells and 3.9 (±0.6)-fold higher plating efficiency on Vero16K cells than on Vero cells. Plating efficiencies on L cells could not be determined using this assay because macroscopic plaques failed to form on L cell monolayers infected with UL16 mutant viruses. To quantify reciprocal complementation of plaque formation, we measured the areas of the plaques produced by SD90e, UL16SΔ10-360, KOS, and UL16KΔ28-359 on monolayers of Vero, Vero16, and Vero16K cells (Fig. 4C) as described in the legend to Fig. 2. Plaque sizes of both UL16 mutant strains were significantly increased on both complementing cell lines. Collectively, these data indicate that the HSV-2 UL16 protein can complement HSV-1 UL16-null strains, and vice versa.
FIG 4.
Reciprocal complementation between HSV-2 and HSV-1 UL16 proteins. (A and B) Monolayers of Vero, Vero16, and Vero16K cells (A) or L, L16, and L16K cells (B) were infected with identical dilutions of each HSV-2 and HSV-1 UL16 mutant. The cells were fixed and stained with 0.5% methylene blue in 70% methanol at 72 hpi. (C) Vero, Vero16, or Vero16K cells were infected with the indicated viruses and fixed, and plaques were stained using antiserum against HSV Us3 by indirect immunofluorescence microscopy at 24 hpi. Plaque sizes were determined as described in Materials and Methods (n = 40 plaques per strain). The error bars represent standard errors of the mean. HSV wild-type strains SD90e and KOS were normalized to 100%. *, P < 0.05; ***, P < 0.0001.
Species-specific requirements for the UL16 protein.
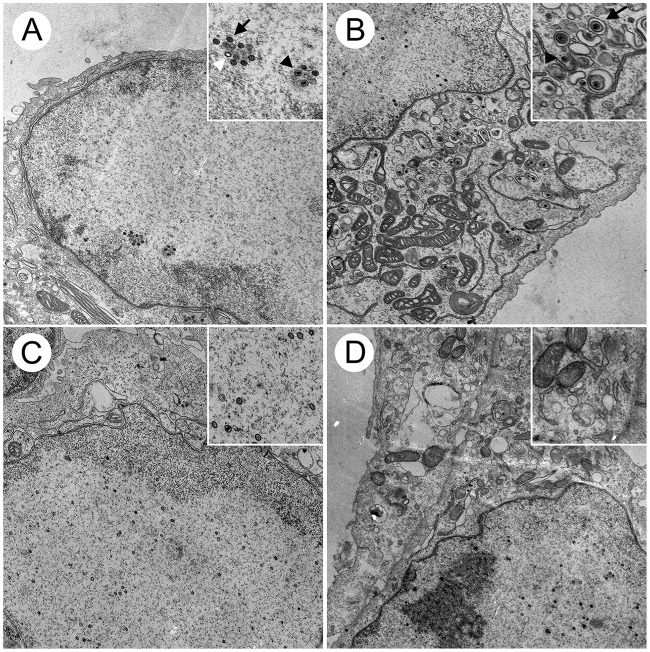
To define and compare the stages at which our HSV-2 and HSV-1 UL16 mutants were blocked in their maturation, transmission electron microscopy (TEM) was performed. Vero cells were infected with HSV-1 and HSV-2 UL16 mutants and the corresponding parental strains, and cells were fixed and processed for TEM at 16 hpi as described in Materials and Methods. A, B, and C capsids were readily observed in the nuclei of parental HSV-2 (SD90e)-infected cells (Fig. 5A) and UL16SΔ10-360-infected cells (Fig. 5C). However, similar to what we reported previously for HSV-2 (186) (6), many more cytoplasmic capsids were observed in HSV-2 (SD90e)-infected cells (Fig. 5B) than in cells infected with its corresponding UL16 mutant, UL16SΔ10-360 (Fig. 5D). Numerous capsids were observed in both the nuclei and cytoplasm of HSV-1 (F)- and UL16FΔ139-359-infected cells (Fig. 6). However, fewer enveloped cytoplasmic capsids were observed in cells infected with the UL16 mutant, UL16FΔ139-359 (Fig. 6D), than in HSV-1 (F)-infected cells (Fig. 6B). These findings are consistent with previous reports indicating that HSV-1 UL16 functions in secondary envelopment (8).
FIG 5.
Ultrastructural analysis of HSV-2-infected cells. Vero cells were infected with HSV-2 SD90e (A and B) and the UL16 deletion mutant, UL16SΔ10-360 (C and D), at an MOI of 3. At 16 hpi, cells were fixed and processed for TEM as described in Materials and Methods. (B) Nonenveloped cytoplasmic capsids and enveloped virions can be observed in the cytoplasm of SD90e-infected Vero cells. (D) These structures were rarely observed in the cytoplasm of UL16SΔ10-360-infected cells. (A and C) Nuclear capsids were readily detected in the nuclei of SD90e-infected (A) and UL16SΔ10-360-infected (C) cells. (Insets) Magnified portions of the images. (A) The white arrowhead identifies an A capsid, whereas a B capsid is identified with a black arrow and a C capsid with a black arrowhead. (B) The black arrow indicates an enveloped capsid and the black arrowhead a nonenveloped capsid.
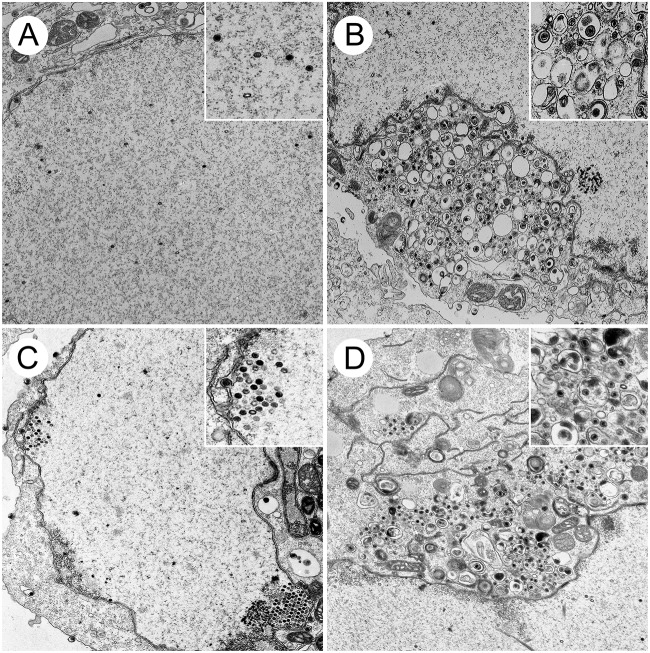
FIG 6.
Ultrastructural analysis of HSV-1-infected cells. Vero cells were infected with HSV-1 F (A and B) and the UL16 deletion mutant, UL16FΔ139-359 (C and D), at an MOI of 3. At 16 hpi, cells were fixed and processed for TEM as described in Materials and Methods. (B) Nonenveloped cytoplasmic capsids and enveloped virions can be observed in the cytoplasm of F-infected cells. (D) Enveloped virions were less frequently observed in the cytoplasm of UL16FΔ139-359-infected cells, where nonenveloped capsids were abundant. (A and C) Nuclear capsids were readily detected in the nuclei of both F-infected cells (A) and UL16FΔ139-359-infected cells (C).
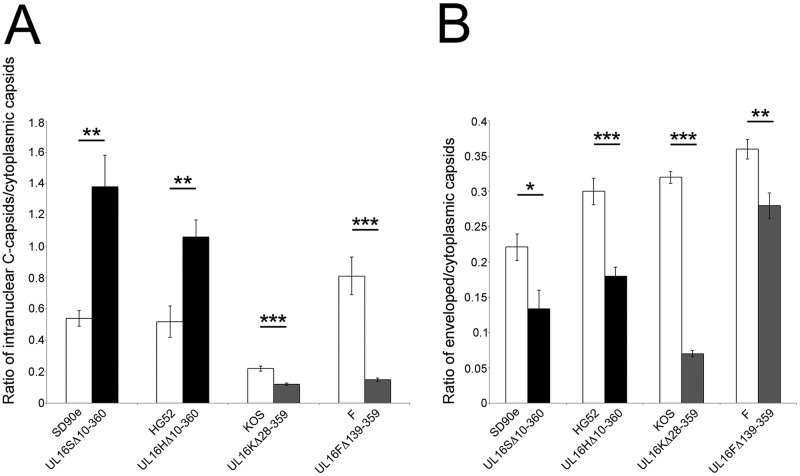
To quantify the distribution of capsids in the presence and absence of UL16, viral particles at various stages of maturation were classified and counted in 10 independent images of Vero cells infected with the strains listed in Table 1, and the ratios of intranuclear C capsids to cytoplasmic capsids and of enveloped capsids to cytoplasmic capsids were analyzed in more detail (Fig. 7). We chose to focus on intranuclear C capsids instead of A, B, and C capsids together, because C capsids are preferentially selected for primary envelopment (13). The ratio of intranuclear C capsids to cytoplasmic capsids was significantly greater in cells infected with HSV-2 UL16 mutants than in cells infected with their parental counterparts (Fig. 7A). By contrast, the ratio of intranuclear C capsids to cytoplasmic capsids was greater for parental HSV-1 strains than for their UL16 mutants, suggesting that HSV-1 UL16 does not play a discernible role in nuclear egress. The fact that the ratios of intranuclear C capsids to cytoplasmic capsids were significantly lower for HSV-1 UL16 mutants than for their parental strains is likely due to defects in secondary envelopment leading to the accumulation of cytoplasmic capsids in cells infected with the UL16 mutant strains (Table 1 and Fig. 6D). The mean ratios of enveloped capsids to cytoplasmic capsids for HSV-2 and HSV-1 UL16 mutant strains were significantly lower than for their parental strains, indicating that UL16 functions in secondary envelopment for both species of HSV (Fig. 7B). Taken together, these data indicate that UL16 has species-specific functions in HSV infection, such that HSV-2 relies strongly on UL16 for nuclear egress whereas both HSV-2 and HSV-1 rely on UL16 for efficient secondary envelopment.
TABLE 1.
Quantification of intracellular capsids in Vero cells infected with UL16 mutant and parental HSV strainsa
| Strain | No. (%) of capsids |
|||||
|---|---|---|---|---|---|---|
| Total | Intranuclear |
Cytoplasmic |
PNSb | |||
| A + B | C | Nonenveloped | Enveloped | |||
| SD90e | 948 | 377 (39.8) | 96 (10.1) | 250 (26.4) | 53 (5.6) | 172 (18.1) |
| UL16SΔ10-360 | 992 | 677 (68.2) | 75 (7.6) | 159 (16) | 13 (1.3) | 68 (6.9) |
| HG52 | 1,429 | 676 (47.3) | 107 (7.5) | 441 (30.9) | 163 (11.4) | 42 (2.9) |
| UL16HΔ10-360 | 990 | 702 (70.9) | 91 (9.2) | 156 (15.7) | 29 (2.9) | 12 (1.2) |
| KOS | 850 | 249 (29.3) | 75 (8.8) | 296 (34.8) | 171 (20.1) | 59 (6.9) |
| UL16KΔ28-359 | 1,058 | 506 (47.8) | 58 (5.5) | 452 (42.8) | 35 (3.3) | 7 (0.7) |
| F | 663 | 217 (32.7) | 77 (11.6) | 175 (26.4) | 126 (19) | 68 (10.3) |
| UL16FΔ139-359 | 1,778 | 892 (50.2) | 84 (4.7) | 671 (37.7) | 99 (5.6) | 32 (1.8) |
Capsids were counted in different cellular compartments from 10 images/strain derived from multiple sections in two independent experiments.
PNS, perinuclear space.
FIG 7.
Analysis of capsid distribution in cells infected with HSV UL16 deletion mutants. (A) Ratios of intranuclear C capsids to cytoplasmic capsids of parental HSV strains and their corresponding UL16 deletion mutants were determined. The values were calculated from 10 independent images per strain. The error bars represent standard errors of the mean. (B) Ratios of enveloped capsids to cytoplasmic capsids of parental HSV strains and their corresponding UL16 deletion mutants were calculated using the same methodology as for panel A. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
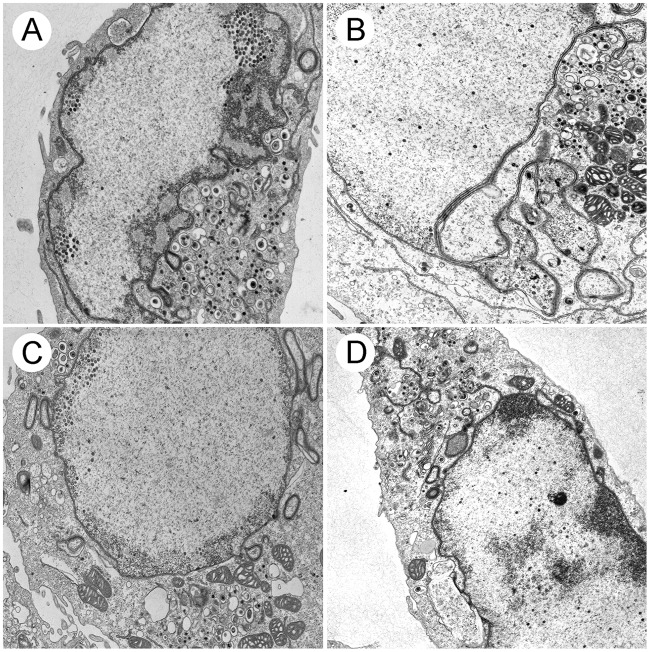
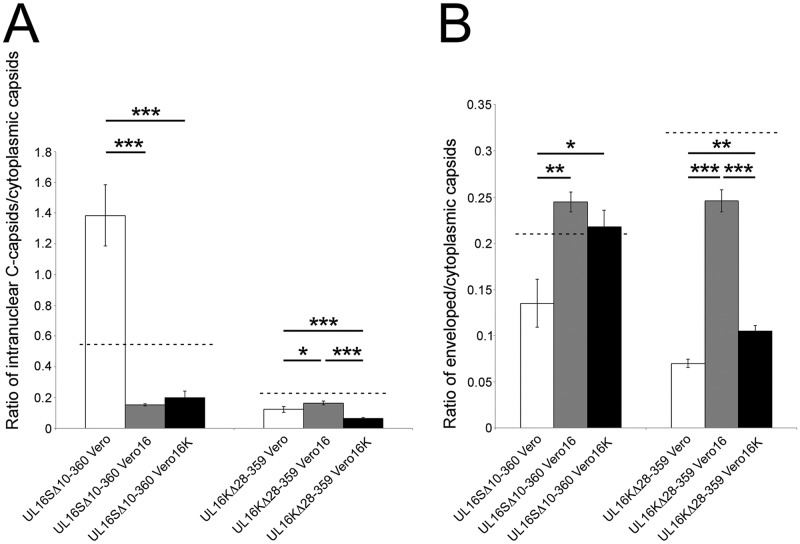
Because HSV-1 did not appear to require UL16 for nuclear egress, we were interested in determining if the HSV-1 UL16 protein had the capacity to promote the nuclear egress of an HSV-2 UL16 mutant. To test this, Vero16 and Vero16K cells were infected with UL16SΔ10-360 and UL16KΔ28-359 and processed for TEM (Fig. 8), and the TEM data were quantified (Fig. 9). Both Vero16 and Vero16K cells were able to support the nuclear egress of UL16SΔ10-360, as evidenced by the appearance of numerous cytoplasmic capsids (Fig. 8A and B). Quantification of these data indicated that complementation of UL16SΔ10-360 in Vero16 and Vero16K cells resulted in more robust nuclear egress than that seen with the parental SD90e strain (Fig. 9A). Furthermore, the abilities of the HSV-2 and HSV-1 UL16 proteins to complement the UL16SΔ10-360 nuclear egress defect were indistinguishable. Expression of HSV-1 UL16, but not HSV-2 UL16, modestly, but significantly, promoted the nuclear egress of the UL16KΔ28-359 strain (Fig. 9A). As expected, both HSV-2 and HSV-1 UL16 proteins were able to complement secondary envelopment of both HSV species (Fig. 9B). Collectively, these data suggest that HSV-1 encodes a function, missing in HSV-2, that can compensate for nuclear egress in the absence of UL16.
FIG 8.
Ultrastructural analysis of trans-complemented UL16 mutants. Vero16 cells expressing HSV-2 UL16 (A and C) and Vero16K cells expressing HSV-1 UL16 (B and D) were infected with UL16SΔ10-360 (A and B) and UL16KΔ28-359 (C and D) at an MOI of 3. At 16 hpi, cells were fixed and processed for TEM as described in Materials and Methods. Numerous nuclear and cytoplasmic capsids can be observed in all the infected cells.
FIG 9.
Quantitative analysis of UL16 trans-complementation. (A) Ratios of intranuclear C capsids to cytoplasmic capsids of representative HSV-2 and HSV-1 UL16 mutants complemented by either HSV-2 (Vero16) or HSV-1 (Vero16K) UL16 protein. The values were calculated from 10 independent images per strain. The error bars represent standard errors of the mean. (B) Ratios of enveloped to cytoplasmic capsids of representative HSV-2 and HSV-1 UL16 mutants complemented by either HSV-2 or HSV-1 UL16 protein were calculated using the same methodology as for panel A. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. The dashed lines indicate values obtained for parental strains on Vero cells, as shown in Fig. 7.
DISCUSSION
Here, we describe the analysis of UL16 deletion mutants derived from four HSV strains. The strategy used to construct these strains utilized CRISPR/Cas9 mutagenesis, which was both efficient and rapid. Our approach utilized two guide RNAs (gRNAs) toward the UL16 locus simultaneously. Cleavage of the UL16 gene at the sites directed by the gRNAs and subsequent repair of the lesion by nonhomologous end joining resulted in the isolation of a variety of mutants, some having in-frame deletions and others with frame shifts after the 5′ gRNA-directed cleavage. For the analysis presented here, we chose to select a variety of UL16 mutants for further study. All the HSV-2 UL16 mutants isolated displayed similar phenotypes. In the case of HSV-1, however, one of the F UL16 mutants, UL16FFS27/3, was an outlier, as its replication was reduced much more severely than that of other HSV-1 UL16 mutants (Fig. 3D). It is not clear why UL16FFS27/3 grows as poorly as it does; however, it is noteworthy that it forms smaller plaques on complementing cells than the other HSV-1 UL16 mutants (Fig. 2C), raising the possibility that additional mutations outside the UL16 locus were introduced during its isolation. Alternatively, the N-terminal fragment of UL16, predicted to be produced by UL16FFS27/3, might act as a dominant-negative protein, resulting in the inhibition of both cell-to-cell spread and virus replication. Clearly, more work is required to determine the cause of the UL16FFS27/3 cell-to-cell spread and replication phenotypes. Because of these caveats, we eliminated the strain from subsequent ultrastructural analyses.
Our kinetic analysis of UL16 mutant replication revealed that HSV-2 UL16 mutants had roughly 50- to 100-fold reductions in virus replication, while HSV-1 UL16 mutants, with the exception of UL16FFS27/3 (see above), had approximately 10-fold reductions (Fig. 3). These results are consistent with previous findings suggesting HSV-2 and HSV-1 have different requirements for UL16 (6–8). Despite replicating better than HSV-2 UL16 deletion mutants (Fig. 3), HSV-1 UL16 mutants consistently formed slightly smaller plaques relative to their parental strains than the HSV-2 UL16 mutants (Fig. 2). These findings may suggest that HSV-1 has a greater reliance on UL16 for cell-to-cell spread of infection than does HSV-2. Along these lines, Yeh and colleagues and Han et al. have documented an interaction between the N terminus of HSV-1 UL16 and the cytoplasmic tail of glycoprotein gE (14) and that a complex formed by UL16, UL11, and UL21 on the gE cytoplasmic tail is important for normal glycosylation of gE, trafficking of gE to the cell surface, and cell-to-cell spread of infection (14, 15). The existence of such interactions and their potential roles in the spread of HSV-2 infection have yet to be determined. Perhaps such interactions are not required for efficient cell-to-cell spread in HSV-2-infected cells and therefore might explain the differences in relative plaque sizes observed. In support of this idea, the N terminus of UL16 is less conserved between HSV-2 and HSV-1 than the remainder of the protein (16), and our preliminary investigations suggest that gE glycosylation is unperturbed in cells infected with HSV-2 Δ16 (data not shown).
Our trans-complementation plaque assays revealed that HSV-1 UL16 can rescue plaque formation of HSV-2 UL16 mutants, and vice versa (Fig. 4). Furthermore, TEM analysis revealed that HSV-1 UL16 can promote the nuclear egress of HSV-2, despite not being required for HSV-1 nuclear egress (Fig. 8 and 9A). Our findings also indicate that both HSV-2 and HSV-1 UL16 proteins function in secondary envelopment and that these proteins are trans-complementary for this process (Fig. 8 and 9B). Perhaps there are similarities in the processes of primary and secondary envelopment and HSV-1 UL16 is able to function in primary envelopment in the context of HSV-2 infection. The observation that HSV-1 and HSV-2 UL16 molecules can complement each other suggests that the genetic basis for the species-specific activities of UL16 lie outside the UL16 locus.
Importantly, these findings do not fully explain the reductions in virus replication observed for all the UL16 mutant strains. The explanation for the magnitude of the replication deficiencies observed for the UL16 mutants is certainly multifactorial. The functions of HSV-1 UL16 in cell-to-cell spread of infection have been well documented (14, 15). Additionally, previous studies on HSV-2 UL16 have implied a role for UL16 in viral DNA packaging into capsids (16), and in support of this idea, we noted that the proportion of intranuclear A and B capsids was greater for all of the UL16 mutants analyzed (Table 1). Moreover, the proportion of perinuclear virions compared to the parental strains was reduced in all the UL16 mutants examined (Table 1). Taken together, these findings suggest that UL16 influences multiple stages of virion morphogenesis.
The goal of this study was to resolve an apparent discrepancy between the functions of the HSV-2 and HSV-1 UL16 proteins during virus maturation. We have conclusively demonstrated that UL16 is important for HSV-2 nuclear egress in multiple strains (186, SD90e, and HG52). Additionally, it is clear that multiple strains of HSV-2 and HSV-1 rely on UL16 for efficient secondary envelopment. Despite important differences in primary and secondary envelopment, such as the well-characterized functions of the nuclear egress complex in primary envelopment, these findings raise the intriguing possibility that some aspects of primary and secondary envelopment may be more similar than previously appreciated.
MATERIALS AND METHODS
Viruses and cells.
HSV-2 strains 186 and SD90e were kind gifts from David Knipe, Harvard University. The construction of the HSV-2 186 strain UL16 knockout (Δ16) was described previously (6). HSV-2 strain HG52 was kindly provided by Aidan Dolan and Duncan McGeoch, University of Glasgow, Glasgow, United Kingdom. HSV-1 strains F and KOS were generously provided by Lynn Enquist, Princeton University. African green monkey kidney (Vero) cells and human embryonic kidney 293T cells were acquired from the ATCC. Phoenix-AMPHO cells were generously provided by Craig McCormick, Dalhousie University, Halifax, Nova Scotia, Canada. The murine L fibroblast line was a kind gift from Frank Tufaro, University of British Columbia, Vancouver, British Columbia, Canada. All the cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% GlutaMax and grown at 37°C in a 5% CO2 environment.
UL16-expressing cell lines were constructed by retroviral transduction using an amphotropic Phoenix-Moloney murine leukemia virus system described previously (17). In brief, plasmid pBMN-IP-UL16 or pBMN-IP-UL16K (see below) was transfected into Phoenix-AMPHO cells to produce the retroviruses. HSV-2 UL16-expressing cell lines (Vero16 and 293T16) and HSV-1 UL16-expressing cell lines (L16K and Vero16K) were isolated by transducing either Vero, 293T, or L cells with the corresponding amphotropic retroviruses and were selected using 2 μg/ml puromycin (InvivoGen) 48 h after transduction. To confirm UL16 expression, cell extracts were prepared and analyzed by Western blotting using HSV-2 or HSV-1 UL16 antiserum (Fig. 1A).
Antibodies.
Chicken polyclonal antiserum against HSV-2 UL16 (6) was used for Western blotting at a dilution of 1:200, and mouse monoclonal antibody against HSV-2 ICP27 (Virusys) was used for Western blotting at a dilution of 1:1,000. Rabbit polyclonal antiserum against HSV-1 UL16 was a kind gift from John Wills, The Pennsylvania State University College of Medicine (18), and was used for Western blotting at a dilution of 1:3,000. Rat polyclonal antiserum against Us3 (19) was used for indirect immunofluorescence microscopy at a dilution of 1:1,000, and mouse monoclonal antibody against β-actin (Sigma) was used for Western blotting at a dilution of 1:2,000. Alexa Fluor 568-conjugated goat anti-rat immunoglobulin G monoclonal antibody (Invitrogen Molecular Probes) was used at a dilution of 1:500 for immunofluorescence microscopy. Horseradish peroxidase-conjugated goat anti-mouse IgG, horseradish peroxidase-conjugated goat anti-chicken IgY, horseradish peroxidase-conjugated rabbit anti-rat IgG, and horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma) were used for Western blotting at dilutions of 1:10,000, 1:30,000, 1:80,000, and 1:5,000, respectively.
Plasmid construction.
pBMN-IP-UL16 encoding HSV-2 UL16 was constructed previously (6). To construct pBMN-IP-UL16K, UL16 KOS sequences were amplified from HSV-1 KOS genomic DNA by PCR using the forward primer 5′-GACTGAATTCATGGCGCAGCTGGGAC-3′ containing an EcoRI restriction site (italics) and the reverse primer 5′-GACTCTCGAGTTATTCGGGATCGCTTG-3′ containing a XhoI restriction site (italics). The PCR product was digested with EcoRI and XhoI and ligated into similarly digested pBMN-IP, a kind gift from Craig McCormick, Dalhousie University, Halifax, Nova Scotia, Canada, to yield pBMN-IP-UL16K.
gRNAs used for producing the UL16 mutant strains were expressed from the guide RNA-Cas9 expression plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9, a gift from Feng Zhang, the Broad Institute of MIT (Addgene plasmid 42230) (20). To construct these gRNA expression plasmids, the top-strand oligonucleotide was annealed to the bottom-strand oligonucleotide (Table 2), and the double-stranded product was cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9, which had been digested with BbsI. Three different UL16 gRNAs were designed, for both HSV-1 and HSV-2, to produce different-size deletions within UL16.
TABLE 2.
Oligonucleotides used to produce HSV-2 and HSV-1 UL16 gRNAs
| gRNA | Predicted nucleotide cleavage site (nt)a | Sequence (5′–3′) |
|
|---|---|---|---|
| Top strand | Bottom strand | ||
| HSV-2 UL16 | 28 | 5′-CACCGCGGGCACTCTGGCGTCCCC-3′ | 5′-AAACGGGGACGCCAGAGTGCCCGC-3′ |
| 177 | 5′-CACCGCGTCGTTCGGGGGGACGAG-3′ | 5′-CACCGCGTCGTTCGGGGGGACGAG-3′ | |
| 1078 | 5′-CACCGAGCTGCCCCGCGGTCGCGC-3′ | 5′-AAACGCGCGACCGCGGGGCAGCTC-3′ | |
| HSV-1 UL16 | 99 | 5′-AAACCCGTTGCCCGGGCCGTTGCC-3′ | 5′-CACCGGCAACGGCCCGGGCAACGG-3′ |
| 430 | 5′-AAACGACCCCGCTCCTGTGCACCC-3′ | 5′-CACCGGGTGCACAGGAGCGGGGTC-3′ | |
| 1095 | 5′-CACCGGGTGCACAGGAGCGGGGTC-3′ | 5′-CACCGGCAACGGCCCGGGCAACGG-3′ | |
Nucleotide (nt) position in the UL16 gene targeted by the gRNA.
CRISPR/Cas9 mutagenesis of the UL16 locus.
An approach similar to that used by Xu and colleagues for the construction of PRV mutants was utilized (21). Viral DNA of each strain (SD90e, HG52, KOS, or F) was purified as described previously (22). 293T16 cells growing in 100-mm dishes were cotransfected with 16 μg of purified viral genomic DNA, along with 1 μg each of two UL16 guide RNA expression plasmids, using a calcium phosphate coprecipitation method (23). Twenty-four hours after transfection, the culture medium was replaced with semisolid medium containing 0.5% methyl cellulose to allow plaque formation. Five to 6 days later, plaques were picked. Viral DNA isolated from a portion of the picked plaque was used for screening for UL16 deletions by PCR. The UL16 loci from viruses bearing UL16 deletions were sequenced in their entirety to determine the precise nature of the UL16 mutations introduced. Roughly 50% of the plaques picked had UL16 deletions or frameshift mutations.
Plaque size determination.
Monolayers of Vero cells were prepared on 35-mm glass bottom dishes (MatTek) and infected with virus at an MOI of 0.005. Plaques were allowed to form for 24 h prior to fixation and processing for indirect immunofluorescence microscopy (6) using antisera against the HSV Us3 protein (19). Images of plaques were captured on a Nikon TE200 inverted epifluorescence microscope using a 10× objective and a cooled charge-coupled-device (CCD) camera. To quantify these results, the pixels in the area of each plaque were counted using Image-Pro 6.3 software. The results shown were derived from 40 distinct plaques per strain.
TEM.
Vero cells growing in 100-mm dishes were infected with virus at an MOI of 3 and processed for TEM at 16 hpi. Infected cells were rinsed with phosphate-buffered saline (PBS) three times before fixing in 1.5 ml of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 60 min. Cells were collected by scraping into fixative and centrifugation at 300 × g for 5 min. Cell pellets were carefully enrobed in an equal volume of molten 5% low-melting-point agarose and allowed to cool. Specimens embedded in agarose were incubated in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 1.5 h and rinsed three times in 0.1 M sodium cacodylate buffer (7.4) and then postfixed in 1% osmium tetroxide for 1 h. The fixed cells in agarose were rinsed with distilled water 3 times and stained in 0.5% uranyl acetate overnight before dehydration in ascending grades of ethanol (30% to 100%). Samples were transitioned from ethanol to infiltration with propylene oxide and embedded in Embed-812 hard resin (Electron Microscopy Sciences). Blocks were sectioned at 50 to 60 nm and stained with uranyl acetate and Reynolds' lead citrate. Images were collected using a Hitachi H-7000 transmission electron microscope operating at 75 kV.
ACKNOWLEDGMENTS
This work was supported by Canadian Institutes of Health Research operating grant 93804, Natural Sciences and Engineering Research Council of Canada Discovery Grant 418719, and Canada Foundation for Innovation award 16389 to B.W.B. J.G. was supported in part by an award from the China Scholarship Council.
We are grateful to Kelsey Won for help with viral-DNA preparation, Laura Ruhge and Greg Smith for advice with sample preparation for electron microscopy, and Renée Finnen for critical reading of the manuscript.
REFERENCES
- 1.Bigalke JM, Heldwein EE. 2017. Have NEC coat, will travel: structural basis of membrane budding during nuclear egress in herpesviruses. Adv Virus Res 97:107–141. doi: 10.1016/bs.aivir.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellberg T, Passvogel L, Schulz KS, Klupp BG, Mettenleiter TC. 2016. Nuclear egress of herpesviruses: the prototypic vesicular nucleocytoplasmic transport. Adv Virus Res 94:81–140. doi: 10.1016/bs.aivir.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 5.Turcotte S, Letellier J, Lippé R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Hay TJM, Banfield BW. 2017. The product of the herpes simplex virus type 2 UL16 gene is critical for the egress of capsids from the nuclei of infected cells. J Virol 91:e00350-17. doi: 10.1128/JVI.00350-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines JD, Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol 65:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klupp BG, Böttcher S, Granzow H, Kopp M, Mettenleiter TC. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J Virol 79:1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. 1998. The genome sequence of herpes simplex virus type 2. J Virol 72:2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgrove R, Diaz F, Newman R, Saif S, Shea T, Young S, Henn M, Knipe DM. 2014. Genomic sequences of a low passage herpes simplex virus 2 clinical isolate and its plaque-purified derivative strain. Virology 450-451:140–145. doi: 10.1016/j.virol.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai W, Chen C, Morse S, Htun Y, Fehler H, Liu H, Ballard R. 2003. Increasing relative prevalence of HSV-2 infection among men with genital ulcers from a mining community in South Africa. Sex Transm Infect 79:202–207. doi: 10.1136/sti.79.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roizman B, Furlong D. 1974. The replication of herpesviruses, p 229–403. In Fraenkel-Conrat H, Wagner RR (ed), Comprehensive virology, vol 3 Plenum Press, New York, NY. [Google Scholar]
- 14.Yeh P-C, Han J, Chadha P, Meckes DG, Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol 85:9425–9436. doi: 10.1128/JVI.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol 143:863–880. doi: 10.1007/s007050050338. [DOI] [PubMed] [Google Scholar]
- 17.Swift S, Lorens J, Achacoso P, Nolan GP. 2001. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems, Curr Protoc Immunol Chapter 10:Unit 10.17C. doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Chadha P, Meckes DG, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol 85:9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finnen RL, Roy BB, Zhang H, Banfield BW. 2010. Analysis of filamentous process induction and nuclear localization properties of the HSV-2 serine/threonine kinase Us3. Virology 397:23–33. doi: 10.1016/j.virol.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu A, Qin C, Lang Y, Wang M, Lin M, Li C, Zhang R, Tang J. 2015. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol Lett 37:1265–1272. doi: 10.1007/s10529-015-1796-2. [DOI] [PubMed] [Google Scholar]
- 22.Sawtell NM, Thompson RL. 2014. Herpes simplex virus mutant generation and dual-detection methods for gaining insight into latent/lytic cycles in vivo. Methods Mol Biol 1144:129–147. doi: 10.1007/978-1-4939-0428-0_9. [DOI] [PubMed] [Google Scholar]
- 23.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]