ABSTRACT
Pulmonary CD4 T cells are critical in respiratory virus control, both by delivering direct effector function and through coordinating responses of other immune cells. Recent studies have shown that following influenza virus infection, virus-specific CD4 T cells are partitioned between pulmonary vasculature and lung tissue. However, very little is known about the peptide specificity or functional differences of CD4 T cells within these two compartments. Using a mouse model of influenza virus infection in conjunction with intravascular labeling in vivo, the cell surface phenotype, epitope specificity, and functional potential of the endogenous polyclonal CD4 T cell response was examined by tracking nine independent CD4 T cell epitope specificities. These studies revealed that tissue-localized CD4 cells were globally distinct from vascular cells in expression of markers associated with transendothelial migration, residency, and micropositioning. Despite these differences, there was little evidence for remodeling of the viral epitope specificity or cytokine potential as cells transition from vasculature to the highly inflamed lung tissue. Our studies also distinguished cells in the pulmonary vasculature from peripheral circulating CD4 T cells, providing support for the concept that the pulmonary vasculature does not simply reflect circulating cells that are trapped within the narrow confines of capillary vessels but rather is enriched in transitional cells primed in the draining lymph node that have specialized potential to enter the lung tissue.
IMPORTANCE CD4 T cells convey a multitude of functions in immunity to influenza, including those delivered in the lymph node and others conveyed by CD4 T cells that leave the lymph node, enter the blood, and extravasate into the lung tissue. Here, we show that the transition of recently primed CD4 cells detected in the lung vasculature undergo profound changes in expression of markers associated with tissue localization as they establish residence in the lung. However, this transition does not edit CD4 T cell epitope specificity or the cytokine potential of the CD4 T cells. Thus, CD4 T cells that enter the infected lung can convey diverse functions and have a sufficiently broad viral antigen specificity to detect the complex array of infected cells within the infected tissue, offering the potential for more effective protective function.
KEYWORDS: CD4 T cell, influenza, lung, specificity
INTRODUCTION
CD4 T effector cells are central to orchestrating protective immune responses to influenza virus (1). In addition to key functions within the local draining lymph node, such as provision of help for antibody responses and enhancement of CD8 T cell expansion, virus control also relies on CD4 T cell responses that localize to the site of infection. Local secretion of cytokines by CD4 T cells promotes recruitment of innate effectors into the lung and facilitate positioning and persistence of CD8 T cells in the airway (2). Additionally, release of cytotoxic mediators has the potential to kill antigen-bearing cells (3). Despite these complexities in functionality, the regulation of CD4 T cell effector function in pulmonary tissue following acute viral infection remains poorly understood.
The lung is a highly vascularized organ containing an extensive network of blood vessels that serves in part to facilitate trafficking of leukocytes into lung under both homeostatic and inflammatory conditions. Intravascular labeling (iv) in vivo is now a well-characterized strategy for clear discrimination of immune cells localized in the pulmonary vasculature and tissue (4), with the latter term comprising the airways, parenchyma, and interstitium. CD4 T cells home to these tissues to deliver their effector function. Currently, our knowledge is quite limited with regard to the consequences of this CD4 T cell compartmentalization following influenza A virus (IAV) infection. T cell trafficking and activity in the lung has largely been studied for CD8 T cell responses, during memory, and/or in chronic infection models (5–7). Two recent reports (8, 9) analyzing effector CD4 T cells after influenza virus infection, however, found that virus-specific, tissue-localized cells were enriched for antigen-experienced, gamma interferon (IFN-γ)-producing CD4 T cells that were CD11ahi CD49d+. Although intriguing, these IAV studies were limited because the antigen and peptide specificity of the CD4 T cells were not identified, and neither were many of the candidate surface markers associated with tissue homing, transendothelial migration (TEM), and lung localization. Because of their multiplicity of antiviral activities and potential for heterosubtypic protection, a better understanding of the cellular signatures underlying tissue recruitment, immunodominance, and functionality of influenza-specific CD4 T cells in pulmonary vasculature and tissue is essential.
One particularly important unresolved issue is whether and how the epitope specificity and functional potential of CD4 T cells primed in the local draining lymph node is altered after they commit to lung homing, leave the lung vasculature, and enter the lung tissue. This issue has not yet been addressed for CD4 T cells. There are reasons to suspect that immunodominance hierarchies or functional potential are altered as cells become established in the lung tissue. First, the microenvironment in the lung following influenza virus infection is highly enriched in inflammatory cytokines and diverse myeloid and lymphoid cells from the innate and adaptive immune response (reviewed in references 10–12). Our recent studies using a novel fluorescent reporter virus (13) have revealed that influenza virus antigens access many different types of antigen-presenting cells (APCs) in the lung, including eosinophils, macrophages, neutrophils, interstitial macrophages, CD11b- and CD103-positive dendritic cells, as well as CD45-negative, nonhematopoietic cells. All of these antigen-bearing cells also express major histocompatibility complex (MHC) class II molecules and might engage infiltrating virus-specific CD4 T cells. None of these class II-positive cells within the infected lung have been evaluated for expression of the key protein cofactors that control MHC class II peptide epitope display. Recent studies have provided evidence that CD4 T cells form contacts with antigen-bearing cells via their T cell receptor at later time points after infection (e.g., day 6 to 8 postinfection) after T cells have entered the lung (14). Also, recent data exploring CD8 T cell immunodominance in viral infections have shown that competition for antigen within the infected tissue shapes the T cell repertoire within the tissue (15), and that for pH1N1 influenza virus infection, monocyte-derived dendritic cells in the lung change CD8 T cell peptide epitope specificity (16). Collectively, these data led us to speculate that disparate epitope display affects CD4 T cell persistence or expansion in the lung or alters their functional capacity.
Because the impact of lung tissue localization on CD4 T cell epitope specificity and evolution of cytokine-mediated effector function during the course of influenza virus infection had not yet been explored experimentally, we designed experiments to comprehensively evaluate this issue. We used a mouse model of IAV infection in combination with intravascular labeling to examine the cellular heterogeneity, specificity, and effector potential of pulmonary CD4 T cells in the lung vasculature and tissue during the influenza-specific immune response. To that end, we simultaneously tracked 9 different peptide specificities from 4 different viral proteins (hemagglutinin [HA], neuraminidase [NA], matrix 1 [M1], and nucleoprotein [NP]) drawn from the polyclonal endogenous repertoire. Comprehensive, multiparameter flow cytometry revealed that compared to cells in the vasculature, the antigen-experienced CD4 T cells in the tissue displayed dramatically different expression patterns for markers associated with trafficking and tissue residency, including CXCR3, functional P- and E-selectin ligands, lymphocyte function-associated antigen-1 (LFA-1), CD49d, very late antigen-1 and -2 (VLA-1 and VLA-2) integrins, and CD69. Despite this evidence that the CD4 T cells engage distinct proteins, cells, and the inflammatory microenvironment of the infected lung, we found that the epitope specificity of influenza-reactive CD4 T cells was not altered during entry into and residence within the lung, and neither was their cytokine potential.
RESULTS
Intravascular labeling in vivo reveals compartmentalization of CD4 T cells during the peak of the response following influenza virus infection.
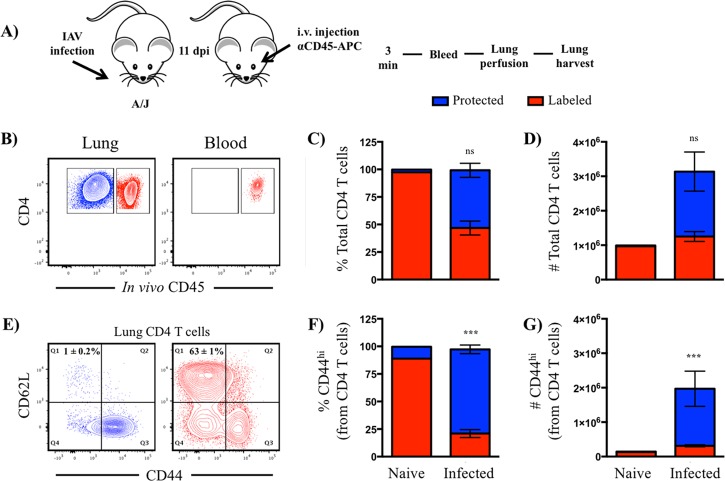
Many tissue and cellular alterations occur in the lung in response to virus infection and inflammation (17, 18). Although proinflammatory mediators and chemokines may localize to the vasculature and tissue, differences in cellular composition, distribution, and activation dramatically alters the microenvironment of CD4 T cells that enter the lung during infection and thus potentially their specificity and functionality. To study infiltrating CD4 T cells in pulmonary vasculature and tissue, we took advantage of a well-characterized iv labeling technique in vivo that enabled clear distinction by flow cytometry. Briefly, A/JCr (H-2a) mice were infected intranasally with a sublethal dose of A/New Caledonia/20/99 (H1N1) virus and analyzed at 10 to 11 days postinfection. This time point was chosen because preliminary experiments indicated that this time frame represented the peak of the adaptive CD4 T cell response in the lung (Fig. 1A), as well as the highest fraction of CD4 T cells within the lung tissue (Fig. 1B). Shortly before sacrifice, mice were injected intravenously with an anti-CD45 antibody conjugated to allophycocyanin to label all circulating hematopoietic cells. Mice were then bled, and the lungs were perfused, harvested, and processed into single-cell suspensions for analytical flow cytometry, as outlined in Fig. 2A. Figure 2B shows two discrete populations of cells, i.e., iv labeled, operationally termed “vasculature,” and cells that were unlabeled or protected from the iv label, which we refer to as “tissue localized.” Figure 2B, right, shows that all of the CD4 T cells in the peripheral blood became labeled, demonstrating that sufficient antibody was delivered. Consistent with earlier studies (4, 8), we observed that many cells in the lung vasculature remained after perfusion, as many of those cells were labeled with the fluorochrome-conjugated antibody (Fig. 2B, left). Although CD4 T cells were distributed equally between pulmonary vasculature and tissue (Fig. 2B to D), over 60% of the CD4 T cells in pulmonary vasculature were naive, as defined by CD44lo CD62L+ (Fig. 2E). Based on CD44 expression as a marker of cells that have previously responded to antigen, 80% of the cells in the tissue were classified as antigen experienced (CD44hi) compared to only 20% in the vasculature. The differences in phenotype in the CD4 T cells within these two compartments differs dramatically in both frequency and absolute abundance (Fig. 2F and G, bars on the right) and from cells isolated from naive mice, analyzed in parallel (Fig. 2F and G, bars on the left).
FIG 1.
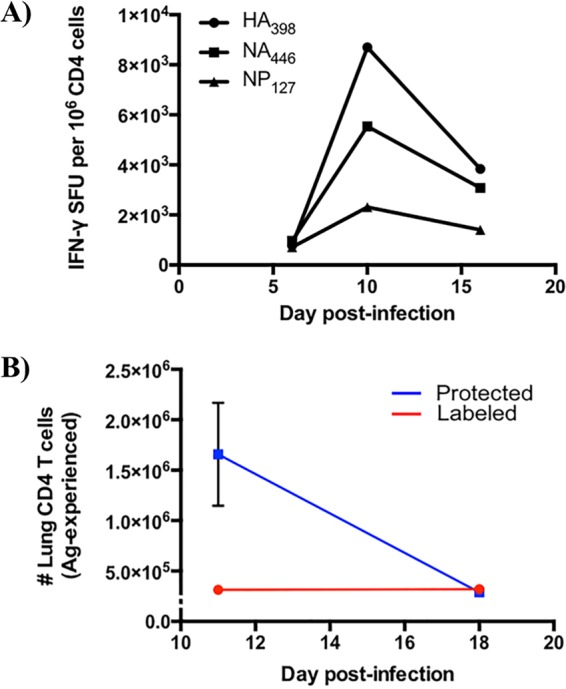
Kinetics of influenza-specific CD4 T cell responses and localization following A/New Caledonia/20/1999 (H1N1) infection. (A) CD4 T cell epitope-specific responses over time after influenza virus infection. CD4 T cell enzyme-linked immunosorbent spot assays were performed at the indicated time points postinfection, where responses of lung CD4 T cells to three different influenza epitopes (HA398, NA446, NP127; Table 1) were enumerated. SFU stands for spot-forming units, which correspond to the number of epitope-specific cells, represented as frequency per million. Data reflect responses from 5 animals whose tissues were pooled prior to analyses. (B) Antigen-experienced CD4 T cells decline dramatically in lung tissue following influenza A virus infection. CD44+, antigen-experienced cells that were iv labeled (red) and protected (blue) were quantified using flow cytometry 11 and 18 days postinfection. Symbols reflect the means, while the error bars represent the standard errors of the means (±SEM). Data reflect responses from 6 to 9 individual animals.
FIG 2.
Tissue compartmentalization of pulmonary CD4 T cells following infection. (A) Schematic representation of experimental design. dpi, day postinfection. (B) Representative contour plots representing iv-labeled (red) and -protected (blue) CD4 T cells. (C and D) Frequency (C) and number (D) of live CD4 T cells, partitioned by iv staining. (E) Representative contour plots representing the relationship between CD44 and CD62L expression in iv-labeled and -protected pulmonary CD4 T cells. (F and G) Frequency (F) and number (G) of live, antigen-experienced (CD44+) CD4 T cells, partitioned by iv staining. For all experiments, n = 5 to 9 mice and error bars reflect standard error of the mean (SEM). ns, not statistically significant, where P > 0.05.
Tissue-localized CD4 T cells are distinct in cell surface phenotype.
To access lung tissue, CD4 T cells undergo a highly orchestrated and progressive process of TEM that involves rolling/tethering, activation, firm adhesion/attachment, and extravasation (17, 19). One critical adhesion molecule involved in this process, LFA-1, binds to intercellular adhesion molecule-1 (ICAM-1), expressed on the surface of the vascular endothelium during inflammation. Here, integrins become further activated by chemokine receptor signaling that increases ligand binding, allowing higher affinity and more stable interactions. The cell surface density of LFA-1 was almost 2.5-fold greater on the tissue-localized CD4 T cells than on the vasculature cells (Fig. 3A), in agreement with an earlier study (8). CD69 has been defined as a prototypic marker associated with tissue residence of T cells, both during acute infections and at memory (7, 20). Cytokines and local antigen are both thought to induce CD69 expression and thus enhance tissue localization of effector T cells, in part through reciprocal antagonism of the S1P receptor (7, 20). We therefore assessed the frequency of CD69-positive cells on antigen-experienced CD4 T cells in pulmonary vasculature and tissue. Using a conservative gating strategy based on a fluorescence-minus-one control (Fig. 3B), we found approximately 50% of the protected cells were CD69 positive, representing an approximately 6-fold change relative to vasculature, highlighting its expression as an indicator of tissue residency in the lung. Collectively, these data regarding cell surface phenotype of cells distinguished by iv labeling indicate that the CD4 T cells in this model system have a phenotype consistent with what has been described for lung-resident CD8 and CD4 T cells identified after respiratory infections (reviewed in references 2, 21, and 22).
FIG 3.
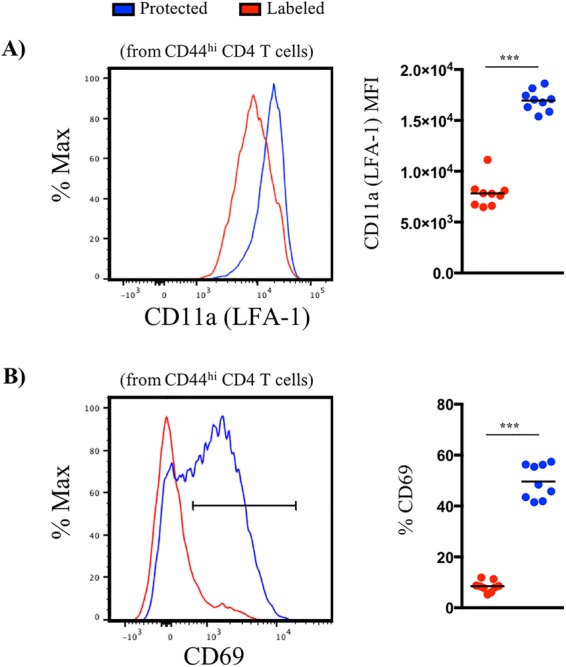
Protected niches of the lung are enriched in LFA-1 and CD69 expression. (A) Representative histogram plots and quantified median fluorescence intensity (MFI) for LFA-1 (A) and CD69 (B) expression by iv-labeled (red) and -protected (blue) cells. Symbols represent responses from individual animals (n = 9), and horizontal lines indicate the means.
Tissue-localized CD4 T cells possess an enhanced capacity for binding selectins, markers associated with inflammation-induced migration, and proteins that control positioning in the lung tissue.
To further identify phenotypic features of CD4 T cells that are compartmentalized between vasculature and lung tissue, we sought to determine if expression of other molecules involved in cellular localization and trafficking were distinct. A number of cell surface proteins have been implicated in homing, extravasation, and positioning of cells within the lung tissue during influenza virus infection (22, 23). However, to our knowledge, CD4 T cells have not yet been evaluated for the expression of these markers as they move from the vasculature and establish residency in the lung during influenza virus infection.
One interesting set of candidates are the selectins (reviewed in reference 24). P- and E-selectins are induced on the vascular endothelium in response to inflammation, and their ligand binding capacity is upregulated on Th1 and cytotoxic T cells (25–27). These molecules are thought to play an important role in regulating T cell migration (28). P-selectin surface ligand expression has been found to be enriched in NKG2C/E-positive cytotoxic CD4 T cells isolated from unfractionated lung cells after PR8 infection in mice (29), suggesting that these molecules are important in extravasation during influenza virus infection. We utilized chimeric fusion proteins that contain the C-type lectin domains fused to the Fc region of a human IgG1 molecule for use in flow cytometry to probe for both P- and E-selectin binding capacity. These studies revealed that effector CD4 T cells in the lung tissue expressed dramatically higher frequency and total abundance of CD4 T cells that displayed high-affinity ligands for both P- and E-selectin receptors (Fig. 4A and B).
FIG 4.
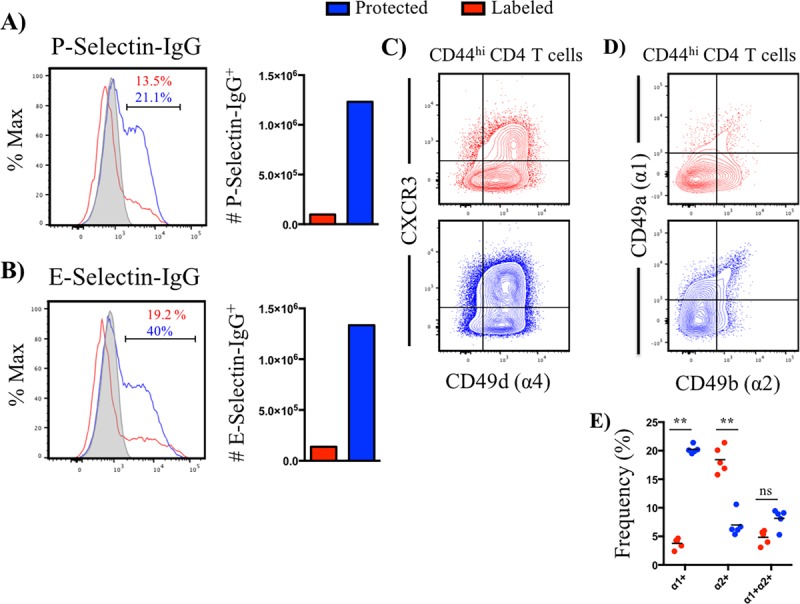
Antigen-experienced, tissue-resident CD4 T cells are enriched in markers related to homing, extravasation, and micropositioning within lung tissue. (A and B) Representative histogram plots depicting frequency of positive cells for P-selectin-IgG (A) and E-selectin-IgG (B) staining and absolute number, partitioned by iv-labeled (red) and -protected (blue) antigen-experienced CD4 T cells. Data are representative of the average from 2 independent experiments, where responses were pooled from 3 to 5 animals per experiment. Gray-filled histogram represents the fluorescence-minus-one control. (C) Representative contour plot illustrating CXCR3 and CD49d expression by iv-labeled (red) and -protected (blue) antigen-experienced CD4 T cells (n = 5). (D and E) Representative contour plots and quantification of the frequency of VLA-1 and VLA-2 single- and double-positive cells, partitioned by iv-labeled (red) and -protected (blue) antigen-experienced (CD44+) CD4 T cells. Symbols represent responses from individual animals (n = 5), and horizontal bars denote the means.
In the context of infection-induced inflammation, increased production of CXCR3-binding ligands CXCL9, CXCL10, and CXCL11 become concentrated in tissue, promoting directional migration via chemotaxis. The chemokine ligands for CXCR3 are IFN-γ inducible (30). Because of the prominence of a diverse set of IFN-γ-producing cells in the lung, including NK cells and CD8 T cells (reviewed in reference 31), we hypothesized that tissue-localized cells display increased expression of CXCR3. Inflammation is also known to cause increased deposition of integrin ligands, such as vascular cell adhesion molecule-1 (VCAM-1), that bind CD49d (α4)-containing molecules, such as VLA-4 (α4β1), promoting firm adhesion during TEM (23). Therefore, expression of CD49d was evaluated. Our analyses confirmed that the tissue-localized population of CD4 T cells was enriched 2-fold in the frequency of double-positive CXCR3+ CD49d+ cells (Fig. 4C), suggesting that high levels of expression enable selection for tissue entry and residence in the lung.
Following TEM, T cells encounter a distinct microenvironment, where a combination of soluble mediators, cell-cell, and cell-extracellular matrix (ECM) interactions influence micropositioning within the tissue (32). While collagen type I is thought to be widely distributed throughout the interstitial space, collagen type IV is predominantly found in the airway epithelium and vascular endothelium (33). Existing data on the expression of collagen binding receptors VLA-1 (α1β1) (collagen type I and IV) and VLA-2 (α2β1) (collagen type I) by influenza-specific CD4 T cells (33) were performed before methods were developed to distinguish cells in vasculature versus tissue. Accordingly, we sought to explicitly examine the distribution of VLA-1 and VLA-2 on pulmonary CD4 T cells. These studies revealed that antigen-experienced (CD44+) CD4 T cells were approximately 4-fold enriched in expression of the integrin subunit α1 (CD49a) (Fig. 4D and E) in tissue- versus vasculature-localized cells. This result suggests that micropositioning within pulmonary tissue depends on cell-ECM interactions, such as those of collagen or other less abundant components known to bind VLA-1 and VLA-2, including laminin.
Unsupervised tSNE analyses reveal the distinct cell surface phenotypes and microheterogeneity of iv-labeled and -protected CD4 T cells after influenza virus infection.
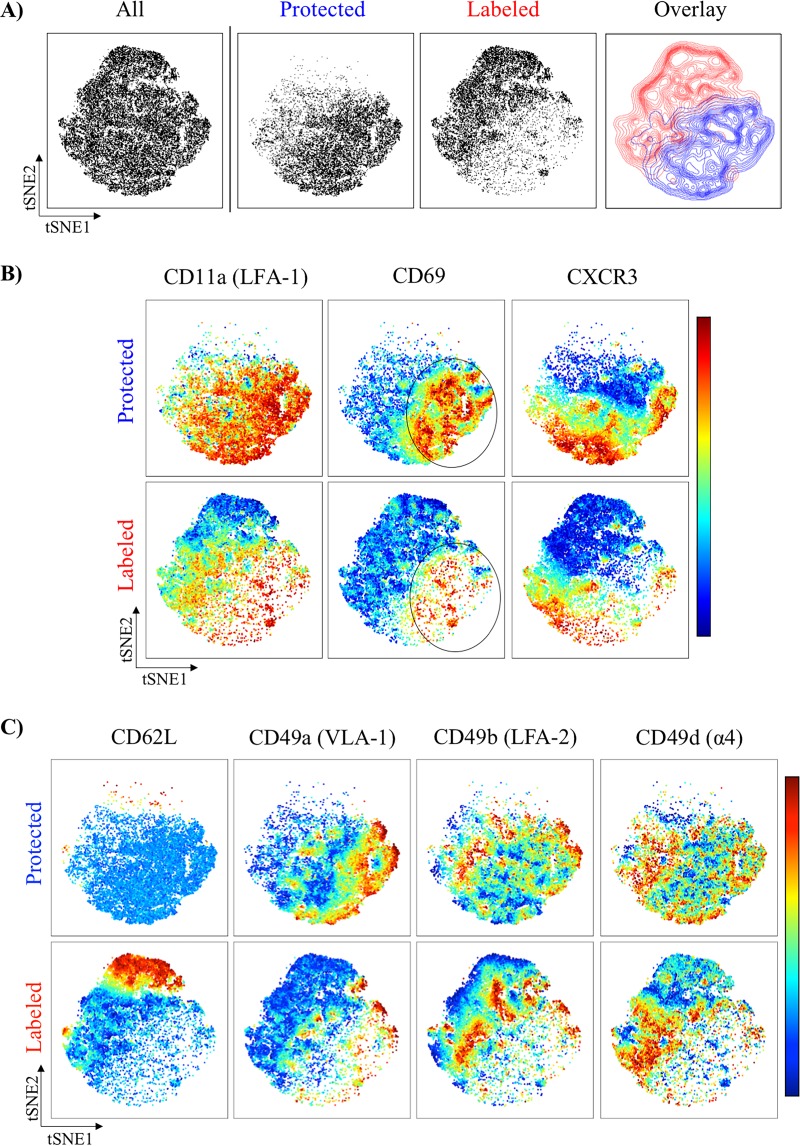
Although multiparameter flow cytometry and hierarchical gating analysis are powerful strategies for the identification and characterization of many cell types, more recently, unsupervised analysis of high-dimensional data sets can be applied using specialized software that eliminates user bias in identification of cell populations between experimental samples. One such analysis tool, t-distributed stochastic neighbor embedding (tSNE), uses an algorithm to reduce high-dimensionality data for visualization in two-dimensional space while maintaining the structure of the data (34). Because of these advantages, we applied tSNE to our flow cytometry data sets to more readily visualize and compare the cell surface profiles of vascular- versus lung tissue-localized CD4 T cells after infection using the following markers: CD62L, CD69, CXCR3, CD49a, CD49b, CD49d, CD11a, and CD107a.
It is clear from the tSNE analyses (Fig. 5A) that the CD4 T cells within these two compartments (demarcated as vasculature in red versus tissue in blue) exhibit profound differences in cell surface phenotype, illustrating that major cellular alterations that occur as cells transition from pulmonary vasculature to the tissue microenvironment. For example, induction of CD69, CD11a, and CXCR3 on antigen-experienced CD4 T cells is enriched in the iv-protected population, as visualized by the distinct island of high expression (Fig. 5B, circled patches) compared to the lower expression observed for vasculature cells within the same regions of the tSNE plots. Strikingly, within these islands of enrichment, dramatic microheterogeneity at the single-cell level is visualized and represented by subclusters of cells with distinct coexpression profiles for the proteins described above. Collectively, these analyses illustrate the unique composition and phenotype of cells within the two microenvironments of the lung and also the degree of heterogeneity within each compartment, likely reflecting kinetic, anatomic, and/or functionally distinct cell subsets.
FIG 5.
tSNE reveals profound distribution of markers between CD4 T cells in vasculature versus tissue after influenza virus exposure. (A) Representative and cumulative tSNE maps from CD44hi pulmonary CD4 T cells 11 dpi with A/New Caledonia/20/1999 (H1N1) virus, partitioned by iv labeling status. (B and C) tSNE pseudocolored maps representing relative expression for the indicated markers by iv-labeled (red) and -protected (blue) cells. The scale bar to the right of panels B and C reflects the relative expression differences with respect to a given marker, where red indicates higher expression and blue indicates lower expression. In all experiments, tSNE analyses were derived from individual mice (at least 5 per experiment). Cells from each mouse were downsampled to 4,880 based on the lowest input value for normalization between mice and then digitally concatenated to produce a single file for each experimental group. tSNE maps were then constructed (perplexity, 50; iterations, 2,000) for visualization of expression profiles of 8 markers in two-dimensional space to compare pulmonary vasculature and tissue-localized cells.
Pulmonary vasculature CD4 T cells are distinct from circulating blood CD4 T cells.
One important issue to understand is whether the CD4 T cells within the pulmonary vasculature contain CD4 T cells specifically en route to the tissue. It has been proposed that cells localized to pulmonary vasculature are largely circulating cells that have been trapped by the narrow diameter of the capillary bed and the larger size of lymphocytes relative to red blood cells (4, 20). If true, we would predict that vascular-localized CD4 T cells isolated from peripheral blood and lung would be largely indistinguishable. To evaluate this, we compared several markers on circulating peripheral blood CD4 T cells with CD4 T cells isolated from the pulmonary vasculature. Pulmonary vasculature cells were approximately 4-fold enriched in CD44+ antigen-experienced cells, suggesting that infection resulted in the selection of lung-homing, antigen-activated cells (Fig. 6A and B). When further analyzed for phenotypic markers, lung vascular cells were enriched for molecules that facilitate tissue entry, such as P- and E-selectin ligands (2.8- and 1.5-fold, respectively), VLA-1 (6.0-fold), and CD69 (3.5-fold) (Fig. 5C). Taken together, these results demonstrate that the pulmonary vasculature CD4 T cells enrich for cells poised to enter the lung, distinct from the CD4 T cells in general peripheral circulation.
FIG 6.
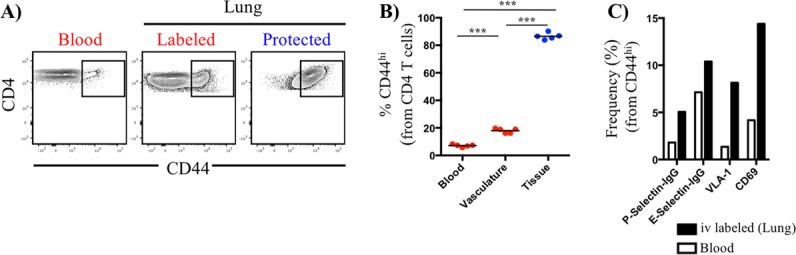
Cellular composition is distinct in pulmonary vasculature and peripheral blood. (A) Representative contour plots illustrating the expression pattern of CD44 by CD4 T cells in blood and lung, partitioned as iv-labeled (vasculature, red) and -protected (tissue, blue) cells. Symbols represent responses from individual animals (n = 5), and horizontal lines indicate the means. (B) Quantification of the frequency of antigen-experienced (CD44+) CD4 T cells, partitioned based on iv labeling status. Statistical significance across multiple comparisons was determined using analysis of variance. ***, P < 0.001. (C) Frequency of CD44hi CD4 T cells in blood and pulmonary vasculature positive for functional selectin ligands, VLA-1, and CD69. Data reflect pooled responses from 3 animals and are representative of 2 independent experiments.
Infection elicits diverse antigen-specific CD4 T cells whose epitope specificity and cytokine potential do not change upon entry into lung tissue.
While some recent studies have assessed the influenza-specific response using major histocompatibility complex (MHC) tetramers (35, 36), T cell receptor (TcR) transgenic systems (37), or coculture with antigen-presenting cells (APCs) and complex antigens (8), few have examined the endogenous polyclonal response using individual peptide epitopes. Study of the viral antigen specificity of the polyclonal response to IAV infection is important for several reasons. First, there is a diversity of APCs that CD4 T cells might encounter in the lung, each of which might display distinct sets of epitopes (13). In addition, there is increasing evidence that CD4 T cell fate decisions can be influenced by specific features of TcR interactions with the peptide-MHC class II complexes (38–40), suggesting that tracking one to two TcR specificities may lead to conclusions that are not generalizable. Therefore, we examined the functional phenotype of influenza-specific CD4 T cells of many different peptide specificities and evaluated whether the change in location from vasculature to tissue modified the effector cytokine profile or the relative representation of epitope-specific cells following infection.
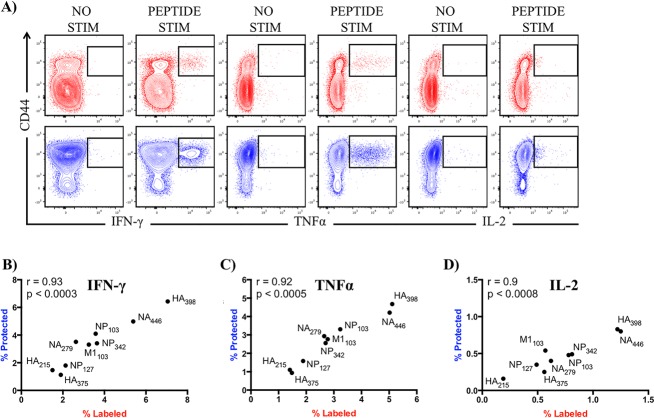
We simultaneously examined CD4 T cell reactivity to 9 different MHC class II-restricted peptide epitopes, including those derived from the viral HA, NA, M1, and NP (Table 1), using peptide restimulation in conjunction with intracellular cytokine staining. It is well established that influenza virus infection preferentially drives a Th1 CD4 T cell response, where release of antiviral cytokines, including gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-2 (IL-2), orchestrate protective immune responses at the site of infection (41). Accordingly, these were the cytokines measured in these assays. Figure 7A shows a representative sample of cytokine reactivity induced by exogenous influenza peptide (HA398) in the pulmonary vasculature and tissue, testing for production of IFN-γ (left), TNF-α (middle), or IL-2 (right) for vascular CD4 cells (top, in red) or tissue CD4 T cells (bottom, in blue). The patterns of expression were similar in each of the other peptide specificities tested (Fig. 7B to D and data not shown). To directly compare the frequency of antigen-experienced cytokine-producing CD4 T cells in the two pulmonary microenvironments, we performed a correlation analysis using all epitope-specific responses and separately represented the data for the different cytokines. As shown in Fig. 7B, among CD44+ antigen-experienced cells, IFN-γ-producing cells were the most dominant. Depending on the epitope used for restimulation, frequencies ranged from 1.1 to 7.1%. TNF-α producers were the second highest in frequency, ranging from 0.9 to 5.1% (Fig. 7C), with IL-2 being the lowest, ranging from 0.2 to 1.3% (Fig. 7D). Despite differences in the range of responses elicited by each cytokine, we found comparable frequencies of antigen-experienced CD4 T cells that were influenza peptide specific in pulmonary vasculature and tissue, with statistically significant relationships in the abundance of CD4 T cells specific for each of the peptides tested and for each cytokine (see P and R values in each panel). The only noticeable change in cytokine profile was a modest decrease in IL-2-producing cells within the tissue (indicated by the diminished slope in panel D), suggesting a lack of recruitment, retention, or persistence of these cells in the tissue. Collectively, the results of these experiments suggest that the functionality of effector CD4 T cells, based on cytokine potential, is comparable within pulmonary vasculature and tissue and that the influenza-specific CD4 T cell repertoire is not remodeled with regard to peptide specificity after accessing lung tissue.
TABLE 1.
MHC class II-restricted epitopes for A/NC/20/99 (H1N1) virus in H-2a mice
| I-Ak/I-Ek-restricted epitope (A/JCr) | Sequence |
|---|---|
| HA215 | 215 VSVVSSHYSRRFTPEIA 231 |
| HA375 | 375 SGYAADQKSTQNAINGI 391 |
| HA398 | 398 VIEKMNTQFTAVGKEFN 414 |
| NA279 | 279 CSCYPDTGTVMCVCRDN 295 |
| NA446 | 446 FCGVNSDTANWSWPDGA 462 |
| M1103 | 103 LKREITFHGAKEIALSY 119 |
| NP103 | 103 KWVRELVLYDKEEIRRI 119 |
| NP127 | 127 DDATAGLTHIMIWHSNL 143 |
| NP342 | 342 RVSSFIRGTRVLPRGKL 358 |
FIG 7.
Infection elicits a broad epitope-specific repertoire of CD4 T cells that do not change epitope-specific abundance or cytokine secretion as they transition from pulmonary vasculature into tissue. (A) Representative contour plots (iv labeled, red; iv protected, blue) depicting influenza-specific IFN-γ-, TNF-α-, and IL-2-producing CD4 T cells following peptide restimulation. Responses from HA398 peptide are shown as an example to illustrate the fold change over background (no peptide) for each cytokine measured. Spearman correlation analyses were performed on epitope-specific responses, as determined by IFN-γ (B), TNF-α (C), and IL-2 (D) production from CD4 T cells. Scales for correlation panels reflect the percentage of cytokine-positive cells pregated on CD44+. All data reflect the average taken from 2 independent experiments using pooled responses from at least 5 mice per experiment.
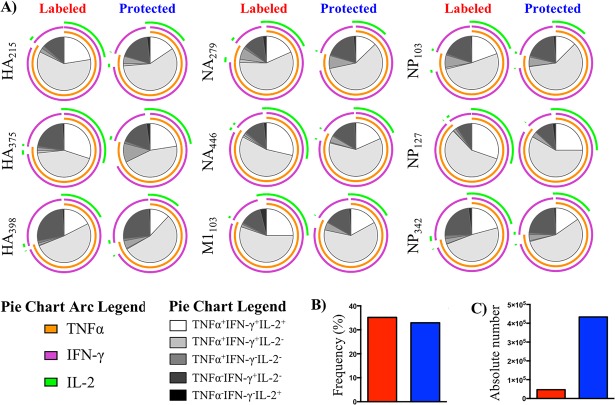
To assess the polyfunctional potential of influenza-specific CD4 T cells following infection and to more readily visualize how cytokine potential compares within the two lung compartments, Boolean gating was applied to the cytokine response from antigen-experienced cells within vasculature versus tissue. This analysis provides quantification for all possible combinations of cytokines. We excluded the triple-negative population from these analyses, as these cells may not be influenza specific. The data for all peptide specificities were then inserted into simplified presentation of incredibly complex evaluations (SPICE) software to construct arced pie charts that enable easy visualization of the fractional response in terms of the cytokine pattern (Fig. 8A). The source of the CD4 T cells (vasculature versus tissue) is identified in the top of the figure. The colored outer arcs, in green, violet, and orange, reflect positive expression for the indicated cytokine for each given peptide specificity. The majority of cytokine-producing, influenza-specific CD4 T cells were IFN-γ and TNF-α coproducers, indicated by the fraction of the pie that contained violet and orange arcs. While nearly all of the TNF-α-producing cells were coproducers, a small fraction of IFN-γ-producing cells were single producers (indicated by the violet-only arc), further illustrating the dominance of IFN-γ-producing CD4 T cells in the lung. IL-2 producers (indicated by the green arc) were the most poorly represented in the lung, mainly detected within triple cytokine producers. Most strikingly for the goals of this study, these experiments revealed that the cytokine patterns measured across 9 specificities were remarkably similar between the pulmonary vasculature and tissue. The cytokine potential of the virus-specific cells was not dramatically modified by residence in the tissue. The interior pie slice shading (in greyscale) represents the distinct cytokine profiles that can be identified with these three cytokines and illustrates the remarkable consistency in cytokine production among influenza-specific cells within the vasculature and tissue. Thus, despite differences in expression of receptors involved in cellular migration and tissue compartmentalization and the unique microenvironment in the lung after influenza virus infection, antigen specificity and cytokine functionality are not remodeled as virus-specific cells accumulate in the tissue. Figure 8B shows that when the frequency of cytokine-producing cells responsive to these 9 epitopes within the antigen-experienced (CD44-positive) cells was summed, they were equivalent between the compartments. The absolute number of cytokine-producing cells was highly enriched in the tissue (Fig. 8C) because of the greater overall abundance of the cells and the enrichment for those that were CD44 positive. We therefore conclude from this that a diverse epitope specificity of CD4 T cells enters the pulmonary vasculature after priming, and the cytokine profile and specificity are not edited, either in a negative or positive manner, as the cells move into the tissue.
FIG 8.
Spice analyses reveal stability of cytokine potential in vasculature and tissue. (A) Spice analysis of cytokine-producing, influenza-specific CD4 T cells, partitioned by iv-labeled (red) and -protected (blue) status to illustrate epitope-specific polyfunctional potential. HA, hemagglutinin; NA, neuraminidase; M1, matrix 1; NP, nucleoprotein. (B and C) Frequency (B) and number (C) of total influenza-specific CD4 T cells based on total cytokine reactivity within antigen-experienced cells following peptide restimulation in culture. Data reflect the average taken from 2 independent experiments using pooled responses from at least 5 mice per experiment.
DISCUSSION
In this report, we examined pulmonary CD4 T cells responding to IAV infection. Tissue-localized CD4 T cells were highly enriched in CD44-positive, antigen-experienced cells and for expression of adhesion molecules and homing receptors implicated in lung trafficking and retention, including previously reported markers associated with tissue residence, LFA-1 and CD69. Striking differences were also noted in expression patterns for other molecules implicated in different stages of transmigration and lung positioning, such as high-affinity P- and E-selectin ligands, CXCR3, CD49d, VLA-1, and VLA-2. Surprisingly, despite these dramatic differences in the cell surface phenotype of vasculature- and tissue-compartmentalized cells, the influenza-specific CD4 T cell response was remarkably similar in fine specificity, frequency, and cytokine polyfunctionality within these two compartments. Results obtained using traditional multiparameter flow cytometry analyses that distinguished vasculature and tissue illustrated both the diversity of cytokine patterns among cells of many specificities and the preservation of phenotype and immunodominance as cells transition from the vasculature into the lung tissue.
Our data are consistent with the following model. After influenza virus infection, CD4 T cells of diverse epitope specificity commit to homing to the lung, expressing a cytokine profile rich in TNF-α and IFN-γ. Vasculature-localized CD4 T cells display a cell surface phenotype that is distinct from that of circulating blood. Compared to the peripheral lymphocytes, the pulmonary vasculature CD4 T cells are enriched for antigen-experienced cells and molecules implicated in extravasation from blood vessels. This finding argues that a subset(s) of recently primed cells in the lung vasculature is poised to enter the lung. Markers expressed on many of the antigen-experienced cells within the lung vasculature enable interactions with proteins expressed on the vascular endothelium, including P- and E-selectins, chemokines, VCAM-1, and ICAM-1. Together, coexpression of selectin ligands, chemokine receptors, and integrins by effector CD4 T cells mediates TEM in pulmonary postcapillary venules. CD4 T cells that accumulate in the tissue display markers associated with tissue residence, such as CD69, as well as the collagen binding receptors VLA-1 and VLA-2, which have the potential to control localization within the respiratory tract. Despite the pronounced changes in cell surface phenotype in cells within vasculature and lung tissue, our studies revealed no changes in the influenza viral epitope specificities of cells within these two compartments. We also did not note any remodeling of the potential of the CD4 T cells to produce the cytokines IFN-γ, TNF-α, and IL-2 as they transition into the tissue, with only modestly diminished abundance of IL-2-producing cells in the tissue, which may reflect diminished potential for extravasation or persistence in the tissue. Overall, our study revealed that much of the functional potential of cells is dictated early, but that CD4 T cells that ultimately enter the lung are exposed to its inflammatory environment and may be further specialized to deliver effector function linked to specific microenvironment or positioning that is not reflected by cytokine potential. In support of this possibility is the conclusion from a recent study suggesting that cytotoxic potential becomes highly enriched as influenza-specific CD4 T cells enter the lung (29).
Our finding that the broad viral epitope specificity detected in vasculature persists in the tissue may have important functional consequences in the protective immune response to influenza. Our previous studies have shown that in general, CD4 T cells responding to influenza recognize a much more diverse set of epitopes than are typically recognized by CD8 T cells (42–45). In this study, we find that this diverse antigen specificity is maintained within the lung tissue. The ability of the population of tissue-homing CD4 T cells to recognize many peptide epitopes presented in the lung may enhance their overall protective capacity because of their ability to recognize a diverse set of antigen-bearing cells whose viral epitope display is selective for a subset of proteins or peptides. Additionally, recognition of different antigen-bearing APCs may allow a diverse repertoire of CD4 cells to expand within the lung or persist, albeit in lower numbers, at memory. It is known that there is a wide variety of MHC class II+ APCs that are positioned differently throughout the lung (13, 46–48). Therefore, a CD4 T cell repertoire encompassing many viral epitopes may ensure optimal recognition of distinct antigen-bearing cells. Additionally, different class II-positive APCs may access viral antigens differently through infection or receptor-mediated uptake and also have unique patterns in expression of intracellular endosomal molecules that participate in antigen processing and presentation, such as proteases, DM, and DO. Each of these endosomal components has the potential to influence the final peptide repertoire expressed at the cell surface (49). Clearly, identification and characterization of MHC class II-positive APCs that encounter infiltrating CD4 T cells will be an important area of future work.
Finally, it is important to gain a more thorough understanding of what virus-specific CD4 T cells, if any, establish long-term memory in lung as the CD4 T cell response contracts postinfection. In our studies, CD4 T cells within the lung tissue decay quite rapidly between day 11 and day 18, preventing a detailed survey of the specificity or functionality of the cells that persist. It has been shown that cognate interactions late in the primary response can be important in establishment of CD4 T cell memory (14, 26), in part through provision of IL-2, and there may be specialized niches where these interactions occur in the lung, leading to persistence of a subset of memory CD4 T cells. Alternatively, or additionally, key protective properties of influenza-specific memory CD4 T cells toward future influenza virus infection may be through rapid recruitment of virus-specific memory cells from the periphery. Several studies have shown that memory CD4 T cells contribute to protective immunity by accelerating the recruitment of early innate effectors into the lung, blunting virus replication (reviewed in reference 50). Our previous studies have shown that a broad diversity of influenza-specific CD4 T cell memory persists in the peripheral lymphoid tissue at memory (45). These results suggest that the recruited pool of memory CD4 T cells established by infection, whether localized to the lung tissue or dispersed in the periphery, has the potential to recognize a diverse array of peptide epitopes generated during a secondary challenge, leading to recognition of infection or antigen-bearing cells in the lung and thus rapidly contributing to protective immunity.
MATERIALS AND METHODS
Animals and infections.
Female A/JCr (H-2a) mice were obtained from Charles River Laboratories (Frederick, MD) and maintained in a specific-pathogen-free facility at the University of Rochester Medical Center according to institutional guidelines. Mice were anesthetized with 2,2,2-tribromoethanol (Avertin) via intraperitoneal injection and infected intranasally with 5 × 104 50% egg infectious doses (EID50) of influenza A/New Caledonia/20/1999 (H1N1) virus. All experimental groups were age matched and used between 12 and 18 weeks of age.
Ethics statement.
All mice were maintained in a specific-pathogen-free facility at the University of Rochester Medical Center according to institutional guidelines. All animal protocols used in this study adhere to the AAALAC International, the Animal Welfare Act, and the PHS guide and were approved by the University of Rochester Committee on Animal Resources, Animal Welfare Assurance Number A3291-01. The protocol under which these studies were conducted was originally approved 4 March 2006 (protocol 2006-030) and has been reviewed and reapproved every 36 months, with the most recent review and approval on 23 January 2018.
Intravascular labeling.
APC anti-mouse CD45 (30-F11; Tonbo) was prepared in sterile Dulbecco's phosphate-buffered saline (DPBS; 3 μg/mouse) and loaded into 0.5-ml insulin syringes. Mice were anesthetized using isoflurane via inhalation, and 100 μl was delivered intravenously per animal via retro-orbital injection.
Lung processing.
Three minutes following intravenous injection, blood was collected in tubes containing heparin sodium followed by intraperitoneal administration of a lethal dose of 2,2,2-tribromoethanol. The lungs were perfused via intracardiac injection of the right ventricle with 5 ml ice-cold lung perfusion medium (1× DPBS supplemented with 0.6 mM EDTA). Lung tissue was excised, dipped in Hanks' balanced salt solution (HBSS), and then placed into lung digestion medium (1 ml/lung) (RPMI 1640 supplemented with l-glutamine, 2.5% fetal bovine serum, 10 mM HEPES, 1 mg/ml collagenase type II, and 30 μg/ml DNase I). Tissues were further processed as previously described (13) to obtain single-cell suspensions. PBLs were isolated from whole blood using lymphocyte separation medium (Corning) according to the manufacturer's instructions.
Flow cytometry.
Single-cell suspensions were stained with Live/Dead aqua viability dye (Thermo Fisher), followed by incubation with purified rat anti-mouse CD16/CD32 (mouse BD Fc block, clone 2.4G2) as previously described (13). Cells were then stained for 25 min at 4°C in the dark using antibodies targeting the following markers: CD69 (H1.2F3; BioLegend), CD4 (RM4-5; BD Biosciences), CXCR3 (220803; R&D Systems), CD11a (2D7; BD Biosciences), CD44 (IM7; Tonbo), CD49a (Ha31/8; BD Biosciences), CD49b (Ha1/29; BD Biosciences), CD49d (R1-2; BD Biosciences), CD62L (MEL-14; BioLegend), CD107a (eBio1D4B; eBiosciences), purified mouse P-selectin-IgG fusion protein (BD Pharmingen), and purified mouse E-selectin Fc chimera protein (R&D Systems). Cells were then washed and prepared for flow cytometry analysis. Data were acquired using a BD LSR-II instrument, configured with 488 (blue)-, 633 (red)-, 407 (violet)-, and 532 (green)-nm lasers. Data were analyzed using FlowJo software (FlowJo, LLC), version 10.
Intracellular cytokine staining.
Cells derived from infected lungs were enriched for CD4 T cells using negative paramagnetic bead selection using magnetically activated cell sorting technology. CD4 T cells were then cocultured in U-bottom 96-well plates (3 × 105 cells/well) with antigen-presenting cells from the spleens of naive, syngeneic donors (5 × 105 cells/well) with or without exogenous influenza peptides. A cocktail of monensin and brefeldin A was then spiked into the cultures and incubated for 6 more hours for a total culture time of 8 h. Plates were then stored at 4°C for up to 12 h. The next day, cells were washed, stained with fixable Live/Dead aqua, washed again, and stained as described above using CD44 (IM7; Tonbo), CD62L (MEL-14; BioLegend), and CD4 (RM4-5; BD Biosciences). Following incubation at 4°C for 25 min, cells were then fixed and permeabilized using the eBioscience FoxP3 transcription factor staining buffer set according to the manufacturer's instructions. Intracellular proteins were then stained using an antibody cocktail of IFN-γ (XMG1.2; BD Biosciences), IL-2 (JES6-5H4; BD Biosciences), and TNF-α (MP6-XT22; BioLegend) and incubated for 45 min at 4°C. Data were acquired using a BD LSR-II instrument, configured with 488 (blue)-, 633 (red)-, 407 (violet)-, and 532 (green)-nm lasers. Data were analyzed using FlowJo software (FlowJo, LLC), version 10. Combination Boolean gating was performed to select cytokine responses (IFN-γ, TNF-α, and IL-2) from single, live, antigen-experienced CD4 T cells. Frequencies for each possible cytokine pattern (23) were tabulated in FlowJo, exported as an Excel file, annotated, and finally converted to a tab-delimited file. This file was then further annotated in Pestle software before importing data into SPICE software, version 5.35, which was used to produce the arced pie charts for data representation.
Synthetic peptides and libraries.
Seventeen-mer peptides overlapping by 11 amino acids to encompass the entire sequences of the viral proteins were obtained from the NIH Biodefense and Emerging Infections Research Repository, NIAID, NIH, and were used for the original epitope mapping as described previously (44). Individual peptides were reconstituted and used at a final concentration of 10 μM.
Statistical analyses.
All statistical analyses were performed using GraphPad Prism software, version 5.0a (GraphPad Software, Inc., La Jolla, CA). Unless otherwise stated, significance comparing iv-labeled and -protected populations was determined by unpaired, nonparametric Mann-Whitney tests, with P values of <0.05 (*), 0.01 (**), and 0.001 (***).
ACKNOWLEDGMENTS
This work was supported by research grants from the NIAID Centers for Excellence for Influenza Research and Surveillance and the National Institutes of Health, including grants HHSN272201400006C, 1R21AI105851, and 5T32AI007285.
We thank the University of Rochester flow cytometry core facility for technical assistance. We also thank Katherine Richards for outstanding editorial assistance with the manuscript. Finally, we thank BEI Resources, NIAID, NIH, for the influenza virus peptide arrays that were instrumental in the elicitation of influenza-specific CD4 T cells in vivo.
We have no conflicts of interest to declare.
REFERENCES
- 1.Strutt TM, McKinstry KK, Marshall NB, Vong AM, Dutton RW, Swain SL. 2013. Multipronged CD4(+) T-cell effector and memory responses cooperate to provide potent immunity against respiratory virus. Immunol Rev 255:149–164. doi: 10.1111/imr.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laidlaw BJ, Craft JE, Kaech SM. 2016. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 16:102–111. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi A, Saito T. 2017. CD4 CTL, a cytotoxic subset of CD4(+) T cells, their differentiation and function. Front Immunol 8:194. doi: 10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilk MM, Misiak A, McManus RM, Allen AC, Lynch MA, Mills KHG. 2017. Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with Bordetella pertussis. J Immunol 199:233–243. doi: 10.4049/jimmunol.1602051. [DOI] [PubMed] [Google Scholar]
- 6.Sakai S, Kauffman KD, Schenkel JM, McBerry CC, Mayer-Barber KD, Masopust D, Barber DL. 2014. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J Immunol 192:2965–2969. doi: 10.4049/jimmunol.1400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner DL, Farber DL. 2014. Mucosal resident memory CD4 T cells in protection and immunopathology. Front Immunol 5:331. doi: 10.3389/fimmu.2014.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner DL, Bickham KL, Thome JJ, Kim CY, D'Ovidio F, Wherry EJ, Farber DL. 2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol 7:501–510. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knudson CJ, Weiss KA, Hartwig SM, Varga SM. 2014. The pulmonary localization of virus-specific T lymphocytes is governed by the tissue tropism of infection. J Virol 88:9010–9016. doi: 10.1128/JVI.00329-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allie SR, Randall TD. 2017. Pulmonary immunity to viruses. Clin Sci 131:1737–1762. doi: 10.1042/CS20160259. [DOI] [PubMed] [Google Scholar]
- 11.Spitaels J, Roose K, Saelens X. 2016. Influenza and memory T cells: how to awake the force. Vaccines 4:33. doi: 10.3390/vaccines4040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braciale TJ, Sun J, Kim TS. 2012. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiPiazza A, Nogales A, Poulton N, Wilson PC, Martínez-Sobrido L, Sant AJ. 2017. Pandemic 2009 H1N1 influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci Rep 7:10857. doi: 10.1038/s41598-017-11313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bautista BL, Devarajan P, McKinstry KK, Strutt TM, Vong AM, Jones MC, Kuang Y, Mott D, Swain SL. 2016. Short-lived antigen recognition but not viral infection at a defined checkpoint programs effector CD4 T cells to become protective memory. J Immunol 197:3936–3949. doi: 10.4049/jimmunol.1600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muschaweckh A, Buchholz VR, Fellenzer A, Hessel C, König P-A, Tao S, Tao R, Heikenwälder M, Busch DH, Korn T, Kastenmüller W, Drexler I, Gasteiger G. 2016. Antigen-dependent competition shapes the local repertoire of tissue-resident memory CD8+ T cells. J Exp Med 213:3075–3086. doi: 10.1084/jem.20160888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz JLG, Pérez-Girón JV, Lüdtke A, Gómez-Medina S, Ruibal P, Idoyaga J, Muñoz-Fontela C. 2017. Monocyte-derived dendritic cells enhance protection against secondary influenza challenge by controlling the switch in CD8+ T-cell immunodominance. Eur J Immunol 47:345–352. doi: 10.1002/eji.201646523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nourshargh S, Alon R. 2014. Leukocyte migration into inflamed tissues. Immunity 41:694–707. doi: 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Wardlaw AJ, Guillen C, Morgan A. 2005. Mechanisms of T cell migration to the lung. Clin Exp Allergy 35:4–7. doi: 10.1111/j.1365-2222.2005.02139.x. [DOI] [PubMed] [Google Scholar]
- 19.Weninger W, Biro M, Jain R. 2014. Leukocyte migration in the interstitial space of non-lymphoid organs. Nat Rev Immunol 14:232–246. doi: 10.1038/nri3641. [DOI] [PubMed] [Google Scholar]
- 20.Schenkel JM, Masopust D. 2014. Tissue-resident memory T cells. Immunity 41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takamura S. 2017. Persistence in temporary lung niches: a survival strategy of lung-resident memory CD8(+) T cells. Viral Immunol 30:438–450. doi: 10.1089/vim.2017.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masopust D, Schenkel JM. 2013. The integration of T cell migration, differentiation and function. Nat Rev Immunol 13:309–320. doi: 10.1038/nri3442. [DOI] [PubMed] [Google Scholar]
- 23.Iijima N, Iwasaki A. 2015. Tissue instruction for migration and retention of TRM cells. Trends Immunol 36:556–564. doi: 10.1016/j.it.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veerman KM, Williams MJ, Uchimura K, Singer MS, Merzaban JS, Naus S, Carlow DA, Owen P, Rivera-Nieves J, Rosen SD, Ziltener HJ. 2007. Interaction of the selectin ligand PSGL-1 with chemokines CCL21 and CCL19 facilitates efficient homing of T cells to secondary lymphoid organs. Nat Immunol 8:532–539. doi: 10.1038/ni1456. [DOI] [PubMed] [Google Scholar]
- 25.Lim Y-C, Henault L, Wagers AJ, Kansas GS, Luscinskas FW, Lichtman AH. 1999. Expression of functional selectin ligands on Th cells is differentially regulated by IL-12 and IL-4. J Immunol 162:3193–3201. [PubMed] [Google Scholar]
- 26.McKinstry KK, Strutt TM, Bautista B, Zhang W, Kuang Y, Cooper AM, Swain SL. 2014. Effector CD4 T-cell transition to memory requires late cognate interactions that induce autocrine IL-2. Nat Commun 5:5377. doi: 10.1038/ncomms6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown DM, Kamperschroer C, Dilzer AM, Roberts DM, Swain SL. 2009. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell Immunol 257:69–79. doi: 10.1016/j.cellimm.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark JG, Mandac-Dy JB, Dixon AE, Madtes DK, Burkhart KM, Harlan JM, Bullard DC. 2004. Trafficking of Th1 cells to lung: a role for selectins and a P-selectin glycoprotein-1-independent ligand. Am J Respir Cell Mol Biol 30:220–227. doi: 10.1165/rcmb.2003-0208OC. [DOI] [PubMed] [Google Scholar]
- 29.Marshall NB, Vong AM, Devarajan P, Brauner MD, Kuang Y, Nayar R, Schutten EA, Castonguay CH, Berg LJ, Nutt SL, Swain SL. 2017. NKG2C/E marks the unique cytotoxic CD4 T cell subset, ThCTL, generated by influenza infection. J Immunol 198:1142–1155. doi: 10.4049/jimmunol.1601297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Braciale TJ. 2013. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol 3:425–429. doi: 10.1016/j.coviro.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaylo A, Schrock DC, Fernandes NRJ, Fowell DJ. 2016. T cell interstitial migration: motility cues from the inflamed tissue for micro- and macro-positioning. Front Immunol 7:858–813. doi: 10.3389/fimmu.2016.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richter M, Ray SJ, Chapman TJ, Austin SJ, Rebhahn J, Mosmann TR, Gardner H, Kotelianski V, deFougerolles AR, Topham DJ. 2007. Collagen distribution and expression of collagen-binding alpha1beta1 (VLA-1) and alpha2beta1 (VLA-2) integrins on CD4 and CD8 T cells during influenza infection. J Immunol 178:4506–4516. doi: 10.4049/jimmunol.178.7.4506. [DOI] [PubMed] [Google Scholar]
- 34.van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J Machine Learning Res 9:2579–2605. [Google Scholar]
- 35.León B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A. 2014. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nat Commun 5:3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Zeng X, Sigal N, Lund PJ, Su LF, Huang H, Chien Y-H, Davis MM. 2016. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc Natl Acad Sci U S A 113:E1890–E1897. doi: 10.1073/pnas.1602488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. 2012. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol 86:6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tubo NJ, Jenkins MK. 2014. TCR signal quantity and quality in CD4(+) T cell differentiation. Trends Immunol 35:591–596. doi: 10.1016/j.it.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol 10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnamoorthy V, Kannanganat S, Maienschein-Cline M, Cook SL, Chen J, Bahroos N, Sievert E, Corse E, Chong A, Sciammas R. 2017. The IRF4 gene regulatory module functions as a read-write integrator to dynamically coordinate T helper cell fate. Immunity 47:481–497. doi: 10.1016/j.immuni.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng N, Weaver JM, Mosmann TR. 2014. Cytokine diversity in the Th1-dominated human anti-influenza response caused by variable cytokine expression by Th1 cells, and a minor population of uncommitted IL-2+ IFNγ- Thpp cells. PLoS One 9:e95986. doi: 10.1371/journal.pone.0095986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiPiazza A, Richards KA, Knowlden ZAG, Nayak JL, Sant AJ. 2016. The role of CD4 T cell memory in generating protective immunity to novel and potentially pandemic strains of influenza. Front Immunol 7:10. doi: 10.3389/fimmu.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. 2007. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol 81:7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nayak JL, Richards KA, Chaves FA, Sant AJ. 2010. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol 23:169–180. doi: 10.1089/vim.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards KA, Chaves FA, Sant AJ. 2011. The memory phase of the CD4 T-cell response to influenza virus infection maintains its diverse antigen specificity. Immunology 133:246–256. doi: 10.1111/j.1365-2567.2011.03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penberthy KK, Juncadella IJ, Ravichandran KS. 2014. Apoptosis and engulfment by bronchial epithelial cells. Implications for allergic airway inflammation. Ann Am Thorac Soc 11(Suppl 5):S259–S262. doi: 10.1513/AnnalsATS.201405-200AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.GeurtsvanKessel CH, Lambrecht BN. 2008. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol 1:442–450. doi: 10.1038/mi.2008.39. [DOI] [PubMed] [Google Scholar]
- 48.Guilliams M, Dutertre C-A, Scott CL, McGovern N, Sichien D, Chakarov S, Van Gassen S, Chen J, Poidinger M, De Prijck S, Tavernier SJ, Low I, Irac SE, Mattar CN, Sumatoh HR, Low GHL, Chung TJK, Chan DKH, Tan KK, Hon TLK, Fossum E, Bogen B, Choolani M, Chan JKY, Larbi A, Luche H, Henri S, Saeys Y, Newell EW, Lambrecht BN, Malissen B, Ginhoux F. 2016. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roche PA, Cresswell P. 2016. Antigen processing and presentation mechanisms in myeloid cells. Microbiol Spectr 4(3):microbiolspec.MCHD-0008-2015 http://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.MCHD-0008-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sant AJ, Richards KA, Nayak J. 2018. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr Opin Immun 50:13–21. doi: 10.1016/j.coi.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]