ABSTRACT
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease caused by a tick-borne phlebovirus of the family Bunyaviridae, SFTS virus (SFTSV). Wild-type and type I interferon (IFN-I) receptor 1-deficient (IFNAR1−/−) mice have been established as nonlethal and lethal models of SFTSV infection, respectively. However, the mechanisms of IFN-I production in vivo and the factors causing the lethal disease are not well understood. Using bone marrow-chimeric mice, we found that IFN-I signaling in hematopoietic cells was essential for survival of lethal SFTSV infection. The disruption of IFN-I signaling in hematopoietic cells allowed an increase in viral loads in serum and produced an excess of multiple inflammatory cytokines and chemokines. The production of IFN-I and inflammatory cytokines was abolished by deletion of the signaling molecules IPS-1 and MyD88, essential for retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) and Toll-like receptor (TLR) signaling, respectively. However, IPS-1−/− MyD88−/− mice exhibited resistance to lethal SFTS with a moderate viral load in serum. Taken together, these results indicate that adequate activation of RLR and TLR signaling pathways under low to moderate levels of viremia contributed to survival through the IFN-I-dependent antiviral response during SFTSV infection, whereas overactivation of these signaling pathways under high levels of viremia resulted in abnormal induction of multiple inflammatory cytokines and chemokines, causing the lethal disease.
IMPORTANCE SFTSV causes a severe infectious disease in humans, with a high fatality rate of 12 to 30%. To know the pathogenesis of the virus, we need to clarify the innate immune response as a front line of defense against viral infection. Here, we report that a lethal animal model showed abnormal induction of multiple inflammatory cytokines and chemokines by an uncontrolled innate immune response, which triggered the lethal SFTS. Our findings suggest a new strategy to target inflammatory humoral factors to treat patients with severe SFTS. Furthermore, this study may help the investigation of other tick-borne viruses.
KEYWORDS: SFTSV, inflammation, innate immunity, interferons, viral pathogenesis
INTRODUCTION
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease caused by a tick-borne phlebovirus of the family Bunyaviridae, SFTS virus (SFTSV) (1, 2). SFTS has been reported to be endemic in China, South Korea, and Japan (2–4). The major clinical manifestations and laboratory findings for SFTS are high fever, gastrointestinal symptoms, thrombocytopenia, leukopenia, and multiple-organ dysfunction, with a fatality rate of 12 to 30% (1, 2). In SFTS patients, levels of inflammatory cytokines and chemokines in serum are dramatically increased. In particular, granulocyte colony-stimulating factor (G-CSF), interferon alpha (IFN-α), IFN-γ, C-C motif ligand 3 (CCL3), interleukin-6 (IL-6), and C-X-C motif ligand 10 (CXCL10) are increased to a greater extent in sera of patients with severe disease than in patients with mild disease, demonstrating that the severity of SFTS is correlated with the levels of these cytokines (5–8). Cytokine expression is tightly regulated by the innate immune system, which controls the host antiviral response and promotes subsequent activation of an adaptive immune system. However, an uncontrolled innate immune system causes excessive production of cytokines and a massive inflammatory response, called a cytokine storm, that leads to vascular leakage and hemorrhage, liver damage, and multiple-organ dysfunction (9). Currently, it is hypothesized that the cytokine storm leads to lethal SFTS (5–8).
SFTSV is a single-stranded RNA virus possessing large (L), medium (M), and small (S) segments (1, 2). The L segment encodes an RNA-dependent RNA polymerase, and the M segment encodes glycoproteins. The S segment is an ambisense RNA consisting of negative- and positive-sense genes, which encode a nucleocapsid (N) protein and a nonstructural S segment (NSs) protein, respectively. SFTSV NSs protein is reported to form granular structures in the cytoplasm of infected cells and to interact with host molecules to suppress antiviral innate immune responses, as described below.
Infection by RNA viruses is sensed by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) and Toll-like receptors (TLRs), which activates an antiviral response by producing type I interferon (IFN-I) and inflammatory cytokines (10–13). RLRs, including RIG-I and melanoma differentiation-associated gene 5 (MDA5), recruit IFN-β promoter stimulator 1 (IPS-1, also known as MAVS, VISA, and Cardif), which relays the signal to TANK-binding kinase 1 (TBK1) to induce IFN-I by activation of interferon regulatory factor 3 (IRF-3) (14–19). Among TLRs, TLR3 and -7 have been identified as viral-RNA sensors. TLR3 and TLR7 recruit TIR domain-containing adaptor-inducing IFN-β (TRIF) and myeloid differentiation factor 88 (MyD88), respectively (12, 20–24). TRIF controls IFN-I induction through TBK1, but MyD88 triggers IFN-I induction in an IRAK (IL-1 receptor-associated kinase)/IRF-7 axis-dependent manner. Secreted IFN-I can bind to its receptor (IFNAR1) and activate signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2) and IRF-9, inducing antiviral genes (25–27).
It has been reported that SFTSV NSs protein interacts with TBK1 (28–30), suggesting that IFN-I production by TBK1 activation could be abolished in SFTSV-infected cells. A recent study demonstrated that recombinant SFTSV genetically devoid of NSs protein strongly induces IFN-I (31). Thus, NSs protein is one of the viral factors evading the host innate immune response to promote efficient viral replication.
To establish an animal model for the study of human SFTS, several laboratory animals have been challenged with SFTSV. Adult immunocompetent animals were reported to show resistance to lethal SFTS (32–36). On the other hand, immunocompromised animals, such as IFNAR1-deficient (IFNAR1−/−) mice and STAT2-deficient (STAT2−/−) hamsters, were shown to exhibit lethal SFTS (34, 36–39). Immunohistochemical examination demonstrated that in SFTSV-infected IFNAR1−/− mice, SFTSV N protein-positive cells could be detected in several tissues, including the spleen, lymph nodes, liver, and kidneys, and that they coexpressed the host cellular markers of macrophages, B lymphocytes, and reticular cells, i.e., positivity for IbaI, Pax5, and gp36, respectively (36). In the spleens and lymph nodes, rather than other tissues, of infected IFNAR1−/− mice, SFTSV N protein-expressing cells are frequently observed, and destruction of the lymphoid architecture by decreasing the number of B lymphocytes can be observed, suggesting that secondary lymphoid tissues are the primary sites of viral replication. These findings from animal models suggest that IFN-I signaling is essential for protection against lethal SFTS and that suppression of IFN-I production by SFTSV NSs protein is a viral strategy to secure replication. However, the signaling pathway and the mechanisms of the IFN-I-dependent antiviral response during SFTSV infection have not been elucidated in vivo.
In this study, we revealed that hematopoietic cells are essential for the IFN-I-dependent antiviral response and that the expression of IFN-I and inflammatory cytokines is regulated by the IPS-1 and MyD88 signaling pathways upon SFTSV infection. We also found that large amounts of inflammatory cytokines and chemokines were produced in the sera of SFTSV-infected IFNAR1−/− mice, suggesting that these inflammatory mediators play key roles in the lethal disease. Our study revealed that IPS-1 and MyD88 signaling pathways are connected to the pathogenesis of SFTSV infection by leading to a severe inflammation called a cytokine storm but that they also contribute to the host defense against lethal SFTSV infection by an IFN-I-dependent antiviral response.
RESULTS
Type I interferon signaling in hematopoietic cells is required for protection from lethal SFTSV infection.
IFNAR1−/− mice have been established as a lethal animal model of SFTSV infection (34, 36–38). To confirm this, wild-type (WT) and IFNAR1−/− mice were intravenously inoculated with 2 × 106 50% tissue culture infective doses (TCID50) of SFTSV. Consistent with previous reports, IFNAR1−/− mice, but not WT mice, died within 7 days postinfection (dpi) (see Fig. 5B). At 48 h postinfection (hpi), the spleen, liver, small intestine, large intestine, stomach, kidneys, heart, lungs, inguinal lymph nodes, and Peyer's patch were collected and subjected to immunohistochemical analysis. The frozen tissue sections were stained with anti-SFTSV N protein antibody (Fig. 1). Immunohistochemical analysis revealed that IFNAR1−/− mice allowed viral replication in many of their organs compared with WT mice, and SFTSV replicated more efficiently in secondary lymphoid tissues, such as the spleen and inguinal lymph nodes, than in other tissues. These results suggest that SFTSV initially targets the hematopoietic cells of secondary lymphoid tissues, but not nonhematopoietic cells, and that the lack of an IFN-I-dependent antiviral response allows viral spread in hematopoietic cells of several tissues.
FIG 5.
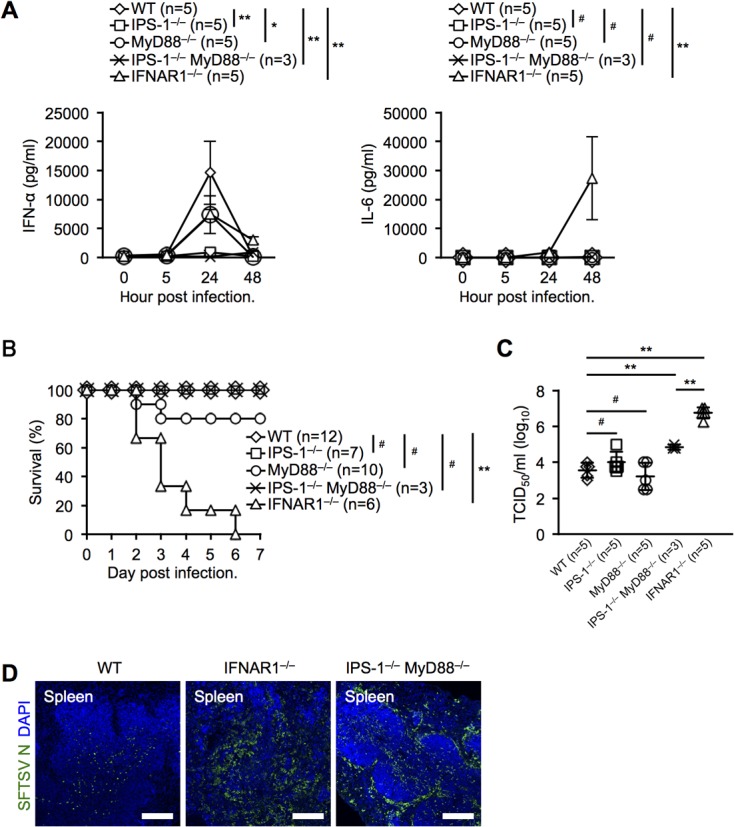
Systemic production of IFN-α and IL-6 is dependent on IPS-1 and MyD88 during SFTSV infection in vivo. (A) IFN-α and IL-6 production levels in sera from SFTSV-infected mice at the indicated time points. (B) Survival rates of WT, IPS-1−/−, MyD88−/−, IPS-1−/− MyD88−/−, and IFNAR1−/− mice after SFTSV challenge. (C) SFTSV titers in peripheral blood of infected mice at 2 dpi. (D) Immunohistochemical examination of SFTSV N in the spleens of WT, IFNAR1−/−, and IPS-1−/− MyD88−/− mice at 2 dpi. Spleen sections were costained for SFTSV N (green) and DAPI (blue) and analyzed by confocal microscopy. Scale bars, 250 μm. The results are representative of 3 mice. (A and C) The data are representative of the results of two independent experiments and are plotted as means ± SD. (A) Unpaired Student t tests were used to determine significant differences in IFN-α and IL-6 production between samples at 24 hpi and 48 hpi, respectively. *, P < 0.05; **, P < 0.01; #, not significant.
FIG 1.
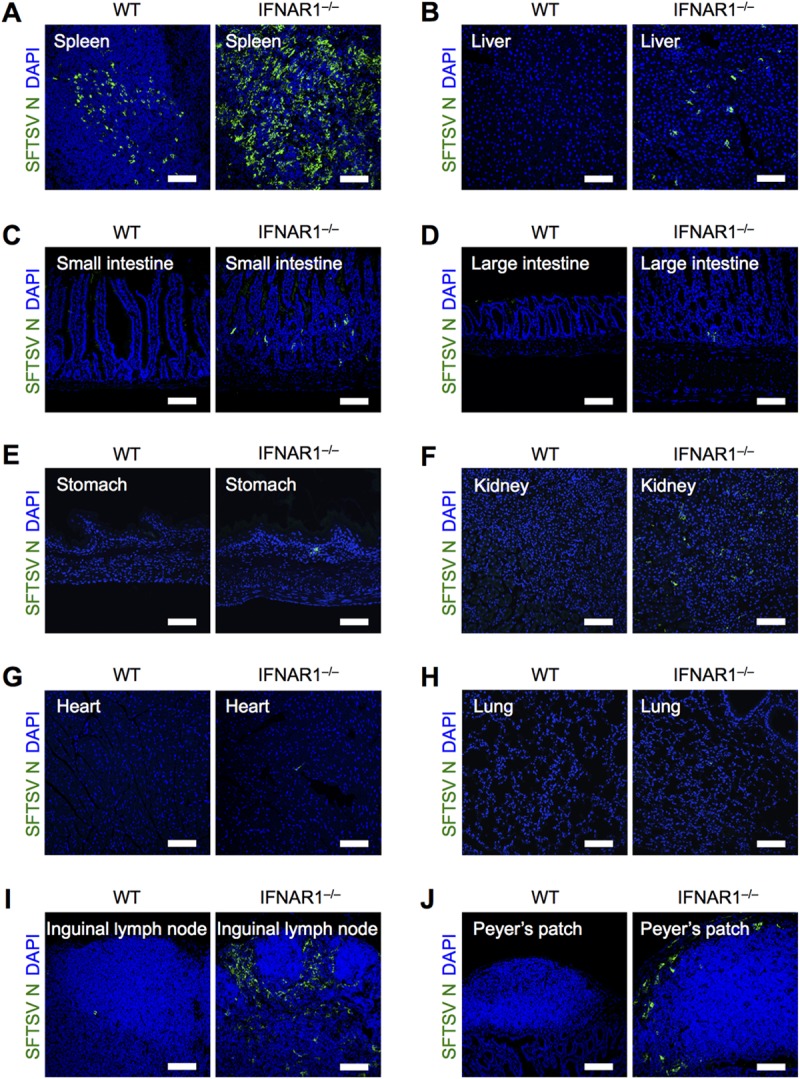
Deficiency of IFN-I signaling promotes SFTSV replication in secondary lymphoid tissues. Shown is immunohistochemical analysis of SFTSV N protein in tissues from WT and IFNAR1−/− mice at 2 dpi. Spleen (A), liver (B), small intestine (C), large intestine (D), stomach (E), kidney (F), heart (G), lung (H), inguinal lymph node (I), and Peyer's patch (J) sections were costained for SFTSV N (green) and DAPI (blue). All the sections were analyzed by confocal microscopy. Scale bars, 100 μm. Representative images from 3 mice are shown.
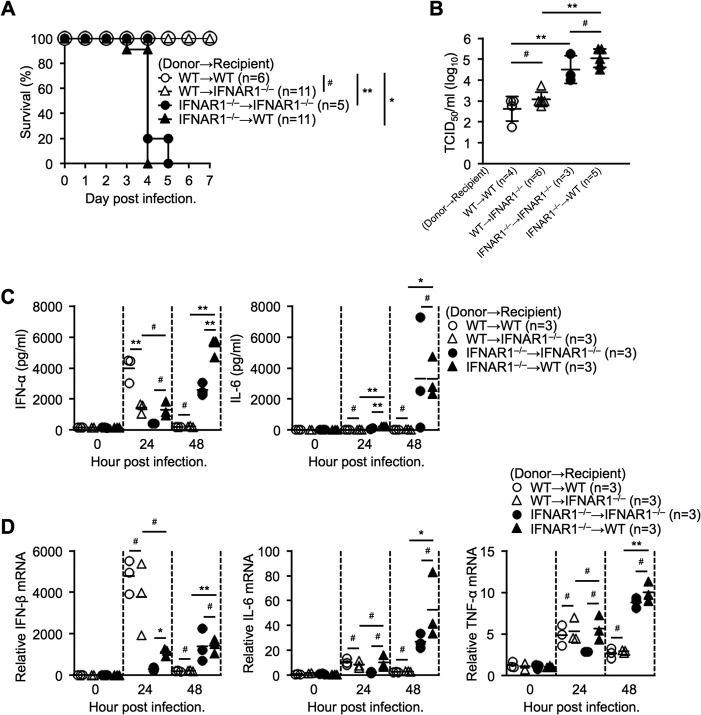
To further investigate the importance of IFN-I signaling in hematopoietic cells, we generated bone marrow (BM)-chimeric mice that selectively lacked IFNAR1 in hematopoietic cells, nonhematopoietic cells, or both cell types by transplantation of bone marrow prepared from WT or IFNAR1−/− donor mice into irradiated WT or IFNAR1−/− recipient mice. After 8 weeks of bone marrow reconstitution, the BM-chimeric mice were intravenously inoculated with 2 × 106 TCID50 of SFTSV. BM-chimeric mice lacking IFNAR1 in hematopoietic cells showed high susceptibility to SFTSV infection (Fig. 2A). In contrast, BM-chimeric mice expressing IFNAR1 in hematopoietic cells exhibited resistance to lethal SFTS (Fig. 2A). We focus on the factors leading to the lethality of SFTSV infection; therefore, it was important to analyze mice just before the infected mice started to die. To collect samples at 3 dpi was manageable; however, there was selection bias, since some susceptible mice started to die as early as 3 dpi. Therefore, we decided to collect samples at 2 dpi in order to provide data on a group of mice. Peripheral blood was harvested from BM-chimeric mice at 48 hpi to determine the viral load in sera. Consistent with the survival, IFNAR1 deficiency in hematopoietic cells significantly increased the viral loads in serum (Fig. 2B). These results clearly demonstrate that IFN-I signaling in hematopoietic cells, but not nonhematopoietic cells, is critical for the antiviral response to protect from lethality. Next, we determined IFN-I and inflammatory cytokine levels in sera of BM-chimeric mice at different time points during SFTSV infection (Fig. 2C). WT chimeric mice with BM cells transferred intravenously (BM-transferred mice) expressed large amounts of IFN-α, reaching a peak at 24 hpi. IFNAR1−/− BM-transferred chimeric mice expressed IFN-α with delayed kinetics. The inflammatory cytokine IL-6 was undetectable up to 48 hpi in WT BM-transferred chimeric mice (Fig. 2C). In contrast, IFNAR1−/− BM-transferred chimeric mice expressed high levels of IL-6 at 48 hpi, coinciding with active viral replication. (Fig. 2C). We also determined the expression levels of IFN-β, IL-6, and tumor necrosis factor alpha (TNF-α) mRNAs in the spleens of BM-chimeric mice at the indicated time points (Fig. 2D). IFN-β induction in IFNAR1−/− BM-transferred chimeric mice was delayed and attenuated, and also, the results confirmed that IFNAR1 deficiency leads to production of large amounts of the inflammatory cytokines IL-6 and TNF-α at 48 hpi. These results suggest that the lack of IFN-I signaling results, due to high viremia status, in hyperproduction of inflammatory cytokines, causing the severe disease phenotype.
FIG 2.
IFN-I signaling in hematopoietic cells is essential for surviving SFTSV infection. (A) Effect of bone marrow cell transplantation on the survival curve of SFTSV-infected mice. (B) Virus titers in peripheral blood of infected bone marrow-chimeric mice at 2 dpi. (C and D) Cytokine levels in serum (C) and relative expression of cytokine mRNA in the spleen (D) in SFTSV-infected chimeric mice at the indicated time points. Representative data from two independent experiments are shown (B to D) and plotted as means ± standard deviations (SD) (B). *, P < 0.05; **, P < 0.01; #, not significant.
F4/80- or sialoadhesin-positive macrophages are targeted by SFTSV in BM-chimeric mice.
Iba1-positive macrophages and Pax5-positive immature B cells have been reported to be SFTSV-susceptible cell types in IFNAR1−/− mice (36). To examine the SFTSV-replicating cell type in our BM-chimeric mouse model, we performed immunohistochemical analysis. The spleens and livers of infected BM-chimeric mice were harvested at 48 hpi and stained for SFTSV N protein and cellular markers. IFNAR1−/− BM-transferred chimeric mice appeared similar to IFNAR1−/− mice and had higher numbers of viral-antigen-positive cells than WT BM-transferred chimeric mice (Fig. 3A to C). SFTSV N protein was observed in some F4/80-positive red pulp macrophages (Fig. 3D) and sialoadhesin-positive marginal metallophilic macrophages (Fig. 3E) in the spleens of IFNAR1−/− BM-transferred WT mice. In the livers of IFNAR1−/− BM-transferred WT mice, most SFTSV N-positive cells expressed F4/80, a marker for Kupffer cells (Fig. 3F) (40). Collectively, our results demonstrate that F4/80- or sialoadhesin-positive macrophages are potentially SFTSV-targeting cells in the chimeric mice, consistent with the results of the previous study (36). Since macrophages reside in different tissues and potentially participate in inflammation, their involvement in SFTS pathogenesis is strongly suggested.
FIG 3.
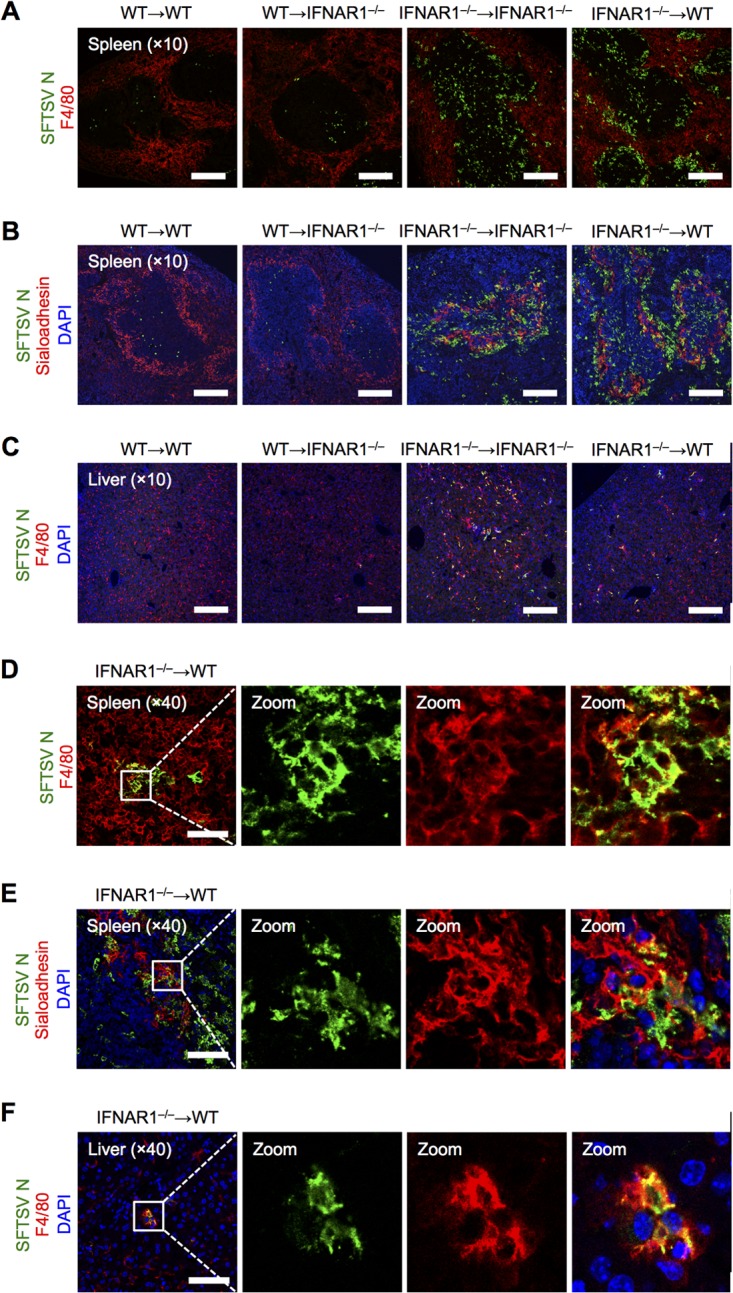
F4/80- or sialoadhesin-positive macrophages are potentially SFTSV-targeting cells in BM-chimeric mice. Shown is immunohistochemical examination of SFTSV N protein expression in the spleens and livers of bone marrow-chimeric mice at 2 dpi. Spleen sections were costained for SFTSV N (green) and F4/80 (red) (A and D) or SFTSV N (green), sialoadhesin (red), and DAPI (blue) (B and E). (C and F) Liver sections were costained for SFTSV N (green), F4/80 (red), and DAPI (blue). Zoom, enlargement of the boxed areas in the leftmost images. All the sections were analyzed by confocal microscopy. Scale bars, 250 μm (A to C) and 75 μm (D to F). The results are representative of 3 mice.
Production of IFN-I and inflammatory cytokines by BMDCs upon SFTSV infection.
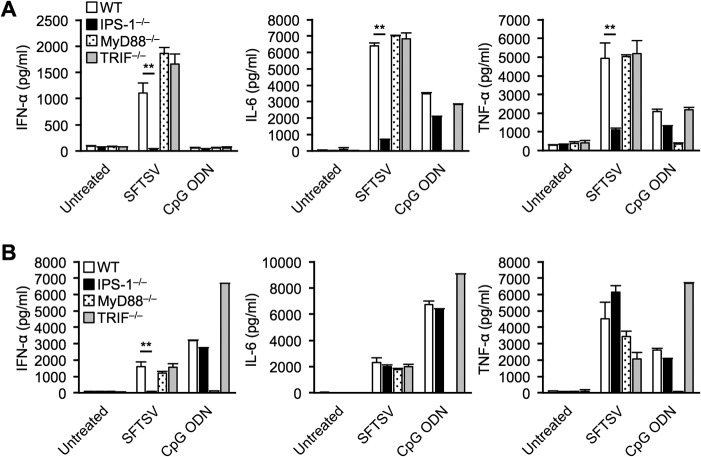
Although the IFN-I system plays a key role in the antiviral response against SFTSV, the precise signaling mechanism remains unclear. To address this, we examined the involvement of IPS-1, MyD88, and TRIF in conventional (cDCs) and plasmacytoid (pDCs) dendritic cells. We cultured BM derived from WT, IPS-1−/−, MyD88−/−, and TRIF−/− mice in the presence of granulocyte macrophage colony-stimulating factor (GM-CSF) or Fms-like tyrosine kinase 3 ligand (Flt3L). GM-CSF generates cDCs, and Flt3L generates a mixture of cDCs and pDCs. These cells were infected with SFTSV or stimulated with a synthetic oligodeoxynucleotide (CpG ODN), a ligand activating the TLR9/MyD88 signaling pathway, and cytokine production was monitored by enzyme-linked immunosorbent assay (ELISA). As expected, GM-CSF-induced cDCs and Flt3L-induced pDCs stimulated by CpG ODN produced the cytokines through MyD88 signaling pathways, but not other signaling pathways (Fig. 4). GM-CSF-induced WT cDCs secreted large amounts of IFN-α, IL-6, and TNF-α in response to SFTSV infection (Fig. 4A). On the other hand, IPS-1 deficiency, but not MyD88 and TRIF deficiency, dramatically decreased the production of these cytokines. The lack of IPS-1 in Flt3L-induced bone marrow-derived dendritic cells (BMDCs) also significantly decreased the production of IFN-α; however, production of IL-6 and TNF-α remained unchanged (Fig. 4B). These results suggest that BMDCs, especially cDCs, respond to SFTSV infection and secrete massive amounts of cytokines in an IPS-1-dependent manner. In contrast, production of IL-6 and TNF-α by Flt3L-induced BMDCs was independent of either IPS-1 or MyD88 (Fig. 4B). This was unexpected, as pDCs utilize MyD88, but not IPS-1, to produce IFN-I and inflammatory cytokines via TLR7 and TLR9. These results suggest the occurrence of SFTSV-specific cytokine-regulatory mechanism(s) in pDCs (see Discussion).
FIG 4.
Production of IFN-I and inflammatory cytokines by BMDCs upon SFTSV infection. (A and B) Bone marrow cells were purified from WT, IPS-1−/−, MyD88−/−, and TRIF−/− mice and cultured with GM-CSF or Flt3L to induce BMDCs. GM-CSF-BMDCs (A) and Flt3L-BMDCs (B) were infected with 2 × 105 TCID50 SFTSV or stimulated with 0.5 μM CpG ODN. After 24 h, production of IFN-α, IL-6, and TNF-α in the culture supernatant was determined by ELISA. The data are representative of the results of two independent experiments and are plotted as means and SD. **, P < 0.01.
To further investigate the signaling pathway for IFN-I and inflammatory cytokine production, we intravenously infected WT, IPS-1−/−, MyD88−/−, IPS-1−/− MyD88−/−, and IFNAR1−/− mice with 2 × 106 TCID50 of SFTSV and measured serum IFN-α and IL-6. Consistent with the results of in vitro infection of BMDCs, IPS-1 deficiency abolished IFN-α production (Fig. 5A). In contrast to the in vitro results, MyD88 deficiency significantly attenuated IFN-α production, and deficiencies of both IPS-1 and MyD88 dramatically abrogated IFN-α production, suggesting that viral single-stranded RNA might be sensed by TLR7 and RLRs. To our surprise, the lack of IFNAR1 dramatically stimulated IL-6 production. To further analyze these mice, we monitored their survival and measured the serum viral titer at 48 hpi. All IFNAR1-deficient mice died within 7 days after SFTSV challenge (Fig. 5B). Although both IPS-1−/− and IPS-1−/− MyD88−/− mice had inhibited IFN-α production upon SFTSV infection (Fig. 5A), they survived (Fig. 5B). Most MyD88-deficient mice survived, suggesting its contribution was minor. The viral titers in both IPS-1- and MyD88-deficient mice were comparable to that in WT mice (Fig. 5C). IPS-1−/− MyD88−/− mice exhibited significantly higher viral titers than WT mice; however, IFNAR1−/− mice exhibited a 10-fold-higher viral titer than IPS-1−/− MyD88−/− mice. Immunohistochemical analysis demonstrated that the viral spread in the spleens of IPS-1−/− MyD88−/− mice was comparable to that in IFNAR1−/− mice (Fig. 5D).
Inflammation-related genes are augmented in the SFTSV-infected IFNAR1-deficient mouse spleen.
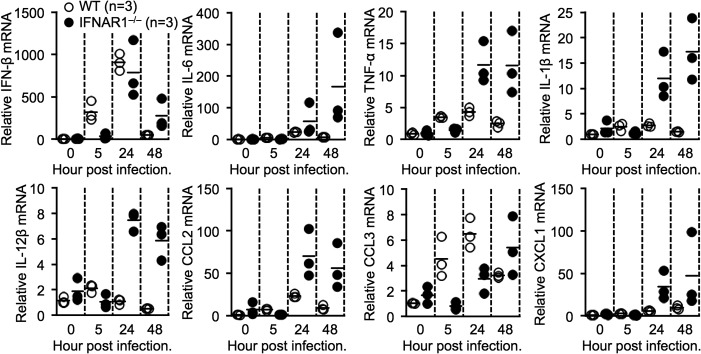
SFTSV infection in humans results in high mortality, and it has been assumed that the severe inflammation known as a cytokine storm is a causative factor (1, 2). Previous studies have demonstrated that levels of specific inflammatory cytokines and chemokines are correlated with the seriousness of SFTS in patients (5–8). Here, to clarify the relationship between inflammation-related gene induction and the lethality of SFTSV infection in more detail in a mouse model, we examined the expression levels of multiple genes, including IFN-β, IL-6, TNF-α, IL-1β, IL-12β, CCL2, CCL3, and CXCL1 genes, in the spleens of WT and IFNAR1−/− mice after SFTSV challenge. The induction of IFN-β in the spleens of both SFTSV-infected WT and IFNAR1−/− mice peaked at 24 hpi (Fig. 6). Infected WT mice exhibited higher CCL3 induction at 24 hpi than infected IFNAR1−/− mice, although this result was reversed at 48 hpi. IFN-I signaling is related to the positive regulation of CCL3 induction (41), and therefore, early and strong induction of CCL3 was observed in infected WT mice. Every inflammation-related gene tested was augmented in the spleens of SFTSV-infected IFNAR1−/− mice compared with WT mice. In particular, SFTSV-infected IFNAR1−/− mice exhibited 100-fold-enhanced IL-6 and CCL2 mRNA levels compared with SFTSV-infected WT mice. This result suggests that induced inflammatory cytokines and chemokines cooperate to cause severe inflammation.
FIG 6.
Enhanced production of multiple inflammatory cytokines and chemokines in SFTSV-infected IFNAR1−/− mice. Shown is relative expression of the indicated cytokine mRNAs in the spleens of infected WT and IFNAR1−/− mice at different time points. The data are representative of the results of two independent experiments.
DISCUSSION
In vitro studies have shown that NSs protein, a nonstructural protein of SFTSV, strongly inhibits innate immunity by targeting TBK1, which activates IRF-3 and -7 (28–30). In contrast, our mouse study demonstrated that systemic production of IFN-I is induced by RLR/IPS-1 signaling, including TBK1 activation (Fig. 5A). It is important to point out that strong inhibition of IFN-I production by NSs protein is observed in NSs protein-overexpressing cell lines, but not SFTSV-infected primary cells (28–31). A possible explanation for this discrepancy could be that the expression level of NSs protein in SFTSV-infected cells is insufficient to block IFN-I production and/or NSs protein is less functional in cell types such as macrophages and B lymphocytes. Bunyavirus NSs proteins play an important role in hijacking host cells, although NSs proteins have low homology of amino acid sequences among bunyaviruses (42). Indeed, NSs proteins from different viruses exhibited varying degrees of IFN-I inhibition, and their cellular localizations were distinct (data not shown). Rift Valley fever virus (RVFV), in the genus Phlebovirus of the family Bunyaviridae, expresses NSs protein that is well known to block IFN-I induction and to lead to proteasomal degradation of protein kinase R (PKR), an IFN-I-inducible antiviral protein (43). The naturally attenuated RVFV strain clone 13 has a truncated defective NSs protein and exhibits lower virulence than the virulent RVFV strain ZH548, with intact NSs protein. These findings indicate that PKR plays a key role in RVFV infection and that RVFV NSs protein is closely related to virulence (44). Although we did not focus on the function of PKR upon SFTSV infection in this study, further studies in the future will provide deep insights into the virulence of SFTSV.
Phleboviruses, such as Rift Valley fever virus, possess panhandle genomic RNAs with a triphosphate at the 5' end (5'ppp) that are sensed by RIG-I (45–50). Since each SFTSV genomic-RNA segment contains complementary sequences at the 5′ and 3′ ends (51), it is probable that SFTSV genomic RNAs form panhandle structures with 5′ppp and are recognized by RIG-I. Upon SFTSV infection, similar to IFN-I, production of IL-6 and TNF-α was also IPS-1 dependent in cDCs (Fig. 4A). However, the induction mechanisms of these cytokines in pDCs are more complicated. With the TLR9 ligand, CpG ODN, induction of IFN-α/IL-6/TNF-α was exclusively dependent on MyD88 (Fig. 4B). However, in SFTSV-infected pDCs, deletion of MyD88 had little effect on the production of these cytokines, except that IFN-α induction appeared to be IPS-1 dependent. This is in sharp contrast to the Newcastle disease virus, which activates IFN-α and IL-6 in a MyD88-dependent but RIG-I-independent manner (52). These results suggest that SFTSV is capable of activating unique signaling in pDCs to produce inflammatory cytokines.
As IFNAR1 deficiency dramatically sensitizes mice to SFTSV infection, we initially thought that blockade of IFN-I production also provokes enhanced pathogenesis. We showed that IPS-1−/− MyD88−/− mice did not produce serum IFN-α, while IFNAR1−/− mice produced levels of IFN-α comparable to those of WT mice (Fig. 5A). However, IPS-1−/− MyD88−/− mice did not exhibit lethality of SFTSV infection (Fig. 5B). Abrogation of both IPS-1 and MyD88 showed a 10-fold increase in viral titer compared to WT mice; however, this enhancement was not accompanied by lethality (Fig. 5B and C). It can be speculated that local IFN-I production by IPS-1- and MyD88-independent mechanisms may contribute to the survival of IPS-1−/− MyD88−/− mice through an IFNAR1-dependent mechanism.
Our bone marrow transfer experiments suggest that the lack of IFNAR1 in hematopoietic cells caused enhanced production of IL-6 and TNF-α (Fig. 2C and D). SFTSV was observed to infect F4/80- or sialoadhesin-positive macrophages (Fig. 3). Taking these observations together, we hypothesize that the infected macrophages lacking IFNAR1 contribute to the disease by producing inflammatory cytokines. The lack of functional IFNAR1 may abrogate induction of interferon-stimulated genes, which promotes viral replication; however, it is not clear how this deficiency results in enhanced production of inflammatory cytokines and chemokines by SFTSV (Fig. 6).
Our in vivo experiments showed that IL-6, TNF-α, IL-1β, IL-12β, CCL2, CCL3, and CXCL1 were strongly induced in SFTSV-infected IFNAR1−/− mice, suggesting that these cytokines/chemokines may play critical roles in lethal SFTS (Fig. 6). The cytokines and chemokines are well-known inflammation mediators and are transiently induced in response to viral infection, contributing to host defense through innate and adaptive immunity (53). Although the induction of these inflammatory mediators is tightly regulated, their strong and sustained production triggers severe inflammatory diseases. The adenosine deaminase acting on RNA 1 (ADAR1) gene p150 isoform, an IFN-I-inducible gene, acts as a negative regulator in innate immune response because loss of function of ADAR1 causes severe autoimmune disease associated with aberrant IFN-I and inflammatory cytokine production (54, 55). A previous report showed that ADAR1 is a negative regulator of MDA5/IPS-1 signaling (56). Furthermore, ADAR1 negatively regulates RIG-I signaling by competing with RIG-I to recognize double-stranded RNA (dsRNA) (57). We speculate that depletion of IFNAR led to dysfunction of IFN-I-inducible negative regulation and resulted in severe inflammation. Among tested cytokines/chemokines, there was obvious induction of IL-6 and CCL2 mRNAs (Fig. 6). CCL2 is predominantly produced by monocytes and macrophages in response to viral infection and cytokines, including IL-6 (53). CCL2 promotes migration and infiltration of monocytes into the inflammation site via its receptor (CCR2). The recruited monocytes further express inflammatory cytokines and chemokines, including IL-6 and CCL2, resulting in amplification of the inflammation. During SFTSV infection, the recruitment of monocytes could contribute to disease progression. Therefore, neutralizing IL-6 or CCL2 in serum or blocking its receptors (58) is potentially beneficial for SFTS patients. Furthermore, the expression of inflammatory cytokines is controlled by nuclear factor κB (NF-κB) (59); in practice, dexamethasone as a chemical compound can bind to glucocorticoid receptors, and the ligand-bound receptor directly blocks NF-κB, resulting in attenuated inflammation (60). Thus, NF-κB is one of the therapeutic targets for inflammation in SFTS.
MATERIALS AND METHODS
Virus strain.
SFTSV strain SPL010, an isolate from a Japanese SFTS patient (4), was used after propagation in Vero cells.
Mice.
All mouse lines were bred under specific-pathogen-free conditions at the Institute for Frontier Life and Medical Sciences, Kyoto University. All animal experiments were conducted in compliance with regulations approved by the Committee for Animal Experiments of the Institute for Frontier Life and Medical Sciences and the National Institute of Infectious Diseases (no. 114049, 116027, 116034, 117003, 117004, and 215021). Among the mouse lines used in our study, C57BL/6 mice and IFNAR1−/− mice on a C57BL/6 background were purchased from Shimizu Laboratory Supplies and B&K Universal, respectively. IPS-1−/−, MyD88−/−, and TRIF−/− mice on a C57BL/6 background were kindly provided by Shizuo Akira (Osaka University). IPS-1−/− mice were crossed with MyD88−/− mice to generate IPS-1−/− MyD88−/− mice.
Generation of bone marrow-chimeric mice.
To generate bone marrow-chimeric mice, recipient CD45.1 C57BL/6 and IFNAR1−/− mice were lethally irradiated with 9 Gy. Donor bone marrow cells were purified from femurs and tibias of CD45.2 C57BL/6, IFNAR1−/−, and CD45.1 C57BL/6 mice and filtered through a 70-μm nylon filter. Purified BM cells were transferred intravenously to the irradiated mice. The chimeric mice were treated with water supplemented with antibiotics for 2 weeks. After 8 weeks of BM transfer, mice were taken for infection experiments.
SFTSV infection in vivo.
When we started this project, only a few studies on SFTSV pathogenesis using a mouse model had been reported. We were not sure how large a dose of SFTSV was sufficient to kill mice and also to monitor detectable immune responses. Therefore, we performed intravenous inoculations with several doses of virus (103 to 106 TCID50) in mice and found that all IFNAR1−/− mice infected with 2 × 106 TCID50 of virus died, but not at other doses. Although subcutaneous injection with the same dose also killed mice, the mice survived much longer than the mice injected intravenously; the survival periods varied (1 to 2 weeks), and lethality was not 100% stable. Considering that we focus on lethal factors in SFTSV infection, it is important to have rigid and stable settings for lethal infection and to analyze the mice just before they die, which might reflect the viremia condition in SFTS patients. Thus, we chose the following infection route and virus dose: mice were infected intravenously with 2 × 106 TCID50 of SFTSV strain SPL010. From the infected mice, blood and sera were collected to quantify SFTSV titers and cytokine production, respectively. For real-time quantitative PCR (qPCR) analysis and immunohistochemistry examination, tissues were harvested from infected mice. All of the experiments involving the handling of infectious SFTSV were performed in a biosafety level 3 (BSL3) facility at the National Institute of Infectious Diseases.
Preparation of bone marrow-derived dendritic cells.
Femurs and tibias were harvested from C57BL/6, IPS-1−/−, MyD88−/−, and TRIF−/− mice, and bone marrow was isolated. To generate BMDCs, bone marrow cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin mixed solution (100 U/ml and 100 μg/ml, respectively), 100 μM 2-mercaptoethanol, and 10 ng/ml murine GM-CSF (Peprotech) or 100 ng/ml human Flt3L (Peprotech) for 6 to 8 days. In the case of GM-CSF-induced BMDCs, the medium was replaced with fresh medium every 2 days. GM-CSF- and Flt3L-induced BMDCs were infected with 2 × 105 TCID50 of SFTSV strain SPL010 or stimulated with 0.5 μM CpG ODN (InvivoGen). After 24 h of stimulation, the culture supernatant was collected, and IFN-α and IL-6 were measured by ELISA.
SFTSV titration.
Titers were determined as previously described (61). SFTSV was serially diluted and inoculated into Vero cells in quadruplicate. After 3 days of culture, the cells were fixed with 10% formalin and permeabilized with 0.1% Triton X-100. The cells were subjected to incubation with rabbit anti-SFTSV N protein (4), followed by further incubation with Alexa Fluor 488-labeled anti-rabbit IgG(H+L) (Life Technologies). Observation was performed under a fluorescence microscope (BZ-9000; Keyence), and viral titers were calculated as the median TCID50 per milliliter by the Reed-Muench method (62).
Immunohistochemistry.
Harvested tissues from infected mice were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 2 to 3 h. The fixed tissues were embedded in OCT compound (Sakura Finetek) and frozen in liquid nitrogen. The embedded tissues were sectioned at 5 μm. The frozen sections were stained with a rabbit polyclonal antibody against SFTSV N protein as a primary antibody to detect the viral antigen. Anti-F4/80 and anti-sialoadhesin antibodies and DAPI (4′,6-diamidino-2-phenylindole) were used for identification of the different types of cells.
Cytokine quantification.
Sera from infected mice and culture supernatant from stimulated BMDCs were collected and mixed with 0.1% NP-40 and irradiated with UV light for 10 min to inactivate the viruses. These samples were subjected to analysis with commercial ELISA kits for mouse IFN-α (PBL Assay Science), IL-6 (eBioscience), and TNF-α (eBioscience) according to the manufacturers' instructions.
RNA isolation and real-time qPCR.
Total RNA was harvested from spleens using TRIzol regent (Invitrogen) and treated with DNase I (Roche Diagnostics). cDNA was generated with a High-Capacity cDNA reverse transcription kit (Applied Biosystems) or ReverTra Ace qPCR RT master mix (Toyobo) according to the manufacturer's instructions. Real-time qPCR was performed with the Step One plus real-time PCR system (Applied Biosystems) using TaqMan fast universal PCR master mix (Applied Biosystems), Thunderbird probe qPCR mix (Toyobo), and Fast SYBR green master mix (Applied Biosystems) according to the manufacturers' instructions.
Statistical analysis.
Unpaired Student t tests were used to determine significant differences between samples. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This study was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Innovative Areas Infection Competency (24115004) from the Japan Agency for Medical Research and Development; the Research Program on Emerging and Reemerging Infectious Diseases (17fk0108202h0902); the Japan Society for the Promotion of Science Core to Core Program; and Grants-in-Aid for Scientific Research A (23249023).
REFERENCES
- 1.Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, Wang P, Tang X, Wang H, Kang K, Zhang S, Zhao G, Wu W, Yang Y, Chen H, Mu F, Chen W. 2011. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog 7:e1002369. doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, Choe PG, Kim NJ, Lee JK, Oh MD. 2013. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis 19:1892–1894. doi: 10.3201/eid1911.130792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M. 2014. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis 209:816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YZ, He YW, Dai YA, Xiong Y, Zheng H, Zhou DJ, Li J, Sun Q, Luo XL, Cheng YL, Qin XC, Tian JH, Chen XP, Yu B, Jin D, Guo WP, Li W, Wang W, Peng JS, Zhang GB, Zhang S, Chen XM, Wang Y, Li MH, Li Z, Lu S, Ye C, de Jong MD, Xu J. 2012. Hemorrhagic fever caused by a novel bunyavirus in China: pathogenesis and correlates of fatal outcome. Clin Infect Dis 54:527–533. doi: 10.1093/cid/cir804. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Jin C, Zhan F, Wang X, Liang M, Zhang Q, Ding S, Guan X, Huo X, Li C, Qu J, Wang Q, Zhang S, Zhang Y, Wang S, Xu A, Bi Z, Li D. 2012. Host cytokine storm is associated with disease severity of severe fever with thrombocytopenia syndrome. J Infect Dis 206:1085–1094. doi: 10.1093/infdis/jis452. [DOI] [PubMed] [Google Scholar]
- 7.Deng B, Zhang S, Geng Y, Zhang Y, Wang Y, Yao W, Wen Y, Cui W, Zhou Y, Gu Q, Wang W, Shao Z, Li C, Wang D, Zhao Y, Liu P. 2012. Cytokine and chemokine levels in patients with severe fever with thrombocytopenia syndrome virus. PLoS One 7:e41365. doi: 10.1371/journal.pone.0041365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu MM, Lei XY, Yu H, Zhang JZ, Yu XJ. 2017. Correlation of cytokine level with the severity of severe fever with thrombocytopenia syndrome. Virol J 14:6. doi: 10.1186/s12985-016-0677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens EM, Koretzky GA. 2017. Review: cytokine storm syndrome: looking toward the precision medicine era. Arthritis Rheumatol 69:1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, ten Oever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 17.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 18.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol 169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 22.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol 4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 23.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 24.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs A. 1962. Antiviral action of interferon. Br Med J 2:353–355. doi: 10.1136/bmj.2.5301.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestka S, Krause CD, Walter MR. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol Rev 202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 27.Ank N, West H, Paludan SR. 2006. IFN-lambda: novel antiviral cytokines. J Interferon Cytokine Res 26:373–379. doi: 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 28.Qu B, Qi X, Wu X, Liang M, Li C, Cardona CJ, Xu W, Tang F, Li Z, Wu B, Powell K, Wegner M, Li D, Xing Z. 2012. Suppression of the interferon and NF-kappaB responses by severe fever with thrombocytopenia syndrome virus. J Virol 86:8388–8401. doi: 10.1128/JVI.00612-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning YJ, Wang M, Deng M, Shen S, Liu W, Cao WC, Deng F, Wang YY, Hu Z, Wang H. 2014. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. J Mol Cell Biol 6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Zheng B, Wang T, Li A, Wan J, Qu J, Li CH, Li D, Liang M. 2017. NSs protein of severe fever with thrombocytopenia syndrome virus suppresses interferon production through different mechanism than Rift Valley fever virus. Acta Virol 61:289–298. doi: 10.4149/av_2017_307. [DOI] [PubMed] [Google Scholar]
- 31.Brennan B, Rezelj VV, Elliott RM. 2017. Mapping of transcription termination within the S segment of SFTS phlebovirus facilitated generation of NSs deletant viruses. J Virol 91:e00743-17. doi: 10.1128/JVI.00743-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen XP, Cong ML, Li MH, Kang YJ, Feng YM, Plyusnin A, Xu J, Zhang YZ. 2012. Infection and pathogenesis of Huaiyangshan virus (a novel tick-borne bunyavirus) in laboratory rodents. J Gen Virol 93:1288–1293. doi: 10.1099/vir.0.041053-0. [DOI] [PubMed] [Google Scholar]
- 33.Jin C, Liang M, Ning J, Gu W, Jiang H, Wu W, Zhang F, Li C, Zhang Q, Zhu H, Chen T, Han Y, Zhang W, Zhang S, Wang Q, Sun L, Liu Q, Li J, Wang T, Wei Q, Wang S, Deng Y, Qin C, Li D. 2012. Pathogenesis of emerging severe fever with thrombocytopenia syndrome virus in C57/BL6 mouse model. Proc Natl Acad Sci U S A 109:10053–10058. doi: 10.1073/pnas.1120246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Wu B, Paessler S, Walker DH, Tesh RB, Yu XJ. 2014. The pathogenesis of severe fever with thrombocytopenia syndrome virus infection in alpha/beta interferon knockout mice: insights into the pathologic mechanisms of a new viral hemorrhagic fever. J Virol 88:1781–1786. doi: 10.1128/JVI.02277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin C, Jiang H, Liang M, Han Y, Gu W, Zhang F, Zhu H, Wu W, Chen T, Li C, Zhang W, Zhang Q, Qu J, Wei Q, Qin C, Li D. 2015. SFTS virus infection in nonhuman primates. J Infect Dis 211:915–925. doi: 10.1093/infdis/jiu564. [DOI] [PubMed] [Google Scholar]
- 36.Matsuno K, Orba Y, Maede-White K, Scott D, Feldmann F, Liang M, Ebihara H. 2017. Animal models of emerging tick-borne phleboviruses: determining target cells in a lethal model of SFTSV infection. Front Microbiol 8:104. doi: 10.3389/fmicb.2017.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada S, Posadas-Herrera G, Aoki K, Morita K, Hayasaka D. 2015. Therapeutic effect of post-exposure treatment with antiserum on severe fever with thrombocytopenia syndrome (SFTS) in a mouse model of SFTS virus infection. Virology 482:19–27. doi: 10.1016/j.virol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tani H, Fukuma A, Fukushi S, Taniguchi S, Yoshikawa T, Iwata-Yoshikawa N, Sato Y, Suzuki T, Nagata N, Hasegawa H, Kawai Y, Uda A, Morikawa S, Shimojima M, Watanabe H, Saijo M. 2016. Efficacy of T-705 (favipiravir) in the treatment of infections with lethal severe fever with thrombocytopenia syndrome virus. mSphere 1:e00061-15. doi: 10.1128/mSphere.00061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gowen BB, Westover JB, Miao J, Van Wettere AJ, Rigas JD, Hickerson BT, Jung KH, Li R, Conrad BL, Nielson S, Furuta Y, Wang Z. 2017. Modeling severe fever with thrombocytopenia syndrome virus infection in golden syrian hamsters: importance of STAT2 in preventing disease and effective treatment with favipiravir. J Virol 91:e01942-16. doi: 10.1128/JVI.01942-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krenkel O, Tacke F. 2017. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 41.Fujita H, Asahina A, Tada Y, Fujiwara H, Tamaki K. 2005. Type I interferons inhibit maturation and activation of mouse Langerhans cells. J Investig Dermatol 125:126–133. doi: 10.1111/j.0022-202X.2005.23803.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu F, Chen H, Travassos da Rosa AP, Tesh RB, Xiao SY. 2007. Phylogenetic relationships among sandfly fever group viruses (Phlebovirus: Bunyaviridae) based on the small genome segment. J Gen Virol 88:2312–2319. doi: 10.1099/vir.0.82860-0. [DOI] [PubMed] [Google Scholar]
- 43.Ly HJ, Ikegami T. 2016. Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virol J 13:118. doi: 10.1186/s12985-016-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol 83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 46.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 47.Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennan B, Li P, Zhang S, Li A, Liang M, Li D, Elliott RM. 2015. Reverse genetics system for severe fever with thrombocytopenia syndrome virus. J Virol 89:3026–3037. doi: 10.1128/JVI.03432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 53.Deshmane SL, Kremlev S, Amini S, Sawaya BE. 2009. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W Jr, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O'Connell MA, Lovell SC, Crow YJ. 2012. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 Independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43:933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang S, Deng P, Zhu Z, Zhu J, Wang G, Zhang L, Chen AF, Wang T, Sarkar SN, Billiar TR, Wang Q. 2014. ADAR1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J Immunol 193:3436–3445. doi: 10.4049/jimmunol.1401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka T, Narazaki M, Kishimoto T. 2016. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 59.Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 60.Ray A, Prefontaine KE. 1994. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc Natl Acad Sci U S A 91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimojima M, Fukushi S, Tani H, Yoshikawa T, Fukuma A, Taniguchi S, Suda Y, Maeda K, Takahashi T, Morikawa S, Saijo M. 2014. Effects of ribavirin on severe fever with thrombocytopenia syndrome virus in vitro. Jpn J Infect Dis 67:423–427. doi: 10.7883/yoken.67.423. [DOI] [PubMed] [Google Scholar]
- 62.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]