ABSTRACT
Antibody and receptor binding are key virus-host interactions that control host range and determine the success of infection. Canine and feline parvovirus capsids bind the transferrin receptor type 1 (TfR) to enter host cells, and specific structural interactions appear necessary to prepare the stable capsids for infection. Here, we define the details of binding, competition, and occupancy of wild-type and mutant parvovirus capsids with purified receptors and antibodies. TfR-capsid binding interactions depended on the TfR species and varied widely, with no direct relationship between binding affinity and infection. Capsids bound feline, raccoon, and black-backed jackal TfRs at high affinity but barely bound canine TfRs, which mediated infection efficiently. TfRs from different species also occupied capsids to different levels, with an estimated 1 to 2 feline TfRs but 12 black-backed jackal TfRs binding each capsid. Multiple alanine substitutions within loop 1 on the capsid surface reduced TfR binding but substitutions within loop 3 did not, suggesting that loop 1 directly engaged the TfR and loop 3 sterically affected that interaction. Binding and competition between different TfRs and/or antibodies showed complex relationships. Both antibodies 14 and E competed capsids off TfRs, but antibody E could also compete capsids off itself and antibody 14, likely by inducing capsid structural changes. In some cases, the initial TfR or antibody binding event affected subsequent TfR binding, suggesting that capsid structure changes occur after TfR or antibody binding and may impact infection. This shows that precise, host-specific TfR-capsid interactions, beyond simple attachment, are important for successful infection.
IMPORTANCE Host receptor binding is a key step during viral infection and may control both infection and host range. In addition to binding, some viruses require specific interactions with host receptors in order to infect, and anti-capsid antibodies can potentially disrupt these interactions, leading to neutralization. Here, we examine the interactions between parvovirus capsids, the receptors from different hosts, and anti-capsid antibodies. We show that interactions between parvovirus capsids and host-specific TfRs vary in both affinity and in the numbers of receptors bound, with complex effects on infection. In addition, antibodies binding to two sites on the capsids had different effects on TfR-capsid binding. These experiments confirm that receptor and antibody binding to parvovirus capsids are complex processes, and the infection outcome is not determined simply by the affinity of attachment.
KEYWORDS: antibody competition, bio-layer interferometry, canine parvovirus, conformational change, feline panleukopenia virus, infection, neutralization, receptor binding, transferrin receptor
INTRODUCTION
Interactions of animal viruses with host cell receptors are key events that control cell infection, tissue tropism, and host range. Specific virus-receptor interactions lead to cellular attachment and initiate cell entry and infection, which generally occur through receptor-mediated endocytosis. However, in addition to the tethering process, receptor binding may also result in structural changes to the capsid that are important for allowing binding to secondary receptors or for controlling later intracellular steps in infection (1–5). Variation in different host receptors also creates host range barriers, and virus adaption to those receptors may be associated with host range expansion (6–8). Other important virus-host interactions occur through antibody binding, where host antibodies bind viral proteins and can directly neutralize viral infectivity or protect against infection through a variety of different mechanisms (9–12). Despite the importance of these different interactions, we still lack a detailed understanding of how receptor or antibody binding dynamics control cell binding and infection, cause disease, or influence transmission to new hosts.
Canine parvovirus (CPV) and feline panleukopenia virus (FPV) have single-stranded DNA genomes of about 5,100 bases that encode at least 4 genes, including the major capsid protein VP2 and the minor capsid protein VP1. The genomes are packaged into small, nonenveloped, and stable capsids consisting of 60 subunits, ∼90% of which are VP2 and the remaining ∼10% of which are VP1. CPV and FPV show natural variations in host range that are primarily controlled by differences in viral capsids and their interactions with the host receptor, the transferrin receptor type 1 (TfR) (13–16). The TfR is the natural cellular receptor for iron-loaded transferrin (Tf) and the hemochromatosis protein (HFE), which regulates transferrin binding and iron uptake in the intestine (17, 18). Once bound to the TfR, diferric Tf or viral capsids are efficiently transported into cells via clathrin-mediated endocytosis (19, 20). While Tf recycles to the cell surface after releasing iron, the virus-TfR complex appears to be retained in the endosomal system for a prolonged period before entering the cytoplasm and releasing viral DNA into the nucleus (21). Both Tf and HFE bind on the side or underneath the TfR protease-like and helical domains (22, 23), while CPV binds to the apical domain (15, 24), making it unlikely that either Tf or HFE binding to the TfR would affect CPV binding.
Canine parvovirus type-2 (CPV-2) arose in the mid-1970s as a variant of a virus similar to but distinct from FPV (25, 26). In 1978, CPV spread worldwide, causing a pandemic of disease among dogs, wolves, and coyotes, and during 1979 and 1980 a virus variant, designated CPV-2a and containing 5 mutations on or near the capsid surface, arose and replaced the original CPV-2 strain worldwide (27–29). In previous studies, we have shown that CPV acquired the canine host range as the result of mutations to a small number of residues on the capsid surface (30, 31). Two of those mutations (VP2 residues 93Lys to Asn and 323Asp to Asn) acted together to give CPV the ability to bind to the canine TfR and infect canine cells and dogs (13, 31), although CPV has been shown to bind to the canine TfR with much lower affinity than to the feline TfR (13, 14). The canine TfR differs from other host receptors both in amino acid sequence and in having an additional (fourth) glycosylation within the apical domain (attached to Asn384 of the canine TfR sequence), which blocked the binding of FPV-like viruses (15, 32). Other mutations that blocked the infection of canine cells were observed in VP2 surface loop 3, around residue 300 (33, 34). The CPV-derived viruses in dogs have continued to evolve since the emergence of the CPV-2a common ancestor, and many different studies have reported VP2 sequences from infected dogs or other carnivore hosts from different regions of the world (35–38). Those studies show that a variety of mutations have become widespread in the virus population, including a number of changes in the capsid surface that fall close to or within the predicted receptor binding site or antigenic epitopes.
The CPV-like viruses have broad host ranges and naturally infect many different animals within the order Carnivora, with infections and sometimes disease being commonly observed in dogs, mink, raccoons, foxes, domestic cats, large cats, and skunks (26, 37, 39, 40). However, these hosts differ naturally in their susceptibility to infection by FPV, CPV, and closely related viruses, and infection of different hosts, including raccoons, mink, and foxes, was often associated with selection of additional mutations that alter residues on the capsid protein structure (37, 39). Similar mutations also arose after passaging viruses in cultured cells from different hosts, showing that the selection occurs at the level of the host cell, most likely the receptor (37).
Those previous studies therefore show that TfR-capsid interactions are highly specific, and that capsid mutations affect the functional interactions between capsids and the TfR. The variant interactions resulting from virus and TfR differences may involve both the affinity of capsid binding to the TfR as well as additional structural interactions essential for virus infectivity of cells. This was seen most clearly in studies where we showed that some capsid variants bound efficiently to the TfR and were endocytosed into cells but did not result in infection (41). However, the details of the TfR-capsid interactions are not yet well understood, including the stoichiometry of receptor binding, the structures involved in attachment, and whether the number of bound receptors is important for successful infection. For CPV and FPV infection, the number of receptors that mediate binding during cell infection appears to be very low. The T=1 parvovirus capsid should display 60 equivalent receptor binding sites, with an estimated maximum occupancy of between 12 and 24 TfRs per capsid after accounting for steric interference between neighboring receptors (42). However, it was seen that single CPV particles bound less than 5 feline TfR molecules on the surface of cells prior to endocytosis and infection (43), and incubating CPV capsids with purified feline TfR ectodomains resulted in only 1 to 4 TfR being copurified with each capsid on average (44).
Antibodies bind to a number of overlapping epitopes on parvovirus capsids, and for CPV and FPV, monoclonal antibodies have been classified into two broad groups recognizing two different structures on the capsids (45). Eight different antigen binding fragments (Fabs) derived from monoclonal antibodies were examined for their interactions with the CPV and FPV capsids using cryo-electron microscopy (cryoEM), and their combined footprints covered more than 60% of the viral surface (46). Three of those antibodies recognized the “A” site and bound close to the top of a raised region surrounding the 3-fold axis of the capsid (the 3-fold spike), while the other five antibodies tested recognized the “B” site and attached to the side of the 3-fold spike (also called the “shoulder”) (46, 47). In contrast to the TfRs, most of the Fabs tested appeared to fully or nearly fully occupy the 60 possible binding sites on the capsids when added in excess (46). However, those Fabs differed significantly in their neutralization efficiencies, with some being poorly neutralizing even when present in excess of the binding sites and others neutralizing the virus with as few as 5 to 10 Fabs per capsid (48). The reasons for the large differences in neutralization efficiency are not yet fully understood, but the high-resolution structure of a complex between the Fab of one highly neutralizing antibody (FabE) and the CPV capsid showed structural variation of nonbound regions of the capsid, suggesting that the Fab triggered allosteric changes that interfered with infection (47).
Other parvoviruses and adeno-associated viruses (AAV) use a variety of different receptors for binding and infection of cells. The AAVs use a protein receptor (the AAV receptor), which they bind through different domains, in addition to a variety of other glycoprotein or glycan receptors, including heparin sulfate proteoglycans and sialic acids (49–51). Those distinct interactions determine the tissue tropism of the AAVs and the gene delivery vectors derived from them (52). Cell infection and the tissue tropisms of the minute virus of mice (MVM) and H1 viruses are in some cases determined by variation in the specific interactions of the capsids with the host sialic acids (53, 54). Complex virus-receptor interactions also occur in many other systems. For example, some New World arenaviruses differ in their cell and host tropism due to variation in their interactions with host TfRs (55–57). Picornavirus receptors may trigger structural changes in the capsid after binding, either at high occupancy or when groups of receptors bind to one region of the capsid (58). HIV gp120 also undergoes conformational changes after binding CD4 that reveal the binding site for the second receptor, either CXCR4 or CCR5 (2, 4, 5). Likewise, antibody binding has been shown to induce changes in the structures of some viral proteins (9, 10, 12, 59).
Here, we further explore this key area in the natural history of animal viruses by examining the specific interactions of the parvovirus capsids and their receptors involved in infection and in determining viral host range. We use a variety of approaches to reveal the dynamic interactions between parvovirus variants and TfRs from different hosts, as well as with antibodies against defined capsid epitopes. These interactions proved to be surprisingly variable and were influenced by mutations in both capsids and TfRs, which altered the binding affinity and occupancy of TfR on the capsids.
RESULTS
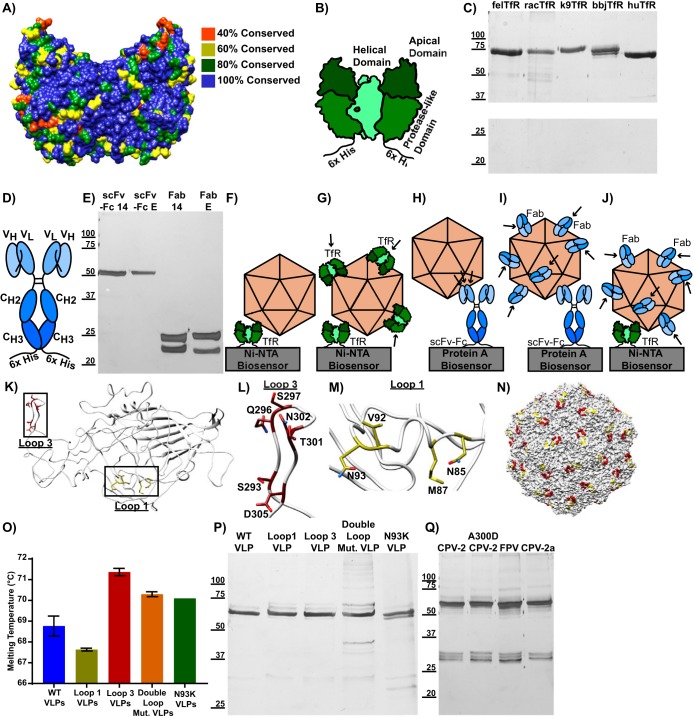
Here, we define the specific interactions of TfRs from various animal hosts with capsids of viruses from different hosts, or with engineered virus-like particles (VLPs), in order to explain how those interactions alter binding and control infection. The receptors examined were derived from cats (Felis catus) (felTfR), dogs (Canis lupus familiaris) (k9TfR), raccoons (Procyon lotor) (racTfR), black-backed jackals (Canis mesomelas) (bbjTfR), and humans (Homo sapiens) (huTfR). These receptors have all been shown to have distinct interactions with capsids when expressed on cells (14, 16, 32, 39, 60), including the canine TfR, which acts as a host range barrier to FPV binding and infection in dogs (60). The protein sequences of these TfRs are largely conserved, but loops with low sequence conservation exist in the apical domain, where CPV has been shown to bind (Fig. 1A). Antibodies against two different sites on the capsids were also examined, both as Fabs and after cloning and expression as single-chain variable fragments fused to antibody constant fragments (scFv-Fcs). The soluble TfR, an scFv-Fc, and the engineered VLPs are diagrammed in Fig. 1, along with an outline of how they were used in the binding studies carried out here, and Table 1 lists the differences between the different parvoviruses and VLPs used in this study. Figure 1 also contains silver stains of proteins to show purity (Fig. 1C, E, P, and Q) and a differential scanning fluorimetry assay on engineered VLPs to show that they are intact and at least as stable as wild-type VLPs (Fig. 1O).
FIG 1.
Composition of ligands and VLPs used in this study. (A) Structure of the human TfR (PDB entry 1suv) with residues color coded by sequence conservation among the canine, feline, raccoon, black-backed jackal, and human TfRs. (B) Soluble TfR, consisting of the transferrin receptor type 1 ectodomain fused to a 6×His tag. Helical, apical, and protease-like domains are labeled. (C) Silver stain showing purified TfRs. (D) scFv-Fc structure, with variable heavy (VH), variable light (VH), heavy constant 2 (CH2), and heavy constant 3 (CH3) domains labeled. (E) Silver stain showing purified scFv-Fcs and Fabs. (F to J) Design of the bio-layer interferometry binding experiments used throughout the study. (F) A capsid binding to soluble TfR, which is attached to a Ni-NTA biosensor via a 6×His tag. (G) Soluble TfRs with His tags removed are shown binding to capsids that are attached to a Ni-NTA biosensor via a TfR with an intact His tag. (H) A capsid binding to an scFv-Fc, which is attached to a protein A biosensor via its Fc domain. (I) Fabs are shown binding to a capsid that is attached to a protein A biosensor via an scFv-Fc. (J) Fabs binding to a capsid that is attached to a Ni-NTA biosensor via a TfR with an intact His tag. (J to N) CPV capsid mutations used in the study. (K) A canine parvovirus VP2 (PDB entry 2CAS) subunit is shown, with mutated capsid surface residues in loop 1 (yellow) and loop 3 (red) highlighted. (L) Magnification of panel H, focusing on residues in loop 3. (M) Magnification of panel H, focusing on residues in loop 1. (N) An intact CPV-2 capsid, showing the positioning of loop 1 and loop 3 after capsid assembly. (O) Differential scanning fluorimetry experiment showing average capsid melting temperature as a measure of stability. Results are from three independent experiments, except for N39K VLPs, for which limited sample quantity permitted only one measurement. Error bars show ± standard error. (P) Silver stain showing purified VLPs. (Q) Silver stain showing purified empty capsids.
TABLE 1.
Differences in VP2 protein sequences for viruses and VLPs relative to CPV-2
| Virus | Difference(s) from CPV-2 |
|---|---|
| CPV-2 | |
| FPV | R80N, N93K, I101T, A103V, I232V, F303Y, N323D, N375D, S564N, G568A |
| A300D CPV-2 | A300D |
| CPV-2a | M87L, I101T, A300G, D305Y, N426D |
| N93K CPV-2 VLPs | N93K |
| Loop 1 mutant VLPs | N85A, M87A, V92A, N93A |
| Loop 3 mutant VLPs | S293A, Q296A, S297A, T301A, N302A, D305A |
| Double loop mutant VLPs | N85A, M87A, V92A, N93A, S293A, Q296A, S297A, T301A, N302A, D305A |
Capsid-TfR binding.
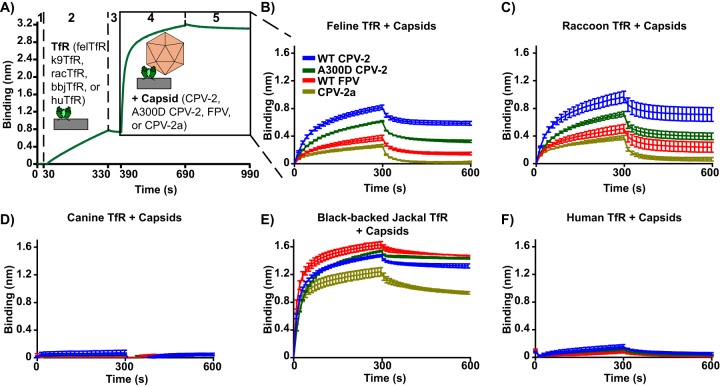
Binding studies were carried out using bio-layer interferometry, where the ligands (TfRs or scFv-Fcs) were bound to nickel-nitrilotriacetic acid (Ni-NTA) biosensors (His-tagged TfRs) or protein A biosensors (scFv-Fcs) (Fig. 1F to J). When TfRs from different species were immobilized on Ni-NTA biosensors and incubated with purified capsids, the results depended on the combinations of capsid and TfR examined, although the results of each combination were quite reproducible (Fig. 2). After attachment, a proportion of the capsids appeared to be loosely bound and were quickly released from the TfRs when washed in buffer, while the remaining capsids stayed attached to receptors for long periods (Fig. 2). The feline TfR bound to all capsids, as expected from previous studies (13, 14, 41), and the raccoon TfR bound capsids to similar levels (Fig. 2B and C). For the feline and raccoon TfRs, CPV-2 capsids bound to the highest levels, while FPV and a point mutant of CPV-2 with a VP2 Ala300Asp mutation, which occurs during natural infection of raccoons or (more rarely) during prolonged passage in cultured feline cells (27, 39, 61), bound to lower levels. The more recent strain of CPV (CPV-2a) bound to the lowest level. The canine TfR showed very low levels of binding to all capsids tested in this assay, close to the background (Fig. 2D), as has been observed previously (13, 14). The black-backed jackal TfR was included in these studies because the black-backed jackal is the closest relative to dogs that lacks the canine TfR's FPV-blocking glycosylation site (32). The black-backed jackal TfR showed the highest level of binding of any of the TfRs tested, regardless of the capsid form examined (Fig. 2E). Compared to the feline, raccoon, and black-backed jackal TfRs, the human TfR bound all of the capsids tested to very low levels, only slightly higher than that of the canine TfR (Fig. 2F).
FIG 2.
Binding kinetics of parvovirus capsids to TfRs from different species. Analysis of the binding of capsids (CPV-2, A300D CPV-2, FPV, or CPV-2a) to the TfR ectodomains from different species (cat, dog, raccoon, black-backed jackal, or human). (A) A sample bio-layer interferometry experiment. Ni-NTA biosensors were washed (step 1), incubated with TfR ectodomains to 0.8 nm of measured binding (step 2), washed again (step 3), incubated with capsids at 240 μg/ml to measure TfR/capsid association (step 4), and washed to measure TfR/capsid disassociation (step 5). Results are measured as the change in wavelength (in nm) over time. Results of capsids binding to feline TfR (B), raccoon TfR (C), canine TfR (D), black-backed jackal TfR (E), and human TfR (F) are shown. Error bars show ± standard error.
Infectivity of parvoviruses on TRVb cells expressing different TfRs.
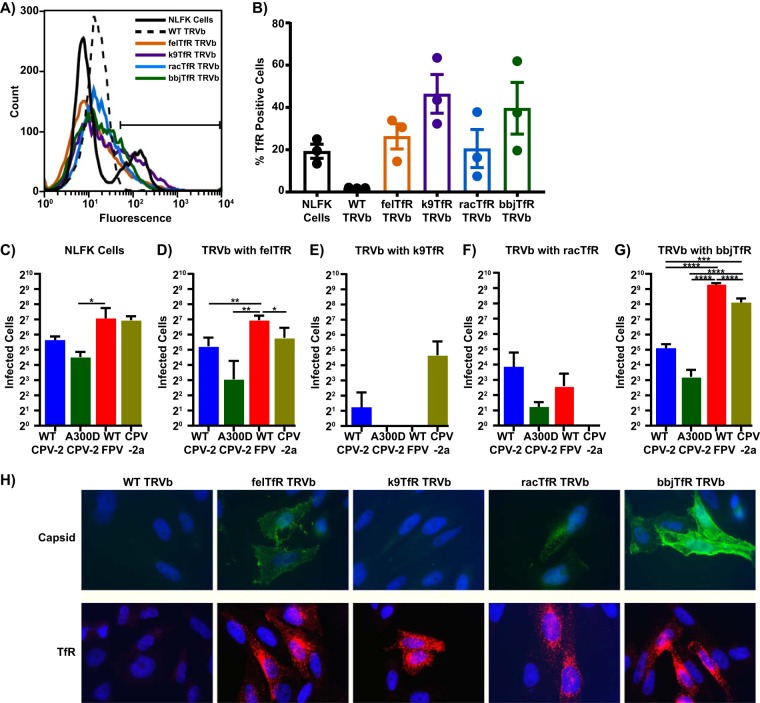
TRVb cells, modified Chinese hamster ovary (CHO) cells without endogenous transferrin receptor, were transfected with plasmids to stably express the feline, canine, raccoon, or black-backed jackal TfRs (Fig. 3A and B) and were then inoculated with parvoviruses. While TfR-free TRVb cells completely resisted infection as expected (16), all of the receptors transfected into TRVb cells added some susceptibility to infection, with the level of infection varying significantly depending on the specific virus and expressed TfR tested (Fig. 3C to G). There was also no direct connection between the affinity of capsid binding to the TfR (Fig. 2) and the level of infection. When CPV-2 capsids were incubated with fixed TRVb cells expressing different TfRs (Fig. 3H), binding approximated the results observed via bio-layer interferometry. Previously published studies examining CPV binding to TRVb cells expressing the TfR or canine or feline cell lines showed similar results (13, 14), further validating the results shown in Fig. 2.
FIG 3.
Relative infectivity of CPV and FPV on cells transfected to express different TfRs. TRVb cells (CHO-derived cells that lack endogenous TfR) were transfected with TfR cDNA from different species. (A) A sample flow cytometry plot showing expression of TfR on wild-type NLFK cells and transfected TRVb cells. (B) Percentage of cells positive for TfR expression, averaged over three independent experiments. (C to G) Cells were seeded at 2 × 104 cells/cm2 in 96-well plates and inoculated with 200 TCID50 of CPV-2, A300D CPV-2, FPV, or CPV-2a. Cultures were incubated for 48 h, fixed, and stained with an anti-NS1 antibody to determine how many cells were infected. Infected cells in each well of wild-type NLFK cells (C) or TRVb cells expressing feline (D), canine (E), raccoon (F), or black-backed jackal (G) TfR were counted as a measurement of infectivity. (H) Binding assay showing attachment of 10 μg/ml CPV-2 capsids to fixed TRVb cells expressing different TfRs (top) and staining of permeabilized TRVb cells with an anti-TfR antibody to visualize TfR expression (bottom). Error bars show ± standard error. *, P < 0.5; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Antibody and TfR binding kinetics and competition.
To better understand the capsid binding of different ligands in these studies, we also compared the binding of two well-characterized monoclonal anti-capsid antibodies and the feline and black-backed jackal TfRs. The antibodies Mab14 and MabE bind to the A and B sites, respectively, of the capsid surface (46). Mab14 and MabE were either purified from hybridomas and cleaved to form Fabs or expressed as scFv-Fcs in insect cells. The cloned scFv-Fcs have the binding properties of the original antibodies (62), can be modified easily, and bind to protein A bio-layer interferometry probes with the same high affinity as the human IgG1 Fc.
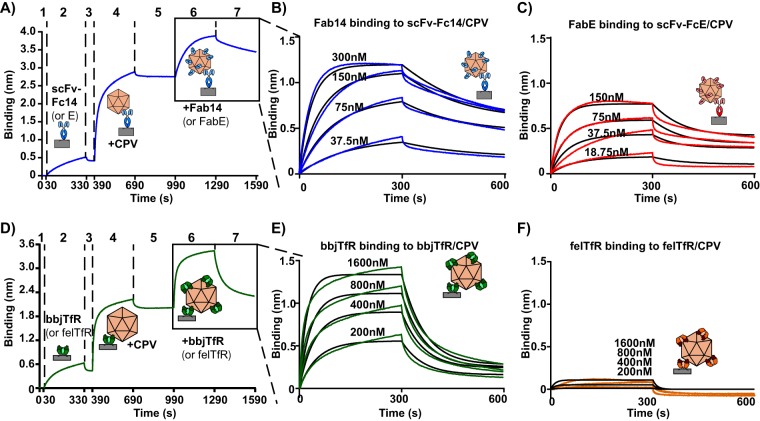
When scFv-Fc14 or E was attached to biosensors and incubated with CPV-2 capsids, capsids showed high on-rates and very low off-rates after binding to the scFv-Fc (Fig. 4A). After binding capsids to scFv-Fcs and washing, we incubated the scFv-Fc/capsid complexes with four concentrations of Fabs derived from the same antibody that had been used to generate the scFv-Fc, allowing us to calculate the affinity of Fab binding. Graphs were globally fit assuming a 1:1 Fab/binding site ratio and showed binding affinities of 2.74 × 10−8 M (association constant [ka] of 8.99 × 104 1/M · s; dissociation constant [kd] of 2.47 × 10−3 1/s) for Fab 14 and 2.16 × 10−8 M for Fab E (ka of 2.28 × 105 1/M · s; kd of 4.91 × 10−3 liters/s) (Fig. 4B and C). Fab14 fit the 1:1 model well in this assay, but the FabE fit was poorer, likely due to a conformational change it has previously been shown to induce in capsids after binding (47).
FIG 4.
Binding kinetics of Fabs or TfRs. Level of binding of Fabs or TfRs to CPV-2 empty capsids. (A) A sample graph showing binding of multiple Fabs to capsids that had been attached to biosensors via scFv-Fcs derived from the same antibody (e.g., FabE and scFv-FcE or Fab14 and scFv-Fc14). Protein A probes were washed (step 1), incubated with scFv-Fc (step 2), washed again (step 3), incubated with CPV-2 empty capsids (step 4), and washed (step 5), resulting in capsids attached to probes by an scFv-Fc. scFv-Fc/CPV complexes were then incubated with Fabs (step 6), followed by a wash (step 7). (B) Fab14 binding to CPV/scFv-Fc14 complexes, with Fab14 concentrations ranging from 37.5 nM to 300 nM. (C) FabE binding to CPV/scFv-FcE complexes, with FabE concentrations ranging from 18.75 nM to 150 nM. (D) A sample graph where CPV-2 capsids are attached to Ni-NTA probes via soluble TfR (steps 1 to 5), followed by incubation of TfR/CPV complexes with purified TfRs with cleaved His tags (step 6) and a wash (step 7). (E) Black-backed jackal TfR binding to CPV/bbjTfR complexes, with concentrations of purified, His tag-cleaved TfR ranging from 200 nM to 1,600 nM. (F) Feline TfR binding to felTfR/CPV complexes, with concentrations of purified, His tag-cleaved TfR ranging from 200 nM to 1,600 nM.
We also examined the binding of the different TfRs to CPV-2 capsids. In this study, feline or black-backed jackal TfRs were attached to Ni-NTA biosensors via a His tag, incubated with CPV-2 capsids, washed, and incubated with four concentrations of soluble TfR of the same type (with cleaved His tags) (Fig. 4D). Additional black-backed jackal TfRs bound efficiently to tethered capsids (Fig. 4E), yielding a binding affinity of 5.32 × 10−7 M (ka of 2.16 × 104 1/M · s; kd of 1.15 × 10−2 1/s). However, the additional feline TfRs showed very low levels of binding (Fig. 4F), even though the initial affinity of CPV for the His-tagged feline TfR on biosensors was high (Fig. 2B).
Competition for binding between antibodies or TfRs and antibodies.
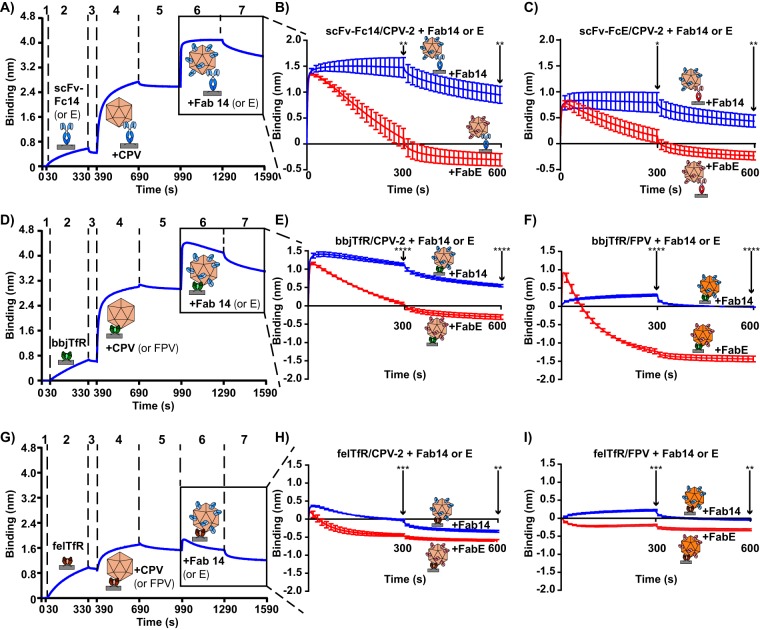
We used bio-layer interferometry to measure how Fab binding affected capsid attachment to either scFv-Fcs or TfRs. Capsids were attached to biosensors with scFv-Fc14, scFv-FcE, black-backed jackal TfR, or feline TfR and then incubated with a very high concentration (4,800 nM) of Fab14 or FabE. There was significant variation in the abilities of the two Fabs to compete CPV or FPV capsids off the biosensor when they were tethered through either scFv-Fc14, scFv-FcE, the black-backed jackal TfR, or the feline TfR (Fig. 5). The species of TfR holding capsids to biosensors (black-backed jackal or feline) and the type of parvovirus tethered to biosensors (CPV or FPV) also affected these results (Fig. 5E, F, H, and I). FabE was particularly efficient in competing capsids off all ligands used to attach it to the biosensor, while Fab14 showed only a mild effect on capsid-TfR binding (Fig. 5B to I). Fab14 also showed the lowest competitive effect and level of binding to the FPV capsid (Fig. 5F and I), as expected since it is largely specific for CPV capsids, compared to levels for FPV (45). Overall, the data strongly support the model where binding of the highly neutralizing FabE induces changes in the capsid that result in capsid detachment from different ligands, including TfRs and other antibodies.
FIG 5.
Competition between scFv-Fcs and Fabs or TfRs and Fabs for capsid binding. Capsids were attached to Ni-NTA probes or protein A probes with TfR or scFv-Fc, respectively. Attached capsids were then incubated with Fabs at high concentration (4,800 nM) to determine whether the Fabs could compete capsids off the ligand that bound them to the probe (scFv-Fc14, scFv-FcE, felTfR, or bbjTfR). (A) A sample graph where capsids are attached to protein A probes via an scFv-Fc (steps 1 to 5), incubated with Fabs (step 6), and washed (step 7). (B) Fab14 or FabE incubation with scFv-Fc14/CPV-2 complexes. (C) Fab14 or FabE incubation with scFv-FcE/CPV-2 complexes. (D) A sample graph where capsids are attached to Ni-NTA probes via the black-backed jackal TfR (steps 1 to 5), incubated with Fabs (step 6), and washed (step 7). (E) Fab14 or FabE incubated with bbjTfR/CPV-2 complexes. (F) Fab14 or FabE incubated with bbjTfR/FPV complexes. (G) A sample graph where capsids are attached to probes via the feline TfR (steps 1 to 5), incubated with Fabs (step 6), and washed (step 7). (H) Fab14 or FabE incubated with felTfR/CPV-2 complexes. (I) Fab14 or FabE incubated with felTfR/FPV complexes. Error bars show ± standard error. Statistics were calculated for the 300-s and 600-s time points using the Student's t test. *, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001.
TfR occupancy on capsids.
Saturating capsids with Fabs and/or TfRs also provided information about the level of occupancy of the different TfRs on capsids. TfR binding levels were calibrated by comparison with Fab binding (Fig. 5B), as Fab14 and FabE are known to fully occupy the capsids when Fab-capsid complexes were examined via cryoEM (46). In this assay, steric blocking by the tethering scFv-Fc and the proximity to the biosensor tip may reduce the number of available binding sites, so we conservatively estimated that around 50 Fabs would be bound to the capsid at saturating levels. We assumed that the 4,800 nM concentration of Fab14 resulted in complete saturation of available binding sites (∼50 sites) on scFv-Fc/CPV-2 complexes in Fig. 5B, and that the 1,600 nM incubation of black-backed jackal TfR with black-backed jackal TfR/CPV-2 complexes in Fig. 4E also resulted in saturation. Comparing the level of Fab14 or black-backed jackal TfR binding to CPV-2 to the amount of CPV-2 bound to biosensors allowed us to estimate that approximately 12 black-backed jackal TfRs bound to CPV-2, assuming that TfR dimers were complexed with two transferrin proteins, as has been observed by biochemical analysis and by cryo-electron microscopy of CPV complexed with the black-backed jackal TfR (H. Lee and S. L. Hafenstein, unpublished data). Using the same calculations and assuming that 1,600 nM feline TfR saturated felTfR/CPV-2 complexes in Fig. 4F, we estimated that approximately 1 to 2 feline TfRs bound to CPV-2.
Influence of initial capsid binding event on attachment of additional TfRs.
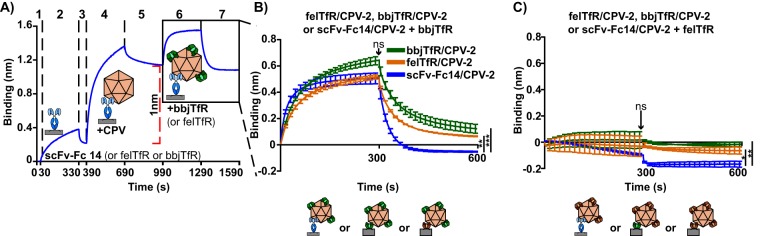
When felTfR/CPV-2, black-backed jackal TfR/CPV-2, or scFv-Fc14/CPV-2 complexes were incubated with soluble feline or black-backed jackal TfRs, we saw that the initial ligand binding event tethering CPV-2 capsids to biosensors affected subsequent attachment of soluble TfR, even though the same amount of capsid was loaded onto biosensors (Fig. 6). Black-backed jackal TfR/CPV-2 complexes bound the highest levels of black-backed jackal or feline TfR, with felTfR/CPV-2 complexes showing slightly lower levels of binding (Fig. 6B and C). However, while scFv-Fc14/CPV-2 complexes bound black-backed jackal TfR to levels similar to those of felTfR/CPV-2 complexes during the association phase, there was a significantly greater drop-off of black-backed jackal TfR during the disassociation phase (Fig. 6B). Significantly higher drop-off during the disassociation phase also occurred with the feline TfR and scFv-Fc14/CPV-2 complexes (Fig. 6C).
FIG 6.
Effects of the initial CPV-ligand interaction on subsequent bbjTfR or felTfR binding. bbjTfR, felTfR, or scFv-Fc14 was used to attach CPV-2 capsids to probes, so that the total amount of CPV attached to the probe, regardless of the ligand attaching it, was equal to 1 nm after washing (steps 1 to 5). Bound capsids were then incubated with 1,600 nM bbjTfR or felTfR with cleaved His tags (step 6) and washed (step 7). (A) A sample binding curve, showing capsids loaded to 1 nm of binding. (B) Incubation of bbjTfR (with cleaved His tag) with bbjTfR/CPV-2, felTfR/CPV-2, or scFv-Fc14/CPV-2 complexes. (C) Incubation of felTfR (with cleaved His tag) with bbjTfR/CPV-2, felTfR/CPV-2, or scFv-Fc14/CPV-2 complexes. Error bars show ± standard error. Statistics were calculated for the 300-s and 600-s time points using ANOVA. *, P < 0.05; **, P < 0.01, ***, P < 0.001.
Capsid residues influencing the TfR binding interaction.
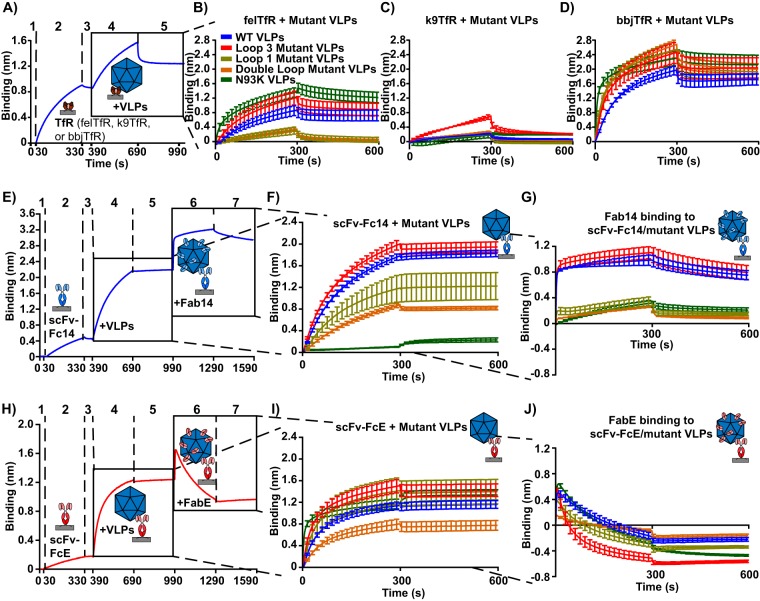
To examine the capsid surface for structures involved in the interactions with the TfRs and antibody 14 or E, we mutated groups of surface-exposed residues in the VP2 structure, four residues in loop 1 (VP2 residues N85A, M87A, V92A, and N93A), six residues in loop 3 (VP2 residues S293A, Q296A, S297A, T301A, N302A, and D305A), or all 10 mutations together (Fig. 1J to N and Table 1). When CPV-2-derived VLPs containing the four surface residue changes within loop 1 were used in binding studies, we saw greatly reduced binding to the feline TfR (Fig. 7B). In contrast, CPV-2-derived VLPs with six substitutions in surface loop 3 showed similar or even increased binding to the feline TfR compared to that of wild-type CPV-2 VLPs (Fig. 7B). Loop 3 mutant VLPs also bound the canine TfR to higher levels than any of the other VLPs (Fig. 7C), and all VLPs bound strongly to the black-backed jackal TfR (Fig. 7D). We did not test the loop 3 mutant for infectivity due to concerns about a possible host range expansion of that virus.
FIG 7.
Binding kinetics of mutant VLPs with TfRs, scFv-Fcs, and Fabs. Mutant VLPs were incubated with feline TfR, canine TfR, black-backed jackal TfR, scFv-Fc14, scFv-FcE, Fab14, and FabE to determine how capsid surface loops affect ligand binding interactions. (A) A sample graph where Ni-NTA probes are washed (step 1), incubated with TfR (step 2), washed (step 3), incubated with VLPs (step 4), and washed again (step 5). (B to D) Results of mutant VLPs binding to the feline (B), canine (C), and black-backed jackal (D) TfRs. (E) A sample graph where VLPs are attached to protein A probes via scFv-Fc14 (steps 1 to 5), incubated with Fab14 (4800 nM) (step 6), and washed (step 7). (F) The initial binding interaction between scFv-Fc14 and VLPs (steps 4 to 5). (G) Binding of 4,800 nM Fab14 to scFv-Fc14/VLP complexes (steps 6 to 7). (H to J) As in panels E to G, but with scFv-FcE replacing scFv-Fc14 and FabE replacing Fab14. Error bars show ± standard error.
scFv-Fc14 and Fab14 bound to all of the mutant capsids except for those with substitutions within its footprint, which included loop 1 mutant VLPs, double loop mutant VLPs, and Asn93Lys VLPs (Fig. 7F and G). In this experiment, Fab14 binding was more affected by these substitutions than scFv-Fc binding, likely due to the monomeric binding of the Fab forms (Fig. 7G). The loop 3 mutant VLPs bound to scFv-FcE as well as wild-type VLPs, even though the antibody E binding footprint overlapped that group of mutations (Fig. 7I) (46, 47), and FabE competed all the different VLPs off scFv-FcE (Fig. 7J).
DISCUSSION
These studies further explain the underlying interactions between CPV capsids and different host TfRs that resulted in the emergence and pandemic spread of CPV in dogs during 1978, as well as the subsequent spread and evolution of that virus within dogs and other hosts during the last 40 years. These specific capsid-TfR contacts and interactions are clearly key to the process of cell infection, as they control the virus's natural host range and also overlap the antibody binding sites on capsids, so that many antibody and receptor interactions are affected by the same mutations (13, 39, 41, 47, 63).
Capsid-TfR interactions.
These studies reveal new molecular details about the interactions between closely related parvovirus capsids and the receptors from susceptible hosts, and in particular they show the dynamic nature of these interactions (Fig. 8). They confirm that there is no direct connection between the affinity of capsid binding to the TfR and the success of cell infection. This was most clearly seen in a comparison of the feline and canine TfRs, which mediate CPV-2a infection to similar levels (Fig. 3D and E), even though the canine TfR shows extremely low binding affinity (Fig. 2B and D). This effect has also been previously observed in studies of a CPV-2 mutant with lysine substitutions of VP2 residues 299 and 300, which bound the feline TfR but did not infect cells (41).
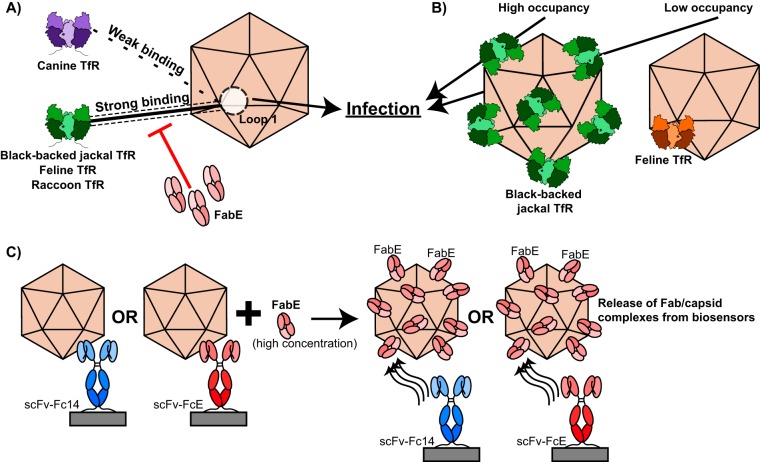
FIG 8.
Summary of major findings. (A) Both strong and weak binding interactions between the capsid and the TfR at capsid surface loop 1 result in infection. Antibody binding can disrupt the capsid-TfR binding interaction and block infection. (B) Both the black-backed jackal TfR, which binds to the capsid at high occupancy, and the feline TfR, which binds to the capsid at low occupancy, facilitate successful infection. (C) When high concentrations of FabE are incubated with scFv-Fc/capsid complexes, FabE competes with scFv-Fcs for capsid binding and FabE/capsid complexes are released from biosensors.
We also revealed more details about the binding site on the capsid and the ways in which viral residues control both the specific interactions with the TfR and the infectious process. Comparing the naturally occurring viruses showed that the capsids of CPV-2 (the first virus strain that spread worldwide in dogs in 1978 [27]) bound to TfR with higher affinity than the CPV-2a capsids, which had substitutions of VP2 residues 87, 300, and 305 on the capsid surface (27, 29, 64). CPV-2a arose naturally during 1979 and completely replaced CPV-2 within dogs worldwide by the end of 1980, again showing that the lower receptor binding affinity was widely selected in nature. We were surprised to find that CPV-2 capsids with 6 mutations in VP2 loop 3 bound the feline and other TfRs to similar or greater levels than wild-type VLPs (Fig. 7). This capsid surface loop was previously implicated in TfR binding because growth in different hosts or host cells may readily select for changes in loop 3 residues (including VP2 residues 297, 299, 300, 301, and 302) (27–29, 33, 37, 39). However, the strong TfR binding of the loop 3 mutant VLP suggests that most of loop 3 is not directly involved in TfR binding but may instead sterically interfere with TfR binding or control the structural transitions required for capsid infectivity. In contrast, loop 1 appears to play a direct role in TfR binding, as mutating 4 residues in that loop greatly reduced attachment, as has been suggested by previous structural and mutational studies and by its role in controlling canine host range (13, 31, 65).
The occupancy of TfR on the capsid was another feature of the capsid-receptor interaction revealed here. It was previously reported that CPV capsids engage fewer than five feline TfRs when they attach to the cell surface (43), and incubating capsids with soluble feline TfR ectodomains prepared from insect cells showed binding of fewer than 5 TfRs on average (44). The binding of fewer than 5 feline TfRs per capsid was confirmed using bio-layer interferometry. However, we also showed that receptor occupancy was controlled at least in part by the receptor form tested, as many more black-backed jackal TfRs bound per CPV capsid tethered to biosensors (Fig. 4). Modeling the occupancy of the TfR on the capsid indicates the maximum is 12 or 24 ectodomain dimers, depending on how the TfR binds to the capsid, due to the large size of the TfR and steric interference between adjacent binding sites. In our study, some binding sites on the capsid would have been blocked by the tethering TfR or scFv-Fc and biosensor, so that the black-backed jackal TfR may be occupying close to the maximum number of sites on the bound capsid. Possible explanations for the low levels of binding of the feline TfR include a limited number of preexisting binding sites on the capsid or that binding of the feline TfR to one or two positions changes the capsid to prevent the binding of additional TfRs. Here, we show that feline TfR binding may block the attachment of at least some additional TfRs, as fewer black-backed jackal TfRs associated with the capsids that were attached to biosensors via the feline TfR compared to capsids that were attached via the black-backed jackal TfR (Fig. 6).
Antibody binding and neutralization.
We also examined potential mechanisms of neutralization by antibodies, using Fabs derived from whole IgGs and cloned versions expressed as scFv-Fcs. Both antibodies we tested bound to similar high affinities, as expected, with both high ka and low kd rates, and capsids remained attached to scFv-Fcs with little or no detectable release over 5 min of washing (Fig. 4). However, FabE efficiently competed with scFv-Fcs for capsid binding, resulting in release of FabE/capsid complexes from biosensors and indicating that FabE was actively changing the capsid to cause release from the scFv-Fc tethering it to the biosensor. Similarly, both Fabs were able to compete the capsids off the feline and black-backed jackal TfRs, suggesting that receptor detachment is a mechanism of virus neutralization.
Summary.
These results provide a clearer view of the complex and dynamic processes of receptor and antibody binding to these superficially simple capsids. In different combinations, the variability of capsid interactions with receptors from different hosts shows that there is variation in the affinity, occupancy, and infection-mediating interactions, or in all of those at once. This highlights the finely tuned processes that mediate efficient infection and host range variability. For these viruses, variations in these processes control the natural host range, including the changes in the capsid that allowed functional canine TfR binding, which resulted in infection of dog cells and the emergence of CPV as a pandemic virus that continues to circulate today. These types of refined virus-receptor interactions are likely the rule rather than the exception, and a better understanding of the rules that apply in general and in particular cases will allow us to both develop antiviral strategies and better anticipate the emergence of viruses with new host ranges or pathogenic properties mediated by host receptors.
MATERIALS AND METHODS
Cells.
Norden Laboratory feline kidney (NLFK) cells (Norden Laboratories) were derived as a single-cell clone of the Crandell Rees feline kidney (CRFK) cell line (66) and were received directly from Norden Laboratories in 1980. TRVb cells (obtained from the laboratory of Timothy McGraw) are Chinese hamster ovary (CHO) cells without endogenous TfR (67). BHK (baby hamster kidney) cells (obtained from the laboratory of Anne Mason), Sf9 (Spodoptera frugiperda) cells (Invitrogen), and High Five (Trichoplusia ni) cells (clone BTI-TN-551-4; Boyce Thompson Institute) were also used in this study. NLFK cells were used to prepare infectious viruses and capsids and were grown in 1:1 McCoy’s/Leibovitz’s L15 medium (Corning) with 5% fetal calf serum (FCS). TRVb cells were grown in Ham's F12 medium (Corning) with 5% FCS. TRVb cells were transfected with pcDNA3.1(−)-derived plasmids containing cDNAs of the feline (Felis catus), canine (Canis lupus familiaris), raccoon (Procyon lotor), or black-backed jackal (Canis mesomelas) TfR using Transit-X2 (Mirus) according to the manufacturer's protocol. Cells were selected for plasmid transfection and TfR expression with G418 (Corning) and were used to measure infectivity of viruses on different TfRs expressed in the same cell background.
BHK cells were used for production of soluble TfR (described below) and were grown in Dulbecco's modified Eagle's medium (DMEM) and F-12 (Lonza) with 10% FCS. Sf9 and High Five cells were used for growth of baculovirus stocks and for production of proteins from baculoviruses, respectively. Sf9 cells were grown in Grace's supplemented insect medium (Gibco) with 10% FCS at 28°C, while High Five cells were grown in Express5 serum-free medium (Gibco) at 28°C.
Quantification of TfR expression on transfected TRVb cells.
TRVb cells transfected to express TfRs from different species were fixed with 4% paraformaldehyde, permeabilized with phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin and 0.5% Triton X-100, and stained with a mouse anti-TfR cytoplasmic tail antibody (QB213080; Life Technologies) and a goat anti-mouse Alexa 488-conjugated antibody (A11029; Life Technologies). Cell staining was then quantified using a Guava EasyCyte plus flow cytometer (Millipore). Three biological replicates were performed, and the percentage of TfR-positive cells was quantified using FlowJo (FlowJo, LLC, v.7.6.5).
Viruses and virus-like particles.
CPV-2, A300D CPV-2, CPV-2a (containing all of the mutations described in references [29 and 68] as well as VP2 426Asp), and FPV were each produced from infectious plasmid clones (69, 70). Viruses were either left in tissue culture supernatant for infectivity assays or purified for binding assays using standard methods involving banding on sucrose gradients to separate the empty and full (DNA-containing) capsids (30, 42). Titers of virus in supernatant were determined via TCID50 assays on NLFK cells and TRVb cells expressing the feline TfR.
For testing infectivity on TRVb cells, TRVb cells stably expressing the feline, canine, raccoon, or black-backed TfR were seeded at 2 × 104 cells/cm2 in 96-well plates (0.32 cm2 per well). Two hundred TCID50 units (measured on TRVb cells expressing the feline TfR) of CPV-2, FPV, A300D CPV-2, or CPV-2a were added to each TfR-expressing TRVb cell line. After inoculation, cells were incubated for 2 days at 37°C, when cells were fixed and stained with an anti-NS1 monoclonal antibody that recognizes an epitope that is identical on all four parvovirus strains tested (71). Viral growth was determined by counting the number of infected cells per well. Three independent biological replicates were performed, and the results were analyzed via analysis of variance (ANOVA).
Virus-like particles (VLPs) were prepared by expression of CPV-2 VP2 from baculovirus vectors. Mutant VLPs that contained groups of substitutions within capsid surface loops 1 and 3, which have been associated with the control of TfR binding (13, 31, 33, 37, 65), were also prepared (Fig. 1J to N). One mutant included six substitutions in VP2 loop 3 (Ser293Ala, Gln296Ala, Ser297Ala, Thr301Ala, Asn302Ala, and Asp305Ala) (Fig. 1L), another included four substitutions in the surface-exposed region of VP2 loop 1 (Asn85Ala, Met87Ala, Val92Ala, and Asn93Ala) (Fig. 1M), and a third variant contained all 10 surface substitutions in both loops 1 and 3. Wild-type VP2, Asn93Lys VP2, and the three mutant forms were prepared as VLPs by expression of VP2 in High Five cells and purified as described previously (41).
Expression and purification of TfR ectodomains.
A plasmid clone containing the soluble human TfR ectodomain (residues 121 to 760 [the C terminus]) fused to a 6×His tag on its N terminus was obtained from Ann Mason, University of Vermont (72) (Fig. 1B). The plasmid was stably transfected into BHK cells, and the TfR ectodomain was secreted into the culture medium. Equivalent clones that expressed the feline, canine, black-backed jackal, and raccoon TfR ectodomains were produced after cloning those sequences in place of the human TfR. Plasmids were transfected into BHK cells using Transit-X2 (Mirus) according to the manufacturer's protocol, and positive cells were selected with 500 μg/ml methotrexate. Cells were incubated with Pro-293a chemically defined medium (Lonza) with 1 mM butyric acid and 2 mM l-glutamine in order to produce protein. Supernatants were collected after 2 to 3 days of culture and stored at 4°C. TfRs were purified by incubating supernatants with Ni-NTA beads (ThermoFisher) for at least 1 h and then washing them three times with 25 mM imidazole. Bound TfR was cleaved on Ni-NTA beads overnight with Factor Xa protease (New England BioLabs) according to the manufacturer's protocol. Protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce).
Preparation of scFv-Fcs and Fabs.
The heavy and light variable domains of two previously characterized monoclonal antibodies, a mouse IgG-2a anti-CPV antibody, Mab14 (73), and a rat IgG-2b anti-FPV antibody, MabE (45), were joined by a linker sequence of three repeats of Gly-Gly-Gly-Ser to prepare an scFv. The baculovirus gp68 signal sequence was added to the N terminus, the scFv was joined through its C terminus to the Fc domain of human IgG1 with a flexible linker containing a thrombin cleavage site, and a 6×His tag was added at the C terminus (Fig. 1D). Clones were expressed in High Five cells using the Bac-to-Bac expression system (Invitrogen) according to the manufacturer's protocol, and protein was collected after 2 to 3 days of incubation of baculovirus with High Five cells at 28°C. scFv-Fc was isolated from supernatant with protein G affinity chromatography, eluted at pH 3.0, and immediately neutralized with Tris-HCl to pH 7.0. Eluted fractions were pooled and buffer exchanged into PBS with a 10-kDa Amicon Ultra centrifugal filter (Millipore).
Fabs were produced by digesting purified MabE or Mab14 with immobilized papain beads (ThermoFisher) according to the manufacturer's instructions. The antibody constant region (Fc) was separated from antibody 14 Fabs by incubating cleaved Mab14 with protein A CL-4B Sepharose beads (Pharmacia Biotech) in 50 mM Tris, pH 7.0. The MabE Fc was removed by running cleaved MabE through a DEAE-Sephadex A25 (Sigma) column in 0.01 M phosphate buffer, pH 7.8, and collecting Fabs in the column flowthrough. Incompletely cleaved proteins were removed via size exclusion chromatography on a Sephadex G100 (Pharmacia) column in PBS. Fabs were then concentrated using an Amicon 10-kDa centrifugal filter (Millipore).
Determination of protein purity and VLP stability.
Protein purity for TfRs, scFv-Fcs, Fabs, VLPs, and parvovirus empty capsids was determined by silver staining samples run on 10% SDS-PAGE gels with a Pierce silver stain kit (Thermo Scientific) according to the manufacturer's protocol. Stability of mutant VLPs was determined by differential scanning fluorimetry of purified VLPs, as previously described (41). Three replicates were performed for each sample except for the N93K VLPs, where limited sample quantity allowed only one measurement.
Measurement of TfR and scFv binding using bio-layer interferometry.
To examine the specific interactions involved in antibody and TfR binding, we bound the capsids and ligands in different combinations, as diagrammed in Fig. 1F to J and in figures throughout. Binding kinetics and dynamics were measured using a BLItz bio-layer interferometer (ForteBio). Ni-NTA biosensors were used to bind TfRs, and protein A biosensors were used to bind scFv-Fcs. To determine the affinities of capsids and TfRs or capsids and scFv-Fcs, biosensors were first blocked and hydrated in kinetics buffer (PBS with 0.02% ovalbumin and 0.02% Tween 20). Basic binding experiments to determine capsid-TfR or capsid–scFv-Fc affinity (Fig. 1F and H) were performed with the following protocol: 30-s baseline in kinetics buffer, 300 s of loading TfR or scFv-Fc, 60-s wash in kinetics buffer, 300 s of association with capsids (240 μg/ml) in PBS, and 300 s of disassociation in kinetics buffer. TfRs in supernatant (with uncleaved His tags) were loaded onto Ni-NTA biosensors to a consistent binding level of 0.8 nm, while purified scFv-Fcs were loaded to approximately 0.4 nm (0.83 μg/ml of protein).
Comparison of TfR affinity between different CPV strains or CPV-2 VLPs.
We compared the relative binding affinities of feline, canine, raccoon, black-backed jackal, and human TfRs by loading TfR in supernatant to 0.8 nm of binding, as described above, followed by incubation with CPV-2, CPV-2a, FPV, or A300D CPV-2 capsids or CPV-2 VLPs at 240 μg/ml. Three independent replicates were performed for each TfR-capsid combination.
Binding of CPV-2 to transfected TRVb cells.
Wild-type TRVb cells and TRVb cells transfected to express different TfRs were seeded on glass coverslips at a density of 2 × 104 cells/cm2 and grown overnight. Cells were incubated in DMEM with 0.1% bovine serum albumin (BSA) for 30 min at 37°C and then washed with PBS and fixed in 4% paraformaldehyde for 10 min. Cells were then incubated with 10 μg/ml CPV-2 empty capsids diluted in DMEM with 0.1% BSA for 1 h at 37°C, stained with anti-capsid monoclonal antibody 8, which does not detach capsids from fixed cells (14, 46, 73), and mounted on slides using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Additional coverslips were permeabilized in PBS with 0.5% BSA and 0.5% Triton X-100 after fixation and stained with a mouse anti-TfR cytoplasmic tail antibody (QB213080; Life Technologies) and a goat anti-mouse Alexa 594-conjugated antibody (A11005; Life Technologies).
Determination of Fab-CPV and TfR-CPV affinity constants and approximation of number of TfRs bound per capsid.
Affinity constants for Fab-CPV and TfR-CPV (cleaved His tag) interactions were determined through multistep binding experiments (Fig. 1G and I), where CPV capsids were first immobilized on biosensors with either scFv-Fcs or TfRs, exactly as in the basic binding experiments described above and in Fig. 1G and I and 2A. These complexes then underwent two additional steps to give 1:1 capsid-ligand binding interactions: 300 s of incubation with Fabs in PBS or His tag-cleaved TfRs, followed by 300 s of disassociation in kinetics buffer. Four different concentrations of Fab or TfR were used to generate four graphs per combination, which were then fitted using 1:1 global fitting in the BLItz Pro software (ForteBio). Experiments were performed in duplicate, and one replicate was shown and analyzed.
The number of copies of TfR that bound to each capsid was approximated by comparing binding of TfR to TfR/CPV complexes with binding of Fab14 to scFv-Fc14/CPV complexes, under conditions where ligand binding was at or near saturation (1,600 nM TfR at 300 s [Fig. 4E and F] and 4,800 nM Fab14 at 300 s [Fig. 5B]). Levels of ligand binding (His tag-cleaved TfR or Fab14) were normalized relative to the loading of CPV onto biosensors and adjusted to account for the difference in molecular weight between the TfR and Fab14. We conservatively estimated that Fab14 saturated our system at 50 Fabs/capsid due to steric hindrance of scFv-Fc14 holding capsids to biosensors and multiplied 50 Fabs/capsid by the normalized and adjusted ratio of TfR versus Fab14 binding to capsids to make our estimate of TfR copies bound per capsid.
Competition between scFv-Fcs and Fabs or TfRs and Fabs for capsid binding.
To determine whether or not antibodies could affect capsid interactions with the TfR or other antibodies, we designed multistep binding experiments where capsids were attached to biosensors with one ligand and then incubated with either the same ligand or a different ligand. scFv-Fc14, scFv-FcE, feline TfR, or black-backed jackal TfR was used to attach CPV-2 capsids to biosensors as described above. After a 300-s wash step, complexes were incubated with either 4,800 nM FabE or Fab14 for 300 s, followed by an additional 300-s wash step. Three independent replicates were performed, and binding at 300 s and 600 s was analyzed via ANOVA.
Effects of the initial CPV-ligand binding interaction on subsequent TfR binding.
To examine whether the ligand holding CPV to biosensors affected subsequent binding of TfR, we bound scFc-Fc14, feline TfR, or black-backed jackal TfR to biosensors, as described above for basic binding experiments. The concentrations of these ligands were adjusted so that all three ligands bound 1 nm of CPV-2 capsids to biosensors (Fig. 6A). Ligand-CPV complexes were then incubated with either black-backed jackal TfR (cleaved His tag, 1,600 nM) or feline TfR (cleaved His tag, 1,600 nM) and washed in kinetics buffer for 300 s. Three independent replicates were performed, and binding at 300 s and 600 s was analyzed via ANOVA.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI092571 to C.R.P. and S.L.H. H.M.C. was supported by NSF-GRFP DGE-1650441.
The funders had no role in study design, data collection and interpretation, or decision to submit the work for publication.
REFERENCES
- 1.Bönsch C, Zuercher C, Lieby P, Kempf C, Ros C. 2010. The globoside receptor triggers structural changes in the B19 virus capsid that facilitate virus internalization. J Virol 84:11737–11746. doi: 10.1128/JVI.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. 2009. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure 17:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattentau QJ, Moore JP. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med 174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry (Mosc) 38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- 6.Li F. 2013. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res 100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohishi K, Suzuki R, Maeda T, Tsuda M, Abe E, Yoshida T, Endo Y, Okamura M, Nagamine T, Yamamoto H, Ueda M, Maruyama T. 2014. Recent host range expansion of canine distemper virus and variation in its receptor, the signaling lymphocyte activation molecule, in carnivores. J Wildl Dis 50:596–606. doi: 10.7589/2013-09-228. [DOI] [PubMed] [Google Scholar]
- 8.Xiong X, McCauley JW, Steinhauer DA. 2014. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr Top Microbiol Immunol 385:63–91. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez R, Paredes A, Brown DT. 2008. Sindbis virus conformational changes induced by a neutralizing anti-E1 monoclonal antibody. J Virol 82:5750–5760. doi: 10.1128/JVI.02673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. A cryo-electron microscopy study identifies the complete H16. V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol 89:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reading SA, Dimmock NJ. 2007. Neutralization of animal virus infectivity by antibody. Arch Virol 152:1047–1059. doi: 10.1007/s00705-006-0923-8. [DOI] [PubMed] [Google Scholar]
- 12.Wien MW, Filman DJ, Stura EA, Guillot S, Delpeyroux F, Crainic R, Hogle JM. 1995. Structure of the complex between the Fab fragment of a neutralizing antibody for type 1 poliovirus and its viral epitope. Nat Struct Biol 2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]
- 13.Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish CR. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J Virol 77:10099–10105. doi: 10.1128/JVI.77.18.10099-10105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palermo LM, Hafenstein SL, Parrish CR. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J Virol 80:8482–8492. doi: 10.1128/JVI.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo LM, Hueffer K, Parrish CR. 2003. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J Virol 77:8915–8923. doi: 10.1128/JVI.77.16.8915-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol 75:3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason AB, Byrne SL, Everse SJ, Roberts SE, Chasteen ND, Smith VC, MacGillivray RTA, Kandemir B, Bou-Abdallah F. 2009. A loop in the N-lobe of human serum transferrin is critical for binding to the transferrin receptor as revealed by mutagenesis, isothermal titration calorimetry, and epitope mapping. J Mol Recognit 22:521–529. doi: 10.1002/jmr.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trinder D, Olynyk JK, Sly WS, Morgan EH. 2002. Iron uptake from plasma transferrin by the duodenum is impaired in the Hfe knockout mouse. Proc Natl Acad Sci U S A 99:5622–5626. doi: 10.1073/pnas.082112299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, Parrish CR. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J Virol 74:1919–1930. doi: 10.1128/JVI.74.4.1919-1930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao N, Enns CA. 2012. Iron transport machinery of human cells: players and their interactions. Curr Top Membr 69:67–93. doi: 10.1016/B978-0-12-394390-3.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harbison CE, Lyi SM, Weichert WS, Parrish CR. 2009. Early steps in cell infection by parvoviruses: host-specific differences in cell receptor binding but similar endosomal trafficking. J Virol 83:10504–10514. doi: 10.1128/JVI.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett MJ, Lebrón JA, Bjorkman PJ. 2000. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. 2004. Structure of the human transferrin receptor-transferrin complex. Cell 116:565–576. doi: 10.1016/S0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 24.Goodman LB, Lyi SM, Johnson NC, Cifuente JO, Hafenstein SL, Parrish CR. 2010. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J Virol 84:4969–4978. doi: 10.1128/JVI.02623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 26.Truyen U, Gruenberg A, Chang SF, Obermaier B, Veijalainen P, Parrish CR. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J Virol 69:4702–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish CR, Have P, Foreyt WJ, Evermann JF, Senda M, Carmichael LE. 1988. The global spread and replacement of canine parvovirus strains. J Gen Virol 69(Part 5):1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- 28.Parrish CR, O'Connell PH, Evermann JF, Carmichael LE. 1985. Natural variation of canine parvovirus. Science 230:1046–1048. doi: 10.1126/science.4059921. [DOI] [PubMed] [Google Scholar]
- 29.Stucker KM, Pagan I, Cifuente JO, Kaelber JT, Lillie TD, Hafenstein S, Holmes EC, Parrish CR. 2012. The role of evolutionary intermediates in the host adaptation of canine parvovirus. J Virol 86:1514–1521. doi: 10.1128/JVI.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155–171. doi: 10.1002/prot.340160204. [DOI] [PubMed] [Google Scholar]
- 31.Chang SF, Sgro JY, Parrish CR. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol 66:6858–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelber JT, Demogines A, Harbison CE, Allison AB, Goodman LB, Ortega AN, Sawyer SL, Parrish CR. 2012. Evolutionary reconstructions of the transferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog 8:e1002666. doi: 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison AB, Organtini LJ, Zhang S, Hafenstein SL, Holmes EC, Parrish CR. 2016. Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J Virol 90:753–767. doi: 10.1128/JVI.02636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llamas-Saiz AL, Agbandje-McKenna M, Parker JS, Wahid AT, Parrish CR, Rossmann MG. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65–71. doi: 10.1006/viro.1996.0575. [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Zeng W, Zhang X, Li S. 2017. The genetic evolution of canine parvovirus–a new perspective. PLoS One 12:e0175035. doi: 10.1371/journal.pone.0175035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda C, Thompson G. 2016. Canine parvovirus: the worldwide occurrence of antigenic variants. J Gen Virol 97:2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 37.Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, Parrish CR. 2014. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog 10:e1004475. doi: 10.1371/journal.ppat.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maya L, Calleros L, Francia L, Hernández M, Iraola G, Panzera Y, Sosa K, Pérez R. 2013. Phylodynamics analysis of canine parvovirus in Uruguay: evidence of two successive invasions by different variants. Arch Virol 158:1133–1141. doi: 10.1007/s00705-012-1591-5. [DOI] [PubMed] [Google Scholar]
- 39.Allison AB, Harbison CE, Pagan I, Stucker KM, Kaelber JT, Brown JD, Ruder MG, Keel MK, Dubovi EJ, Holmes EC, Parrish CR. 2012. Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus. J Virol 86:865–872. doi: 10.1128/JVI.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinel A, Parrish CR, Bloom ME, Truyen U. 2001. Parvovirus infections in wild carnivores. J Wildl Dis 37:594–607. doi: 10.7589/0090-3558-37.3.594. [DOI] [PubMed] [Google Scholar]
- 41.Callaway HM, Feng KH, Lee DW, Allison AB, Pinard M, McKenna R, Agbandje-McKenna M, Hafenstein S, Parrish CR. 2017. Parvovirus capsid structures required for infection: mutations controlling receptor recognition and protease cleavages. J Virol 91:e01871-16. doi: 10.1128/JVI.01871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson CDS, Minkkinen E, Bergkvist M, Hoelzer K, Fisher M, Bothner B, Parrish CR. 2008. Detecting small changes and additional peptides in the canine parvovirus capsid structure. J Virol 82:10397–10407. doi: 10.1128/JVI.00972-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cureton DK, Harbison CE, Cocucci E, Parrish CR, Kirchhausen T. 2012. Limited transferrin receptor clustering allows rapid diffusion of canine parvovirus into clathrin endocytic structures. J Virol 86:5330–5340. doi: 10.1128/JVI.07194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hafenstein S, Palermo LM, Kostyuchenko VA, Xiao C, Morais MC, Nelson CDS, Bowman VD, Battisti AJ, Chipman PR, Parrish CR, Rossmann MG. 2007. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc Natl Acad Sci U S A 104:6585–6589. doi: 10.1073/pnas.0701574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parrish CR, Carmichael LE. 1983. Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology 129:401–414. doi: 10.1016/0042-6822(83)90179-4. [DOI] [PubMed] [Google Scholar]
- 46.Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J Virol 83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Organtini LJ, Lee H, Iketani S, Huang K, Ashley RE, Makhov AM, Conway JF, Parrish CR, Hafenstein S. 2016. Near-atomic resolution structure of a highly neutralizing Fab bound to canine parvovirus. J Virol 90:9733–9742. doi: 10.1128/JVI.01112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson CDS, Palermo LM, Hafenstein SL, Parrish CR. 2007. Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology 361:283–293. doi: 10.1016/j.virol.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 50.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, Deng X, Wosen JE, Davulcu O, Yan Z, Engelhardt JF, Brown KE, Chapman MS, Qiu J, Carette JE. 5 July 2017. AAV serotypes have distinctive interactions with domains of the cellular receptor AAVR. J Virol doi: 10.1128/JVI.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava A. 2016. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol 21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.López-Bueno A, Rubio M-P, Bryant N, McKenna R, Agbandje-McKenna M, Almendral JM. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J Virol 80:1563–1573. doi: 10.1128/JVI.80.3.1563-1573.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pittman N, Misseldine A, Geilen L, Halder S, Smith JK, Kurian J, Chipman P, Janssen M, Mckenna R, Baker TS, D'Abramo A, Cotmore S, Tattersall P, Agbandje-McKenna M. 2017. Atomic resolution structure of the oncolytic parvovirus LuIII by electron microscopy and 3D image reconstruction. Viruses 9:E321. doi: 10.3390/v9110321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham J, Kwong JA, Albariño CG, Lu JG, Radoshitzky SR, Salazar-Bravo J, Farzan M, Spiropoulou CF, Choe H. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog 5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarute N, Ross SR. 2017. New World arenavirus biology. Annu Rev Virol 4:141–158. doi: 10.1146/annurev-virology-101416-042001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H, Shingler KL, Organtini LJ, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2016. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv 2:e1501929. doi: 10.1126/sciadv.1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazear E, Whitbeck JC, Ponce-de-Leon M, Cairns TM, Willis SH, Zuo Y, Krummenacher C, Cohen GH, Eisenberg RJ. 2012. Antibody-induced conformational changes in herpes simplex virus glycoprotein gD reveal new targets for virus neutralization. J Virol 86:1563–1576. doi: 10.1128/JVI.06480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hueffer K, Parker JSL, Weichert WS, Geisel RE, Sgro J-Y, Parrish CR. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol 77:1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hueffer K, Parrish CR. 2003. Parvovirus host range, cell tropism and evolution. Curr Opin Microbiol 6:392–398. doi: 10.1016/S1369-5274(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 62.Yuan W, Parrish CR. 2000. Comparison of two single-chain antibodies that neutralize canine parvovirus: analysis of an antibody-combining site and mechanisms of neutralization. Virology 269:471–480. doi: 10.1006/viro.2000.0230. [DOI] [PubMed] [Google Scholar]
- 63.Strassheim ML, Gruenberg A, Veijalainen P, Sgro JY, Parrish CR. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 64.Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol 65:6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Govindasamy L, Hueffer K, Parrish CR, Agbandje-McKenna M. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J Virol 77:12211–12221. doi: 10.1128/JVI.77.22.12211-12221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crandell RA, Fabricant CG, Nelson-Rees WA. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185. [DOI] [PubMed] [Google Scholar]
- 67.McGraw TE, Greenfield L, Maxfield FR. 1987. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J Cell Biol 105:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parrish CR, Aquadro CF, Carmichael LE. 1988. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink, and raccoon parvoviruses. Virology 166:293–307. [DOI] [PubMed] [Google Scholar]
- 69.Parker JS, Parrish CR. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol 71:9214–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parrish CR. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195–205. [DOI] [PubMed] [Google Scholar]
- 71.Yeung DE, Brown GW, Tam P, Russnak RH, Wilson G, Clark-Lewis I, Astell CR. 1991. Monoclonal antibodies to the major nonstructural nuclear protein of minute virus of mice. Virology 181:35–45. [DOI] [PubMed] [Google Scholar]
- 72.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RTA, Kaltashov IA, Mason AB. 2006. Effect of glycosylation on the function of a soluble, recombinant form of the transferrin receptor. Biochemistry (Mosc) 45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 73.Parrish CR, Carmichael LE, Antczak DF. 1982. Antigenic relationships between canine parvovirus type 2, feline panleukopenia virus and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch Virol 72:267–278. [DOI] [PubMed] [Google Scholar]