Abstract
Classical cadherins are a family of transmembrane proteins that mediate cell–cell adhesion at adherens junctions. A complex chain of cis- and trans-interactions between cadherin ectodomains establishes a cadherin adhesive cluster. A principal adhesive interaction in such clusters is an exchange of β strands between the first extracellular cadherin domains (EC1). The structure of cadherin adhesive clusters can be modified by other adherens junction proteins including additional transmembrane proteins, nectins and various intracellular proteins that directly or indirectly interact with the intracellular cadherin region. These interactions determine the dynamics and stability of cadherin adhesive structures.
5.1 Introduction
The assembly of the vast majority of multiprotein structures includes two distinct steps—nucleation and elongation. The latter step is often based on cooperative interactions between the structure’s subunits. The assembly process is followed by the reverse process of structure disassembly. The balance between these two opposite processes determines the size of the structure and its dynamics. It is highly likely that adherens junctions are not an exception and that the same principles underlie their homeostasis. Assembly of adherens junctions is likely to be initiated at specific sites of cell–cell contacts by a nucleation reaction, the nature of which remains to be determined. Recent advances in the field have shown that junction assembly is based on a set of cooperative trans and cis interactions between cadherin ectodomains. These binding reactions produce adhesive clusters in which cadherin molecules are arranged in specific linear arrays. These reactions of cadherin adhesive cluster self-assembly are, perhaps, the most ubiquitous and currently the best understood event in formation of adherens junctions. Importantly, these reactions are specific to vertebrate classic cadherins; invertebrate cadherins employ another, much less studied, set of extracellular interactions (Harrison et al. 2011).
It is not known how these adhesive clusters arrange themselves into mature adherens junctions. The junctions are not static: they continuously loose and gain cadherin. The reorganization of the adhesive clusters into the adherens junctions and their subsequent disassembly are, perhaps, regulated by diverse intracellular signaling pathways and the cytoskeleton. The complexity and redundancy of these intracellular mechanisms are likely key reasons for the morphological and structural pleomorphism of adherens junctions, which can be classified by a number of subtypes (zonulae adhaerentes, fasciae adhaerentes, puncta adhaerentia and many others, see Franke 2009). This diversity of adherens junctions reflects the varying requirements for cell–cell junction positioning, their strength and their dynamics in different types of cells.
In this chapter we will discuss some basic principles of cadherin–cadherin interactions resulting in the assembly and disassembly of adherens junctions.
5.2 From Cadherin Monomer to Cadherin Adhesive Clusters
Adherens junctions are formed as a result of two independent but coordinated cellular activities. The first one is cadherin adhesiveness, which, as we discuss below, is based on cis- and trans-interactions between cadherin molecules. The second one is the activity of actin cytoskeleton controlling protrusion-retracting cycles of plasma membranes of the contacting cells. In cell culture of MDCK epithelial cells, the initial junction contact is established by lamellipodia of two adjacent cells (McNeill et al. 1993; Adams and Nelson 1998). In mouse keratinocytes, the initial contact is made by interdigitating filopodia that form transient point contacts, which then zipper into a continuous mature junction (Vasioukhin and Fuchs 2001). In both cases, formation of adherens junctions coincides with extensive reorganization of the actin cytoskeleton. Inactivation of this reorganization by inhibitors of actin polymerization or actomyosin contractility affects junction formation. How these two activities are coordinated is one of the key unknown aspects of cadherin adhesion. Detailed understanding of cadherin adhesive interactions is essential for unraveling how the actin cytoskeleton regulates cadherin adhesion.
5.2.1 Cadherin Strand-Swapping is at the Core of Cadherin-Based Cell–Cell Adhesion
Different experimental approaches have compellingly shown that the cadherin adhesive site is localized to the EC1 domain. This was first indicated by domain shuffling experiments (Nose at al. 1990). This work showed that cells expressing an E/P-cadherin chimera with a P-cadherin-derived EC1 domain co-aggregate with P-cadherin-expressing cells. Similar experiments, but based on a co-immunoprecipitation assay, confirmed the key role of the EC1 domain in binding specificity (Klingelhöfer et al. 2000). These biochemical data have been corroborated by an electron microscopy study that showed intercadherin interactions through the EC1 domain (Tomschy et al. 1996). Two independent cryo-electron-tomography studies of desmosomes (He et al. 2003; Al-Amoudi et al. 2007) also documented the aminoterminal location of the adhesive sites. Finally, two recent FRET-based studies, which used elegantly designed cadherin molecules bearing fluorescent tags at different locations of the E-cadherin extracellular region, also showed that cadherin adhesion is established by the EC1 domain (Zhang et al. 2009; Kim et al. 2011).
Point mutagenesis of the EC1 domain in conjunction with co-immunoprecipitation and zonal sedimentation (Chitaev and Troyanovsky 1998; Tamura et al. 1998; Shan et al. 2000; Kitagawa et al. 2000; Laur et al. 2002) provided strong evidence that the cadherin adhesive site corresponds to the strand-swap dimer interface detected first in N-cadherin three-dimensional structure by Shapiro et al. (1995) and then in many other type I cadherins (see Posy et al. 2008). The involvement of this site in adhesion was further indicated by cross-linking experiments performed with engineered cadherin cysteine mutants (Troyanovsky et al. 2003; Harrison et al. 2005) and later documented by two independent FRET studies (Zhang et al. 2009; Kim et al. 2011).
Strand-swap cadherin dimerization is based on the exchange of N-terminal β strands of the EC1 domains (A* strand) between pairing cadherin molecules (Fig. 5.1, see below). Since the amino-terminal amino group stabilizes strand swapping by the salt bridge with Glu89, strand swapping is destroyed by the prodomain present in the unprocessed cadherin or by extra aminoterminal amino acids in recombinant cadherins (Troyanovsky 2005). Proteolytic removal of the prodomain is a key event activating cadherin adhesiveness (Häussinger et al. 2004).
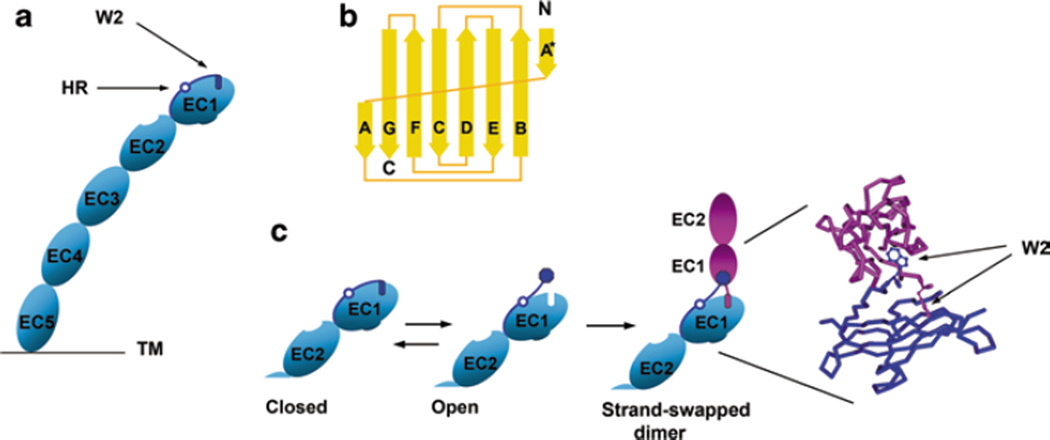
Fig. 5.1.
Cadherin dimerization using the strand-swap interface. a Schematic representation of the cadherin ectodomain. It consists of five homological cadherin-like domains (EC1–EC5). The A* and A strands of EC1 are dark blue. They are separated by the hinge region (open circle, HR). The most important residue of the A* strand is Trp2 (dark blue rectangle, W2). The cadherin molecule shown is in the closed conformation—its Trp2 residue is inserted into its own core. b Topology diagram of the classical cadherin EC1 domain. Note that the domain consists of seven β strands. The first strand is broken into two parts, strands A* and A. Strand A* forms a contact with strand B. This interaction can be intra- or inter-molecular. c Schematic representation of the strand-swapping process. Only EC1 and EC2 domains are shown. In the presence of calcium ions, the closed cadherin conformation is unstable and is in equilibrium with the open conformation in which Trp2 is exposed to solvent. Two cadherin molecules in open conformation produce a strand-swap cadherin dimer. The structural model of the strand-swapped dimer (only EC1 domains of both molecules are shown) is on the right. Note that the W2 residues of both molecules in the dimer are in nearly perpendicular planes
Structural analysis of the full-size cadherin ectodomain shows that the cadherin rod is bent, so that the long axes of the EC1 and EC5 domains are at a nearly right angle (Boggon et al. 2002; Harrison et al. 2011). Such curvature of the cadherin ectodomain presents EC1 domain in a way that it can swap its A* strand with the cadherin from the adjacent cell more efficiently than with cadherin located at the same plasma membrane. The curved structure of the cadherin ectodomain had been shown by EM in 1989, and it was proposed that such a conformation of cell adhesion receptors may represent a general evolutionary solution to the specific problems of cell–cell adhesion: the flexible bent may absorb the stress when adjoining cell surfaces are in motion (Becker et al. 1989).
Biochemical examination of cadherin–cadherin interactions in cell culture is completely consistent with the structural data: it has shown that strand swapping results in the formation of both lateral (or cis) and adhesive (or trans) cadherin dimers on the cell surface (Chitaev and Troyanovsky 1998; Harrison et al. 2005). Importantly, from structural stand-point, these lateral and adhesive dimers are the same: the only difference between them is that cadherins in the dimers originate either from the same or from opposite cell surfaces. Inactivation of calcium-binding interface or placing cells in low-calcium media attenuates trans, but has no effect on cis strand-swapping (Klingelhöfer et al. 2002). Apparently, the loss of correct ectodomain curvature impedes trans and promotes cis dimer formation. As we will discuss below, it is not quite clear how strand-swapping proceeds at low calcium. It is possible that some amount of cis strand-swap dimers can be assembled even at high calcium, but no available data suggests that these strand-swap cis dimers play any role in adhesion.
In conclusion, adherens junction homeostasis is a process of strand-swap dimer formation, clustering of these dimers, and stabilization and disintegration of the resulting clusters. These subjects will be discussed below. In addition, the formation of strand-swap dimers and concomitant processes should trigger signaling effects informing the cells about cell–cell contact formation. These outside-in signaling pathways make up an important but weakly explored area of cadherin adhesion that lies outside the focus of our review.
5.2.2 Unique Features of Cadherin Strand Swapping
Crystallographic, biophysical, and computational studies provide a clear understanding of the cadherin strand-swap process (Shapiro et al. 1995; Posy et al. 2008; Vendome et al. 2011; Vunnam and Pedigo 2011a). A key player in this binding reaction is the amino-terminal β strand (the A*/A strand) of the EC1 domain. This strand is followed by the residue Glu11 that anchors this strand to the EC1-EC2 interface through calcium ions. The A*/A strand contacts B and G strands of the seven β strands that constitute the core of the EC1 domain. A conserved N-terminal segment of the A*/A strand (the A* strand), which comprises residues 1–3, including Trp2, forms β-sheet hydrogen bonds with the B strand. In addition, the A* strand is linked to the rest of the molecule by a salt bridge between the N-terminal amino-group and a conserved Glu89. Trp2 is inserted in the core of the EC1 domain where it forms multiple hydrophobic bonds. In monomeric cadherin all of these contacts are intramolecular. Upon strand-swapping, the A* strand is swapped to another EC1 domain with which it forms exactly the same contacts (Fig. 5.1).
The second segment of the A*/A strand (residues 7–10, the A strand) is immobile; it is locked in place by the hydrogen bonds with the G strand. The mobile A* and immobile A strands are separated by a three-residue-long hinge region which, with few exceptions, contains two consecutive Pro residues in positions 5 and 6. This region does not make any hydrogen bonds with either strand (Vendome et al. 2011).
The two Pro residues conformationally strain the A strand between Trp2 and Glu11 (Vendome et al. 2011; Vunnam and Pedigo 2011b). Since in the presence of calcium ions, the A strand is tightly fixed to the rest of the EC1 domain, the strain can be relieved only by releasing Trp2 from its pocket. Therefore, the strain imposed by Pro5/Pro6 residues prevents the stable anchorage of Trp2 to its own EC1. Once the A* strand is relocated to another EC1, the resulting intermolecular contact is much more stable than the intramolecular one because the strain is released. Specific mutations that release the strain in the cadherin monomer, thereby stabilizing the A* strand anchorage to its own protomer, significantly reduce the affinity of strand swap binding. Thus, strand swapping is based on the instability of the A* strand that is imposed by the A strand and Ca2+-binding.
This mechanism of cadherin strand-swapping has two important consequences for the assembly of adherens junctions. First, strand-swapping is a relatively slow binding process and, therefore, depends on the duration of the cadherin–cadherin encounter. Second, extracellular conditions (like temperature or ion concentrations) or interactions with other proteins that increase the A* strand instability can facilitate the strand swapping.
Regardless of the large binding interface, the strand-swap dimers are unstable. For example, the KD of E-cadherin strand-swap dimerization is about 100 µM (Harrison et al. 2010). It suggests that lifetime of the dimers should be in the millisecond range. It has been originally proposed that the instability of strand swap dimers is based on their competition with intramolecular anchorage of the A* strand (Chen et al. 2005). However, more recent experiments clearly show that the main reason for strand-swap dimer instability is the competition with another type of cadherin dimer, the X-dimer. The inability to produce X-dimers increases the dimer’s lifetime almost indefinitely (Harrison et al. 2011; Vunnan et al. 2011). As we discuss below, the X dimer requirement for the disassembly of strand-swap dimers is a very important feature of adherens junctions.
5.2.3 Cadherin X-Dimerization Maintains Strand-Swap Dimer Dynamics
One of the remarkable features of strand-swap dimers is that despite their low affinity, they are detectable by a co-immunoprecipitation assay, which typically requires much stronger interactions. Recent examinations of the strand-swap dimerization kinetics provided a clue in understanding this obvious paradox.
In addition to strand-swap dimers, another type of cadherin dimer has been reported for two-domain (EC1–EC2) E-cadherin fragments (Nagar et al. 1996; Pertz et al. 1999). The paired molecules in this dimer contact each other via interdomain calcium-binding interfaces leading to X-shaped arrangement of two molecules. Initially, this “X” mode of dimerization has been regarded as a crystal-packing artifact (Häussinger et al. 2004) since cadherin forms such dimers only upon blocking its natural amino-terminus by an N-terminal extension. However, recently obtained data unraveled the important functional significance of the cadherin X-dimer.
It was found that this extremely unstable dimer (KD ~ 900 µM) serves as a kinetically important intermediate in strand-swap dimerization (Harrison et al. 2010; Vunnam et al. 2011). Cadherin bearing a compromised X-dimer interface exhibits a slowly exchanging monomer-dimer equilibrium: monomers have very slow kinetics of strand-swap association but, once formed, dimers have extremely slow kinetics of dissociation. These experiments definitively showed that the X-dimer represents an initial encounter complex in a strand-swap binding reaction, the requirements for which had also been proposed based on the results of single molecule tracking experiments (Sivasankar et al. 2009), and also revealed a role in disassembly.
Our examination of X-dimer mutants expressed in A-431 cells suggests, however, that X-dimerization might not be so essential in cadherin strand-swapping in real cell–cell junctions (Hong et al. 2011). We have proposed that two factors enhance the production of strand-swap dimers in living cells thereby lifting the X-dimer requirement. The first factor is cadherin “presentation”: in cell–cell junctions two encountering EC1 domains may be presented such that they are set for swapping. The second factor is a slow diffusion of cadherin molecules on the cell surface: each cadherin–cadherin encounter has a long enough duration to allow two A* strands to swap. Importantly, the experiments with X-dimer mutants clearly showed that strand-swap adhesive bond cannot be disassembled without its reconfiguration into X-dimer. This observation suggests that in order to disassemble adherens junctions, cadherin has to change its adhesive bond from a strand-swap to an “X” configuration. How this strand-swap-to-X-dimer transition works and whether cells can regulate this transition is an exciting avenue for future research.
The X-dimer requirement for strand-swap dimer dissociation changes our understanding of adherens junction disassembly in calcium-switch assays, which are widely used in cadherin adhesion studies. Since X-dimer formation requires the calcium-binding interface, the disruption of this interface by EDTA or other calcium chelators locks cadherin into the strand-swap configuration (Harrison et al. 2010; Vunnam et al. 2011). The strand-swap trans dimers may still dissociate because cell rounding triggered by calcium switch can physically disrupt the strand-swap trans bond. In contrast, strand-swap cis bonds are stabilized. This explains the fast accumulation of cadherin cis dimers in low calcium conditions. Furthermore, the inability of strand-swap dimers to dissociate explains another inconsistency in the field—the stability of strand-swapped dimers in co-immunoprecipitation assays. Indeed, the lysis buffers used for these experiments (Chitaev and Troyanovsky 1998; Shan et al. 2000; Ozawa 2002; Troyanovsky et al. 2007) typically contain EDTA or other calcium chelators to prevent cadherin proteolysis. The absence of calcium ions would lock cadherin into the strand-swap dimer conformation and allow dimer detection.
5.2.4 A Specific Form of Cis Interaction Reinforces and Clusters Strand-Swap Dimers
Theoretical studies show that at a KD of about 100 µM, cadherin cannot self-assemble adhesive clusters (Kusumi et al. 1999; Wu et al. 2010): at such low affinity, some specific intracellular mechanisms have to assist cadherin recruitment into the adhesive clusters. However, live-imaging experiments with the tailless cadherin mutant clearly showed that the extracellular cadherin region alone can produce cadherin clusters (Hong et al. 2010). In order to do so, the cadherin extracellular region has to participate in some type of cis interactions that stabilizes strand-swapping and promotes clustering (Wu et al. 2010).
It had long been proposed that cadherin forms cis dimers and that the cis dimers are essential for cadherin adhesion. However, neither biochemical approaches—including cross-linking or co-immunoprecipitation assays (Troyanovsky 2005)—nor a FRET study of the recombinant cadherin ectodomains (Zhang et al. 2009), have presented compelling evidence for cadherin cis dimerization. The only cis cadherin dimers that have been detected are the strand-swap lateral dimers. But these dimers, especially prominent in low calcium conditions, apparently play no role in adhesion (Ozawa 2002).
Current data suggests that precursory cadherin cis dimers, if they do form, are very weak and transient. They may be maintained through the cadherin transmembrane domain (Huber et al. 1999) or through unknown intracellular interactions. Such transient cis dimers may be important for increasing local cadherin density. Obviously, more work remains to be done to identify such transient forms of cadherin cis dimerization and to assess their roles in adhesion.
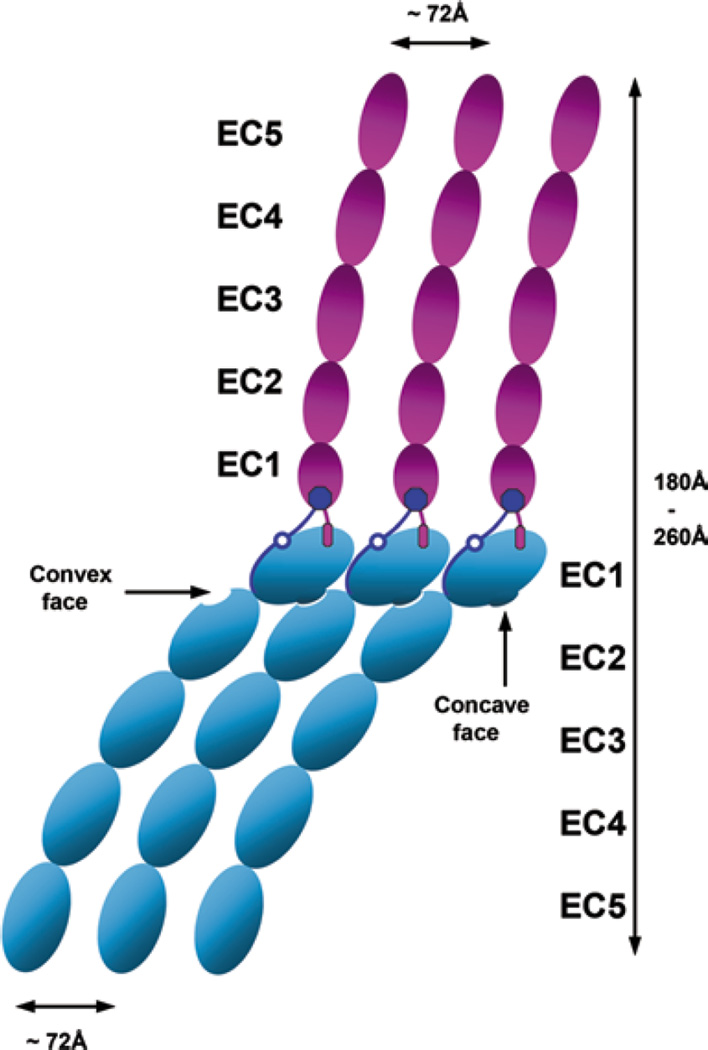
While the quest for stable, precursory cadherin cis dimers has not yet produced any definitive results, structural analysis of the crystal packing interactions in the E-, N-, and C-cadherin crystals has revealed that strand-swapped cadherin trans dimers do form cis contacts (Boggon et al. 2002; Harrison et al. 2011). This cis interface comprises a nonsymmetrical interaction between the concave face of the EC1 domain of one molecule and the convex face of the EC2 domain of the partner cadherin. The EC1 cis binding surface is opposite to the trans dimer interface. The interaction is stabilized by a small hydrophobic core and several intermolecular hydrogen bonds. Each cadherin molecule can provide simultaneously both its concave EC1 surface and its convex EC2 surface for two identicalcis interactions. Thus, the cis interface arranges cadherin molecules into linear arrays (Fig. 5.2). Each cadherin in such a cis array also has a single trans bond with the cadherin located at the opposing plasma membrane. Importantly, since trans bonded cadherin molecules are nearly perpendicular to each other, the linear arrays of cadherin molecules on the opposing surfaces crisscross at right angles.
Fig. 5.2.
Schematic representation of the cadherin adhesive cluster formed by cis and trans intercadherin interactions. Blue molecules are organized in a linear array through cis interactions. The periodicity of the array is 72 Å. Each molecule in the array is engaged in strand-swapped trans interactions with magenta molecules, which belong to the opposite cell. Each of these molecules is part of its own array. Note that the opposing arrays are at right angles
Computational analyses suggest that the formation of such perpendicular trans interacting linear arrays could be a driving force for self-assembly of cadherin adhesive clusters (Wu et al. 2011). To demonstrate this experimentally, two point mutations (V81D/L175D) that destroy the hydrophobic core of the cis interface were introduced into E-cadherin and the molecular structure of the resulting mutant and its recruitment into adherens junctions were studied (Harrison et al. 2011). This work showed that the cis interface is functional—its inactivation completely abolished adherens junction assembly. Importantly, the mutant is still able to produce trans dimers, but the resulting dimers are much less stable. Because of this, the cadherin cis mutant can be recruited into cell–cell contacts by a “diffusion trap” mechanism (Perez et al. 2008). However, the junctions formed by the cis mutant are extremely transient and unstable.
Taken together, these data illuminate the important role of cis interactions in junction formation. The cis interactions are too weak to be detected in solution and are not expected to produce stable cis dimers on an extrajunctional cell surface. However, in cooperation with trans interactions, they produce stable and ordered adhesive structures. Importantly, strand-swap trans bonds are stable only once they are interconnected by cis interactions. For cadherin to exit such structures the strand-swap trans bond must be exchanged for the X-dimer trans bond. Intercadherin cis interactions can also be significantly distorted by cadherin interactions with cytoplasmic proteins, other transmembrane proteins and the cytoskeleton. In theory, these additional components can increase or decrease stability of the adhesive bonds by adding new levels of cadherin–cadherin cis interactions or by preventing the ectodomain cis bond formation, respectively. The contribution of these elements to adherens junction assembly is discussed in the next section.
5.3 From Adhesive Clusters to Adherens Junctions
5.3.1 Evidence for Reorganization
The data discussed above shows that cadherin adhesive clusters may self-assemble through a combination of trans and cis interactions. In the resulting clusters cadherin molecules are organized in linear arrays. The intercadherin distance in an array is about 7.2 nm. Indeed, electron microscopy examination of desmosomes, which consist of close relatives of classical cadherins, desmosomal cadherins, revealed periodical organization of intercellular rod-like structures, approximately 7 nm apart (Al-Amoudi et al. 2007). Paradoxically, no signs of such an organized structure have been detected in EM studies of adherens junctions (Hirokawa and Heuser 1981; Miyaguchi 2000).
A study of adherens junctions of chicken retinal pigment epithelium, using quick-freeze, deep-etch electron microscopy (Miyaguchi 2000), revealed no periodicity in adherens junction organization. Instead, intramembrane particles, approximately 7.7 nm in diameter, were irregularly packed within the inner face of the membrane. While it is impossible to exclude that the linear cadherin arrangement was lost in this study during EM sample preparation, strikingly, the average density of the intramembrane particles was only 700 per µM2. The number of the rod-like intermembrane structures that apparently corresponds to cadherin molecules was even smaller (approximately 100 rods per µM2). Similar low density of the rod-like intermembrane structures was evident in the study of cell–cell junctions in intestinal epithelium using a quick-freeze, deep-etch, rotary-replication technique (Hirokawa and Heuser 1981). Such densities are much less that the predicted density of cadherin molecules in the cadherin adhesion clusters (~17,000 molecules per µM2) (Harrison et al. 2011). Even in desmosomes, the dense and ordered cadherin organization was found only in their specific “hyperadhesive” state that is maintained by intracellular signaling (Garrod and Kimura 2008, see below).
To reconcile these EM observations with the structural data described above, one may propose that the formation of the ordered adhesive clusters is a transient process, which is immediately followed by their internal reconfiguration into more loose structures. Another possibility is that other transmembrane or intracellular proteins associated with cadherin distort the assembly of organized cadherin clusters. In any scenario, the reconfiguration of cadherin adhesive clusters into adherens junctions should include multiple cycles of cadherin adhesive dimer assembly and disassembly that, as discussed above, require strand-swap to X-dimer transitions. The requirement of this transition for cluster remodeling may explain why X-dimer interface cadherin mutants induce a dramatic dominant negative effect on cadherin adhesion in epithelial cells (Harrison et al. 2010; Hong et al. 2011). Cadherin cluster remodeling may also explain a very rapid turnover of cadherin molecules in adherens junctions (de Beco et al. 2009; Hong et al. 2010). Another piece of circumstantial evidence of the reconfiguration of cadherin clusters is the high pleomorphism of adherens junctions with respect to their morphology and protein composition (Meng and Takeichi 2009). The most prominent is the difference between apical adherens junctions (zonulae adhaerentes) and spot-like adherens junctions (puncta adhaerentia) present on the lateral (bounded) cell surfaces. The apical adherens junctions typically associate with a group of cytosolic actin-binding proteins such as vinculin, VASP, and EPLIN (Meng and Takeichi 2009). Their transmembrane adhesive domain also co-associates with another transmembrane adhesion receptor, nectin (Okabe et al. 2004; see also below). In contrast, lateral spot-like junctions do not exhibit association with these proteins, while they are also interconnected to the actin cytoskeleton. This interconnection is important for their basal to apical flow (Kametani and Takeichi 2007). Collectively, this evidence, while circumstantial, suggests that the self-assembly of the cadherin adhesive clusters is only a first step in adherens junction assembly.
The reconfiguration of the adhesive clusters or their assembly modifications could be, in theory, very important to how adherens junctions mature. This process could reconfigure uniformly packed cadherins into cell type-specific clusters. One may propose that cadherin cluster reconfiguration is mediated through additional types of intercadherin interactions, anchorage of cadherin to the cytoskeleton, and, finally, via interactions with other adhesion proteins, such as nectins or JAMs. While currently too little information is available to describe detailed mechanisms of cluster reconfiguration, we briefly outline the main possible driving forces of this process below.
5.3.2 Potential Role of Catenins
A linear array of cadherin molecules, which is formed during cadherin clustering, brings the intracellular cadherin tails into proximity. Such specific arrangement of the cadherin-catenin complexes on the intracellular face of plasma membrane may initiate new binding reactions that are too weak to be detected in solution using regular in vitro binding assays. Moreover, these inter-catenin interactions may induce specific conformational changes in catenin molecules, which, in turn, may open or establish new binding interfaces.
One such potentially important interaction resulting in α-catenin dimerization was observed in the crystal lattice of the α-catenin VH2 domain (Yang et al. 2001; Pokutta et al. 2002). The binding interface of this dimer is localized within the C-terminal four-helix bundle (residues 507–632) of this domain. The dimer is formed by the perpendicular packing of helices E and H against their counterparts. Dimerization of VH2 domain in solution mediated by this interface was also detected using a cross-linking assay. Importantly, the α-catenin region involved in this dimerization exactly corresponds to the adhesion modulation domain, which had been mapped by experiments with cadherin-α-catenin chimeric proteins (Imamura et al. 1999). The authors of this work showed that the chimera consisting of a β-catenin-uncoupled mutant of E-cadherin and an α-catenin VH2 domain mediates aggregation of cadherin-deficient L cells. It has been proposed that this adhesion modulation domain is involved in cadherin clustering. Importantly, since the paired catenin molecules in the dimer are in an antiparallel orientation, this dimerization is unlikely to occur in the parallel arrays of catenins that could be formed in the process of cadherin cluster self-assembly described above. Therefore, for a VH2 domain dimerization interface to be used, the linear cadherin arrays need to be broken and the entire cluster must be reorganized in a particular way.
Another potential α-catenin-dependent mechanism for remodeling the cluster is the binding of α-catenin VH3 domain to the actin filaments. A similar mechanism has been shown to be important in focal adhesions. It was shown that the α-catenin relative, vinculin, forms dimers through its VH3 domain (Bakolitsa et al. 1999; Johnson and Craig 2000; Janssen et al. 2006). Importantly, the dimerization of the VH3 domain of vinculin is proposed to be triggered by its binding to F-actin. The model suggests that actin filaments may be directly involved in molecular organization of vinculin-containing structures. Therefore, α-catenin-mediated reconfiguration of adherens junctions may also involve the actin cytoskeleton.
Finally, very interesting intermolecular interactions were detected in the p120 crystal lattice (Ishiyama et al. 2010). Here, the cadherin-p120 complexes were found to be arranged into linear head-to-tail oligomers with ~6 nm periodicity, which is close to periodicity of cadherin in the self-assembled arrays. Interestingly, residues of both, E-cadherin and p120, are involved in this interaction. Its most crucial feature is the conserved p120 residue W363: it is positioned within the paired p120 molecule, in a hydrophobic cleft between Arm repeats six and seven.
This secondary, catenin-based lateral ligation of cadherin molecules may have two consequences. First, it may reinforce the cadherin cluster if the ligation is compatible with cis interactions between cadherin ectodomains. Alternatively, if they are not, such interactions may change the position of cadherins in the cluster, thereby disengaging extracellular cis interactions. In both cases, the formation of such intracellular layers of cadherin–cadherin bonds can lift the requirement for extracellular cis interactions for cadherin cluster stability: strand-swapped adhesive bonds can be reinforced in the remodeled clusters by catenin-dependent inter-cadherin associations. Therefore, instead of a cis interface, cadherin positioning in the remodeled clusters can be determined by catenin conformations and the cytoskeleton. The advantage of these new cis bonds is that they can be directly regulated by a cell signaling network.
5.3.3 Nectins
Another obvious mechanism of cadherin adhesive cluster remodeling is cadherin interaction with other transmembrane proteins that can interfere with trans or cis intercadherin interactions. Once bound to cadherin, such transmembrane proteins may induce unspecific steric clashes into the process of cadherin cluster assembly. They also may specifically target cadherin cis-binding interfaces, thereby weakening strand-swap trans bonds. In both cases, cadherin molecules bound to such transmembrane proteins would be excluded from adherens junction assembly. A possible example of this mechanism is a cadherin interaction with a large trans-membrane proteolytic enzyme, γ-secretase: the complex consisting of E-cadherin and γ-secretase is mostly present within the extrajunctional lateral surface of epithelial cells (Kiss et al. 2008).
The most interesting example is another group of proteins that, through interactions with cadherin, can mediate specific distortion in the cadherin cluster assembly. The most promising candidates for such a role are the transmembrane immunoglobulin-like cell adhesion receptors, nectins. These proteins form calcium-independent adhesive clusters by their own in cadherin-deficient cells (Takahashi et al. 1999). While the mechanism of nectin clustering and adhesion is far from being clear, similar to cadherin clustering, it is, apparently, based on the self-assembly mechanism. It is suggested by two observations; (i) nectins can form cis and trans bonds (Momose et al. 2002; Narita et al. 2011), and (ii) nectin binding to the large cytosolic scaffolding protein, afadin, the only known intracellular nectin-binding partner, is not essential for nectin junction formation (Takahashi et al. 1999; Krummenacher et al. 2003).
Importantly, upon co-expression with cadherin, this nectin/afadin complex loses its independence and co-localizes with adherens junctions (Takahashi et al. 1999; Asakura et al. 1999). Whether nectin molecules produce the same trans and cis contacts in the adherens junctions, as in the cadherin-free adhesive clusters, remains to be determined. Furthermore, it is not known how and at what step these two adhesive systems interact. The function of the association between these two adhesive systems, particularly with respect to the structure of cadherin adhesion, is also unknown.
One of the possible mechanisms of interactions between cadherin and nectin adhesive systems is their intracellular association through α-catenin and afadin. This possibility is suggested by a number of observations: (i) It was shown that the intracellular C-terminal region of nectins forms a stable complex with the PDZ domain of afadin. Afadin-uncoupled mutants of nectin form adhesive clusters, which are not integrated into adherens junctions (Takahashi et al. 1999; Krummenacher et al. 2003). (ii) Afadin binds with a low affinity to the VH2 domain of α-catenin (Tachibana et al. 2000; Pokutta et al. 2002). (iii) Experiments with cadherin-α-catenin chimeras definitively showed that this VH2 domain of α-catenin is essential for cadherin-nectin co-clustering (Tachibana et al. 2000).
It was suggested that nectins are crucial for the nucleation of cadherin adhesion (Takai et al. 2008; Sato et al. 2006). However, several observations are not consistent with this point of view. While afadin was shown to be required for the general organization of cell–cell contacts in epithelial cells, it is not essential for the assembly of the individual adherens junctions (Zhadanov et al. 1999; Ikeda et al. 1999). Vice versa, α-catenin-deficient cells can recruit cadherin into nectin-deficient, adherens junction-like structures (Tachibana et al. 2000; Troyanovsky et al. 2011). A chimeric protein consisting of a β-catenin-uncoupled cadherin mutant and the α-catenin VH3 domain, which is unable to interact with afadin, still can form junctions upon expression in cadherin-deficient L cells (Imammura et al. 1999). Finally, tailless cadherin mutants rapidly form junctions in the calcium-switch assay (Hong et al. 2010). Taken together, these observations demonstrate that nectin association with cadherin via the intracellular domain is not a key step in the formation of cadherin adhesive clusters.
The data described above show that nectin can be co-recruited with cadherin into adherens junctions and that α-catenin-afadin interactions play a role in this process. A very important and still open question is whether the interaction between these adhesive systems occurs at the level of nectin and cadherin monomers or at the level of independently pre-assembled cadherin and nectin clusters. This question is important because the entire process of cadherin cluster assembly can be distorted if adherens junctions can be assembled from cadherin-nectin cis dimers. For example, cis interactions between nectin’s extracelluar domains can provide an alternative mechanism for the reinforcement of cadherin strand-swap trans dimers. The possibility that extrajunctional, free cadherin molecules can interact with nectin is suggested by experiments with dominant negative cadherin mutants: they destroy both cadherin and nectin adhesion (Tanaka et al. 2003). Alternatively, nectins may only be able to recognize and interact with preassembled cadherin clusters. In this scenario, independently formed cadherin and nectin clusters would associate along their periphery. In this case nectins would play a role in organizing small cadherin clusters into mature adherens junctions.
Adding even more complexity to the problem of cadherin-nectin interactions is a recent work that, using a Xenopus developmental model, suggested that cadherin and nectin molecules can interact through their extracellular regions (Morita et al. 2010). Again, whether this interaction is specific to some oligomeric forms of cadherin, remains to be studied.
5.3.4 Intercellular Distance
As discussed above, the cadherin/catenin complex interacts with a number of other molecules and structures. These interactions can significantly change not only the global distribution of cadherin clusters but also their internal organization. The changes can be cell type-specific or can be specific to the type of adherens junctions—for instance, the zonula adherens or the puncta adherens. Through regulation of the lateral alignment of cadherin molecules in the clusters, a junction can change its strength and its signaling potentials. In addition to transmembrane proteins and to intracellular bridging by catenins, the lateral alignment of cadherin in the junctions can also be controlled by the junctional intercellular distance. This variable largely depends on two opposite forces, stretching the junction by actomyosin contraction and compressing the junction by actin polymerization. Indeed, experiments performed in several laboratories demonstrate that junctional tension controls different parameters of adherens junctions including their protein composition (Ladoux et al. 2010; le Duc et al. 2010; Yonemura et al. 2010; Taguchi et al. 2011). It was proposed, therefore, that the cadherin-catenin complex is a mechanosensor that transmits force between F-actin and the cadherin adhesive bond (Yonemura et al. 2010; le Duc et al. 2010). However, it is also possible that a change in cadherin lateral alignment induced by junctional tension controls different properties of adherens junctions.
Electron microscopy shows that the distance between two adjoining plasma membranes in the junctions varies from 15–30 nm (McNutt and Weinstein 1973; Hirokawa and Heuser 1981; Drenckhahn and Franz 1986; Miyaguchi 2000). The distance between the opposite cadherin C-termini of the strand-swap cadherin dimer is 37–38 nm (Harrison et al. 2011). To be accommodated in the narrow intermembrane space, cadherin dimers, therefore, must transverse this space at an angle. Indeed, such a configuration of cadherin trans dimers in crystal lattice narrows the distance between the presumptive membranes to 18–25 nm. By changing the angle between cadherin and membrane, cells can potentially change cis and trans binding interfaces. Such structural changes can be crucial. Indeed, a cadherin inclination that is compatible with cis and strand-swap trans interactions would ultimately stabilize adherens junctions. In contrast, an angle that is incompatible with these interactions would result in junction disassembly. Therefore, the angle between cadherin and membrane can govern the strength of cadherin adhesion as well as the junction assembly-disassembly process.
Clear evidence for cadherin reorganization within particular adhesive structures, desmosomes, was obtained in the Garrod laboratory (Garrod and Kimura 2008). Desmosomal cadherins and classic cadherins share the same strand-swap trans dimerization binding site (Posy et al. 2006). However, desmosomal cadherins lack a classic cadherin-like cis interface, suggesting that desmosomal cadherin trans dimers have a specific lateral alignment (Harrison et al. 2011). Nevertheless, because of extensive structural similarities, the major principles of adhesion in adherens junctions and desmosomes may be similar. It was shown that desmosomal cadherins in desmosomes have, at least, two types of arrangements. The mature or “hyperadhesive” desmosomes are calcium-independent and exhibit a dense midline. The adjoining membranes in these desmosomes are 30 nm apart. Cryo-electron tomography of rapidly frozen epidermal desmosomes (Al-Amoudi et al. 2007) and computer modeling (Garrod et al. 2005) showed that cadherin molecules in mature desmosomes form arrays with a periodicity of 7.5 nm. Such an arrangement is very similar to that of classic cadherin in crystal lattices. In migrating cells, however, desmosomes become calcium-dependent and lose their midline and cadherin periodicity, and their intercellular space narrows to about 27 nm. This dramatic change in desmosome organization is regulated by PKCα-dependent signaling pathways (Garrod et al. 2005). Therefore, the rearrangement of cadherin molecules within adhesive structures can be a general mechanism regulating junctional dynamics and functions. Future works should address this important issue.
5.4 From Adherens Junctions to Cadherin Monomer
Cadherin-mediated adherens junctions are not static. Live imaging experiments have shown that they are in constant and directional motion (Kametani and Takeichi 2007; Hong et al. 2010). Spot-like adherens junctions are assembled in the basal area of the lateral cell surface and move in the apical direction. Reaching the apical surface, these junctions integrate into the zonula adherens. Such basal-to-apical movement of adherens junctions suggests that the adhesive bonds cementing the junction are strong enough to sustain the stress induced by this motion, which is unlikely to be completely synchronized in two neighboring cells. FRAP (fluorescence recovery after photobleaching) experiments showed that adherens junctions continuously loose and gain cadherin molecules (Yamada et al. 2005; Stehbens et al. 2006; Thoumine et al. 2006; Hong et al. 2010). This exchange of cadherin has been traditionally regarded as a result of dynamic equilibrium between junctional and extrajunctional cadherin (Kusumi et al. 1999). However, experiments performed at both cellular and molecular levels indicate that the mechanism of cadherin exchange in adherens junctions is far more complex: certain active processes continuously remove cadherin molecules from the junction. Active removal of cadherin from junctions has been suggested by the fact that ATP depletion completely stalls cadherin strand-swap dimer dynamics and rapidly blocks dimer disassembly in calcium-switch assay (Troyanovsky et al. 2006). Photoconversion of Dendra2-tagged cadherin in adherens junctions further demonstrated that cadherin molecules are locked in the adherens junctions of ATP-depleted cells (Hong et al. 2010). Importantly, both, live-cell imaging and biochemical approaches, have shown that ATP depletion does not interfere with the recruitment of the plasma membrane exposed cadherin into the junctions (Troyanovsky et al. 2006; Hong et al. 2010). This imbalance between cadherin recruitment and its release rapidly traps nearly all available cadherin in intercellular junctions. This data suggests that adhesive and lateral interactions between cadherin molecules in adherens junctions are strong enough to immobilize cadherin. To unlock cadherin from such a stable immobile state, some specific, energy-consuming processes are required. The active processes disassembling the junctions are far from clear. They can range from ATP-dependent conformational changes that destroy particular catenin-dependent intercadherin cis bonds, discussed in the previous section, to a more complex active process that physically removes cadherin from the junctions.
Among the possible mechanisms of the removal of cadherin molecules from the junctions is clathrin-dependent endocytosis. Indeed, broad inhibition of endocytosis by 0.4 M sucrose in A431 cells (Troyanovsky et al. 2006; Hong et al. 2010) as well as the inactivation of clathrin-dependent endocytosis by more specific inhibitors, dynasore or MiTMAB, in MDCK cells (de Beco et al. 2009) were shown to block cadherin exchange in adherens junctions. However, the process that unlocks cadherin and removes it from the junctions is clearly much more complex. For example, our attempts to prevent a release of cadherin from the junctions by clathrin depletion (Troyanovsky unpublished) or by point mutations of cadherin endocytic motifs (Hong et al. 2010) in A431 cells failed: both maneuvers blocked cadherin endocytosis but were ineffective in slowing down cadherin dynamics in the junctions. Similarly, the same inhibitors, dynasore and MiTMAB that blocked cadherin junctional turnover in MDCK cells produce little effect in MCF7 cells (de Beco et al. 2009).
As we showed recently, cadherin undergoes a strand-swap-to-X-dimer transition before exiting the adherens junction (Hong et al. 2011). This suggests that the mechanism unlocking cadherin from the junction includes the reconfiguration of the main adhesive bonds. Such reconfiguration could be the same ATP-consuming process that is detected in the ATP-depletion experiments. Apparently, the intra-cellular mechanisms that participate in the maturation of adherens junctions after initial cadherin clustering may play the leading role in disengagement of cadherin from the junction. How exactly this strand-swap-to-X-dimer transition is initiated and performed remains to be studied.
5.5 Perspectives and Future Directions
In our review we have highlighted recent progress in understanding the molecular mechanisms of adherens junction assembly. The data we discussed show that the initial formation of adhesive contact is based on cadherin trans dimerization via a strand-swapping mechanism. The resulting strand-swap dimers are unstable unless they are clustered through cis interactions. Despite some advances, we still have no answers to many outstanding questions. For example, virtually no data suggests whether any specific nucleation process triggers this initial cadherin clustering. Little is also known about how these initial clusters are organized into mature adherens junctions and how structural and morphological diversity of the junctions is achieved. Finally, what is the mechanism of adherens junction dynamics and disassembly? Answering these questions is a critical step in our understanding of various pathologies that are associated with abnormalities in cell–cell adhesion.
References
- Adams CL, Nelson WJ. Cytomechanics of cadherin-mediated cell-cell adhesion. Curr Opin Cell Biol. 1998;10:572–577. doi: 10.1016/s0955-0674(98)80031-8. [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A, Díez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- Asakura T, Nakanishi H, Sakisaka T, Takahashi K, Mandai K, Nishimura M, Sasaki T, Takai Y. Similar and differential behaviour between the nectin-afadin-ponsin and cadherin-catenin systems during the formation and disruption of the polarized junctional alignment in epithelial cells. Genes Cells. 1999;4:573–581. doi: 10.1046/j.1365-2443.1999.00283.x. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Becker JW, Erickson HP, Hoffman S, Cunningham BA, Edelman GM. Topology of cell adhesion molecules. Proc Natl Acad Sci U S A. 1989;86:1088–1092. doi: 10.1073/pnas.86.3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev NA, Troyanovsky SM. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J Cell Biol. 1998;142:837–846. doi: 10.1083/jcb.142.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beco S, Gueudry C, Amblard F, Coscoy S. Endocytosis is required for E-cadherin redistribution at mature adherens junctions. Proc Natl Acad Sci U S A. 2009;106:7010–7015. doi: 10.1073/pnas.0811253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn D, Franz H. Identification of actin-, alpha-actinin-, and vinculin-containing plaques at the lateral membrane of epithelial cells. J Cell Biol. 1986;102:1843–1852. doi: 10.1083/jcb.102.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW. Discovering the molecular components of intercellular junctions—a historical view. Cold Spring Harb Perspect Biol. 2009;1:a003061. doi: 10.1101/cshperspect.a003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D, Kimura TE. Hyper-adhesion: a new concept in cell-cell adhesion. Biochem Soc Trans. 2008;36:195–201. doi: 10.1042/BST0360195. [DOI] [PubMed] [Google Scholar]
- Garrod DR, Berika MY, Bardsley WF, Holmes D, Tabernero L. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J Cell Sci. 2005;118:5743–5754. doi: 10.1242/jcs.02700. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Corps EM, Kilshaw PJ. Cadherin adhesion depends on a salt bridge at the N-terminus. J Cell Sci. 2005;118:4123–4130. doi: 10.1242/jcs.02539. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, Shapiro L. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, Troyanovsky RB, Ben-Shaul A, Frank J, Troyanovsky SM, Shapiro L, Honig B. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Heuser JE. Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. J Cell Biol. 1981;91:399–409. doi: 10.1083/jcb.91.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci U S A. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Cadherin exits the junction by switching its adhesive bond. J Cell Biol. 2011;192:1073–1083. doi: 10.1083/jcb.201006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber O, Kemler R, Langosch D. Mutations affecting transmembrane segment interactions impair adhesiveness of E-cadherin. J Cell Sci. 1999;112:4415–4423. doi: 10.1242/jcs.112.23.4415. [DOI] [PubMed] [Google Scholar]
- Ikeda W, Nakanishi H, Miyoshi J, Mandai K, Ishizaki H, Tanaka M, Togawa A, Takahashi K, Nishioka H, Yoshida H, Mizoguchi A, Nishikawa S, Takai Y. Afadin: a key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J Cell Biol. 1999;146:1117–1132. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol. 1999;144:1311–1322. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. Actin activates a cryptic dimerization potential of the vinculin tail domain. J Biol Chern. 2000;275:95–105. doi: 10.1074/jbc.275.1.95. [DOI] [PubMed] [Google Scholar]
- Kametani Y, Takeichi M. Basal-to-apical cadherin flow at cell junctions. Nat Cell Biol. 2007;9:92–98. doi: 10.1038/ncb1520. [DOI] [PubMed] [Google Scholar]
- Kim SA, Tai CY, Mok LP, Mosser EA, Schuman EM. Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc Natl Acad Sci U S A. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A, Troyanovsky RB, Troyanovsky SM. p120-catenin is a key component of the cadherin-gamma-secretase supercomplex. Mol Biol Cell. 2008;19:4042–4050. doi: 10.1091/mbc.E08-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Natori M, Murase S, Hirano S, Taketani S, Suzuki ST. Mutation analysis of cadherin-4 reveals amino acid residues of EC1 important for the structure and function. Biochem Biophys Res Commun. 2000;271:358–363. doi: 10.1006/bbrc.2000.2636. [DOI] [PubMed] [Google Scholar]
- Klingelhöfer J, Troyanovsky RB, Laur OY, Troyanovsky S. Amino-terminal domain of classic cadherins determines the specificity of the adhesive interactions. J Cell Sci. 2000;113:2829–2836. doi: 10.1242/jcs.113.16.2829. [DOI] [PubMed] [Google Scholar]
- Klingelhöfer J, Laur OY, Troyanovsky RB, Troyanovsky SM. Dynamic interplay between adhesive and lateral E-cadherin dimers. Mol Cell Biol. 2002;22:7449–7458. doi: 10.1128/MCB.22.21.7449-7458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacher C, Baribaud I, Eisenberg RJ, Cohen GH. Cellular localization of nectin-1 and glycoprotein D during herpes simplex virus infection. J Virol. 2003;77:8985–8999. doi: 10.1128/JVI.77.16.8985-8999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Suzuki K, Koyasako K. Mobility and cytoskeletal interactions of cell adhesion receptors. Curr Opin Cell Biol. 1999;11:582–590. doi: 10.1016/s0955-0674(99)00020-4. [DOI] [PubMed] [Google Scholar]
- Ladoux B, Anon E, Lambert M, Rabodzey A, Hersen P, Buguin A, Silberzan P, Mège RM. Strength dependence of cadherin-mediated adhesions. Biophys J. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laur OY, Klingelhöfer J, Troyanovsky RB, Troyanovsky SM. Both the dimerization and immunochemical properties of E-cadherin EC1 domain depend on Trp(156) residue. Arch Biochem Biophys. 2002;400:141–147. doi: 10.1006/abbi.2002.2774. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Ryan TA, Smith SJ, Nelson WJ. Spatial and temporal dissection of immediate and early events following cadherin-mediated epithelial cell adhesion. J Cell Biol. 1993;120:1217–1226. doi: 10.1083/jcb.120.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt NS, Weinstein RS. Membrane ultrastructure at mammalian intercellular junctions. Prog Biophys Mol Biol. 1973;26:45–101. doi: 10.1016/0079-6107(73)90017-5. [DOI] [PubMed] [Google Scholar]
- Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaguchi K. Ultrastructure of the zonula adherens revealed by rapid-freeze deep-etching. J Struct Biol. 2000;132:169–178. doi: 10.1006/jsbi.2000.4244. [DOI] [PubMed] [Google Scholar]
- Momose Y, Honda T, Inagaki M, Shimizu K, Irie K, Nakanishi H, Takai Y. Role of the second immunoglobulin-like loop of nectin in cell-cell adhesion. Biochem Biophys Res Commun. 2002;293:45–49. doi: 10.1016/S0006-291X(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Morita H, Nandadasa S, Yamamoto TS, Terasaka-Iioka C, Wylie C, Ueno N. Nectin-2 and N-cadherin interact through extracellular domains and induce apical accumulation of F-actin in apical constriction of Xenopus neural tube morphogenesis. Development. 2010;137:1315–1325. doi: 10.1242/dev.043190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Narita H, Yamamoto Y, Suzuki M, Miyazaki N, Yoshida A, Kawai K, Iwasaki K, Nakagawa A, Takai Y, Sakisaka T. Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions. J Biol Chem. 2011;286:12659–12669. doi: 10.1074/jbc.M110.197368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–155. doi: 10.1016/0092-8674(90)90222-z. 1990. [DOI] [PubMed] [Google Scholar]
- Okabe N, Ozaki-Kuroda K, Nakanishi H, Shimizu K, Takai Y. Expression patterns of nectins and afadin during epithelial remodeling in the mouse embryo. Dev Dyn. 2004;230:174–186. doi: 10.1002/dvdy.20033. [DOI] [PubMed] [Google Scholar]
- Ozawa M. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J Biol Chem. 2002;277:19600–19608. doi: 10.1074/jbc.M202029200. [DOI] [PubMed] [Google Scholar]
- Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:954–968. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148(3):579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Fujita N, Yamada A, Ooshio T, Okamoto R, Irie K, Takai Y. Regulation of the assembly and adhesion activity of E-cadherin by nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J Biol Chem. 2006;281:5288–5299. doi: 10.1074/jbc.M510070200. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grübel G, Legrand JF, AlsNielsen J, Colman DR, Hendrickson WA. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens SJ, Paterson AD, Crampton MS, Shewan AM, Ferguson C, Akhmanova A, Parton RG, Yap AS. Dynamic microtubules regulate the local concentration of E-cadherin at cell-cell contacts. J Cell Sci. 2006;119:1801–1811. doi: 10.1242/jcs.02903. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Nakanishi H, Mandai K, Ozaki K, Ikeda W, Yamamoto Y, Nagafuchi A, Tsukita S, Takai Y. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol. 2000;150:1161–1176. doi: 10.1083/jcb.150.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Ishiuchi T, Takeichi M. Mechanosensitive EPLIN-dependent remodeling of adherens junctions regulates epithelial reshaping. J Cell Biol. 2011;194:643–656. doi: 10.1083/jcb.201104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Ikeda W, Ogita H, Rikitake Y. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol. 2008;24:309–342. doi: 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nakanishi H, Kakunaga S, Okabe N, Kawakatsu T, Shimizu K, Takai Y. Role of nectin in formation of E-cadherin-based adherens junctions in keratinocytes: analysis with the N-cadherin dominant negative mutant. Mol Biol Cell. 2003;14:1597–1609. doi: 10.1091/mbc.E02-10-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoumine O, Lambert M, Mège RM, Choquet D. Regulation of N-cadherin dynamics at neuronal contacts by ligand binding and cytoskeletal coupling. Mol Biol Cell. 2006;17:862–875. doi: 10.1091/mbc.E05-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky S. Cadherin dimers in cell-cell adhesion. Eur J Cell Biol. 2005;84:225–233. doi: 10.1016/j.ejcb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Troyanovsky RB, Sokolov E, Troyanovsky SM. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23:7965–7972. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Sokolov EP, Troyanovsky SM. Endocytosis of cadherin from intracellular junctions is the driving force for cadherin adhesive dimer disassembly. Mol Biol Cell. 2006;17:3484–3493. doi: 10.1091/mbc.E06-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Laur O, Troyanovsky SM. Stable and unstable cadherin dimers: mechanisms of formation and roles in cell adhesion. Mol Biol Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Klingelhöfer J, Troyanovsky SM. α-Catenin contributes to the strength of E-cadherin-p120 interactions. Mol Biol Cell. 2011;22:4247–4255. doi: 10.1091/mbc.E11-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Fuchs E. Actin dynamics and cell-cell adhesion in epithelia. Curr Opin Cell Biol. 2001;13:76–84. doi: 10.1016/s0955-0674(00)00177-0. [DOI] [PubMed] [Google Scholar]
- Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, Honig B. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18:693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vunnam N, Flint J, Balbo A, Schuck P, Pedigo S. Dimeric states of neural- and epithelial-cadherins are distinguished by the rate of disassembly. Biochemistry. 2011;50:2951–2961. doi: 10.1021/bi2001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vunnam N, Pedigo S. Calcium-induced strain in monomer promotes dimerization in neural-cadherin. Biochemistry. 2011a;50(39):8437–8444. doi: 10.1021/bi200902s. [DOI] [PubMed] [Google Scholar]
- Vunnam N, Pedigo S. Prolines in βA-sheet of neural cadherin act as a switch to control the dynamics of the equilibrium between monomer and dimer. Biochemistry. 2011b;50:6959–6965. doi: 10.1021/bi2007788. [DOI] [PubMed] [Google Scholar]
- Wu Y, Jin X, Harrison O, Shapiro L, Honig BH, Ben-Shaul A. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc Natl Acad Sci U S A. 2010;107:17592–17597. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. Alpha-catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Zhadanov AB, Provance DW, Jr, Speer CA, Coffin JD, Goss D, Blixt JA, Reichert CM, Mercer JA. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr Biol. 1999;9:880–888. doi: 10.1016/s0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci U S A. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]