SUMMARY
Zinc finger protein ZFP24, formerly known as ZFP191, is essential for oligodendrocyte maturation and CNS myelination. Nevertheless, the mechanism by which ZFP24 controls these processes is unknown. We demonstrate that ZFP24 binds to a consensus DNA sequence in proximity to genes important for oligodendrocyte differentiation and CNS myelination, and we show that this binding enhances target gene expression. We also demonstrate that ZFP24 DNA binding is controlled by phosphorylation. Phosphorylated ZFP24, which does not bind DNA, is the predominant form in oligodendrocyte progenitor cells. As these cells mature into oligodendrocytes, the non-phosphorylated, DNA-binding form accumulates. Interestingly, ZFP24 displays overlapping genomic binding sites with the transcription factors MYRF, SOX10, and OLIG2, which are known to control oligodendrocyte differentiation. Our findings provide a mechanism by which dephosphorylation of ZFP24 mediates its binding to regulatory regions of genes important for oligodendrocyte maturation, controls their expression, and thereby regulates oligodendrocyte differentiation and CNS myelination.
In Brief
Elbaz et al. describe a mechanism by which dephosphorylation of the zinc finger protein ZFP24 mediates its binding to the regulatory regions of genes important for oligodendrocyte maturation, controls their expression, and thereby regulates oligodendrocyte differentiation and CNS myelination.

INTRODUCTION
Myelin, which is produced by oligodendrocytes in the CNS, is a multilayer lipid membrane structure that ensheathes and insulates axons, allowing the efficient propagation of action potentials (Simons and Nave, 2015). Myelination begins before birth in humans and is mainly completed by early adulthood. During development, a majority of oligodendrocyte progenitor cells (OPCs) originate in the ventricular zone and migrate along vasculature to their target axons (Tsai et al., 2016). There, they terminally differentiate to mature oligodendrocytes, express myelin-related genes, produce myelin, and provide metabolic support to the axons (Simons and Nave, 2015).
Oligodendrocytes differentiation is under tight transcriptional control (Mitew et al., 2014). Several regulatory factors have been shown to participate in the differentiation of OPCs into mature, myelinating oligodendrocytes. NKX2.2, for example, is required for the generation of post-mitotic oligodendrocytes (Qi et al., 2001). Transient downregulation of Nkx2.2 prompts oligodendrocytes to re-express Nkx6.2, which in turn promotes myelination (Cai et al., 2010). OLIG1 is expressed along the lineage of oligodendrocytes and is important at the onset of myelination (Dai et al., 2015; Xin et al., 2005). OLIG2 promotes differentiation of oligodendrocytes by recruiting the chromatin-remodeling enzyme BRG1 to regulatory elements of key genes during differentiation (Yu et al., 2013). SOX10 is also required for the maturation of oligodendrocytes and is a direct activator of several myelin genes (Li et al., 2007), including the transcription factor MYRF (Hornig et al., 2013). Following its induction, MYRF mediates the progression of premyelinating oligodendrocytes to a mature, myelinating state (Emery et al., 2009). Although MYRF and SOX10 bind many of the same myelin gene transcription control regions (Bujalka et al., 2013; Hornig et al., 2013), they also appear to be able to bind individual enhancers independently.
In a mouse forward genetics screen, we discovered that the gene encoding zinc finger protein Zfp24 (previously known as Zfp191) is required for oligodendrocyte maturation and developmental CNS myelination (Howng et al., 2010). ZFP24 is a Cys2-His2 (C2H2) zinc finger protein that contains highly conserved linker domains that intersperse four putative DNA-binding zinc finger domains (Mertins et al., 2013, 2016), suggesting that it may act as a transcription factor. Here, we sought to identify the molecular mechanism by which ZFP24 controls CNS myelination.
RESULTS
ZFP24 Binds in Proximity to Genes Involved in Oligodendrocyte Development and Function and Mediates Their Expression
To identify ZFP24 target genes in the oligodendrocyte lineage, we performed chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) analysis by immunoprecipitating the endogenously expressed ZFP24 and associated chromatin from cultured primary mouse OPCs and mature oligodendrocytes using an anti-ZFP24 antibody. OPCs and oligodendrocytes derived from the Zfp24-null mice served as a negative control. All peak sequences were submitted to multiple expectation maximization for motif elicitation (MEME) (Bailey et al., 2006) for de novo motif analysis, which revealed a consensus sequence composed of six repeats of the sequence TCAT as the strongest candidate (Figure 1A). Binding of ZFP24 to several of the peaks identified by next generation sequencing was further validated by PCR (Figure S1A). ZFP24 peaks were found to be concentrated around known transcription start sites (TSSs), suggesting that ZFP24 binding in the genome is not random (Figure S1B). Binding of ZFP24 to the identified consensus sequence was further validated by an electrophoretic mobility shift assay (EMSA), which revealed that ZFP24 bound to the identified consensus sequence (herein TCAT probe) but did not bind a scrambled DNA probe, suggesting that this binding is DNA sequence specific (Figure S1C).
Figure 1. ZFP24 Binds in Proximity to Genes that Are Crucial for Myelination and Mediates Their Expression.

(A) Chromatin immunoprecipitation was performed on both OPCs and mature oligodendrocytes from the brain of WT and Zfp24-null mice. The eluted DNA was subjected to next-generation sequencing. ZFP24 peaks were subjected to motif finding by MEME. We identified the motif 5′-cattcattcattcattcattc-3′ with an E value of 3.0e–118 as the binding motif of ZFP24.
(B) The identified ZFP24 peaks were subjected to GREAT analysis for biological functions associated with genes that are adjacent to peaks. We found that the foremost enriched categories are “glial cell development,” “regulation of gliogenesis,” “oligodendrocyte development,” “regulation of glial cell differentiation,” and “regulation of oligodendrocyte differentiation,” with a p value less than 1.26 × 10−5.
(C) An example of the binding of ZFP24 upstream to the TSS of Sox10 is highlighted.
(D) ZFP24 was overexpressed in HEK293 cells, and its ability to bind its identified binding site upstream to the TSS of Mbp and Sox10 was assessed by EMSA. ZFP24 was incubated with biotinylated DNA composed of the identified binding site upstream to Mbp or Sox10, or with the same probe with an excess of non-biotinylated probe, or with biotinylated TCAT probe (which we identified as the binding motif of ZFP24) (n = 3).
(E) The identified ZFP24-binding sites upstream to the TSS of Mbp and Sox10 were cloned into pGL3 luciferase reporter assay plasmids (herein pGL3-MBP and pGL3-Sox10). The pGL3-control (pGL3-c) plasmid (with SV40 enhancer) served as a positive control. The expression of these plasmids was enforced in OPCs derived from WT (blue) or Zfp24-null mice (red). The Mbp peak region was placed in a forward (pGL3-MBP-For) and reverse (pGL3-MPB-Rev) orientation (n = 3). Error bars represent the SD.
In order to understand the biological functions in which the identified peaks are involved, the OPC peak coordinates were submitted to Genomic Regions Enrichment of Annotations Tool (GREAT). ZFP24 putative target genes were highly enriched for biological functions (Gene Ontologies) related to glial cell and oligodendrocyte development, as well as regulation of oligodendrocyte differentiation (Figure 1B). This result was based on binding of ZFP24 upstream to genes known to play a role in oligodendrocyte differentiation, among them Sox10 (Figure 1C). To verify the identified ZFP24-binding sites, we focused on two sites upstream to the TSS of Mbp and Sox10, genes that are crucial for CNS myelination. As determined by EMSA analysis, ZFP24 specifically bound to both sites (Figure 1D).
The identified binding sites of ZFP24 upstream to Mbp and Sox10 may indicate that ZFP24 binding is required for their expression. In order to test this, the ZFP24-binding sites that we identified upstream to the TSS of Mbp and Sox10 were cloned into luciferase reporter vectors, which were introduced into oligodendrocyte lineage cells derived from wild-type (WT) or Zfp24-null mice. ZFP24 expression was necessary to induce luciferase expression from these vectors (Figure 1E).
Phosphorylation State of ZFP24 Determines Its DNA-Binding Capacity and Activity
ZFP24 contains a SCAN domain and four DNA-binding C2H2 zinc finger domains that are interspersed by linkers that contain potential phosphorylation sites (Figure 2A). Linker phosphorylation is a mechanism that potentially results in the deactivation of the C2H2 proteins: phosphorylation introduces a negative charge to the DNA-binding domain of the protein and hence reduces its affinity to DNA (Dovat et al., 2002; Rizkallah et al., 2011). We confirmed that ZFP24 linkers undergo phosphorylation in vivo by subjecting ZFP24 immunoprecipitated from 16-day-old mouse brain to western blot analysis using an antibody that specifically recognizes the phosphorylated form of the linkers (Figure 2B). In order to study the effect of linker phosphorylation on the affinity of ZFP24 to its DNA consequence sequence, we tested the ability of phosphorylated and non-phosphorylated ZFP24 to bind to the consensus DNA sequence. Protein samples that were enriched for the phosphorylated form of ZFP24 had a lower capability to bind the ZFP24 target DNA sequence than samples enriched for the non-phosphorylated ZFP24 isoform (Figures 2C and 2D). To confirm the effect of the phosphorylation state of ZFP24 on its DNA-binding capacity, we mutated the phosphorylation sites within the three linkers to either alanine, to mimic a non-phosphorylated form of ZFP24, or to glutamate, to mimic constant phosphorylation (Zhao et al., 1994). We enforced the expression of these mutants in HEK293 cells and measured the ability of nuclear extracts to bind the ZFP24 target DNA sequence by EMSA. The alanine mutant that resembles the non-phosphorylated form of ZFP24 was able to bind the ZFP24 target DNA sequence comparable with WT ZFP24. In contrast, the glutamate mutant that resembles the fully phosphorylated form of ZFP24 failed to bind the ZFP24 target DNA sequence (Figure 2E). In addition, we have generated phosphomimetic mutants that represent all eight potential states of phosphorylation and tested their capability to bind the TCAT target sequence. We found that as long as one linker is not phosphorylated, ZFP24 has the capacity to bind to the ZFP24 target DNA sequence (Figures S1D and S1E).
Figure 2. Phosphorylation State of ZFP24 Determines Its DNA-Binding Capacity and Activity.
(A) Schematic diagram of ZFP24 protein structure showing the relative positions of the SCAN domain (green), zinc fingers (ZF; yellow), linkers, and potential phosphorus-acceptor sites within the linker regions (red).
(B) ZFP24 was immunoprecipitated from 16-day-old WT or Zfp24-null mouse brain using an antibody against ZFP24 (Sigma-Aldrich). The eluted protein was subjected to western blot using an antibody that recognized ZFP24 (top) or an antibody that recognized the phosphorylated linker (anti-HpTGEKP antibody) (n = 3).
(C) ZFP24 was expressed in HEK293 cells, and nuclear extracts from untransfected (U) and transfected (T) cells were subjected to western blot using antibodies against ZFP24, the phosphorylated linker (anti-HpTGEKP antibody), and actin (used as equal loading control). To enrich for phosphorylated ZFP24, we treated the enriched for the phosphorylated form of ZFP24 has a lower capability to bind the ZFP24 target DNA sequence.
(D) The ratio between DNA-binding capacity in DMSO-treated (non-phosphorylated) and nocodazole-treated (phosphorylated) cells is shown (n = 3). Error bars represent the SEM.
(E) The alanine mutant (S274A, T302A, T330A) that resembles the non-phosphorylated ZFP24 is able to bind the identified DNA motif, whereas the glutamate mutant (S274E, T302E, T330E) that resembles phosphorylated ZFP24 cannot (n = 3).
(F) ZFP24 was immunoprecipitated from primary OPCs and from mature oligodendrocytes (OLGs) derived from rat. The eluted proteins were subjected to isoelectric focusing followed by western blot. In OPCs the majority of the protein migrated to the acidic side, whereas in mature OLGs the protein shifted to the basic side, suggesting that the protein loses acidic phosphate groups.
(G) Phosphorylation was verified using an antibody that recognizes the phosphorylated linker only. Four forms of the ZFP24 protein appear to be represented in this figure: in the first form, all three linkers are phosphorylated; the second form is demonstrated by a shift to the basic side, consistent with the loss of one phosphate group; the third is represented by a larger shift to the basic side, consistent with the loss of two phosphate groups. The most basic fourth form is non-phosphorylated and hence was not recognized as the antibody recognizes the phosphorylated form only.
(H) The ratio between non-phosphorylated to fully phosphorylated ZFP24 in OPCs and OLGs is shown (n = 3). Error bars represent the SEM.
In order to better understand how this phosphorylation may be relevant to myelin-related gene expression, we immunoprecipitated ZFP24 from OPCs as well as mature oligodendrocytes, then subjected the eluted protein to isoelectric focusing followed by western blot (two-dimensional [2D] gel electrophoresis). We found that as OPCs differentiate to mature oligodendrocytes, ZFP24 shifts from the acidic side of the gel to the basic side, suggesting that ZFP24 loses acidic phosphates as OPCs differentiate to mature oligodendrocytes (Figure 2F). Changes in the phosphorylation state were further confirmed using the phosphorylated linker antibody (Figures 2G and 2H).
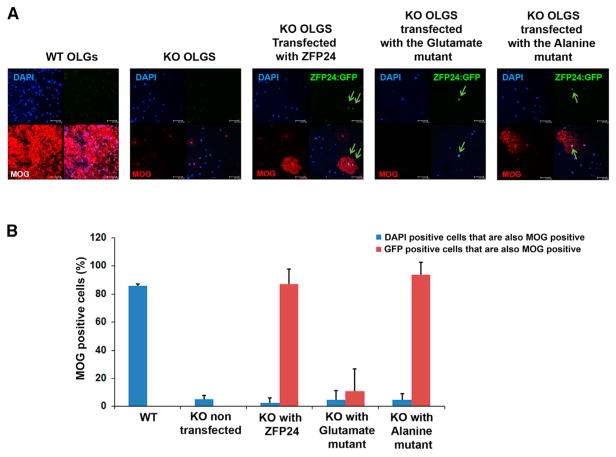
To study the role of the ZFP24 phosphorylation state on oligodendrocyte lineage cell differentiation, we enforced the expression of the glutamate and alanine mutants separately in Zfp24-null-derived oligodendrocyte lineage cells and measured the ability of each to restore MOG expression. MOG is a mature myelinating oligodendrocyte marker that is not expressed by oligodendrocytes derived from Zfp24-null mice (Howng et al., 2010). WT ZFP24 was able to restore MOG expression in Zfp24-null cells (Figure 3). In contrast, the glutamate mutant that resembles the fully phosphorylated form of ZFP24 failed to restore MOG expression (Figure 3). Meanwhile, the alanine mutant was able to restore MOG expression (Figure 3), indicating that the non-phosphorylated form of the protein is functional.
Figure 3. Expression of an Alanine Mutant (S274A, T302A, T330A) that Resembles the Non-phosphorylated ZFP24 Rescues MOG Expression, Whereas the Glutamate Mutant (S274E, T302E, T330E) that Resembles Phosphorylated ZFP24 Does Not.
(A) OPCs isolated from WT and Zfp24-null mice were transfected with a plasmid encoding ZFP24 or its mutants fused to GFP. The cells were allowed to differentiate to mature oligodendrocytes (OLGs), fixed, and immunostained with anti-MOG antibody, a mature myelinating oligodendrocyte marker. WT-derived oligodendrocytes express MOG, whereas Zfp24-null-derived cells do not. Reintroduction of WT ZFP24 to the Zfp24-null-derived cells is able to restore MOG expression (cells that express the ZFP24:GFP protein are green). The alanine mutant has a similar effect, whereas the glutamate mutant failed to restore MOG expression. More than 700 cells were counted per sample from three different experiments. Green arrows highlight GFP+ cells.
(B) The percentage of GFP-expressing cells that also stain for MOG. Error bars represent the SEM.
ZFP24 Targets Genes in Oligodendrocyte Lineage Cells
In order to characterize ZFP24 target genes, we examined the genes with perturbed expression in Zfp24-null oligodendrocyte lineage cells (Aaker et al., 2016) for ZFP24-binding sites in proximity to their TSSs (on the basis of our ChIP data). We identified 700 potential ZFP24 target genes (Figure 4A), which are listed in Table S1.
Figure 4. ZFP24 Targets Genes in Oligodendrocyte Lineage Cells.
(A) The genes that have both a ZFP24-binding site in proximity of 50 kb to their TSSs (on the basis of our ChIP-seq data, in blue) and expression that is affected by ZFP24 loss (on the basis of our RNA sequencing [RNA-seq] data, in purple) (Aaker et al., 2016) are highlighted.
(B) OPCs were isolated from WT and Zfp24-null mice and transfected with a plasmid encoding Flag-tagged MYRF or GFP-fused SOX10. The cells were allowed to differentiate to mature oligodendrocytes, fixed, then immunostained with anti-MBP antibody, a myelinating oligodendrocyte marker. More than 850 cells were counted per sample from three different experiments. Green arrows highlight transfected or GFP+ cells.
(C) The percentage of MYRF- or SOX10-expressing cells that also stain for MBP. Error bars represent the SEM.
(D) One hundred eighty-four ZFP24-binding sites overlap with the identified binding sites of OLIG2, 149 overlap with the identified binding sites of SOX10, and 51 overlap with the identified binding sites of MYRF.
(E) A scheme describing the relation between these factors. The lines (arrowheads) indicate the presence of binding site for one factor in vicinity of the promoter of the other factor (marked by the arrowhead). Factors also bind their own promoters.
The identified ZFP24 target genes include transcription factors that control oligodendrocyte differentiation and affect myelination, most notably Sox10 and Myrf. We therefore hypothesized that the effect of ZFP24 loss is mediated, at least in part, by the reduced expression of its downstream target genes Sox10 and Myrf. In order to test this possibility we enforced the expression of SOX10 and MYRF in Zfp24-null-derived oligodendrocyte lineage cells and measured the ability of each to restore expression of MBP, an important component of myelin, which is reduced approximately 4-fold in Zfp24-null oligodendrocytes. We found that enforced expression of MYRF, but not SOX10, in Zfp24-null oligodendrocyte lineage cells can rescue MBP expression in these cells, suggesting that MYRF mediates the expression of myelin genes downstream to ZFP24 (Figures 4B and 4C).
Our data suggest that ZFP24 directly mediates the expression of Sox10 and Myrf. We therefore sought to determine whether the ZFP24 DNA-binding sites overlapped with the ChIP-seq identified binding sites of the other three transcription factors that are known to mediate oligodendrocyte differentiation and myelination: OLIG2 (Yu et al., 2013), SOX10 (Lopez-Anido et al., 2015), and MYRF (Bujalka et al., 2013). As a control, we compared the intersection of ZFP24, OLIG2, SOX10, and MYRF with the identified binding sites of the transcription factor JunD in leukemia cells (Yue et al., 2014) and which is also expressed in the brain (Zhang et al., 2014). Interestingly, we found that 184 of the ZFP24-binding sites overlap with the binding sites of OLIG2, 149 overlap with the binding sites of SOX10, and 52 overlap with the binding sites of MYRF (Figure 4D). This degree of overlap is statistically significant (HOMER, hypergeometric test; Table S2), while for JunD no significant overlap was found (Table S2). Interestingly, we found 13 regions that were occupied by all four transcription factors. The list of the overlapping binding sites and the genes in proximity to the 13 regions that were occupied by all four transcription factors is provided in Table S3. Analyzing binding sites near the promoters of these four transcription factors, we also found that they bind their own promoter and often bind the promoters of each other (Figure 4E).
DISCUSSION
The human genome contains roughly 700 C2H2-containing zinc finger proteins (Vaquerizas et al., 2009; Weirauch and Hughes, 2011). Nevertheless, the DNA-binding sequences and the biological functions of the vast majority of these proteins remain unknown. Here, we elucidate the molecular mechanisms by which ZFP24 acts to drive oligodendrocyte differentiation, myelin protein gene expression, and CNS myelination.
Interestingly, although expression of the putative ZFP24 target genes changes as oligodendrocyte lineage cells progress toward maturation, the ZFP24 mRNA and protein are expressed at all phases of oligodendrocyte development. ZFP24 cells with nocodazole, which is known to induce phosphorylation of C2H2 zinc finger proteins (Experimental Procedures). It can be seen that the sample that was contains four zinc finger domains that are interspersed by highly conserved linker domains, which have been shown previously to be potential phosphorylation targets (Mertins et al., 2013, 2016). Strikingly, the phosphorylated status of ZFP24 changes as oligodendrocyte lineage cells differentiate. In mitotic OPCs, ZFP24 is phosphorylated, whereas it appears to lose its phosphate groups in more mature oligodendrocytes. Using the ZFP24 consensus DNA-binding domain as bait, we demonstrated that non-phosphorylated ZFP24 has an increased capacity to bind the ZFP24 target DNA sequence, and phosphomimetic substitution abolishes ZFP24’s ability to restore myelin protein gene expression in Zfp24-null oligodendrocytes. Therefore, the role ZFP24 plays in oligodendrocyte maturation is likely controlled post-translationally through phosphorylation.
Oligodendrocyte transcription factor mouse mutants (e.g., Olig2, Myrf, Sox10) do not generate detectable numbers of mature oligodendrocytes, indicating that these proteins work early in the oligodendrocyte lineage (Emery et al., 2009; Lu et al., 2002; Stolt et al., 2002; Takebayashi et al., 2002). We and others speculated previously that ZFP24, in contrast, plays a role in the final stages of oligodendrocyte maturation (Emery, 2010; Howng et al., 2010). This suggestion was based on our analysis of Zfp24-mutant mice, which, despite having normal oligodendrocyte numbers, fail to myelinate (Howng et al., 2010). Our present studies, however, raise the possibility that ZFP24 works in collaboration with SOX10, OLIG2, and MYRF (Figure 4E) to establish a transcriptional network that is critical for proper oligodendrocyte lineage development. ZFP24 binds in the vicinity of the transcription initiation sites of the genes that encode SOX10, OLIG2, and MYRF, and Zfp24-mutant oligodendrocytes display reduced expression of Sox10 and Myrf, although Olig2 expression appears unaltered in the absence of ZFP24 (Aaker et al., 2016). The ability of MYRF to rescue the maturation phenotype of Zfp24-mutant oligodendrocytes (Figure 4B) suggests that Myrf is a downstream target of ZFP24. Moreover, a number of the ZFP24 DNA-binding sites in genes essential to oligodendrocyte function overlap with the binding sites of one or more of these oligodendrocyte transcription factors (Figure 4D). Therefore, ZFP24 might have a quantitative effect on the expression of genes critical for oligodendrocyte maturation and function. The reduced expression of ZFP24 target genes, along with the resulting reduced expression of the downstream targets of key oligodendrocyte transcription factors, likely combine to contribute to the hypomyelinating phenotype of Zfp24-mutant mice. Further studies are required to elucidate how ZFP24, MYRF, SOX10, and OLIG2 cooperate to regulate oligodendrocyte development.
Interestingly, it has recently been reported that patients hemizygous for 18q chromosomal deletions that include the region that contains the Zfp24 human ortholog, ZNF24, display seizures and tremors, suggestive of myelin abnormalities (Cody et al., 2015). This raises the possibility that a subset of human disorders with 18q deletions might be due, at least in part, to ZNF24 haploinsufficiency.
Taken together, our findings provide a direct molecular mechanism by which dephosphorylation of ZFP24 mediates its binding to regulatory regions of genes critical for oligodendrocyte differentiation and myelination, mediates their expression, and as a result, controls oligodendrocyte differentiation and CNS myelination. Further examination of this critical component of CNS myelination could unveil potential therapeutic targets for enhanced myelination and remyelination.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J mice (The Jackson Laboratory), Zfp24-null mice (Howng et al., 2010), and Rattus norvegicus rats (Harlan) were housed according to the University of Chicago’s Institutional Animal Care and Use Committee (IACUC) guidelines.
Primary Rat and Mouse Cell Cultures
Mouse OPCs were isolated from C57BL/6J mice and from Zfp24-null mice (Howng et al., 2010) by immunopanning as previously described (Emery and Dugas, 2013). Rat OPCs were similarly isolated from rat pups by immunopanning as previously described (Dugas and Emery, 2013). For OPC isolations, 7-day-old pups were used, both males and females. OPCs were transfected by electroporation using Amaxa cell nucleofector II device. Transfection efficiency was ~40% (as determined by transfection of a GFP-encoding plasmid).
ChIP-Seq
OPCs were isolated from WT and Zfp24-null animals by immunopanning as previously described (Emery and Dugas, 2013). Cells were then cultured for 5 days in differentiative conditions (−PDGF, +40 ng/mL triiodothyronine; Sigma-Aldrich). ChIP was performed on formaldehyde cross-linked chromatin (~50 μg/sample) using an anti-ZFP24 antibody (Sigma-Aldrich) by Active Motif. Briefly, ChIP samples were used for Illumina library construction, and libraries were submitted to 75-base Illumina sequencing (>20 million reads/sample). The 75 nt sequence reads were mapped to the mouse genome (mm10) assembly using the BWA algorithm (default settings) (Kofler et al., 2016). Peak calling was performed with HOMER (Heinz et al., 2010) using only uniquely mapped reads and PCR duplicates removed (makeTagDirectory -tbp 1). The peaks for OPC and OLG were calculated twice (HOMER findPeaks –style factor): against ChIP data derived from the Zfp24-null as control and against input DNA as control. The final list contains only peaks that were identified against both controls.
Subsequent Analysis of ChIP-Seq Data
For de novo motif discovery, sequences of identified ZFP24 peaks from HOMER were submitted to the MEME-ChIP suite (Machanick and Bailey, 2011). GREAT (McLean et al., 2010) was used to find enriched annotations with gene association rule set to basal + up to 100 kb. Occurrence of overlapping peaks (Table S3) for MYRF, SOX10, and OLIG2 peaks were identified using mergePeaks from HOMER package and plotted on the University of California, Santa Cruz (UCSC), browser. For this purpose, previously reported binding sites for OLIG2 (Yu et al., 2013), SOX10 (Lopez-Anido et al., 2015), and MYRF (Bujalka et al., 2013) were converted to mm10 using UCSC liftover. The rat ChIP-seq peak coordinates were converted to the mouse NCBI37/mm10 genomic assembly using the UCSC browser liftover function and the coordinates submitted to GREAT using a 100 or 1,000 kb regulatory region setting. Overlapping peaks for MYRF, SOX10, and OLIG2 peaks were identified using mergePeaks from the HOMER package (Heinz et al., 2010).
ChIP-Real-Time PCR
Rat OPCs were isolated from rat pups by immunopanning as previously described (Dugas and Emery, 2013). The cells were transfected with a plasmid encoding the FLAG-tagged ZFP24. Immunoprecipitation was performed using anti-Flag antibody (Sigma-Aldrich). Immunoprecipitation and isolation of the ZFP24-bound DNA was performed using the DNA ChIP-IT Express kit (Active Motif) according to the manufacturer’s instructions. The immunoprecipitated DNA was amplified using iQ SYBR Green Supermix (Bio-Rad) in a Bio-Rad C1000 thermocycler. OLIG2 is known to bind upstream to p21 (Ligon et al., 2007). We used this known protein-DNA interaction as a control. OLIG2 was immunoprecipitated using OLIG2 antibody (R&D). The following set of primers were used: Sox10: distal: forward: 5′-GCAGAGGACATGGTTGTGGA-3′, reverse: 5′-CTGCCTCAGACTGCTGACAA-3′. Sox10: no TCAT: forward: 5′-CA TACCCAAGGGCTCTCTGC-3′, reverse: 5′-TCCACAACCATGTCCTCTGC-3′. Sox10: TCAT: forward: 5′-GGGTAGGTCACAACCCACTG-3′, reverse: 5′-TG CCATCCTTTGTGTGTGGT-3′. Zfp536: forward: 5′-CGGCTCCTCTAACGT GACTG-3′, reverse: 5′-CCTCGCCGATGTCTGAAGAA-3′. Elovl7: forward: 5′-TGCTTATACAGCGTGCACCA-3′, reverse: 5′-TGGACTGAGGATTGAACT CAGA-3′. Smad7: forward: 5′-AGGATCTTGTCCCCGAGCAG-3′, reverse: 5′-CTGCGTCTCAGGCAGCTCTC-3′. p21: forward: 5′-AGGTGTCTAGACTC CAGATT-3′, reverse: 5′-AAAATCAAGGCTTTGCTGG-3′.
Luciferase Assays
The identified ZFP24-binding sites upstream to the TSSs of Mbp and Sox10 were amplified by PCR from mouse genomic DNA and cloned into pGL3 luciferase reporter assay plasmids (pGL3-promter; Promega). Luciferase assays were performed using the Promega luciferase reporter assay kit according to the manufacturer’s instructions. OPCs were co-transfected with both the test pGL3 luciferase construct and a plasmid encoding GFP, and luciferase readings were normalized to the GFP levels. Samples were normalized to the pGL3-p plasmid that does not contain an enhancer. Luciferase results are shown as the average and SD of at least three independent experiments. The following set of primers were used to clone the identified ZFP24-binding sites: Sox10 forward 5′-GGA CAT ATC CAA GGG CTC TG-3′, Sox10 reverse 5′-TGC CAT GCA CAT CAG TAT CC-3′; Mbp forward 5′-TGT GCC TTT CCA GCA CTG T-3′, Mbp reverse 5′-CTG AGG TGC TGT GGA AAG GT-3′; MBPshort forward 5′-ctctcctcatctgcgttcgtt-3′, MBPshort reverse 5′-aggaacagtgccagcaaaca-3′. Insertions sequence was verified by PCR and DNA sequencing.
EMSA
EMSA was performed using the LightShift Chemiluminescent EMSA kit (Pierce). Nuclear extract derived from HEK cells transfected with a ZFP24 encoding plasmid was incubated with biotinylated probe and run on 5% TBE PAGE gel. The gels were then transferred to nylon membranes (Thermo Fisher Scientific). Biotin-labeled probes were detected by primary streptavidin antibody attached to horseradish peroxidase. Biotinylated probes used were as follows: TCAT probe: 5′-TCATTCATTCATTCATTCATTCATCAT-3′; scramble probe: 5′-AGAGTGGTCAATACCCCCTCTG-3′; Mbp probe: 5′-GTT TTG TTA ATG CAT TTA ATT CCT CAC AGT ATA GCT GTT GTC TTC TCT CCT CAT CTG CGT TCG TTT CTG ACA GCT ACA GAA TTT AT-3′; Sox10: 5′-GGT AGT TGC TTC ATT CAT TCA TTC ACT CAT TCA TTC ATT CAT TCA TTC ATT CAT TCA TTC ATC TTG GAG TTT CCT TTA GTA CA-3′. For reactions that contained the unlabeled “cold” probe, the same unlabeled probe was added to the reaction in 200-fold excess.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using a site-directed mutagenesis kit (Agilent) according to the manufacturer’s guidelines. Mouse Zfp24 coding sequence was cloned into pCMV-AC-GFP mailman expression vector (Origene). For phosphomimetic substitution, serine 274 in the first linker, threonine 302 in the second linker, and threonine 330 in the third linker were replaced by glutamate. For the mutant that mimics the non-phosphorylated state, serine 274, threonine 302, and threonine 330 were replaced by alanine.
DNA-Binding Assay
The expression of ZFP24 was enforced in HEK293 cells, and its ability to bind to its target DNA was tested. To enrich for phosphorylated ZFP24, we treated the cells with nocodazole, a small molecule that interferes with the polymerization of microtubules (Lee et al., 1980). Nocodazole-treated cells enter mitosis but cannot form metaphase spindles, because their microtubules can no longer polymerize. It has been shown that cell-cycle arrest at mitosis induces phosphorylation of C2H2 zinc finger proteins (Rizkallah et al., 2011).
We therefore treated the cells with either nocodazole to induce cell-cycle arrest or with equivalent amounts of DMSO as a negative control, and we found that nocodazole treatment increased phosphorylation of ZFP24 (Figure 2). The ability of phosphorylated and non-phosphorylated ZFP24 to bind to the consensus DNA sequence was tested using biotinylated TCAT probe as bait. The biotinylated DNA (and the protein bound to it) was immunoprecipitated using streptavidin-coated beads, then subjected to western blot analysis.
Immunocytochemistry
Cells were fixed for 10 min in 4% paraformaldehyde, washed in PBS, and subjected to immunostaining. The antibodies used were anti-Flag antibody (Sigma-Aldrich), anti-MBP, and anti-MOG antibody (Millipore).
Immunoprecipitation and Western Blotting
ZFP24 was immunoprecipitated from the brain of P16 pups of Zfp24-null mice (Howng et al., 2010) and littermate WT using anti-ZFP24 antibody (Sigma-Aldrich). The eluted proteins were subjected to western blot analysis using polyclonal anti-ZFP24 (Sigma-Aldrich) antibody and the anti-phospho linker antibody (Sigma-Aldrich).
Isoelectric Focusing
ZFP24 was immunoprecipitated from OPCs and from mature oligodendrocytes using anti-ZFP24 antibody (Sigma-Aldrich). The eluted proteins were subjected to isoelectric focusing followed by western blot. Two-dimensional electrophoresis was performed according to the carrier ampholine method of isoelectric focusing (Burgess-Cassler et al., 1989; O’Farrell, 1975).
Statistical Analysis
Data are presented as mean ± SEM unless otherwise noted. Multiple comparisons were statistically evaluated using mixed-results ANOVA or two-way ANOVA; single comparisons were performed using two-sided unpaired t tests. A p value < 0.05 was considered to indicate statistical significance.
Supplementary Material
Highlights.
ZFP24 binds to the regulatory regions of genes important for CNS myelination
The DNA-binding capability of ZFP24 is controlled by phosphorylation
The phosphorylation state of ZFP24 changes along the oligodendrocyte lineage
ZFP24 displays overlapping genomic binding sites with MYRF, SOX10, and OLIG2
Acknowledgments
We would like to thank Rejani B. Kunjamma, Ani Solanki, and Gloria Wright for their technical assistance. This work was supported by grants to B.P. from the Myelin Repair Foundation, the NIH (R01NS067550), the National Multiple Sclerosis Society (RG-1501-02797), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. S.I. and A.E. are supported by the Israel Science Foundation (ISF; grant 1361/14).
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the ChIP-seq data reported in this paper is GEO: GSE101535.
Supplemental Information includes one figure and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.089.
AUTHOR CONTRIBUTIONS
B.E. and B.P. conceived the study and experimental design, performed the experiments, analyzed the data, and wrote the manuscript. J.D.A. and A.K. performed the experiments and analyzed data. S.I. and A.E. analyzed the ChIP-seq experiments. P.B. designed and analyzed the EMSA experiments.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Aaker JD, Elbaz B, Wu Y, Looney TJ, Zhang L, Lahn BT, Popko B. Transcriptional Fingerprint of Hypomyelination in Zfp191null and Shiverer (Mbpshi) Mice. ASN Neuro. 2016;8:8. doi: 10.1177/1759091416670749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013;11:e1001625. doi: 10.1371/journal.pbio.1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Cassler A, Johansen JJ, Santek DA, Ide JR, Kendrick NC. Computerized quantitative analysis of coomassie-blue-stained serum proteins separated by two-dimensional electrophoresis. Clin Chem. 1989;35:2297–2304. [PubMed] [Google Scholar]
- Cai J, Zhu Q, Zheng K, Li H, Qi Y, Cao Q, Qiu M. Co-localization of Nkx6.2 and Nkx2.2 homeodomain proteins in differentiated myelinating oligodendrocytes. Glia. 2010;58:458–468. doi: 10.1002/glia.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody JD, Sebold C, Heard P, Carter E, Soileau B, Hasi-Zogaj M, Hill A, Rupert D, Perry B, O’Donnell L, et al. Consequences of chrom-some18q deletions. Am J Med Genet C Semin Med Genet. 2015;169:265–280. doi: 10.1002/ajmg.c.31446. [DOI] [PubMed] [Google Scholar]
- Dai J, Bercury KK, Ahrendsen JT, Macklin WB. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J Neurosci. 2015;35:4386–4402. doi: 10.1523/JNEUROSCI.4962-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002;16:2985–2990. doi: 10.1101/gad.1040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Emery B. Purification of oligodendrocyte precursor cells from rat cortices by immunopanning. Cold Spring Harb Protoc. 2013;2013:745–758. doi: 10.1101/pdb.prot070862. [DOI] [PubMed] [Google Scholar]
- Emery B. Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20:601–607. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Emery B, Dugas JC. Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb Protoc. 2013;2013:854–868. doi: 10.1101/pdb.prot073973. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J, Fröb F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, Wegner M. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet. 2013;9:e1003907. doi: 10.1371/journal.pgen.1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA, Popko B. ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 2010;24:301–311. doi: 10.1101/gad.1864510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Langmuller AM, Nouhaud P, Otte KA, Schlotterer C. Suitability of different mapping algorithms for genome-wide polymorphism scans with Pool-Seq data. G3 (Bethesda) 2016 doi: 10.1534/g3.116.034488. Published online September 12, 2016. https://doi.org/10.1534/g3.116.034488. [DOI] [PMC free article] [PubMed]
- Lee JC, Field DJ, Lee LL. Effects of nocodazole on structures of calf brain tubulin. Biochemistry. 1980;19:6209–6215. doi: 10.1021/bi00567a041. [DOI] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Anido C, Sun G, Koenning M, Srinivasan R, Hung HA, Emery B, Keles S, Svaren J. Differential Sox10 genomic occupancy in myelinating glia. Glia. 2015 doi: 10.1002/glia.22855. Published online May 14, 2015. https://doi.org/10.1002/glia.22855. [DOI] [PMC free article] [PubMed]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. NCICPTAC. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitew S, Hay CM, Peckham H, Xiao J, Koenning M, Emery B. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2014;276:29–47. doi: 10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Rizkallah R, Alexander KE, Hurt MM. Global mitotic phosphorylation of C2H2 zinc finger protein linker peptides. Cell Cycle. 2011;10:3327–3336. doi: 10.4161/cc.10.19.17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Nave KA. Oligodendrocytes: myelination and axonal support. Cold Spring Harb Perspect Biol. 2015;8:a020479. doi: 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Niu J, Munji R, Davalos D, Chang J, Zhang H, Tien AC, Kuo CJ, Chan JR, Daneman R, Fancy SP. Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science. 2016;351:379–384. doi: 10.1126/science.aad3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell Biochem. 2011;52:25–73. doi: 10.1007/978-90-481-9069-0_3. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. Mouse ENCODE Consortium. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hawes J, Popov KM, Jaskiewicz J, Shimomura Y, Crabb DW, Harris RA. Site-directed mutagenesis of phosphorylation sites of the branched chain alpha-ketoacid dehydrogenase complex. J Biol Chem. 1994;269:18583–18587. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.