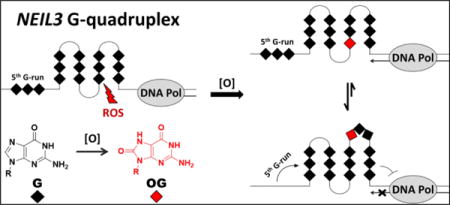
Abstract
DNA oxidation is an inevitable and usually detrimental process but the cell is capable of reversing this state due to possessing a highly developed set of DNA repair machineries, including the DNA glycosylase NEIL3 that is encoded by the NEIL3 gene. In this work, the G-rich promoter region of the human NEIL3 gene was shown to fold into a dynamic G-quadruplex (G4) structure under near physiological conditions using spectroscopic techniques (e.g., NMR, circular dichroism, fluorescence, and UV-vis) and DNA polymerase stop assays. The presence of 8-oxo-7,8-dihydroguanine (OG) modified the properties of the NEIL3 G4 and entailed the recruitment of the 5th domain to function as a “spare tire,” in which an undamaged 5th G-track is swapped for the damaged section of the G4. The polymerase stop assay findings also revealed that owing to its dynamic polymorphism, the NEIL3 G4 is more readily bypassed by DNA polymerase I (Klenow Fragment) compared to well-known oncogene G4s. The present study identifies the NEIL3 promoter possessing a G-rich element that can adopt a G4 fold, and that when OG is incorporated, the sequence can lock into a more stable G4 fold via recruitment of the 5th track of Gs.
Graphical abstract
INTRODUCTION
Damage to DNA can result from oxidative stress due to chronic exposure to exogenous oxidants (e.g., ionizing radiation, UV light, and genotoxic agents) and endogenous reactive oxygen species (ROS) generated by metabolic and inflammatory processes. Oxidation of genomic DNA can lead to the acceleration of aging and the development of chronic diseases such as cancer, neurodegenerative disorders, immune deficiencies, cardiovascular and pulmonary ailments, and diabetes.1–5 Sequences of DNA bearing electron-rich guanine (G), the nucleobase that has the lowest reduction potential (E°= 1.29 V),6 are especially prone to undergo oxidation and yield 8-oxo-7,8-dihydroguanine (OG) as a major oxidation product.7,8 It has been estimated that up to 100,000 OGs are formed per cell daily,9–11 and it is the most commonly measured biomarker of ROS-induced DNA oxidation to date.12–19 The longstanding view regarding cellular ROS is that they interfere with normal cellular processes and are generally believed to be detrimental to the viability of the cell; however, it was pointed out early on that ROS can function as signaling molecules inside cells.20–22
Likewise, OG lesions in DNA have been largely documented to have a negative impact on cellular activities such as transcription. The presence of OG was shown to reduce the binding affinity of the transcription factors SP1,23 NF-κB,24 and CREB25 when located in the recognition sequences for these proteins, inhibit RNA polymerase elongation,26 and can cause transcriptional gene inactivation.27 Recently, there is a growing consensus among our laboratory and others suggesting that OG can also serve as a regulatory element in the genome.28–31 For instance, the gene expression levels of BCL2,32 SIRT1,33 VEGF,34 and TNF-α31 are enhanced in correlation with increased OG present in the promoter regions. Recent work in our laboratory incorporated OG into specific sites of the G-rich VEGF promoter sequence regulating a reporter gene and demonstrated that when OG was present in this sequence gene expression increased in mammalian cells.35,36 A comprehensive PCR-based gene expression profiling assay in liver and colon tissues from mice with infection-induced colitis divulged numerous genes involved in cell cycle checkpoint, oxidative stress response, and DNA repair that were upregulated or downregulated concurrently with high genomic OG content.37 Furthermore, an interplay between OGG1, the glycosylase responsible for excision of OG, and the KRAS promoter sequence that contains a G-rich region has been recently reported.38
One of the DNA repair genes identified to be activated by ROS is NEIL3 that is known to have an expression pattern that is cell cycle regulated.37,39–41 This gene codes for the human enzyme NEIL3 (Nei-like or Endonuclease VIII-like 3) belonging to a class of glycosylases homologous to the bacterial Fpg/Nei family that recognize oxidatively damaged DNA.42,43 It was found that NEIL3 expression commences in the early S phase and is sustained through the G2/M phase by the Ras-dependent ERK-MAPK pathway,40 and it is mostly limited to occur in proliferating cells, including cancer cells. For instance, NEIL3 expression in humans has been reported in the thymus and spleen,44,45 while in mice it was detected in thymus, spleen, blood cells, bone marrow and in the developing brain.45–47 Knockout mice experiments revealed that NEIL3 expression is involved in neurogenesis48,49 and atherogenesis.50 Interestingly, experiments using the Drosophila midgut revealed that stem cell proliferation is induced by ROS via activation of the Ras/MAPK pathway.51 Moreover, NEIL3 is overexpressed in various tumor tissues,47 as well as in primary malignant melanoma associated with metastatic progression.52 Based on these observations, it is apparent that NEIL3 is expressed in cells that are in the proliferative state under oxidative conditions; however, not all cell lines have been tested.
A closer look at the GC-rich but TATA-less promoter of the NEIL3 gene uncovers a classical sequence motif in the coding strand that is a potential G-quadruplex sequence (PQS) capable of intramolecular G-quadruplex (G4) folding (Figure 1). The folding rule would propose that a sequence of the format 5`-d(G3+N1-7G3+N1-7G3+N1-7G3+)-3`, wherein the loop N refers to any nucleotide, has the potential to fold into a G4 structure near physiological conditions (100 mM KCl at pH 7.4, 10 mM Tris-HCl).53 Recently, this widely accepted consensus has been broadened to include G4 sequences that contain even larger loops as well as bulges in the G-tracks.54–56 DNA G4 structures have been detected in cells predominantly in the telomeric and regulatory regions of the human genome.57–59 High-throughput sequencing data revealed that 716,310 G-rich sequences in the human genome may adopt G4 folds during high-throughput sequencing,60 and more than 40% of human genes are predicted to carry at least one PQS in the promoter region.56,61 It is proposed that the enrichment of gene promoters with PQSs suggests that G4 structures may regulate vital biological processes such as DNA replication, transcription, translation, and overall genome stability.62,63 A high-resolution genome-wide mapping of human chromatin discovered around 10,000 PQSs folded to G4s primarily in regulatory, nucleosome-depleted genes linked to elevated transcription.59 We have recently demonstrated that G4-regulated genes can be activated when OG is present in a coding strand G4,36 and we also discovered that the average density of PQSs in human DNA repair genes was found to be double the amount of what is detected in all coding and non-coding genes.64 Coupled with the observation that NEIL3 is activated by ROS and has a promoter PQS in the coding strand, this suggests a possibility that G oxidation in the NEIL3 PQS may function to activate the gene under oxidative stress condition. Thus, it is important to find out whether the NEIL3 promoter forms an alternative structure, in particular a G4, as a crucial step in understanding its fundamental role in cellular processes.
Figure 1.
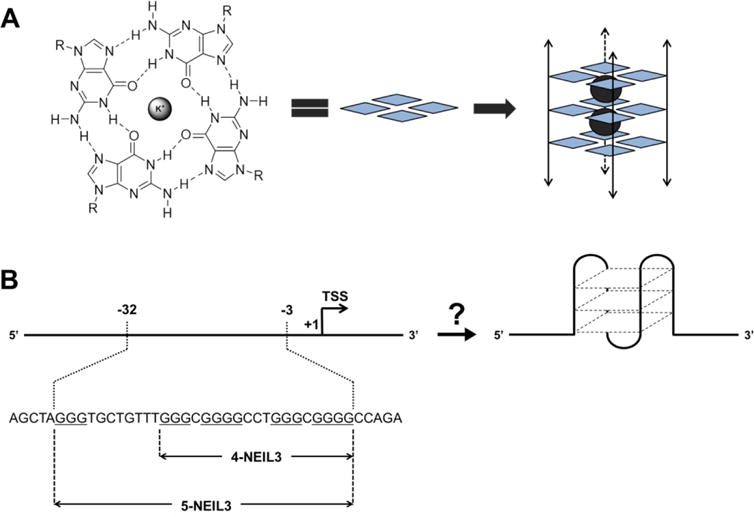
(A) Hoogsteen H-bonding interactions between guanines in a G-tetrad and its graphical representation (guanine = blue diamond, K+ = gray sphere) assembling into a G4 structure. The double-headed arrows aligning the G-tetrads symbolize either intermolecular or intramolecular folding with undefined strand polarities. (B) The promoter region of the human NEIL3 gene containing a G-rich coding strand that may switch to an intramolecular G4 fold. Oligonucleotides including four (4-NEIL3) and five (5-NEIL3) G-tracks were analyzed in this study.
Here, we report characterization of the NEIL3 PQS using spectroscopic methods for G4 formation such as nuclear magnetic resonance (NMR), circular dichroism (CD), fluorescence, and UV melting curve analysis (Tm), as well as a DNA polymerase stop assay. An increasing number of studies have confirmed the formation of G4s from the promoter sequences of DNA repair genes,36,64,65 and the present work aims to expand this category.
It is known that NEIL3 excises spiroiminodihydantoin (Sp) and 5-guanidinohydantoin (Gh) from single- and double-stranded DNAs66 but has a preference for removal of the hydantoins from telomeric and promoter G4 sequences.67,68 These NEIL3 substrates, which are the two-electron oxidation products of OG, are highly mutagenic,69,70 and therefore must be removed and initiate DNA repair. The NEIL3 gene has a regulation pattern indicating that NEIL3 is implicated in replication-associated repair of DNA lesions in proliferating cells.40 These observations underline the association of NEIL3 with cellular proliferation, oxidation and repair and emphasize the need to study what happens when the G-rich promoter sequence of the NEIL3 gene is exposed to oxidative stress. Thus, we also examined how the NEIL3 G4 fold responds to the synthetic incorporation of OG in different positions and monitored the changes in its properties via spectroscopy and a DNA polymerase stop assay.
Our laboratory has hypothesized that PQSs in the genome may function as sensors of oxidative stress involving OG formation as an epigenetic-like factor.36 The key feature that optimized gene induction of VEGF and NTHL1 promoters is the possession of the 5th G-track or the “spare tire” domain. The impact of the 5th G-track on the G4 fold has been explored when the presence of lesions, such as an OG or an abasic site, led to the alteration of the G4 configuration that depends on the location of the base lesions.38,68,71,72 Our laboratory in collaboration with the Wallace laboratory has successfully demonstrated a relevance for the 5th G-track to function as a “spare tire” when it recreated the G4 fold and facilitated the extrusion of a base lesion-containing G-run that subsequently became a substrate for base excision repair (BER) enzymes.73 The NEIL3 PQS also bears a 5th G-track (Figure 1B); thus, we investigated whether the “spare tire” hypothesis also applies to this gene of interest. Whether the NEIL3 5th domain is required to maintain the G4 fold when OG is present is also among the topics we have addressed.
MATERIALS AND METHOD
Oligonucleotide preparation
All oligonucleotide sequences used in this study were synthesized and deprotected at the University of Utah DNA/Peptide Core Facility using standard protocols, followed by HPLC purification. An anion-exchange HPLC column was used and the mobile phases were as follows: solution A = 1:9 acetonitrile:ddH2O, solution B = 1.5 M lithium acetate in 1:9 acetonitrile:ddH2O (pH 7). The purification was accomplished using a linear gradient of 5% to 100% of solution B over 30 min while the absorbance was measured at 260 nm. The purified DNAs were desalted by dialysis against ddH2O for 48 h then lyophilized using a centrifugal evaporator. The freeze-dried samples were resuspended in ddH2O and their concentrations were calculated using the values obtained from their respective absorbance at 260 nm and extinction coefficients derived from the nearest neighbor method. Annealing of the duplexes and the G4s was achieved at the specified concentrations and buffers by heating the samples at 90 °C for 5 min, and then cooling them to room temperature for about 3 hours. The samples were stored at 4 °C for at least 24 h before analysis.
1H-NMR Spectroscopy
The sample (300 μM) was annealed in 20 mM potassium phosphate buffer (pH 7) containing 50 mM KCl and 10% D2O. The samples were analyzed on an 800-MHz NMR spectrometer at 24 °C running a Watergate solvent suppression pulse sequence that allowed observation of the G:G Hoogsteen imino peaks in G-tetrads at 10-12 ppm.
CD Spectroscopy
The CD spectra were collected at 25 °C using a 1.0-mm quartz cuvette containing the samples (20 μM) in 20 mM cacodylic acid (pH 7.4) with 140 mM KCl and 12 mM NaCl. For the K+-dependent studies, the samples were annealed in potassium phosphate buffer (pH 7) with the K+ concentration ranging from 1-200 mM. Plots of molar ellipticity were generated by normalizing the background-subtracted data.
Melting Temperature Analysis
The melting temperature (Tm) values were determined for the samples (5 μM) in buffered solutions (20 mM cacodylic acid, pH 7.4) containing 140 mM KCl and 12 mM NaCl. The samples were thermally equilibrated at 20 °C for 3 min before being heated to 100 °C then cooled back down to 20 °C at a rate of 1 °C/min. Absorbance readings at 295 nm were measured in triplicate samples. The melting curves were plotted from the absorbance at 295 nm vs. temperature, and the Tm values were determined using the instrument’s software. To calculate the number of coordinated K+ ions in the G4 fold (ΔnK+), the Tm studies were performed on samples (5 μM) that were annealed in the presence of 10, 50, 100, 150 and 200 mM K+ ions in potassium phosphate buffer (pH 7) following a method previously described.74
Thioflavin T Fluorescence Enhancement Assay
Experiments were performed at 25 °C using the method developed by Renaud de la Faverie et al.,75 with some modifications. Triplicate samples (1 μM) were mixed with 0.5 μM thioflavin T (ThT) in Tris-HCl buffer (25 mM Tris, pH 7.4) containing 140 mM KCl and 12 mM NaCl. Fluorescence emission of highly fluorescent 1:1 complexes was measured at 490 nm after excitation at 425 nm using Hitachi F-7000 fluorescence spectrophotometer.
DNA Polymerase Stop Assay
This assay is a modification of a method described elsewhere.76,77 The 15-mer primer used for the reaction was 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase, and then purified using a PD SpinTrap™ G-25 chromatography column (GE Healthcare Life Sciences, UK) following the manufacturer’s protocol. The radiolabeled primer was then annealed to a 60-mer sequence carrying the NEIL3 PQS (1:2 primer:template) in Tris-HCl buffer (25 mM Tris, pH 7.4) with 140 mM KCl, 12 mM NaCl and 5 mM MgCl2. The extension reaction (20 μL) was initiated by introducing 2′-deoxyribonucleotide triphosphates (final concentration 100 μM) to the primer-annealed template sample (100 nM) that was pre-equilibrated with Klenow Fragment deficient in exonuclease activity (2.5 U/reaction; New England Biolabs) at 37 °C. After a 30 min incubation period, an equal volume of the stop buffer (95% formamide, 10 mM EDTA, 10 mM NaOH, 0.1% bromophenol blue and 0.1% xylene cyanol) was added into the mixture to quench the reaction and then heated to 90 °C for 5 minutes. For the control experiment, the assay was performed on samples in buffered solution containing 152 mM LiCl instead. The products were separated on a denaturing 20% polyacrylamide gel that was electrophoresed for about 3 hours. The bands were then visualized using storage-phosphor autoradiography. The relative signal intensities were calculated using Imagequant software.
RESULTS AND DISCUSSION
The guanine-rich promoter sequence of the human NEIL3 gene forms a dynamic mixture of G-quadruplexes
The G-rich promoter of the human NEIL3 gene carries a G4 sequence motif that is 3 nucleotides (nt) upstream of the transcription start site (TSS), and contains five consecutive G-runs wherein the fifth domain is at the 5′ end separated 8 nt away from the 4 G-runs (Figure 1B). For this study, we analyzed the NEIL3 PQSs that include and exclude the 5th G-run, hereby labeled as 5-NEIL3 and 4-NEIL3, respectively. To determine whether the native NEIL3 PQS folds into a G4, different spectroscopic techniques were utilized.
The 1D 1H-NMR spectra of the native NEIL3 sequences (i.e., 5-NEIL3 native and 4-NEIL3 native) showed the presence of imino protons diagnostic of G:G Hoogsteen base pairs in a G-tetrad that aid in confirming the formation of a G4 fold (Figure 2A). The broad nature of the peaks supports a complex mixture of interconverting G4 species, consistent with the ability of these sequences to adopt many possible folds in solution on the basis of the sequences having two G tracks with 4 Gs (Figure 1B). Comparison of the imino regions of native 5-NEIL3 and 4-NEIL3 sequences showed that the loss of the 5th G-run leads to more defined peaks probably as a consequence of limiting the possible number of G4 conformations in solution. To further evaluate this, we obtained a 1H-NMR spectrum of a 5-NEIL3 variant wherein the 5th G-run was replaced by a thymine repeat or T-run, and found that it also produced a more defined imino region compared to that of the native 5-NEIL3 (Figure S1). This suggests that the 5th G-run affects the equilibrium mixture of folded conformations and makes the NEIL3 G4 more dynamic. Conformational dynamics is a distinctive aspect of many G4 structures as is the case for known promoter G4s such as c-MYC,78 VEGF,79 and BCL-2.80 It is evident that the NEIL3 G4 also exhibits dynamic polymorphism.
Figure 2.
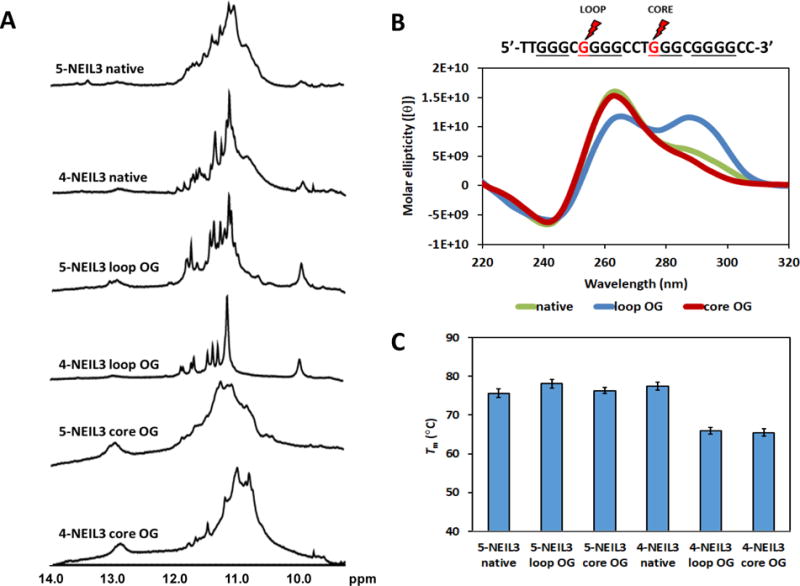
Characterization of the 5- and 4-track G4 motif in the promoter sequence of the human NEIL3 gene by (A) 1H-NMR, (B) CD, and (C) Tm with and without OG. Positions of the loop and core OGs in the primary sequence are marked with the red lightning symbols and are red. Only the CD spectra for the 4-track sequences, which share similar profiles with the 5-track sequences, are shown here for brevity. Error bars correspond to 95% confidence intervals derived from triplicate trials.
The high degree of structural polymorphism in the native NEIL3 G4s is also supported by their CD spectra (Figure 2B). Analogous CD profiles were obtained for native 5-NEIL3 and 4-NEIL3 sequences with slight differences in their intensities (Figure S2), which means that the presence or the absence of the 5th G-run does not affect the topology of the native NEIL3 G4s; thus, only the spectra for the 4-NEIL3 sequences are displayed in the text for brevity and those for the 5-NEIL3 sequences can be found in Figure S2. In the presence of physiologically relevant cationic concentrations that mimic intracellular conditions, i.e., 140 mM K+ and 12 mM Na+, the native NEIL3 G4s form a mixture of parallel and anti-parallel topologies. The claim of a mixture of folds for NEIL3 G4s is based on the similarity in CD profiles recorded for the hTERT and BCL-2 G4s.80,81 The similarities include a dominant positive band at 260 nm with a broad shoulder around 290 nm and a negative band at 240 nm indicate G4 structures with a mixture of (3+1) and parallel folds, as elucidated by multi-dimensional NMR methods.
To determine the thermal stabilities of the native 4 and 5 G-track NEIL3 G4s, we performed UV melting studies wherein the denaturation of the G4 was monitored via the absorbance at 295 nm across a temperature gradient (Figure 2C and Figure S3). The Tm studies found both native NEIL3 sequences had Tm values >70 °C. These values are relatively high suggesting they would be stable under physiological temperatures.
In the next studies, enhancement of ThT fluorescence was used as another predictive assessment tool to further confirm the NEIL3 G4s folded. It was demonstrated that ThT fluorescence is significantly enhanced upon binding to DNA or RNA G4s compared to other single/double-stranded secondary structures.75,82 Therefore, we measured the fluorescence of ThT when bound to the native 5-NEIL3 or 4-NEIL3 sequences and compared them to known oligonucleotide sequences (Table 1). First, the negative controls that include single- and double-stranded DNA sequences, as well as a triplex forming sequence (i.e., a modified human telomeric sequence that will not form a G4 because one of its G-track was replaced by a thymine repeat or T-run) yielded a small enhancement in ThT fluorescence similar to previous studies (Figure 3B).75 The results for the native 4 and 5 G-track NEIL3 G4s showed a fluorescence enhancement of nearly 180-fold that is a value similar to the established G4s c-MYC and the human telomere sequence (Figure 3B). A limitation for this assay is that ThT cannot distinguish between G4 conformations, but it was demonstrated in other studies that ThT can bind both parallel and anti-parallel G4s;83,84 thus, we assume that ThT was also able to recognize both the parallel and anti-parallel folds that constitute the folded mixture observed for the native NEIL3 G4s. All the spectroscopic information gathered in this study presents evidence supporting that the motif of guanines, including the 5th domain, in the promoter region of the human NEIL3 gene can assemble into a G4 fold in vitro.
Table 1.
Oligonucleotide sequences used in the thioflavin T fluorescence enhancement assay.*
| Name of oligonucleotide | Description | Primary Sequence |
|---|---|---|
| hTel G4 | human telomeric G-quadruplex sequence | 5′-TAGGGTTAGGGTTAGGGTTAGGGTT-3′ |
| c-myc G4 | c-myc G-quadruplex sequence (2,3,4,5 G-track) | 5′-TGAGGGTGGGGAGGGTGGGGAA-3′ |
| 4-NEIL3 native | unmodified 4 G-track NEIL3 G-quadruplex sequence | 5′-TTGGGCGGGGCCTGGGCGGGGCC-3′ |
| 5-NEIL3 native | unmodified 5 G-track NEIL3 G-quadruplex sequence | 5′-TAGGGTGCTGTTTGGGCGGGGCCTGGGCGGGGCC-3′ |
| 4-NEIL3 loop OG | 4-NEIL3 with OG in the loop position | 5′-TTGGGC GGGCCTGGGCGGGGCC-3′ |
| 5-NEIL3 loop OG | 5-NEIL3 with OG in the loop position | 5′-TAGGGTGCTGTTTGGGC GGGCCTGGGCGGGGCC-3′ |
| 4-NEIL3 core OG | 4-NEIL3 with OG in the core position | 5′-TTGGGCGGGGCCT GGCGGGGCC-3′ |
| 5-NEIL3 core OG | 5-NEIL3 with OG in the core position | 5′-TAGGGTGCTGTTTGGGCGGGGCCT GGCGGGGCC-3′ |
| hTel triplex | human telomeric triplex (one G-run is replaced with a T-run) | 5′-TAGGGTTAGGGTTATTTTTAGGGTT-3′ |
| 4-NEIL3 C-rich | ssDNA C-rich complementary strand of 4-NEIL3 native | 5′-GGCCCCGCCCAGGCCCCGCCCAA-3′ |
| 5-NElL3 C-rich | ssDNA C-rich complementary strand of 5-NEIL3 native | 5′-GGCCCCGCCCAGGCCCCGCCCAAACAGCACCCTA-3′ |
| DNA duplex | DNA duplex combined from the G- and C-rich complementary strands of 4-NEIL3 native | (a) 5′-TTGGGCGGGGCCTGGGCGGGGCC-3′ (b)3′-AACCCGCCCCGGACCCGCCCCGG-5′ |
The synthetically incorporated OGs are indicated by the red Gs.
Figure 3.
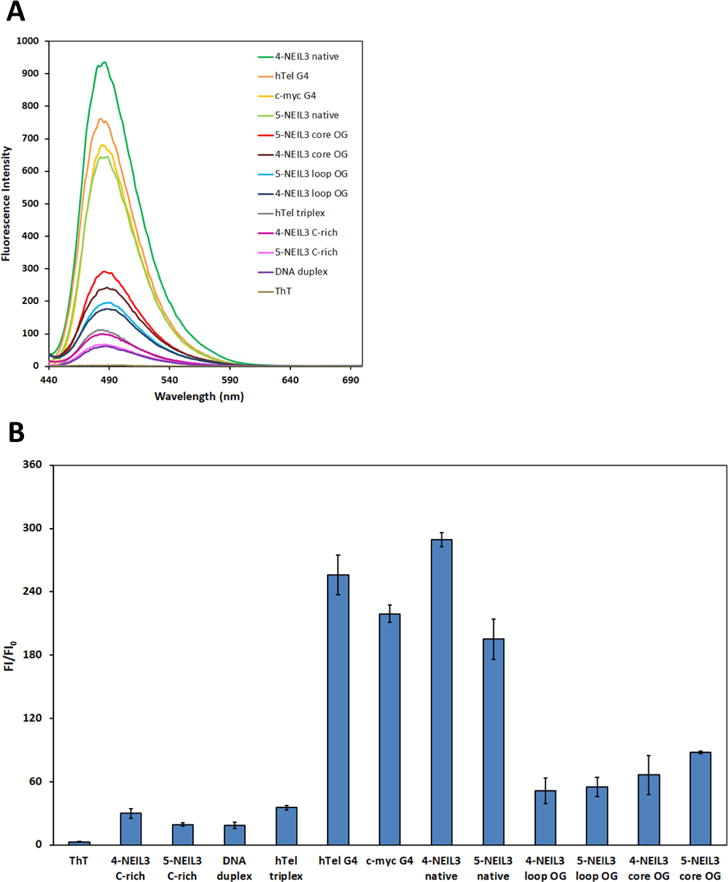
(A) Fluorescence emission spectra of ThT alone and in the presence of various oligonucleotide sequences listed in Table 1. (B) Bar graph of ThT fluorescence enhancement. Error bars correspond to 95% confidence intervals. The 4-NEIL3 C-rich and 5-NEIL3 C-rich sequences are the complementary strands to the G-rich sequences, and they were utilized as single-stranded controls.
Oxidative damage to G destabilizes the NEIL3 G-quadruplex and induces a topological transition as a function of lesion position
After initial characterization of the native NEIL3 sequences via spectroscopic methods and establishing that it can fold into a mixture of G4s, we examined the differences in behavior of the NEIL3 G4 when one of the Gs in the primary sequence was oxidized to OG. We studied this by synthetically incorporating an OG into the NEIL3 sequences and then monitoring the changes in the properties of the spectroscopic signatures as a function of OG position while comparing it to the undamaged native sequences. The OG positions studied in the 5-track and 4-track NEIL3 PQSs were located on the 5` side of a 4-G track or on the 5` side of a 3-G track (Table 1). The OG in the 4-G track is likely positioned in a loop because the other 3 Gs in the track can form G-tetrads, while the OG in the 3-G track causes a disruption to the core of the G4 fold. Previous studies have found that when OG replaces a core OG, the new structure is destabilized.68,85 The positions were selected based on a previous observation form our laboratory that mapped the oxidation-prone sites in the VEGF promoter sequence identified the 5′-most side of each G-run as the most susceptible to oxidation.73
The results showed that the effect of OG in the G4 fold is location dependent. The appearance of sharper 1H-NMR peaks accompanied the substitution of a loop G with an OG (Figure 2A). The 5-NEIL3 sequence with a loop OG carries 4 G-runs that contain 3 Gs each and one G-run that has 4 Gs, and this sequence produced an imino spectrum that is more resolved than that of the native 5-NEIL3 sequence. The profound effect on the 1H-NMR resolution caused by restricting the probable number of NEIL3 G4 conformations via replacement of G with an OG is particularly evident for the 4-NEIL3 loop sequence. The loop OG-containing 4-NEIL3 sequence only contains 3 G-runs with 3 Gs each and one G-run with 4 Gs, and this is predicted to lock the NEIL3 G4 into a more rigid fold manifesting in an 1H-NMR spectrum that has distinct peaks.
In contrast, the substitution of a core G with an OG in either the 4 or 5 track NEIL3 sequence resulted in a drastic loss in the peak resolution observed in the 1H-NMR spectrum. The broadening of the imino regions for these sequences with core OG reflects the highly disruptive impact of OG on the NEIL3 G4 fold (Figure 2A). Whereas the 1H-NMR spectra of the native sequences indicate overlapping imino peaks due to dynamic polymorphism, the spectra of the sequences that contain core OG reveals indistinguishable imino peaks due to destabilization of the G4 fold. This is because OG cannot participate in Hoogsteen base pairing with a G.68 Thus, a loop OG stabilizes the organization of the G4 fold while a core OG weakens it.
The thermal melting analysis revealed that the presence of OG is slightly destabilizing for the 4 G-track sequence but was not disruptive to the stability when the 5th G-track was present (Figure 2C and Figure S3). From the standpoint of human physiology the OG-containing NEIL3 sequences formed stable folds that have Tm values that are well above the homeostatic setting. This may indicate that an oxidative base lesion can possibly exist in a stable G4 fold inside the cell.
The thermal melting behaviors of the 5-NEIL3 G4s with OG are similar to those previously observed in our laboratory with the VEGF G4 and human telomere G4 bearing OG whose 5th domain also raised the thermal stabilities of core damaged sequence back to the level of the undamaged sequence;72,73 additionally, the Xodo laboratory has documented the same feature for the 5th G run when the KRAS G4 possessed an oxidized G site.38
The CD spectra revealed that the OG-containing NEIL3 G4 sequences undergo a conformational change as a function of the OG position. The CD profile of the native NEIL3 G4 sequences show a mixture of parallel and anti-parallel folds but it changes into one predominant fold when there is an OG in the sequence (Figure 2B). On the basis of comparisons to literature reports,81,86 the presence of OG induced a location-dependent shift in the sequence folding preference wherein the loop damaged G4 sequences favored an anti-parallel fold while the core damaged G4 sequence favored a parallel-stranded fold (Figure 2B). The CD spectra of loop and core OG-containing 5-NEIL3 and 4-NEIL3 sequences were similar except for the slight differences in their intensities (Figure S2). A similar impact of OG on the topology of a G4 was also observed in the VEGF G4.73 This differential effect of OG in either the loop or core position of the G4 fold is also observed in a parallel study using NEIL3 sequences with modifications in the other G-runs (Figure 4).
Figure 4.
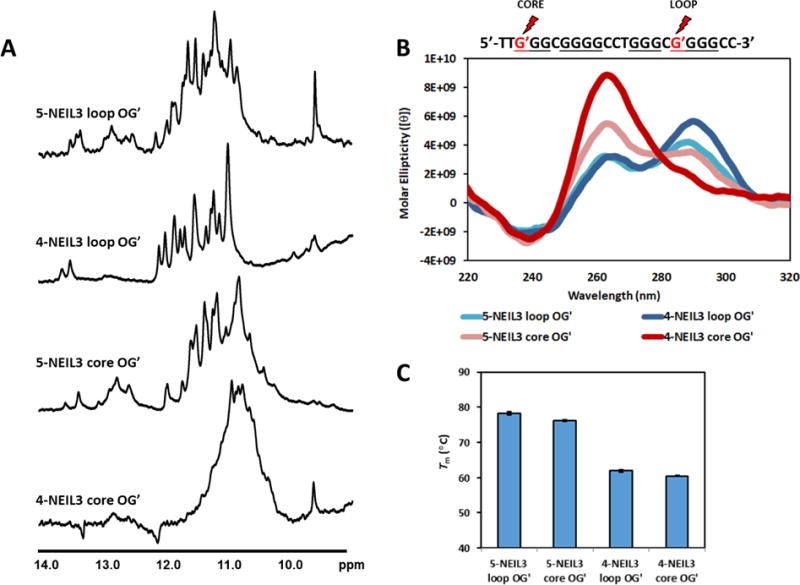
(A) 1H-NMR, (B) CD, and (C) Tm values of NEIL3 G4 sequences with OG in a different putative loop or core (indicated by the a G′ and marked by a red lightning symbol) position in the fold. The primary sequence of the 4-track NEIL3 G4 is shown but the location of OGs are exactly the same as that in the 5-track. Error bars correspond to 95% confidence intervals derived from triplicate trials.
As expected, well defined peaks were observed when the OG position is in the loop while the opposite occurs when the OG is in the core position as seen in the 5- and 4-NEIL3 loop OG’ and core OG’ sequences, respectively (Figure 4A). Moreover, the locking effect of the 5th G-track on the folding of NEIL3 G4 is more pronounced; the imino protons of the 5-NEIL3 core OG’ sequence are greatly enhanced compared to that of the 4-NEIL3 core OG’ sequence. Topological changes were also evident depending on the OG location (Figure 4B). The thermal stabilities of these sequences (Figure 4C) were also found to be consistent with those analyzed in Figure 2C.
All OG-containing NEIL3 sequences were found to be capable of enhancing the ThT fluorescence greater than that observed with other non-G4 DNA secondary structures studied (Figure 3B). However, the ThT fluorescence enhancement was observed at a much lower level compared to the native sequences and known G4s. The reason for this relatively low fluorescence enhancement is not known as the chemical interaction between OG and ThT has not been studied. Nevertheless, the present ThT data provide additional support that OG-containing NEIL3 sequences can adopt G4 folds.
The 5th domain of the NEIL3 promoter sequence is required to form the G-quadruplex in the presence of oxidative damage and to stall replication
A sensitive and specific indicator of G4 formation is the ability of the fold to block or arrest DNA synthesis with dependency on the presence of K+.76 Thus, we determined whether the NEIL3 PQS exhibits this diagnostic property using the DNA polymerase stop assay to confirm NEIL3 G4 formation. In this assay, we performed the DNA polymerase extension reactions in buffers containing physiological cation concentrations mimicking the intracellular environment (140 mM K+, 12 mM Na+, 5 mM Mg2+), and we monitored the formation of gel bands indicating arrest at the G4 site (Figure 5A). The G4 site would be where the replicative enzyme Klenow Fragment will be stalled as it encounters either of the first two guanines along the sequence participating in a stable G4 fold. Control experiments were carried out in buffers containing LiCl of the same ionic strength as the previous studies wherein stable G4 folds will not form,87,88 and thus, arrest bands on the gel would not be observed.
Figure 5.
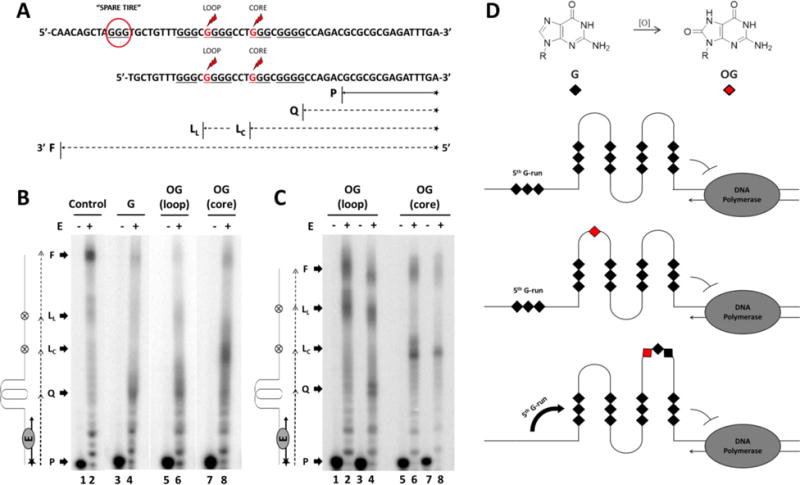
(A) The NEIL3 gene promoter sequence specifying where the “spare tire” domain (enclosed in a red circle) and the OG positions (indicated by the red Gs marked with the red lightning symbols) are placed. Extension of the annealed 15-mer primer P with expected arrest sites either at the G4 site Q, or at the base lesion OG in the loop LL or core position LC are marked while the fully synthesized DNA product is designated as F. (B-C) Autoradiographs of the DNA polymerase stop assay. Primer annealed-template DNAs were incubated at 37 °C for 30 minutes with (+) or without (−) the Klenow Fragment E. (B) DNA polymerase stop assays of five-track NEIL3 G4s with or without OG. Control lanes were reactions using the native strand in LiCl. (C) DNA polymerase stop assays of OG-containing four-track NEIL3 G4s. Lanes 1-2 and 5-6 were control experiments run in buffer with LiCl. (D) Model illustrating the NEIL3 G4 stalling the DNA polymerase and the 5th G-run acting as a “spare tire” that reconstructs the G4 fold in the presence of an oxidative base lesion.
The results showed that G4 formation was detected for the five-track NEIL3 sequences with and without OG as signified by the emergence of arrest bands in the expected G4 site that were not observed in the control (Figure 5B). About 40% of the band intensity distribution constituted the native sequence that stopped at the G4 site. However, even though G4s in the template strand were able to impede the activity of the DNA polymerase, the appearance of a band signifying the fully extended sequence (<10%) may suggest that the enzyme was able to bypass the G4 fold or that some of the conformations are more easily linearized by the enzyme. In the LiCl solution, the band intensity distribution for the fully extended sequence was approximately 35%. When there was a loop OG, the DNA polymerase also paused at the lesion site that constitutes about 10% of the band intensity distribution. The blockage intensity at the G4 site of the loop OG-containing sequence was similar to that of the native sequence. The observation that the loop OG-containing NEIL3 sequence resulted in stalling of the polymerase at the G4 suggests the lesion was well tolerated in the fold.
The susceptibility of the G4 structure to polymerase bypass became most prominent when the OG was placed at the core position of the 5-track NEIL3 sequence (Figure 5B); the band intensity distribution diminished at the G4 site (15%) but increased at the lesion (30%) and remained constant at the full extension site as that of the native sequence (Figure 5B). This suggests that the core damage weakened the G4 structure and made it easier for the DNA polymerase to relax the G4 and process the sequence until it stops at the lesion site or at the end of the sequence. These results reassert that a core OG is more destabilizing to the G4 structure than a loop OG.
Next, we wanted to determine the impact the 5th domain had on stalling the polymerase when OG was present. This would allow us to determine if the 5th domain was replacing the damaged G-track as previously documented for the VEGF G4 sequence.73 This was achieved by observing what happens when the 5th domain was removed while oxidative damage was present during the polymerase stop assay (Figure 5A). Arrest bands at the expected G4 and lesion sites were observed for the OG-containing 4-track sequence wherein both showed roughly equivalent band intensity distribution at 25% (Figure 5C). It is interesting to note that when there was a 5th domain, the G4 site presented a stronger barrier for the DNA polymerase than the OG site, but when it was removed the enzyme stopped equally at the two sites. This signifies that the 5th domain strengthens the G4 fold when a lesion was present. Meanwhile, when the OG was at the core position the arrest band at the G4 site was not detectable (Figure 5C). Instead, only the arrest bands at the lesion (~35%) and fully extended sequences (~20%) were produced that have similar intensity distribution as the control. This finding signifies that the 5th domain was required to form a G4 stable enough to block the DNA polymerase (Figure 5D). According to the “spare tire” hypothesis, in which the 5th G-track exchanges with the damaged G-track and allows the repair process to take place before DNA replication or transcription is resumed with high fidelity.73 Thus, the possession of the 5th domain is especially critical for the NEIL3 gene to ensure faithful DNA replication.
Another noteworthy observation was that the arrest band at the G4 site of the loop OG-containing 4-track sequence showed two distinct arrest bands at the G4 site (Figure 5C). These two bands may correspond to the two possible G4 configurations for this sequence that contain 3 G-runs with 3 Gs each and one G-run with 4 Gs yielding two possible folds. This is in contrast to the diffuse quality of the arrest band at the G4 site produced by the 5-track loop OG-containing sequence (Figure 5B). This might be attributed to the highly dynamic property of the NEIL3 G4 with the 5th domain present.
To further confirm whether the 5th domain of the NEIL3 PQS was utilized as a “spare tire,” we replaced the Gs in the G-run that was presumed to contain a core OG with Ts instead (Figure S4). This experiment works on the assumption that an OG cannot contribute in Hoogsteen H-bonding in a G-tetrad,68 much like a thymine. However, the broad CD spectra of the T-substituted sequences describe a profile that would fit for a mixture of folds (Figure S4). Moreover, their thermal stabilities were much lower with Tm values of 46.1 ± 0.3 °C (5-NEIL3 TGG), 43.4 ± 0.3 °C (5-NEIL3 TTG), and 49.5 ± 0.3 °C (5-NEIL3 T-run) compared to those of the core-damaged sequences at 76.3 ± 0.8 °C (5-track) and at 65.5 ± 0.9 °C (4-track). These results suggest that the 5th G run contributes to the folding of the native NEIL3 G4 sequence and exhibits preferential involvement as a “spare tire” when an OG is present. This study showed that in the oxidative damage context, the 5th G-run served as a “spare tire” and was recruited by the core OG in order to rebuild the destabilized G4 structure, as well as halt the DNA replication process.
The NEIL3 G4 does not arrest DNA synthesis as effectively as established oncogene G4s
We wanted to compare the behavior of the NEIL3 G4 to known promoter G4 sequences such as c-MYC, VEGF, and the hTERT gene promoter G4 sequences. To do this we replaced the NEIL3 G4 motif with the respective motifs from the oncogenes while retaining the original flanking sequences used in the NEIL3 studies (Figure 6A). The results showed that the G4 structures of c-MYC and VEGF obstructed the DNA polymerase more strongly than the NEIL3 G4 did (Figure 6B). However, the hTERT G4 did not block the DNA polymerase as strongly as c-MYC and VEGF, and in fact presented a weak hindrance to bypass similar to that observed for the NEIL3 G4 (Figure 6B). The sensitivity of the DNA polymerase stop assay may also be sequence and G4 topology dependent. The Sugimoto laboratory has shown that the topology of non-canonical G4 structures impact the activity of Klenow Fragment to process a template strand.89 It was established that the VEGF and the c-MYC G4s are parallel stranded,78,79 while the hTERT sequence forms a mixture of parallel and anti-parallel G4,81 much like the NEIL3 G4 studied in this work. This suggests that parallel-stranded G4s present a more solid obstacle to DNA polymerase bypass compared to dynamic G4 structures that equilibrate between parallel and anti-parallel configurations.
Figure 6.
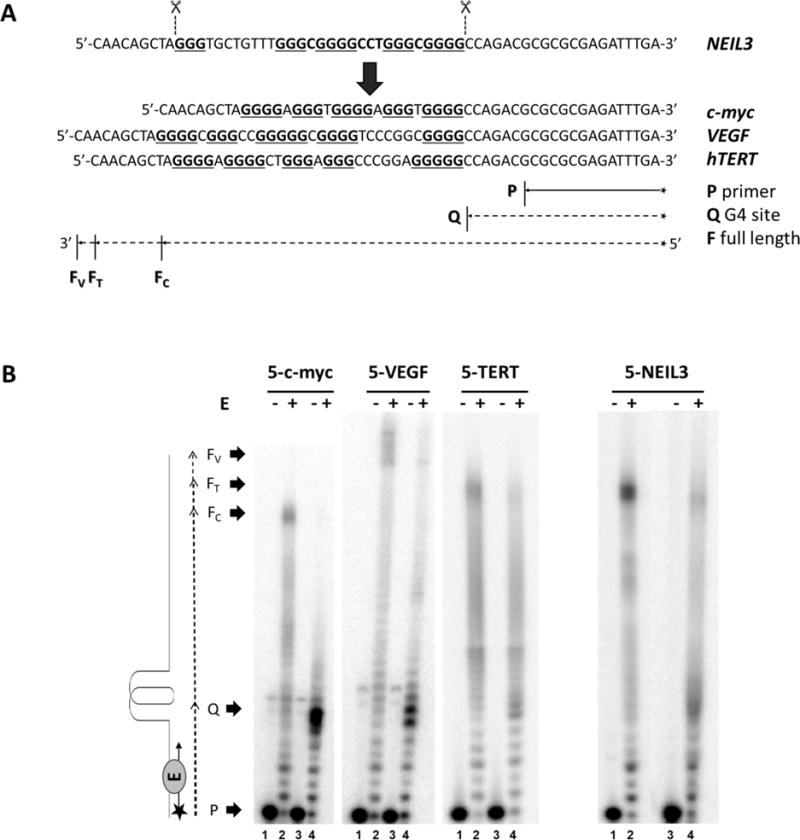
(A) The 5-track G4 sequence of the NEIL3 was replaced with the 5-track G4 sequences of c-MYC, VEGF and hTERT. DNA templates were annealed to a radiolabeled 15-mer primer P. Expected arrest site at the G4 site is marked as Q while the fully synthesized duplex DNA for c-MYC, VEGF and hTERT are designated as FC, Fv and FT, respectively. (B) Autoradiograph of the DNA polymerase stop assay. Primer annealed-template DNAs were incubated at 37 °C for 30 min with (+) or without (−) the Klenow Fragment enzyme E. All even-numbered lanes were reacted in buffered physiological salts while the odd-numbered lanes were assayed in LiCl buffer.
The dynamic property of the NEIL3 G-quadruplex depends on the K+ concentration and the presence of the 5th domain
The G4 stability depends on the coordinately bound K+ ions,88 and the ability to arrest DNA synthesis can be observed even when the KCl concentration is as low as 5 mM.76 However, the native NEIL3 G4 was not able to block the DNA polymerase at 10 mM KCl, unlike the c-MYC G4 (Figure S5), suggesting that the NEIL3 G4 must be highly dynamic and requires very high K+ concentration to stabilize the fold. This led us to use Tm measurements with varying K+ concentration to obtain the van’t Hoff-derived thermodynamic parameters for the folding of NEIL3 G4, as well as to estimate the number of bound K+ ions.73,90 The results revealed that the Tm values of the NEIL3 G4 strands were found to be linearly dependent with increasing K+ concentration (Figure 7A), and all melting curves are displayed in Figure S6. As seen in Figure 7A, the presence of the 5th domain conferred a more pronounced linear dependence as shown by the steeper slopes compared to 4-track sequences. The presence of OG in a core position resulted in the G4 strands to be less sensitive to changes in K+ concentration compared to the loop OG and native NEIL3 sequences.
Figure 7.
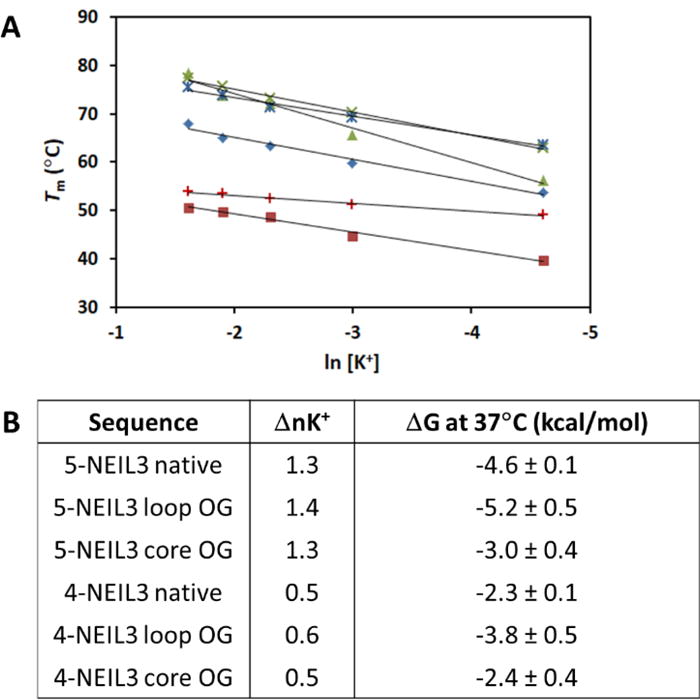
(A) The Tm dependence on K+ concentration.
 : 5-NEIL3 native,
: 5-NEIL3 native,
 : 5-NEIL3 loop OG,
: 5-NEIL3 loop OG,
 : 5-NEIL3 core OG,
: 5-NEIL3 core OG,
 : 4-NEIL3 native,
: 4-NEIL3 native,
 : 4-NEIL3 loop OG,
: 4-NEIL3 loop OG,
 : 4-NEIL3 core OG. (B) Tabulated list of van’t Hoff-derived K+ ion coordination values and favorable ΔG for the formation of each NEIL3 G4 sequences.
: 4-NEIL3 core OG. (B) Tabulated list of van’t Hoff-derived K+ ion coordination values and favorable ΔG for the formation of each NEIL3 G4 sequences.
Favorable ΔG values for the formation of all NEIL3 G4s were obtained. Additionally, the presence of the 5th domain allowed more than twice as much uptake of K+ ions compared to 4-track sequences as indicated by the ΔnK+ values (Figure 7B). Based on the number of Gs in the NEIL3 PQS it should form a typical G4 composed of 3 stacks of G-tetrads that require coordination of 2 K+ ions in the channel, like in the case of the VEGF G4.73 However, the ΔnK+ values obtained were less than 2, which hints at the extremely dynamic polymorphism of the NEIL3 G4 as it equilibrates between different folded states and exchanges K+ ions during the transition. This feature has been previously discussed.90 This property of the native NEIL3 G4 might be one of the reasons why the G4 arrest bands found in the DNA polymerase stop assays have a somewhat indistinct appearance (Figure 5B-C). The complete thermodynamic profile of the NEIL3 G4 sequences is available in the Supporting Information (Table S1).
We also examined how the topology of all the NEIL3 G4s would be influenced by the varying K+ concentration (Figure 8). To this end, we collected the CD profiles for the 5-track and 4-track native and OG-containing sequences as a function of K+ concentration (Figure S7). Both 4-track and 5-track NEIL3 G4 sequences responded the same to changes in K+ concentration; thus, we only show here the CD spectra for the 5-track sequences for brevity. For the native G4 sequence, although the global shapes of the spectra were not altered, the magnitude of the CD band around 260 nm amplified with increasing K+ concentration while those around 240 and 290 nm remained constant (Figure 8A). This increase in the population of the parallel-stranded G4s at higher K+ concentration may explain why the NEIL3 G4 was able to arrest the DNA polymerase bypass more successfully than at lower K+ concentration (Figure S5). Meanwhile, the loop OG-containing sequences did not exhibit K+ concentration dependence as seen in the invariable shape of their CD profiles (Figures 8B). Next, in the core OG-containing sequences, the magnitude of the band near 260 nm also goes up with increasing K+ concentration while the broad band around 290 nm goes down to ultimately fold into a parallel-stranded G4 (Figures 8C). These data suggest that the K+ concentration directs the topology and thus the polymorphic nature of the NEIL3 G4 under normal and oxidatively damaged conditions.
Figure 8.
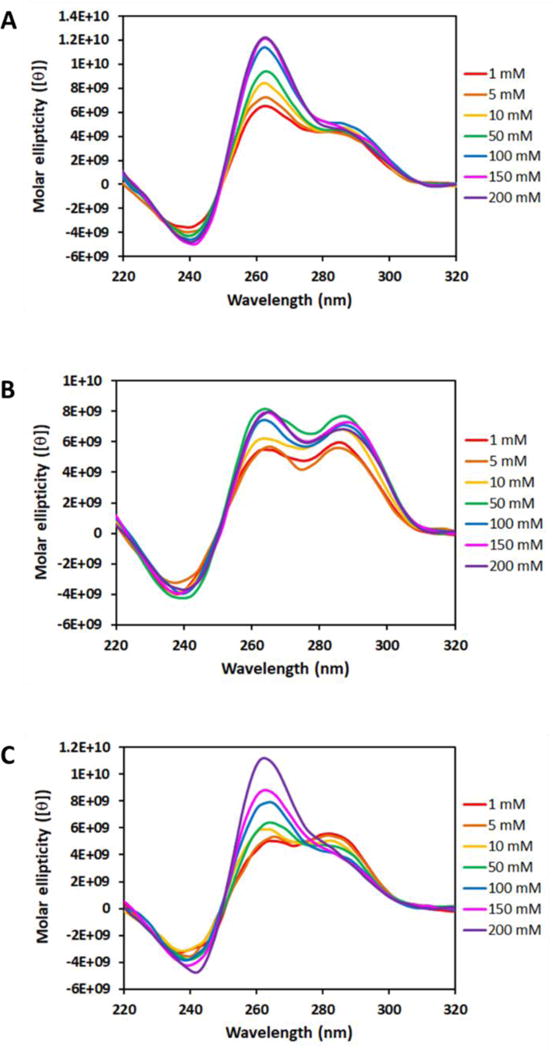
The effect of different K+ concentrations on the CD profiles of NEIL3 PQS without an OG (A) and with an OG in the loop (B) or core (C) position. For conciseness, only the CD spectra for 5-track sequences are presented here due to comparability with those of 4-track sequences.
CONCLUSIONS
Our studies have determined through a series of established methods for characterizing G4 formation that the G-rich promoter sequence of the human NEIL3 gene can adopt a G4 fold. The native NEIL3 G4 sequence was found to be comprised of a mixture of folds with a relatively high Tm value; however, the DNA polymerase bypass study and 1H-NMR results suggest this sequence is highly dynamic (Figures 2 and 4). We have also shown that the presence of OG, a product of oxidative stress damaging the genome, modulates the properties of the NEIL3 G4 when the 5th G track is present to favor a parallel-stranded G4 when OG was located in a core position and an anti-parallel fold when OG is in a loop position. These studies further identify that oxidation of G in a critical G-track for G4 formation can be rescued by recruitment of the 5th G track to lock the G4 structure in a more stable fold, as was determined in a DNA polymerase bypass assay, 1H-NMR, and Tm studies (Figures 2 and 4).
Supplementary Material
Acknowledgments
This research publication was supported by a National Cancer Institute grant (R01 CA090689). The oligonucleotides were provided by the University of Utah Health Sciences Core facilities that are supported in part by a National Cancer Institute Cancer Center Support grant (P30 CA042014).
Footnotes
SUPPORTING INFORMATION
CD spectra, Tm curves, DNA polymerase stop assay results, and thermodynamic values for the oligonucleotide sequences analyzed in this study can be accessed in the Supporting Information that is available free of charge via the internet at http://…
CONFLICT OF INTEREST
The authors declare no competing financial interest.
References
- 1.Yu Y, Cui Y, Niedernhofer LJ, Wang Y. Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem Res Toxicol. 2016;29:2008–2039. doi: 10.1021/acs.chemrestox.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribezzo F, Shiloh Y, Schumacher B. Systemic DNA damage responses in aging and diseases. Semin Cancer Biol. 2016;37–38:26–35. doi: 10.1016/j.semcancer.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 6.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 7.Cadet J, Wagner JR, Shafirovich V, Geacintov NE. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int J Radiat Biol. 2014;90:423–432. doi: 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming AM, Burrows CJ. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic Biol Med. 2017;107:35–52. doi: 10.1016/j.freeradbiomed.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins A, Cadet J, Epe B, Gedik C. Problems in the measurement of 8-oxoguanine in human DNA. Report of a workshop, DNA oxidation, held in Aberdeen, UK, 19-21 January, 1997. Carcinogenesis. 1997;18:1833–1836. doi: 10.1093/carcin/18.9.1833. [DOI] [PubMed] [Google Scholar]
- 13.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Hu CW, Cooke MS, Tsai YH, Chao MR. 8-Oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2′-deoxyguanosine concentrations in various human body fluids: implications for their measurement and interpretation. Arch Toxicol. 2015;89:201–210. doi: 10.1007/s00204-014-1255-1. [DOI] [PubMed] [Google Scholar]
- 15.Andreoli R, Spatari G, Pigini D, Poli D, Banda I, Goldoni M, Riccelli MG, Petyx M, Protano C, Vitali M, Barbaro M, Mutti A. Urinary biomarkers of exposure and of oxidative damage in children exposed to low airborne concentrations of benzene. Environ Res. 2015;142:264–272. doi: 10.1016/j.envres.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Nikitaki Z, Hellweg CE, Georgakilas AG, Ravanat JL. Stress-induced DNA damage biomarkers: applications and limitations. Front Chem. 2015;3:35. doi: 10.3389/fchem.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozalski R, Gackowski D, Siomek-Gorecka A, Starczak M, Modrzejewska M, Banaszkiewicz Z, Olinski R. Urinary 5-hydroxymethyluracil and 8-oxo-7,8-dihydroguanine as potential biomarkers in patients with colorectal cancer. Biomarkers. 2015;20:287–291. doi: 10.3109/1354750X.2015.1068860. [DOI] [PubMed] [Google Scholar]
- 18.Sliwinska A, Kwiatkowski D, Czarny P, Toma M, Wigner P, Drzewoski J, Fabianowska-Majewska K, Szemraj J, Maes M, Galecki P, Sliwinski T. The levels of 7,8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1) - A potential diagnostic biomarkers of Alzheimer’s disease. J Neurol Sci. 2016;368:155–159. doi: 10.1016/j.jns.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Schupp N, Stopper H, Heidland A. DNA damage in chronic kidney disease: evaluation of clinical biomarkers. Oxid Med Cell Longev. 2016;2016:3592042. doi: 10.1155/2016/3592042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halliwell B. Free radicals, proteins and DNA: oxidative damage versus redox regulation. Biochem Soc Trans. 1996;24:1023–1027. doi: 10.1042/bst0241023. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki YJ, Forman HJ, Sevanian A. Oxidants as stimulators of signal transduction. Free Radic Biol Med. 1997;22:269–285. doi: 10.1016/s0891-5849(96)00275-4. [DOI] [PubMed] [Google Scholar]
- 22.Day RM, Suzuki YJ. Cell proliferation, reactive oxygen and cellular glutathione. Dose-Response. 2005;3:425–442. doi: 10.2203/dose-response.003.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramon O, Sauvaigo S, Gasparutto D, Faure P, Favier A, Cadet J. Effects of 8-oxo-7,8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box) Free Radic Res. 1999;31:217–229. doi: 10.1080/10715769900300781. [DOI] [PubMed] [Google Scholar]
- 24.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2‘-deoxyguanosine lesions in the NF-κB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 25.Moore SP, Toomire KJ, Strauss PR. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst) 2013;12:1152–1158. doi: 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Allgayer J, Kitsera N, Bartelt S, Epe B, Khobta A. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016;44:7267–7280. doi: 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleming AM, Burrows CJ. 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst) 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifermann M, Epe B. Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark? Free Radic Biol Med. 2017;107:258–265. doi: 10.1016/j.freeradbiomed.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Antoniali G, Malfatti MC, Tell G. Unveiling the non-repair face of the base excision repair pathway in RNA processing: A missing link between DNA repair and gene expression? DNA Repair (Amst) 2017;56:65–74. doi: 10.1016/j.dnarep.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor appaB-driven gene expression. J Biol Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 33.Antoniali G, Lirussi L, D’Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, Tell G. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol Biol Cell. 2014;25:532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1367–L1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming AM, Zhu J, Ding Y, Burrows CJ. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on-off switch for transcription. ACS Chem Biol. 2017;12:2417–2426. doi: 10.1021/acschembio.7b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci U S A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cogoi S, Ferino A, Miglietta G, Pedersen EB, Xodo LE. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018;46:661–676. doi: 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 40.Neurauter CG, Luna L, Bjoras M. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst) 2012;11:401–409. doi: 10.1016/j.dnarep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Chan J, Lambele M, Yusufzai T, Stumpff J, Opresko PL, Thali M, Wallace SS. NEIL3 repairs telomere damage during S phase to secure chromosome segregation at mitosis. Cell Rep. 2017;20:2044–2056. doi: 10.1016/j.celrep.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 43.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- 46.Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 2008;7:1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci U S A. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Regnell CE, Hildrestrand GA, Sejersted Y, Medin T, Moldestad O, Rolseth V, Krokeide SZ, Suganthan R, Luna L, Bjoras M, Bergersen LH. Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep. 2012;2:503–510. doi: 10.1016/j.celrep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Skarpengland T, Holm S, Scheffler K, Gregersen I, Dahl TB, Suganthan R, Segers FM, Ostlie I, Otten JJ, Luna L, Ketelhuth DF, Lundberg AM, Neurauter CG, Hildrestrand G, Skjelland M, Bjorndal B, Svardal AM, Iversen PO, Hedin U, Nygard S, Olstad OK, Krohg-Sorensen K, Slupphaug G, Eide L, Kusnierczyk A, Folkersen L, Ueland T, Berge RK, Hansson GK, Biessen EA, Halvorsen B, Bjoras M, Aukrust P. Neil3-dependent base excision repair regulates lipid metabolism and prevents atherosclerosis in Apoe-deficient mice. Sci Rep. 2016;6:28337. doi: 10.1038/srep28337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu C, Luo J, He L, Montell C, Perrimon N. Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca(2+) signaling in the Drosophila midgut. eLife. 2017;6:e22441. doi: 10.7554/eLife.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 53.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukundan VT, Phan AT. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J Am Chem Soc. 2013;135:5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 56.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Jr, Sen D, Lansdorp PM. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 60.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 61.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fleming AM, Zhu J, Ding Y, Visser JA, Zhu J, Burrows CJ. Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5`-untranslated regions. Biochemistry. 2018;57:991–1002. doi: 10.1021/acs.biochem.7b01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marcel V, Tran PL, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou MP, Hall J, Mergny JL, Hainaut P, Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 66.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 70.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 71.Beckett J, Burns J, Broxson C, Tornaletti S. Spontaneous DNA lesions modulate DNA structural transitions occurring at nuclease hypersensitive element III(1) of the human c-myc proto-oncogene. Biochemistry. 2012;51:5257–5268. doi: 10.1021/bi300304k. [DOI] [PubMed] [Google Scholar]
- 72.An N, Fleming AM, Burrows CJ. Human telomere G-Quadruplexes with five repeats accommodate 8-oxo-7,8-dihydroguanine by looping out the DNA damage. ACS Chem Biol. 2016;11:500–507. doi: 10.1021/acschembio.5b00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fleming AM, Zhou J, Wallace SS, Burrows CJ. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent Sci. 2015;1:226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olsen CM, Gmeiner WH, Marky LA. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J Phys Chem B. 2006;110:6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]
- 75.Renaud de la Faverie A, Guedin A, Bedrat A, Yatsunyk LA, Mergny JL. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 2014;42:e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weitzmann MN, Woodford KJ, Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J Biol Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 77.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 79.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 82.Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, Wang L, Liu Y, Yu L, Shi Y, Chen H, Tang Y. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci Rep. 2016;6:24793. doi: 10.1038/srep24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI, Bhasikuttan AC. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J Am Chem Soc. 2013;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- 84.Li Y, Xu S, Wu X, Xu Q, Zhao Y, Lou X, Yang X. Thioflavin T as a fluorescence light-up probe for both parallel and antiparallel G-quadruplexes of 29-mer thrombin binding aptamer. Anal Bioanal Chem. 2016;408:8025–8036. doi: 10.1007/s00216-016-9901-5. [DOI] [PubMed] [Google Scholar]
- 85.Lech CJ, Cheow Lim JK, Wen Lim JM, Amrane S, Heddi B, Phan AT. Effects of site-specific guanine C8-modifications on an intramolecular DNA G-quadruplex. Biophys J. 2011;101:1987–1998. doi: 10.1016/j.bpj.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 87.Novy J, Bohm S, Kralova J, Kral V, Urbanova M. Formation and temperature stability of G-quadruplex structures studied by electronic and vibrational circular dichroism spectroscopy combined with ab initio calculations. Biopolymers. 2008;89:144–152. doi: 10.1002/bip.20875. [DOI] [PubMed] [Google Scholar]
- 88.Kim BG, Shek YL, Chalikian TV. Polyelectrolyte effects in G-quadruplexes. Biophys Chem. 2013;184:95–100. doi: 10.1016/j.bpc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Takahashi S, Brazier JA, Sugimoto N. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc Natl Acad Sci U S A. 2017;114:9605–9610. doi: 10.1073/pnas.1704258114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olsen CM, Lee HT, Marky LA. Unfolding thermodynamics of intramolecular G-quadruplexes: base sequence contributions of the loops. J Phys Chem B. 2009;113:2587–2595. doi: 10.1021/jp806853n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


