Abstract
Autism spectrum disorder (ASD) is a heterogeneous group of heritable neurodevelopmental disorders whose symptoms, which include deficits in social interaction skills, impaired communication ability, and ritualistic-like repetitive behaviors, appear in early childhood and continue throughout life. Genetic studies have revealed at least two clusters of genes frequently associated with ASD and intellectual disability: genes encoding for proteins involved in translational control and proteins involved in synaptic function. We hypothesize that mutations occurring in these two clusters of genes interfere with interconnected downstream signaling pathways to cause ASD symptomatology. In this review, we focus on the monogenic forms of ASD caused by mutations in genes encoding for proteins that regulate translation and synaptic proteins. Specifically, we describe the function of these proteins, the intracellular signaling regulated by them, and the current mouse models utilized to characterize synaptic and behavioral features of these mutations. Finally, we summarize recent studies that have established a connection between these two signaling pathways in models of ASD and propose that dysregulation of one pathway has a detrimental impact of the other.
Autism spectrum disorder (ASD) is a complex group of heterogeneous neurodevelopmental disorders categorized by three key behavioral abnormalities: restricted interests accompanied by repetitive behavior, deficits in language and communication skills, and inability to engage in reciprocal social interactions (1–3). These core symptoms often are co-morbid with intellectual disability, epilepsy, motor impairment, anxiety, sleep disorder, attention-deficit hyperactivity disorder, and tics (4–6). The behavioral symptomatology of ASD encompasses a spectrum of wide ranging phenotypes, which span from mild behavioral and personality traits to severe and debilitating impairments (7).
The remarkable clinical heterogeneity that characterizes ASD is paralleled by an equally multifaceted etiological heterogeneity. ASD has been recognized to have a genetic component (twin concordance rate 73–95%) and is extraordinary heritable (>90%) (8, 9). Recently, linkage and association studies have identified numerous susceptibility genes located on multiple chromosomes, especially 2q, 7q, 15q, and the X chromosome. Thus, ASD is considered a polygenic disorder generated by the additive effect of multiple common genetic variants in combination with as of yet unidentified environmental factors (10). These forms of ASD are referred to as non-syndromic ASD (11).
In contrast to non-syndromic ASD, in approximately 10% of the cases, ASD appears as part of a syndrome with a known genetic cause (12–14). These monogenic forms of ASD can result from genomic DNA mutations, de novo copy number variants (CNVs) and chromosomal rearrangements (e.g. deletion and translocation) (11). Even when ASD associated is genetically defined, the clinical symptoms are highly heterogeneous, likely due to differences in genetic background and epigenetic regulation (11). Nevertheless, monogenic forms of ASD have been paramount for understanding key neurobiological processes and complex physiological pathways that when perturbed increase the risk for ASD.
Recent studies of monogenic forms of ASD have focused on at least two different clusters of genes frequently associated with ASD and intellectual disability: genes encoding for structural synaptic proteins and genes involved in the regulation of protein synthesis (reviewed in 3, 11, 15). These studies also suggest that mutations in the same clusters of genes may be pathogenic in non-syndromic forms of ASD. We hypothesize that mutations in these two clusters of genes interfere with interconnected downstream signaling pathways resulting in ASD symptomatology.
Here, we will first review the monogenic forms of ASD caused by mutations in these two clusters of genes, the molecular function of their protein products, and the current mouse models utilized to characterize the neurobiological features of these mutations. Next, we summarize key findings that have established a connection between the protein products of these two clusters of genes in models of ASD.
Genes encoding for proteins that regulate translation
Fragile X mental retardation protein
Nearly all individuals with fragile X syndrome (FXS) present with a trinucleotide (CGG) repeat expansion adjacent to the fragile X mental retardation 1 (FMR1) gene promoter that leads to the transcriptional silencing and subsequent loss of its protein product (16, 17). Recently, it was discovered that the silencing of the FMR1 gene is mediated by the formation of a DNA-mRNA duplex between the gene promoter and the trinucleotide repeat region of the mRNA (18). Epidemiological studies show that FXS is the most common known disorder associated with inherited intellectual disability and ASD (19, 20). It occurs in approximately 1:5000 males and roughly half as many females. Affected males with FXS usually have other neurological and psychiatric conditions in addition to ASD and intellectual disability, including motor abnormalities, speech delay, hyperactivity, and anxiety. Postmortem neuropathological studies have revealed an increase in spine-like protrusions on apical and basal dendrites in the cerebral cortex of individuals with FXS (21, 22).
The fragile X mental retardation protein (FMRP), the product of the FMR1 gene, is an RNA-binding protein that is involved in many aspects of the posttranscriptional regulation of mRNA such as stability, dendritic transport, and translational control. In particular, its function as repressor of protein synthesis has been intensively studied, but the molecular mechanism responsible for this repression remains controversial. Experimental evidence indicate a role for FMRP in the initiation (23–28) and elongation (29–33) steps of protein synthesis. In the initiation model, FMRP inhibits translation initiation by interacting with CYFIP1, a eIF4E-binding protein (4E-BP)-like protein, which is associated with eIF4E, the cap-binding translation factor for mRNAs (27). On the other hand, there is experimental evidence indicating that the function of FMRP as a translation repressor is at the level of the elongation. (29–33). Moreover, ribosomal run-off of these mRNAs demonstrated that FMRP is associated with mRNAs bound to stalled ribosomes (34) and ribosome transit assays indicate that elongation is enhanced in mice that lack FMRP (35). It is possible that FMRP acts by inhibiting both initiation and elongation steps of translation that depends mRNA identity and/or neuronal stimuli.
The most studied animal model for FXS is a mouse line in which the Fmr1 gene has been deleted. Fmr1 null mice display a range of phenotypes that mimic many of the symptoms observed in individuals with FXS. For example, these mice display hyperactivity, altered sensorimotor gating, deficits in learning and memory, increased susceptibility to audiogenic seizures, increased body growth rate, and macroorchidism (36). Fmr1 null mice also exhibit increased density of dendritic spines and numerous filopodia-like spines in the cortex, recapitulating another pathological feature observed in FXS patients (21, 37–39). Fmr1 null mice exhibit high levels of basal brain protein synthesis (40) and using HITS-CLIP, at least 842 FMRP target mRNAs have been identified (34). These mRNAs encode for both pre- and postsynaptic proteins. Importantly, the postsynaptic proteins include SHANK1–3, SAPAP1–4, SynGAP1, and neuroligins, whereas the presynaptic proteins include the neurexins, among others. These findings suggest that synaptic proteins and regulators of protein synthesis may interact to generate the FXS phenotype.
In addition to directly repressing translation, FMRP impacts protein synthesis by acting indirectly on signaling pathways involved in translational control. Elevated mammalian target of rapamycin complex 1 (mTORC1) signaling has been reported in Fmr1 null mice (41), which likely is induced by the elevated expression of PIKE, a GTPase that connects the activation of mGluR5 to the PI3K-mTORC1 signaling pathways in the hippocampus (41, 42). Moreover, several mRNAs targets of FMRP encode for repressors of the mTORC1 signaling pathway, including tuberin (TSC2) and phosphatase and tensin homolog (PTEN) (34). Thus, it is possible that FMRP silencing may have an indirect, secondary effect on protein synthesis by repressing translation of components of the mTORC1 signaling pathway.
Eukaryotic initiation factor 4E
Several studies suggest an association between mutations in the eukaryotic initiation factor 4E (EIF4E) gene and ASD. Genetic variants in chromosome 4q, which contains the EIF4E locus, have been described in patients with ASD (43, 44). Notably, in ASD subjects several of these common genetic variants in the EIF4E gene are associated with a clinical phenotype characterized by repetitive and stereotyped behaviors, but not intellectual disability (45). A de novo chromosomal translocation involving the promoter region of the EIF4E gene in a boy with classic non-syndromic ASD has been described (46). In addition, a nucleotide insertion in the promoter region of the EIF4E gene that increases promoter activity was discovered in two unrelated families with autistic siblings. These genetic studies link mutations in EIF4E to ASD; however further investigations are needed to clearly establish a causal connection.
eIF4E binds to the cap structure at the 5’ terminus of mRNA and regulates the initiation step of cap-dependent translation (47, 48). The main role of eIF4E in translation initiation is in the formation of the eIF4F initiation complex, which brings mRNAs to the ribosome for correct translation initiation. The critical step in the formation of eIF4F is the direct association of eIF4E with eIF4G (49), an mRNA-ribosome bridging factor, and the indirect association with the RNA helicase eIF4A (50). The interaction of eIF4E with eIF4G is regulated by eIF4E-binding proteins (4E-BPs), which repress translation by blocking the interaction of eIF4E with eIF4G (51). Upon stimulation, 4E-BP is phosphorylated and inactivated by mTORC1, thereby allowing eIF4E to associate with eIF4G to form eIF4F (52). eIF4E also is regulated by the extracellular signal-regulated kinase (ERK) signaling pathway via phosphorylation by Mnk1/2, which is a substrate for ERK. In some experimental conditions, eIF4E phosphorylation is correlated with the rate of protein synthesis (53). Thus, eIF4E and cap-dependent protein synthesis can be regulated by both mTORC1 and ERK signaling (54).
The relationship between eIF4E, cap-dependent translation, and ASD has been recently studied by genetically increasing the levels of eIF4E in a transgenic mouse (55). eIF4E transgenic mice showed increased brain protein synthesis and aberrant behaviors reminiscent of ASD, including impairments in social interactions and repetitive/perseverative behaviors. The ASD-like behaviors were corrected by blocking the interaction of eIF4E with eIF4G with the cap-dependent translation inhibitor, 4EGI-1. Notably, mice with a genetic reduction in 4E-BP2, the predominant 4E-BP isoform in the brain, exhibit ASD-like behaviors that mimic those displayed by eIF4E transgenic mice (56). Thus, mice with elevated eIF4E-dependent translation display ASD-like behaviors, strongly suggesting a link between exaggerated protein synthesis and ASD.
TSC1 and TSC2
Tuberous sclerosis complex (TSC) is a multi-system disorder characterized by the presence of benign tumor-like lesions (hamartomas) in many organs, such as brain, skin, eye, kidneys and heart (57). TSC is an autosomal dominant inherited disorder caused by loss-of-function mutations in either TSC1 (encoding hamartin, also referred to as TSC1) or TSC2 (encoding tuberin, also referred to as TSC2) genes. These mutations comprise a mix of missense, nonsense, insertions, and deletions involving nearly all exons present in the TSC1 and TSC2 genes (4, 5, 58). The impact of the different mutations on clinical phenotypes is extremely variable with respect to symptoms and disease severity, and in part is dependent on which TSC gene is affected (59). Seizures are the most common neurological symptom occurring in up to 90% of the patients, whereas intellectual disability and ASD occur in approximately 50% of the patients (57).
TSC1 and TSC2 form a heterodimeric complex that can regulate protein synthesis by controlling mTORC1 activity. TSC1/TSC2 are phosphorylated by many kinases and factors, including Akt, ERK, glycogen synthase kinase-3β, AMP-activated kinase, and cyclin-dependent kinase 1 (60–63). The active TSC1/TSC2 complex inhibits mTORC1 through activation of the small GTPase Ras homolog enriched in the brain (Rheb). Rheb activates mTORC1 when it is bound to GTP. The TSC1-TSC2 complex has GTPase-activity localized in the GAP domain of TSC2. When phosphorylated by Akt, the GAP activity of TSC1/TSC2 is increased, which in turn hydrolyzes GTP bound to Rheb, thereby inhibiting mTORC1 (60–63). Therefore, in the absence of either TSC1 or TSC2, high levels of Rheb-GTP lead to constitutive activation of mTORC1 signaling, thereby resulting in dysregulated protein synthesis and cell growth (63). Several mouse models of TSC have been employed to understand the etiology of this disorder. For example, heterozygous genetic deletion of either Tsc1 or Tsc2 results in cognitive and synaptic impairments consistent with ASD (64–66). Tsc1 and Tsc2 mutant mice display ASD-like phenotypes in absence of neuropathological brain tumors and epilepsy, suggesting that the cognitive dysfunction in TSC arise independently of brain tumors and/or epilepsy. However, it should be noted that specific genetic ablation of Tsc1 in either astrocytes (67) or neurons (65, 68) result in epilepsy and lethality. Recently, either genetic reduction or complete depletion of Tsc1 in cerebellar Purkinje cells (PC) (69) was demonstrated to result in ASD-like behaviors, including impaired social interaction, altered ultrasonic vocalizations, and increased repetitive behaviors that are correlated with decreased PC excitability and changes in the number and morphology of PCs.
Importantly, in the aforementioned mouse models, postnatal and post-development treatment with rapamycin, which inhibits mTORC1 activity, ameliorates multiple behavioral and synaptic phenotypes (65, 68–71). Thus, inhibition of mTORC1 activity in adulthood is sufficient to correct ASD-like phenotypes in TSC model mice, which suggests that these behaviors are caused by ongoing, elevated mTORC1 signaling rather than irreversible pathophysiological changes that occur during brain development.
Phosphatase and tensin homolog
Phosphate and tensin homolog (PTEN), a gene located on chromosome 10q23, is a candidate risk gene for ASD and macrocephaly (72–75). Different studies have suggested a causal role for PTEN mutations in a subset of individuals with ASD. Recently, a novel frameshift variant of PTEN was identified in a patient with extreme macrocephaly, ASD, intellectual disability, and epilepsy, confirming that mutations in this gene are involved in the etiology of ASD and macrocephaly (76). In general, PTEN mutation are more frequent (10–20%) in ASD children that develop macrocephaly (77, 78).
PTEN is a phosphatase with activity directed against 3’ phosphate of the phosphatidylinositol-3,4,5-triphosphate (79). PTEN is a negative regulator of the Akt-mTORC1 signaling pathway. Thus, activation of this phosphatase leads to inhibition of PI3K signaling, thereby inactivating Akt and mTORC1. In contrast, deletion of PTEN results in a constitutively active Akt-mTORC1 signaling pathway. Given the importance of PI3K/AkT/mTORC1 signaling in controlling cell growth, survival and proliferation, it is not surprising that PTEN inactivation leads to human cancers and neurological disorders (80).
Mouse models with Pten deletions have been studied mostly to clarify the role of PTEN in neuronal hypertrophy and number, since the most obvious phenotype in human patients is macrocephaly. Overall, the effect of genetic deletion of Pten during development is dramatic resulting in brain enlargement and gross anatomical abnormalities that are often accompanied by the development of seizures and premature death (81, 82). Several studies have directly addressed the role of PTEN mutations in ASD. These studies bypassed the severe developmental phenotype by deleting Pten in mice either in a specific cell population or at a certain time after development using conditional genetic technology. For example, in one of the mouse models, Pten was ablated in a subset of postmitotic cortical and hippocampal neurons (83). These mice develop macrocephaly and display ASD-like behaviors, including impairments in social interactions, seizures, anxiety, and cognitive deficits (83). Treatment with the mTORC1 inhibitor rapamycin reverses the neuronal hypertrophy and leads to the amelioration of the impairments in social interactions and the seizures (84). Moreover, mice with germline Pten haploinsufficiency (Pten+/−), exhibit an increase in total brain mass and behavioral impairments such as abnormal social behavior and sensorimotor gating (85), increased repetitive behaviors, and depressive-like behaviors (86). These ASD-like behaviors were exacerbated when Pten+/− mice were crossed with serotonin transporter heterozygote mice Slc6a4+/−, which also is considered an ASD susceptibility gene (85). These findings demonstrate that deletion of two ASD risk genes, Pten and Slc6a4, can cooperate to give rise to ASD-like behavioral phenotypes.
Genes encoding for proteins involved in synaptic function
SHANK
Phelan-McDermid Syndrome (PMS) is a genetic disorder characterized by ASD and intellectual disability. Affected patients exhibit impairments in communication skills often accompanied by reduced socialization and stereotypical movements. In addition, patients with PMS also display aggressive behaviors and seizures (87–89). Genetic studies have identified deletions of variable length in the terminal region of the long arm of chromosome 22 as being responsible for the disorder. SHANK3, which is located in this region of chromosome 22, is one of the candidate genes for PMS (89–91). In addition, duplications, copy number variations (CNVs), microdeletions, and mutations in SHANK3 have been described in patients with ASD and intellectual disability (92–97).
The SH3 and multiple ankyrin repeat domains (SHANK) protein family, also known as proline-rich synapse-associated proteins (ProSAPs), is encoded by three genes (SHANK1–3) that share a high degree of homology. The SHANK proteins are expressed abundantly in the central nervous system, are enriched in the postsynaptic density (PSD) of excitatory synapses (98–100), and interact with cytoskeleton and scaffolding proteins, which in turn bind to receptors to create a matrix for the stabilization and organization of the PSD. Indeed, SHANK proteins bind to PSD-95-binding proteins (SAPAP), which interact with PSD-95 proteins associated with glutamate receptors (101). Moreover, SHANK proteins bind to the Homer family of scaffolding proteins, which are associated with metabotropic glutamate receptors (102). Finally, SHANK proteins also are involved in the regulation of cytoskeleton by binding cortactin (103), inositol 1, 4, 5-triphosphate (IP3) receptors, and F-actin (104, 105).
Recently, multiple mouse models with genetic deletions of the Shank genes have been intensively studied. In particular, four different lines of Shank3 mice have been studied, each with a specific deletion of exons encoding for the functional interaction domains of the protein. Overall, the Shank3 mutant mice exhibit behavioral deficits consistent with ASD, including social deficits, communication alterations, repetitive and stereotyped behaviors, and abnormal learning and memory that are accompanied generally by changes in synaptic function and molecular composition of the PSD (106), (107, 108). Notably, Shank3B mutant mice, carrying an ablation of the PDZ domain of the protein, exhibit a particularly severe phenotype. Consistent with the marked expression of Shank3 in the striatum, the Shank3B null-mice groomed so excessively that they exhibited self-inflicted skin lesions and displayed anxiety-like behaviors and impaired social interactions. Genetic deletion of Shank1 results in abnormal grooming behavior and impairments in ultrasonic vocalization, but normal social interactions (109, 110) as well as contextual fear memory and long-term spatial memory (111). Overall, these PMS mouse models suggest that molecular changes perturbing synaptic and structural functions at the PSD of excitatory synapses are likely to generate ASD-like phenotypes.
Neuroligins and Neurexins
Several mutations and deletions in genes encoding for neuroligin3 (NLGN3), neuroligin4 (NLGN4) and neurexin1 (NRXN1) have been associated with ASD and intellectual disability, including frameshift, substitution, and missense mutations, as well as CNVs and gene deletions (112–115) (116–119) (43, 116, 120–123). In addition, a de novo mutation resulting in a base pair substitution A335G in the promoter region of NLGN4 gene has been reported in a boy with autism and intellectual disability. Importantly, this base pair substitution (A335G) results in an increased activity of promoter and subsequently, an increase in mRNA expression (124), suggesting that overexpression of neuroligin4 is detrimental to neuronal functions and results in ASD-like phenotypes similar to deletion or loss-of-function mutations affecting the NLGN4 gene.
Neuroligins and neurexins are synaptic cell adhesion molecules that are critical for synaptic efficacy and plasticity (125–128) (129, 130). Neurexins are type 1-membrane proteins encoded by three genes (NRXN1, 2, 3), which generate larger α - neurexins and shorter β - neurexins from independent promoters (131). Furthermore, each gene undergoes extensive alternative splicing that is capable of generating thousands of neurexin isoforms (132). Neuroligins are endogenous ligands for neurexins (125) and are encoded by four different genes (NLGN1, 2, 3, 4) located on the X-chromosome (133). Neuroligins are type 1-membrane proteins like neurexins, but have a simpler domain structure and less diversity. All neuroligins are enriched in PSD, but neuroligin1 and neuroligin2 are exclusively localized to excitatory and inhibitory synapses, respectively, whereas neuroligin3 may be present in both (134–136) (128).
Mouse models recapitulating the genetic mutations and/or deletions of Nrxns and Nlgns described in ASD patients have been important for understanding the association of human genetic aberrations to the clinical manifestation of the disorder. For example, mice with either a genetic deletion of Nlgn3 (137) or carryng a knockin allele with an R451C substitution (138) in Nlgn3 displayed ASD-like behaviors that were mostly restricted to social and communication domains, such us impairments in ultrasonic vocalization, social interaction and memory. Similarly, mice with a deletion of the Nlgn4 ortholog exhibited impaired social interactions and ultrasonic vocalization (139). This indicates neuroligins are important in the generation of normal social skills and vocalization.
The studies performed on mice with genetic ablation of the genes that encode the neurexins are more difficult to interpret, given the high degree of genetic redundancy. Mice with a genetic deletion that results in a lack of all the neurexin α-isoforms die prenatally, whereas mice with ablation a single genedeletion live, but they are severely compromised and still die postnatally (126). It will be interesting to study the role of neurexins in a specific neuron-type (or postdevelopmental time window) to avoid the lethal phenotype and establish a link with ASD.
SAPAP
DLGAP2, which encodes for SAPAP2, has been identified as a candidate ASD risk gene in a large study aimed to identified genome-wide rare CNVs occurring in ASD patients (140). Moreover, rare genetic variants in DLGAP2, also have been identified in ASD patients (141), suggesting that it could act concomitantly with other genetic mutations and/or environmental factors to contribute to ASD phenotypes in patients. Although not clearly established, a possible involvement of the proteins of the SAPAP family in ASD is intriguing given their demonstrated interaction with the proteins of the SHANK family, which have been more clearly described in ASD (see section, SHANK proteins and signaling) (93–95). However, the involvement of the SAPAP3 gene in obsessive-compulsive spectrum disorders (OCD), trichotillomania, and Tourette syndrome is fairly well established. Indeed, genetic studies have found increased frequency in SAPAP3 gene variants in patients with these disorders (142–145).
The members of the SAP90/PSD-95-associated proteins (SAPAP) family, also referred to as guanylate kinase-associated proteins (GKAPs), are postsynaptic scaffold proteins that are localized in the PSD and are uniquely expressed at the excitatory synapses (146). SAPAP proteins are encoded by a family of four genes that are widely, but differentially, expressed throughout the nervous system. The SAPAP proteins have been proposed to provide a link between the PSD-95 family proteins and the actin cytoskeleton via interactions with the SHANK proteins, which in turn bind the actin-binding protein cortactin. Therefore, in the current model of PSD organization, PSD-95/SAPAP/SHANK interactions play an important role in the constitution of the large postsynaptic signaling complex at glutamatergic synapses (147).
The member of the SAPAP family that has been studied in great detail is SAPAP3, which is highly expressed in the striatum (146, 148). Genetic ablation of Sapap3 caused behavioral abnormalities consisting of extremely high levels of self-grooming accompanied by self-inflicted snout lesions and anxiety-like behaviors. Consistently, Sapap3 null mice display synaptic, morphological, and molecular defects at striatal glutamatergic synapses. The behavioral and synaptic phenotypes of the Sapap3 null mice is similar to those generated by genetic ablation of Shank3 indicating that genetic changes perturbing these synaptic proteins in the striatum results in specific phenotypes that are consistent with ASD.
Based on the genetic link between DLGAP2 and ASD, the role of SAPAP2 in ASD was investigated recently (149). Genetic ablation of DLGAP2 in mice results in elevated aggressive behavior and impairments in social interactions in mice. Moreover, the DLGAP2 null mice exhibit reduced dendritic spines, changes in receptor composition and decrease in PSD length and thickness (149). Overall, these results suggest that deletion of SAPAP2 leads to reduction in synaptic and postsynaptic responses.
SYNGAP1
Genetic studies of ASD patients with intellectual disability have indicated SYNGAP1 as candidate risk gene. In one of these studies, a de novo deletion and a premature stop-codon was discovered in SYNGAP1. In this study, the genetic alterations in SYNGAP1 were described in one child with ASD and intellectual disability and two children with intellectual disability without ASD (out of 30 children observed) (150). Another study identified an extended deletion that included SYNGAP1 together with several other genes in a patient with ASD (140).
The SYNGAP1 gene encodes for a RasGTPase-activating protein (RasGAP) termed synaptic GTPase-activating protein (SynGAP). The SYNGAP1 gene has several alternative start sites and the transcripts can be spliced extensively to generate multiple SynGAP1 isoforms (151, 152). SynGAP1 is a brain-specific protein highly enriched at excitatory synapses that co-localizes and interacts with NMDA receptors and the PDZ domains of PSD-95 via its C-terminal amino acids and (151, 152). It works as a negative regulator of the signaling pathways that control NMDA receptor-mediated synaptic plasticity and AMPA receptor membrane insertion (153–155). It was shown that SynGAP1 links Ca2+ influx to activation of ERK pathway downstream of NMDA receptors (156). Given the multiple isoforms and the possible high degree of redundancy, the impact of deleting SynGAP1 in neurons is not clear. In fact, deletion of SynGAP1 in hippocampal neurons in culture has been reported to both enhance (153, 155) and suppress (154) dendritic spine formation. Mice with a homozygous genetic deletion of Syngap1 die postnatally, but heterozygous mice survive (157). Behavioral analysis of these mice revealed hyperactivity, diminished sensorimotor gating, and enhanced startle response. Moreover, they display a reduction in social memory and tendency toward social isolation. Moreover, Syngap1 mutant mice have enhanced ERK activation and impairment in hippocampal synaptic plasticity (156). Recently, it was demonstrated that mice with a heterozygote deletion of Syngap1 (157) exhibited glutamatergic synapses that mature at an accelerated rate during development with a consequent disruption in the excitation/inhibition balance in hippocampal neurons (158). This study indicates that changes in synapses maturation during the development results in enduring behavioral abnormalities.
Reciprocal Signaling Links Two Clusters of ASD Genes
The studies summarized above are consistent with at least two defined clusters of genes that are involved in ASD and intellectual disability. One cluster encodes for proteins that regulate protein synthesis, a fundamental process for long-lasting changes in synaptic strength and dendritic spine plasticity underlying cognition. The second cluster of genes produces proteins involved in the regulation of synaptic transmission and structure, which are important in the establishment and remodeling of neuronal networks. Currently, there is limited experimental evidence suggesting a direct interaction between the protein products of these two gene clusters. However, their critical and central biological functions strongly suggest that an anomaly in one of these pathways would almost necessarily perturb the other (Figures 1 and 2).
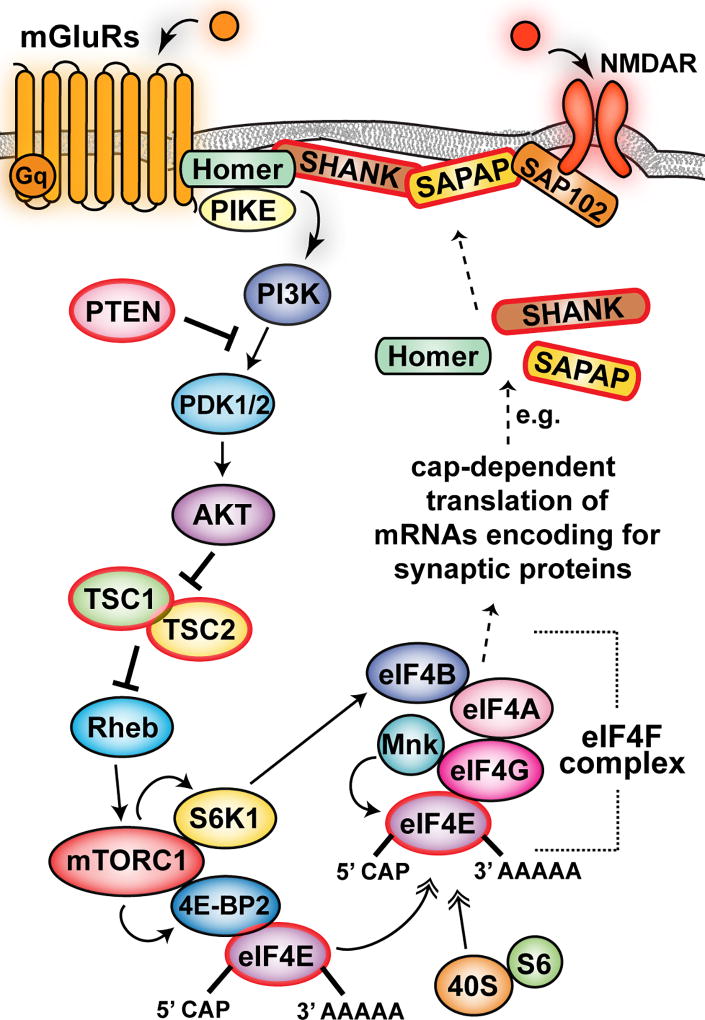
Figure 1. Schematic of the hypothetical connection between protein synthesis and synaptic proteins.
Activation of group I mGluR receptors results in the activation of mTORC1 signaling, which increases protein synthesis. mTORC1 phosphorylates S6K1 and 4E-BP2.; phosphorylation of 4E-BP2 release eIF4E and results in the association of eIF4E with eIF4G to form of the active eIF4F (eIF4E-eIF4G-eIF4A) complex. eIF4F promotes the binding of mRNAs to ribosomes and recruits Mnk, which phosphorylates eIF4E, and eIF4B, which is phosphorylated by S6K1. The eIF4F complex and the poly(A) tail act synergistically together with MnK-dependent phosphorylation of eIF4E to stimulate cap-dependent translation initiation. Cap-dependent protein synthesis translates some mRNAs that encode for synaptic proteins located in the PSD. It is possible that mutation in genes encoding for proteins involved in the regulation of the mTORC1 pathway results in aberrant synthesis synaptic proteins such as neuroligins, SHANK, SAPAP, etc. The altered synthesis of these proteins would generate changes in molecular, structural, and synaptic plasticity, ultimately leading to ASD pathophysiology. The protein products of genes associated with ASD are circled in red.
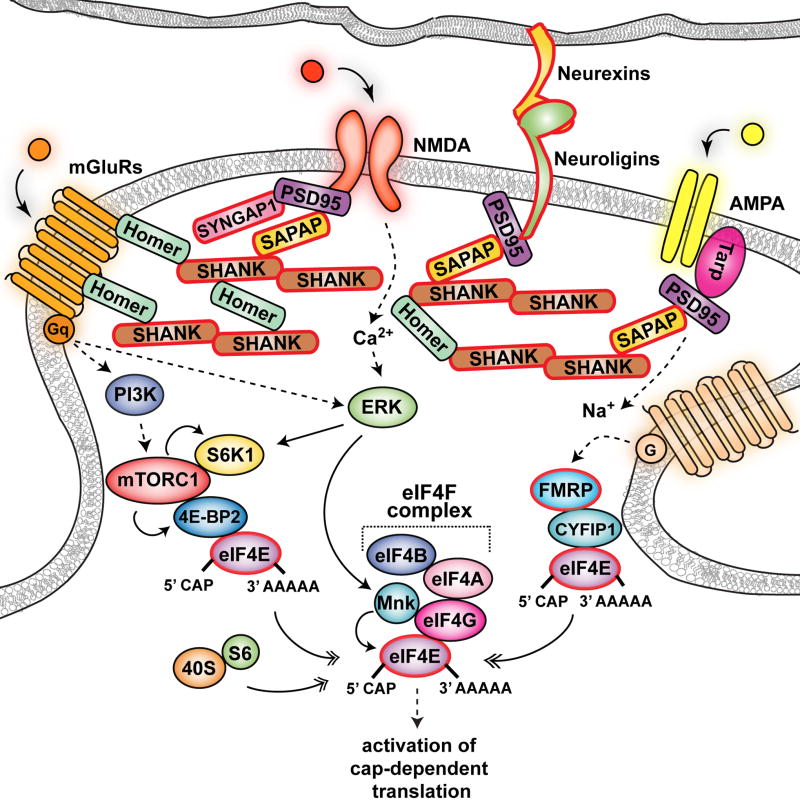
Figure 2. Schematic of the hypothetical connection between synaptic proteins and protein synthesis.
Intracellular signal transduction is initiated by the activation of neurotransmitter receptors that are organized with scaffolding proteins and adhesion molecules in the PSD. Receptor stimulation triggers the activation of intracellular signaling cascades including the mTORC1 and ERK pathways, which results in increased translation (see also Fig1). Given the importance of synaptic proteins in this type of signal transduction, mutations affecting genes encoding for these proteins could result in abnormal signaling that ultimately results in aberrant protein synthesis. The protein products of genes associated with ASD are circled in red.
Activity-dependent changes in PSD composition and/or structure represent molecular mechanisms that drive complex brain functions, including learning and memory. These long-term synaptic and structural changes are critically dependent on dendritic protein synthesis (Figure 1). Indeed, it recently was shown that aberrant protein synthesis driven by overexpression of the cap-binding translation factor eIF4E causes synaptic impairments and ASD-like behaviors (55), indicating that exaggerated translation directly influences synaptic and structural plasticity. Consistent with this idea, a related study revealed that overexpression of neuroligins is likely responsible for the generation of certain ASD-like phenotypes in these mice (56). Moreover, the synaptic, structural, and behavioral abnormalities in mice with exaggerated eIF4E-dependent translation we corrected by reducing protein synthesis and/or diminishing the expression of neuroligins with short interfering RNAs (siRNA) (55, 56).
The examination of FMRP-regulated target mRNAs also supports this idea and demonstrates that both pre- and post-synaptic proteins are part of the transcripts dysregulated in FXS (34), which include, SHANK3, SynGAP1, neuroligin3 and neurexin1 (34), SHANK1, and SAPAPs 1 and 3 (159). This suggests that changes in synaptic and PSD proteins driven by dysregulated protein synthesis may contribute to enduring changes in synaptic plasticity, dendritic morphology, and ASD-like behavioral abnormalities. Another set of mRNA targets of FMRP are proteins directly involved in the regulation of translation, such as TSC2 and PTEN (34, 160), suggesting that synaptic proteins and regulators of mTORC1 activity may interact to give rise to the FXS phenotype.
Conversely, it is possible that the ASD associated mutations that result in changes in the level and function of synaptic and PSD proteins alter protein synthesis and contribute to the generation of ASD (Figure 2). Unfortunately, to our knowledge, there is limited information regarding the activity of translational control pathways in human patients and mouse models of ASD caused by mutations in genes encoding for synaptic proteins (reviewed above). However, a recent study investigating mGluR signaling in mice with a genetic deletion of Fmr1 reveals a fundamental role of the PSD scaffolding protein Homer1a (161, 162). Altered mGluR5-Homer interactions contribute to abnormal mGluR signaling, altered protein synthesis, and other ASD-like phenotypes in FXS. Importantly, genetic deletion of Homer1a restores the normal mGluR5-Homer association and corrects several phenotypes in Fmr1 null mice, including enhanced global protein synthesis (162). Although the effect of mGluR5-Homer interactions on protein synthesis is secondary to the direct role of FMRP in translation, this study indicates the possibility that alteration in synaptic proteins results in aberrant translational control. It is tempting to speculate that ASD linked to SHANK mutations is also associated with alterations in protein synthesis because SHANK directly interacts with Homer (102). Therefore, defects in synaptic protein function could result in aberrant protein synthesis, resulting in abnormal synaptic plasticity and ASD-like behaviors. Future studies are necessary to conclusively address this hypothesis.
Several lines of evidence indicate that loss-of-function mutations, deletions, and overexpression of synaptic and PSD proteins are detrimental and result in ASD-like behavioral phenotypes. A good example of this bidirectional effect is illustrated in patients where deletions and a de novo mutation of neuroligin, which increase the activity of the promoter of NLGN4 gene, result in ASD and intellectual disability (112, 113, 124). Similarly, animal models with genetic deletion (139, 137) or exaggerated expression (56) of NLGNs display behavioral and synaptic phenotypes consistent with ASD. This is in agreement with our hypothesis that increased expression of synaptic proteins generated by alteration in protein synthesis could trigger synaptic abnormalities and behaviors associated with ASD. In contrast, investigations concerning the proteins regulating translation seem to point toward a connection between exaggerated protein synthesis and ASD phenotypes in humans and animal models (55, 56, 163). However, inhibition of de novo protein synthesis impairs long-lasting plasticity and cognition (reviewed in (47, 164)) and likely contributes to cognitive deficits in TSC model mice (165). An intriguing possibility is that excessive translation contributes to aberrant behaviors associated with ASD whereas insufficient translation contributes to impaired cognition associated with intellectual disability, which often accompanies ASD.
To conclude, we have reviewed recent data supporting the hypothesis that proteins involved in the regulation of translation and synaptic function may be interconnected and act in concert to give rise to synaptic and behavioral aberrations associated with ASD. Future genetic studies are necessary to reveal the molecular players that link these two pathways and to understand whether it is possible to intervene therapeutically at the level of these molecular crossroads.
Gloss.
Autism spectrum disorder (ASD) is a heterogeneous group of heritable neurodevelopmental disorders characterized by repetitive behavior, deficits in communication skills and impaired social interactions. Human genetic studies have uncovered at least two clusters of genes associated with ASD and intellectual disability: genes encoding for proteins involved in the regulation of protein synthesis and proteins involved in synaptic function. We hypothesize that mutations in these risk genes impact interconnected intracellular signaling pathways to disrupt synaptic function and behavior. In this review, we describe the protein products of these two clusters of genes and the findings that connect their signaling pathways. Knowledge of these molecular and synaptic abnormalities should generate novel therapeutic targets for the treatment of ASD.
References
- 1.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119:747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fombonne E, Du Mazaubrun C, Cans C, Grandjean H. Autism and associated medical disorders in a French epidemiological survey. J Am Acad Child Adolesc Psychiatry. 1997;36:1561–1569. doi: 10.1016/S0890-8567(09)66566-7. [DOI] [PubMed] [Google Scholar]
- 5.Fombonne E. Epidemiological surveys of autism and other pervasive developmental disorders: an update. J Autism Dev Disord. 2003;33:365–382. doi: 10.1023/a:1025054610557. [DOI] [PubMed] [Google Scholar]
- 6.Belmonte MK, et al. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- 7.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci (Regul Ed) 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozonoff S, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–95. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klei L, et al. Common genetic variants, acting additively, are a major source of risk for autism. Molecular Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persico AM, Napolioni V. Autism genetics. Behav Brain Res. 2013;251:95–112. doi: 10.1016/j.bbr.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Lintas C, Persico AM. Autistic phenotypes and genetic testing: state-of-the-art for the clinical geneticist. Journal of Medical Genetics. 2009;46:1–8. doi: 10.1136/jmg.2008.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Toriello HV. Approach to the genetic evaluation of the child with autism. Pediatr. Clin. North Am. 2012;59:113–28. xi. doi: 10.1016/j.pcl.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pieretti M, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 17.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 18.Colak D, et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffee B, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85:503–514. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010;7:264–274. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comery TA, et al. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 23.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 24.Lacoux C, et al. BC1-FMRP interaction is modulated by 2'-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Research. 2012;40:4086–4096. doi: 10.1093/nar/gkr1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centonze D, et al. Abnormal striatal GABA transmission in the mouse model for the fragile X syndrome. Biological Psychiatry. 2008;63:963–973. doi: 10.1016/j.biopsych.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix J-L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Research. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napoli I, et al. The Fragile X Syndrome Protein Represses Activity-Dependent Translation through CYFIP1, a New 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 28.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 31.Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 32.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamanini F, et al. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 34.Darnell JC, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 37.Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKinney BC, Grossman AW, Elisseou NM, Greenough WT. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:98–102. doi: 10.1002/ajmg.b.30183. [DOI] [PubMed] [Google Scholar]
- 39.Dölen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross C, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Autism Genome Project Consortium et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yonan AL, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waltes R, et al. Common EIF4E variants modulate risk for autism spectrum disorders in the high-functioning range. Journal of neural transmission (Vienna, Austria : (1996) 2014 doi: 10.1007/s00702-014-1230-2. [DOI] [PubMed] [Google Scholar]
- 46.Neves-Pereira M, et al. Deregulation of EIF4E: a novel mechanism for autism. Journal of Medical Genetics. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- 47.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 48.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 49.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 50.Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- 51.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 52.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waskiewicz AJ, et al. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santini E, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gkogkas CG, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curatolo P, Bombardieri R. Tuberous sclerosis. Handb Clin Neurol. 2008;87:129–151. doi: 10.1016/S0072-9752(07)87009-6. [DOI] [PubMed] [Google Scholar]
- 58.Kielinen M, Rantala H, Timonen E, Linna S-L, Moilanen I. Associated medical disorders and disabilities in children with autistic disorder: a population-based study. Autism. 2004;8:49–60. doi: 10.1177/1362361304040638. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Inoki K, Vikis H, Guan K-L. Measurements of TSC2 GAP activity toward Rheb. Meth Enzymol. 2006;407:46–54. doi: 10.1016/S0076-6879(05)07005-9. [DOI] [PubMed] [Google Scholar]
- 60.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 61.Inoki K, Corradetti MN, Guan K-L. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 62.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14(Spec No. 2):R251–8. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 64.Goorden SMI, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- 65.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nie D, et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uhlmann EJ, et al. Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures. Ann Neurol. 2002;52:285–296. doi: 10.1002/ana.10283. [DOI] [PubMed] [Google Scholar]
- 68.Meikle L, et al. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012:1–6. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meikle L, et al. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci. 2008;28:5422–5432. doi: 10.1523/JNEUROSCI.0955-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansen LA, Uhlmann EJ, Crino PB, Gutmann DH, Wong M. Epileptogenesis and reduced inward rectifier potassium current in tuberous sclerosis complex-1-deficient astrocytes. Epilepsia. 2005;46:1871–1880. doi: 10.1111/j.1528-1167.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 72.Orrico A, et al. Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin. Genet. 2009;75:195–198. doi: 10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 73.Conti S, et al. Phosphatase and tensin homolog (PTEN) gene mutations and autism: literature review and a case report of a patient with Cowden syndrome, autistic disorder, and epilepsy. J. Child Neurol. 2012;27:392–397. doi: 10.1177/0883073811420296. [DOI] [PubMed] [Google Scholar]
- 74.Buxbaum JD, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. Journal of Medical Genetics. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marchese M, et al. Autism-epilepsy phenotype with macrocephaly suggests PTEN, but not GLIALCAM, genetic screening. BMC Med. Genet. 2014;15:26. doi: 10.1186/1471-2350-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lainhart JE, et al. Macrocephaly in children and adults with autism. J Am Acad Child Adolesc Psychiatry. 1997;36:282–290. doi: 10.1097/00004583-199702000-00019. [DOI] [PubMed] [Google Scholar]
- 78.Fidler DJ, Bailey JN, Smalley SL. Macrocephaly in autism and other pervasive developmental disorders. Dev Med Child Neurol. 2000;42:737–740. doi: 10.1017/s0012162200001365. [DOI] [PubMed] [Google Scholar]
- 79.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 80.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22:873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 81.Kwon CH, et al. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 82.Backman SA, et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 83.Kwon C-H, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou J, et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Page DT, Kuti OJ, Prestia C, Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proceedings of the National Academy of Sciences. 2009;106:1989–1994. doi: 10.1073/pnas.0804428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014;23:3490–3505. doi: 10.1093/hmg/ddu057. [DOI] [PubMed] [Google Scholar]
- 87.Phelan MC. Deletion 22q13.3 syndrome. Orphanet J Rare Dis. 2008;3:14. doi: 10.1186/1750-1172-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phelan MC, et al. 22q13 deletion syndrome. Am J Med Genet. 2001;101:91–99. doi: 10.1002/1096-8628(20010615)101:2<91::aid-ajmg1340>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 89.Dhar SU, et al. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am. J. Med. Genet. A. 2010;152A:573–581. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderlid B-M, et al. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum. Genet. 2002;110:439–443. doi: 10.1007/s00439-002-0713-7. [DOI] [PubMed] [Google Scholar]
- 91.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. Journal of Medical Genetics. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gauthier J, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proceedings of the National Academy of Sciences. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gauthier J, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 94.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moessner R, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boccuto L, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur. J. Hum. Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waga C, et al. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr. Genet. 2011;21:208–211. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- 98.Boeckers TM, et al. Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J Neurosci. 1999;19:6506–6518. doi: 10.1523/JNEUROSCI.19-15-06506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim S, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 100.Petralia RS, Sans N, Wang Y-X, Wenthold RJ. Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol Cell Neurosci. 2005;29:436–452. doi: 10.1016/j.mcn.2005.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naisbitt S, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 102.Tu JC, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 103.Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol. 1998;18:5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tu JC, et al. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 105.Sala C, et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 106.Bozdagi O, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Molecular Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang X, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Silverman JL, et al. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wöhr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS ONE. 2011;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hung AY, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laumonnier F, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan J, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 115.Talebizadeh Z, et al. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. Journal of Medical Genetics. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lawson-Yuen A, Saldivar J-S, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur. J. Hum. Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 118.Chocholska S, Rossier E, Barbi G, Kehrer-Sawatzki H. Molecular cytogenetic analysis of a familial interstitial deletion Xp22.2-22.3 with a highly variable phenotype in female carriers. Am. J. Med. Genet. A. 2006;140:604–610. doi: 10.1002/ajmg.a.31145. [DOI] [PubMed] [Google Scholar]
- 119.Macarov M, et al. Deletions of VCX-A and NLGN4: a variable phenotype including normal intellect. J Intellect Disabil Res. 2007;51:329–333. doi: 10.1111/j.1365-2788.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 120.Kim H-G, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yan J, et al. Neurexin 1alpha structural variants associated with autism. Neuroscience Letters. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 122.Zahir FR, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. Journal of Medical Genetics. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 123.Feng J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neuroscience Letters. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 124.Daoud H, et al. Autism and nonsyndromic mental retardation associated with a de novo mutation in the NLGN4X gene promoter causing an increased expression level. Biological Psychiatry. 2009;66:906–910. doi: 10.1016/j.biopsych.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 125.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 127.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 128.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Graff JR, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 131.Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- 132.Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 133.Taniguchi H, et al. Silencing of neuroligin function by postsynaptic neurexins. J Neurosci. 2007;27:2815–2824. doi: 10.1523/JNEUROSCI.0032-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci USA. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur. J. Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- 136.Graf ER, Zhang X, Jin S-X, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Radyushkin K, et al. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 2009;8:416–425. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 138.Tabuchi K, et al. A Neuroligin-3 Mutation Implicated in Autism Increases Inhibitory Synaptic Transmission in Mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jamain S, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proceedings of the National Academy of Sciences. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chien W-H, et al. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Molecular Autism. 2013;4:26. doi: 10.1186/2040-2392-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Züchner S, et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol Psychiatry. 2009;14:6–9. doi: 10.1038/mp.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bienvenu OJ, et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:710–720. doi: 10.1002/ajmg.b.30897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Crane J, et al. Family-based genetic association study of DLGAP3 in Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:108–114. doi: 10.1002/ajmg.b.31134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Boardman L, et al. Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Compr Psychiatry. 2011;52:181–187. doi: 10.1016/j.comppsych.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 146.Welch JM, Wang D, Feng G. Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J Comp Neurol. 2004;472:24–39. doi: 10.1002/cne.20060. [DOI] [PubMed] [Google Scholar]
- 147.Ting JT, Peca J, Feng G. Functional consequences of mutations in postsynaptic scaffolding proteins and relevance to psychiatric disorders. Annu Rev Neurosci. 2012;35:49–71. doi: 10.1146/annurev-neuro-062111-150442. [DOI] [PubMed] [Google Scholar]
- 148.Welch JM, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jiang-Xie L-F, et al. Autism-associated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. 2014;5:1–13. doi: 10.1186/2040-2392-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hamdan FF, et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N. Engl. J. Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 152.Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 153.Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 154.Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 155.Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci USA. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Komiyama NH, et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim JH, Lee H-K, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci. 2003;23:1119–1124. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Clement JP, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schütt J, Falley K, Richter D, Kreienkamp H-J, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. Journal of Biological Chemistry. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Giuffrida R, et al. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–8916. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ronesi JA, et al. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci. 2012;15:431–40–S1. doi: 10.1038/nn.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kelleher RJ, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 164.Santini E, Huynh TN, Klann E. Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci. 2014;122:131–167. doi: 10.1016/B978-0-12-420170-5.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ehninger D, de Vries PJ, Silva AJ. From mTOR to cognition: molecular and cellular mechanisms of cognitive impairments in tuberous sclerosis. J Intellect Disabil Res. 2009;53:838–851. doi: 10.1111/j.1365-2788.2009.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]