SUMMARY
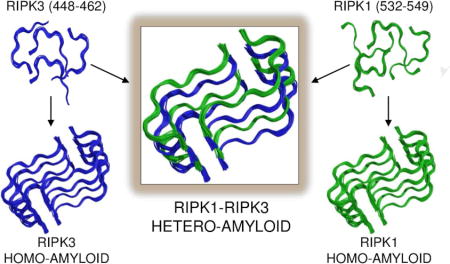
The RIPK1-RIPK3 necrosome is an amyloid signaling complex that initiates TNF-induced necroptosis, serving in human immune defense, cancer and neurodegenerative diseases. RIPK1 and RIPK3 associate through their RIP homotypic interaction motifs with consensus sequences IQIG (RIPK1) and VQVG (RIPK3). Using solid-state nuclear magnetic resonance we determined the high-resolution structure of the RIPK1-RIPK3 core. RIPK1 and RIPK3 alternately stack (RIPK1, RIPK3, RIPK1, RIPK3, etc.) to form heterotypic β-sheets. Two such β-sheets bind together to along a compact hydrophobic interface featuring an unusual ladder of alternating Ser (from RIPK1) and Cys (from RIPK3). The crystal structure of a four-residue RIPK3 consensus sequence is consistent with the architecture determined by SSNMR. The RIPK1-RIPK3 core is the first detailed structure of a hetero-amyloid, and provides a potential explanation for the specificity of hetero- over homo-amyloid formation and a structural basis for understanding the mechanisms of signal transduction.
Graphical abstract
In brief: Solid-state NMR structures of the human necroptosis RIPK1-RIPK3 complex reveal a hetero-oligomeric amyloid signaling complex
INTRODUCTION
Receptor interacting protein kinases 1 and 3 (RIPK1 and RIPK3) are key arbitrators in cell fate regulation, and the decision between TNF-induced cell survival and cell death (Moquin and Chan, 2010). RIPK1 can activate the NF-κB transcription factors leading to cell proliferation and differentiation (Walczak, 2011). Alternatively it can form a cytosolic complex engaging Fas-associated death domain (FADD) and caspase-8 to initiate apoptosis (Wang et al., 2008). In other circumstances, RIPK1 can associate with RIPK3 through their RIP homotypic interaction motifs (RHIM) to form a necrosome and trigger necroptosis. Numerous features distinguish the two cell death pathways, apoptosis vs. necroptosis, including the involvement of kinase activities of RIPK1 and RIPK3 in necrosome formation (Cho et al., 2009; He et al., 2009).
The discovery of small molecule compounds (necrostatins) that block necroptotic cell-death in vitro and in vivo (Degterev et al., 2005) opened new avenues to target necroptosis in pathological scenarios, such as viral infection or neurodegenerative diseases (Vandenabeele et al., 2010). A recent example of particular interest is axonal degeneration by necroptosis, and the link to death of motor neurons in amyotrophic lateral sclerosis (ALS) (Ito et al., 2016). In addition, necroptosis can suppress immune response against cancer and promote tumor growth (Seifert et al., 2016). All these findings highlight the importance of the structure and energetics of the necrosome.
We discovered previously that the RIPK1-RIPK3 necrosome is a functional amyloid required for necroptotic signaling and activation, and RIPK1 or RIPK3 RHIM mutants that were defective for amyloid formation in vitro did not undergo necroptosis upon stimulation with appropriate signals in cells (Li et al., 2012). These findings are consistent with the observation that the murine cytomegalovirus protein M45 suppresses necroptosis through RIPK3–M45 RHIM-mediated interactions (Upton et al., 2010). Similarly, Herpes simplex viruses 1 and 2 unleash their inhibition of necroptosis by disrupting RHIM-dependent RIPK1 and RIPK3 association (Guo et al., 2015), and the enteropathogenic E. coli protease EspL prevents necroptosis signaling by cleavage of the RHIMs of monomeric RIPK1 and RIPK3, but not within the hetero-amyloid structure (Pearson et al., 2017).
A number of studies suggest the existence of multiple amyloidal signalosomes as prominent mediators in signal transduction, since RHIM-containing proteins such as Toll-like receptor signaling adaptor TRIF and DAI, the cytoplasmic DNA sensor involved in interferon response, both interact with RIPK1 and/or RIPK3 to induce programmed cell death or NF-κB activation (Kaiser and Offermann, 2005; Meylan et al., 2004). We previously observed the formation of DAI-RIPK1 fibrillar complexes (Li et al., 2012), and preliminary anti-His resin pulldown assays (not shown) confirm that RIPK1 interacts with RIPK3 and can also interact with DAI. Other support for this hypothesis comes from the discovery of a RHIM-like regulatory amyloid motif in the fungal protein HELLP that signals to a programmed cell death related to necroptosis (Daskalov et al., 2016). Elucidating the association of RIPK1 and RIPK3 can provide insight to the signaling biology mediated by these varied amyloids.
Here we present the high-resolution solid-state nuclear magnetic resonance (SSNMR) structure of the RIPK1-RIPK3 amyloidal signaling complex, featuring sequential and specific stacking to form a hetero-amyloid serpentine fold, stabilized by hydrophobic packing and enriched with interactions along the fibrillar axis such as Asn and Gln ladders, Tyr stacking, and an unusual Cys-Ser ladder. We found that RIPK1 and RIPK3 homo-amyloids possess an analogous fold as supported by the crystal structure of the tetrapeptide VQVG (RIPK3) motif and biochemical validations. These homo-amyloids are, however, less preferred energetically in comparison with the RIPK1-RIPK3 hetero-amyloid. Clearly hetero-amyloids can form highly specific structures with defined folds, analogous to globular proteins and prior homo-amyloids. These results offer insights into how RHIM-mediated interactions provide nuanced control of biological pathways.
RESULTS
A Robust Core Flanked by Flexible Regions
We previously observed that the core of the RIPK1-RIPK3 amyloid comprises the RHIMs of both proteins and flanking segments, and only amino acid residues within these regions were detected in SSNMR spectra collected near room temperature (non cryogenic). This observation was consistent for hydrated samples made from a variety of constructs of different lengths (Li et al., 2012). Extensive portions of the proteins are missing from CPMAS-based spectra, which we interpret in terms of the presence of a rigid amyloid core centered on the critical RHIM motifs, flanked by flexible regions. This is analogous to prior reports regarding the CPEB functional amyloid from Aplysia californica (Raveendra et al., 2013) or the HET-s fungal prion (Wasmer et al., 2008).
Although NMR chemical shifts are very sensitive to structural changes, this fibrillar core exhibited remarkably consistent shifts, including shifts previously reported and confirmed in this work, as well as a handful of additional assignments reported here for the first time (Figure 1 and S1A). This consistent observation of a single set of resonances (Figure S1B) can be contrasted with other amyloids, where polymorphism is evident.
Figure 1. SSNMR Spectra of the RIPK1(532–549)-RIPK3(448–462) Complex.

An expanded view of the DARR spectrum recorded on the U-[13C-15N] sample (Tmix=250 ms) showing selected intermolecular, long-range (blue labels), intra-residue (black), and sequential (red) cross-peaks. See also Figure S1.
SSNMR Data Provide Valuable Long-Range Distance Restraints
We prepared a number of samples including 13C-, 15N-uniformly enriched (U-[13C-15N]) or sparsely 13C enriched (using 2-13C-, or 1,3-13C-glycerol as the carbon source) (Castellani et al., 2002), and recorded a number of SSNMR experiments containing 13C–13C and 15N–13C inter-chain RIPK1-RIPK1, RIPK1-RIPK3 and RIPK3-RIPK3 contacts within the RIPK1-RIPK3 hetero-amyloid. Specifically we utilized: (1) dipolar assisted rotational resonance (DARR) (Takegoshi et al., 2001); (2) 13C-13C proton-assisted recoupling (PAR) (De Paepe et al., 2008); (3) 15N–13C proton-assisted insensitive nuclei (PAIN) (Lewandowski et al., 2007), and (4) 2D transferred-echo double resonance (TEDOR) (Jaroniec et al., 2002). We refer the reader to the Methods section for more details.
From these experiments, we assigned a number medium-range (< 4 residues apart) and long-range (separated by > 5 residues) cross-peaks, which are highlighted in expansions of a homonuclear DARR (250 ms of mixing) spectrum (Figure 1). A parameter n can be used to characterize the number of possible interpretations for a cross-peak. For example, unambiguous cross-peaks are characterized as n = 1 (one unique interpretation for each chemical shift in the two dimensions); for other cross-peaks, one of the atoms is unambiguously identified but two possible assignments are compatible with the chemical shift of the other atom, and we designate them as n = 2. Following this convention, 21 unambiguous (n=1) medium- and long-range cross-peaks were detected in the DARR, PAR, PAIN and TEDOR experiments in total (Table S1, Figure S2), in addition to another 21 medium and long-range low-ambiguity contacts (n=2–4) contacts (Table S2, Figure S2). All the cross-peaks correspond to amino acid residues located in the RHIM regions and directly flanking stretches (Figure 2A).
Figure 2. Assignment, Secondary Chemical shifts, and Considerations for Calculating the RIPK1-RIPK3 Hetero-Amyloid Structure.

(A) Sequence alignment of RIPK1 (520–560) and RIPK3 (439–479), centered around the I(V)QI(V)G RHIM consensus residues. Residues detected by NMR experiments are indicated.
(B) Secondary chemical shifts. The red asterisk indicates a missing resonance assignment for Y453 (RIPK3). Note that several G or N residues (denoted with black asterisks) break the strands into short β-strand segments.
(C) Residues used in structure calculations are indicated. “B” denotes the short β-strand segments. “E” stands for “extended”, that is, segments out of the stacked core not establishing H-bonds.
(D) Selected long-range contacts defining the parallel strands’ register between RIPK1 (green) and RIPK3 (blue).
(E) Wire diagrams establishing the parallel contacts that support the alternation of RIPK1 and RIPK3, as detected by C’-Cα cross-peaks, and their depiction onto a structural bundle. RIPK1 is shown in green and RIPK3 in blue.
See also Figure S2.
Elucidating the Structure of the RIPK1-RIPK3 Hetero-Amyloid: Initial Considerations
A key feature of amyloid structures (and their determination by NMR) is the “registration” or patterns in the backbone hydrogen bonds between neighboring β-strands, for example whether they are parallel or antiparallel. Once the registration isidentified, backbone atom “hypothesis-based restraints” are often used to enforce canonical intra- and inter-β-sheet separations consistent with fibril diffraction patterns (Tuttle et al., 2016). Other protocols include restraints on the inter-strand H-bonds within the β-sheet (Schutz et al., 2015; Walti et al., 2016), or a combination of both kinds of restraints (Colvin et al., 2016). Only if the correct registration is hypothesized will the resulting structure lack conflicts with the data. Although our work broadly reflects the logic and the final validation procedures in this prior work, we utilized a couple of slightly original approaches to identify the registration. In one approach, secondary chemical shifts Δ(δCα−δCβ), were used to locate the β-strand segments (Figure 2B and 2C). The pattern in the backbone conformations of RIPK1 and RIPK3 based on these chemical shifts suggested a parallel registration (Figure 2B), in that the breaks in the β-sheet are well aligned (in-register) if the two peptides are parallel rather than antiparallel. This registration is supported by a number of intermolecular cross-peaks (Figure 2D and 2E). We then show that only this in-register model was compatible with our data, by comparing it to other topologies, as elaborated below. In an independent effort, the H-bond restraints were entirely left out, and only experimental tertiary contacts drove the same pairings to appear in an unbiased computation.
Our calculations (unless otherwise specified) were performed using a building block comprising 4 RIPK1 molecules and 4 RIPK3 molecules, with amino acid sequences: T532IYNSTGIQIGAYNYMEI549 (RIPK1) and P448LVNIYNCSGVQVGD462 (RIPK3) (Figure 2A–C). We consider this to be a minimal relevant complex, and smaller systems were deemed to be incomplete in terms of the distinct interaction surfaces present. The reason is that RIPK1 (RIPK3) interacts with two RIPK3 (RIPK1) molecules, corresponding to the strands above and below in a hetero-amyloid with alternating proteins. A given cross-peak between the two proteins (e.g. S536 (RIPK1) and I452 (RIPK3) (Table S2)) could correspond to either the RIPK1–RIPK3 (above) or the RIPK1–RIPK3 (below) interface. Since there is only one resolved NMR peak per atom, we have no a priori way to know which interface a specific contact arises from. The calculation protocol therefore treats the intermolecular interactions as ambiguous restraints with respect to which interface the contact resides in (even if they are unambiguous in terms of the assigned atoms). In the final structure of the octameric building block, discussed below, typically only one of the two distances is within the upper bounds of the cross-peak. This is consistent with the observation of a single set of spectral resonances, indicating that all monomers have similar conformations within the fibrils and any hypothetical differences are not resolved by our methods (Figure S1B). No symmetry was imposed within the hetero-amyloid in our calculations. Nevertheless, in the final structure the various copies of each peptide were very consistent, as elaborated below, with the observation that only one peak is observed per residue.
Calculation of the RIPK1-RIPK3 Hetero-Amyloid Structure
The 21 unambiguous cross-peaks (Table S1) were converted into upper distance limits of 7.5 Å for restraints obtained from 13C–13C DARR and PAR experiments, and 6.5 Å from those detected in the 15N–13C PAIN and TEDOR spectra, analogous to literature precedents (Colvin et al., 2016). This data set was complemented with 28 dihedral angle restraints obtained using TALOS+ (Shen et al., 2009), and “knowledge-based” information, namely intermolecular H-bond restraints for residues in β-sheet conformation (Figure 2B and 2C, highlighted in yellow) introduced as upper and lower distance limits of 2.0 or 1.8 Å for the H···O distance, and 3.0 or 2.7 Å for the N···O distance, respectively. The combination of H-bond hypothesis driven restraints and experimentally derived restraints was used as an input for our initial CYANA (Guntert, 2004) calculation, and yielded a structure without violations (Figure 3A), with a clear and unique H-bonding strand pairing arrangement. This arrangement explains the presence of both parallel (Figure 2D and 2E) and antiparallel (Figure 3B) contacts between RIPK1 and RIPK3.
Figure 3. Structure of the RIPK1-RIPK3 Hetero-Amyloid (I).
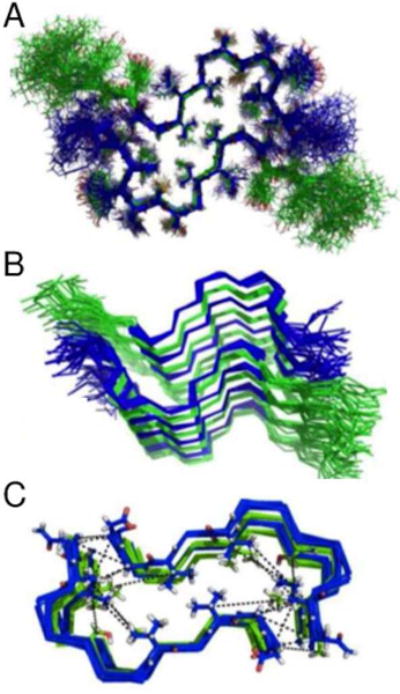
(A) The 20 lowest-energy conformations for the RIPK1-RIPK3 hetero-amyloid using only the 21 unambiguous restraints depicted in Table S1 plus H-bonding restraints. The bundle RMSD based on heavy atom backbone atoms in the core was 1.15 +/− 0.2 Å
(B) Top view of the 20-lowest energy conformers obtained with inclusion of low-ambiguity, resolved restraints listed in Table S2. The bundle RMSD based on heavy atom backbone atoms in the core was 1.01 +/− 0.1 Å (and 0.8 Å after incorporation of ambiguous constraints as described in Figure 4). See also Figure S3.
Evidence supporting this fold comes from the analysis of an additional set of restraints including 21 cross-peaks with low ambiguity or n=2–4 (Table S2); for each of these peaks it is possible to identify an interpretation that is compatible with the proposed structure (Figure S3). A second, independent calculation that included this set of resolved ambiguous restraints combined with all the information from the previous step resulted in the same fold without any restraint violations (Figure 3C), therefore confirming the compatibility of the two data sets with respect to the predicted fold. A list of the experiments from which all these distance restraints were derived is tabulated (Table S3).
Further Computational Validation of the RIPK1-RIPK3 Hetero-Amyloid Structure
To obtain a more refined ensemble of conformers and confirm the validity of the previous structure, peaks in the 250 ms DARR spectrum recorded on the U-[13C-15N] sample were manually picked and included for a full CYANA (Guntert, 2004) structure calculation of the hetero-amyloid consisting of 4 molecules of RIPK1 and 4 of RIPK3, along with all the previous information and beginning from unbiased, extended chains. In other words, we included peaks with a higher degree of ambiguity (n > 4) and they were automatically evaluated by CYANA. Key assignments are displayed on this spectrum (Figure S4). The resulting fibrillar fold (Figure 4) is essentially identical to that obtained in the previous steps, and had no violations, and the core exhibited a pairwise backbone root-mean-square deviation of 0.8 Å. The final structural statistics after AMBER refinement (Case et al., 2005) are shown (Table S4). The resulting structure is deposited in the PDB and described in detail in the next section.
Figure 4. Structure of the RIPK1-RIPK3 Hetero-Amyloid (II).

The hydrophobic interface including residues 532–543 of RIPK1 and 451–462 of RIPK3 is depicted. Hydrophobic residues (Ile and Val) are shown in white and enclose a rectangular hydrophobic core. Asn (red) and Gln (blue) participate in solvent-exposed side chain–side chain H-bonds, and Tyr residues (sand color) and Ser and Thr residues are also stacked out of the nonpolar core. A Cys-Ser ladder is also contained within the hydrophobic core. This structure is deposited as PDB entry 5V7Z.
Among the cross-peaks automatically evaluated by CYANA, a number of carbonyl–alpha carbon (C’–Cα) contacts provide support for presence of a parallel, sequential stack of RIPK1 and RIPK3 around their RHIM motifs. This prompted us to remove the hypothesis derived H-bond restraints in a new, independent round of structure calculations in which the starting point consisted of eight extended, disconnected molecules (4 RIPK1 and 4 RIPK3), and ambiguous restraints were included (and treated as ambiguous), along with the unambiguous restraints, and the TALOS constraints. This information alone allowed us to obtain the RIPK1-RIPK3 fibrillar fold with precision comparable to that obtained when including H-bonding hypothesis derived pseudorestraints (figure S5A–S5D). To further verify our structure, a number of other hypotheses regarding registration and topology were explored computationally and resulted in calculations with numerous violations and poor overall energy (Figure S5E–S5L, and Table S5). These observations in aggregate indicate that our structure correctly describes the fold, and we conclude that only this registration and fold is compatible with our data.
The RIPK1-RIPK3 Hetero-Amyloid Shows a Serpentine Fold
The overall organization of the RIPK1-RIPK3 hetero-amyloid consists of a pair of meandering, parallel β-sheets that are broken into four short β-segments by short turns, which we here term a serpentine fold. The two sheets come together in an antiparallel fashion to create a hydrophobic core. The four copies of RIPK1 are nearly identical (RMSD of all heavy atoms in the core 0.81 Å), and the four copies of RIPK3 are also nearly identical (RMSD of all heavy atoms in the core 0.59 Å), as expected since a single chemical shift is observed per atom. Key interactions in the core involve residues from each chain (532–549 in RIPK1 and 448–462 of RIPK3), near their I(V)QI(V)G RHIM consensus sequence elements. These residues adopt an extended strand–turn/kink1–extended strand–turn/kink2–extended strand–turn/kink3–extended strand conformation defined by T532–I533–Y534–N535–S536–T537–G538–I539–Q540–I541–G542–A543–Y544–N545–Y546–M547–E548–I549 in RIPK1 and P448–L449–V450–N451–I452–Y453–N454–C455–S456–G457–V458–Q459–V460–G461–D462 in RIPK3, with residues in bold corresponding to the kinks or turns (Figure 2B). A central 12-residue core (532-543 in RIPK1 and 451-462 in RIPK3) includes the most ordered part of the structure, consistent with the secondary chemical shift plot (Figure 2B) and the pattern of long-range restraints (Figure S2). Although the NMR data indicate that flanking sequences are also in an extended conformation (Figure 2B), they contain fewer long-range cross-peaks and are less ordered (Figure S2).
An interface formed by the side chains of 12 central residues from each strand constitutes an oblong-shaped hydrophobic core, which stretches beyond the amyloid spine of the consensus RHIM elements of IQIG in RIPK1 and VQVG in RIPK3 (Figure 4). The meandering conformation in the serpentine fold plays a critical role in the structure. In particular, the turns or kinks at N535 and G538 of RIPK1 and N454 and G457 of RIPK3 bring I533 of RIPK1 and I452 of RIPK3 into the hydrophobic core, where they interdigitate with the hydrophobic Ile and Val side chains of the RHIM consensus sequences to make a compact hydrophobic core. These turns or kinks result in an enclosed oblong-shaped hydrophobic core. A number of cross-peaks at the edges of the motif support the conclusion of such a compact core (Table S1 and S2, Figure S2). In addition to the six hydrophobic positions in the core, we detected contacts involving Ser536CB (RIPK1) and Cys455CB (RIPK3) that indicate their burial within the hydrophobic interface as stacked Cys-Ser ladders (Figure 5A). In aggregate, our data show that the interface in the RIPK1-RIPK3 amyloid complex is dependent not only on local β-structure, but also on long-range tertiary and quaternary interactions that are reminiscent of hydrophobic cores in globular proteins.
Figure 5. Parallel Contacts between RIPK1 and RIPK3 Identified by NMR driven CYANA calculations, and biochemical confirmation of Cys-Ser ladder.

(A) Depiction of the alternation of Ser (RIPK1) and Cys (RIPK3) in the Cys-Ser ladder.
(B) SDS-PAGE of RIPK1-RIPK3, RIPK1, and RIPK3 under reducing and non-reducing conditions to detect Cys-Cys contacts. Monomeric bands corresponding to RIPK1 and RIPK3 appear at ~10 and ~15 kDa, respectively. RIPK3 samples show an additional higher molecular weight band near 30 kDa under non-reducing conditions but not under reducing conditions (red asterisk), which is evidence for Cys-linked dimers due to the Cys ladders in homo-amyloids of RIPK3. Such a higher molecular weight band is not detected in the RIPK1 samples, nor the RIPK1-RIPK3 mixed samples, despite the fact that their centrifugation behavior indicted the formation of amyloids; we interpret this as evidence that in place of the Cys ladders these samples have Ser ladders or alternating Cys-Ser ladders, respectively.
(C) SDS-PAGE of RIPK1 and RIPK1(S537C) homo-amyloids reveals that under non-reducing condition the monomer band of RIPK1(S537C) is greatly reduced with concurrent appearance of higher molecular weight oligomeric bands (black asterisks) and material that did not enter the gel. We interpret this result as Cys-cross linking in RIPK1 homo-amyloids, which suggest an analogous structure to that of RIPK3 and RIPK1-RIPK3 amyloids.
(D) SDS-PAGE of RIPK1-RIPK3 and RIPK1(S537C)-RIPK3 hetero-amyloids under reducing and non-reducing conditions are contrasted here. The hetero-amyloid RIPK1(S537C)-RIPK3 exhibits a high molecular weight band (black asterisk) under non-reducing conditions, but only displays a weaker and lower molecular weight oligomeric band (red asterisk) under reducing conditions, suggesting that Cys linkages are formed between neighboring RIPK1 S537C and RIPK3 in the fibrils. We interpret this as evidence for Cys ladders in the RIPK1(S537C)-RIPK3 hetero-amyloid and support for the existence of Cys-Ser ladders in the RIPK1-RIPK3 signaling complex of the necroptotic pathway.
See also Figure S5.
Stabilizing Interactions Facing Out from the Hydrophobic Core
Numerous solvent-exposed stacking interactions outside the hydrophobic core act to stabilize the RIPK1-RIPK3 complex. The structure contains an Asn ladder and a Gln ladder, analogous to ladders that are known to support other amyloid structures through H-bonding among the side chain amide groups along the fibril axis (Nelson et al., 2005). The Asn ladder includes the side chains of Asn535 (RIPK1) and Asn454 (RIPK3), while the Gln ladder includes the side chains of Gln540 (RIPK1) and Gln459 (RIPK3), all of which project their side chains to the amyloid exterior (Figure 4). Another stabilizing chain is formed from the side chains of Tyr534 (RIPK1) and Tyr453 (RIPK3) which protrude and stack along the fibril axis (Figure 4). A chain is formed from the side chains of Thr537 (RIPK1) and Ser456 (RIPK3). These motifs collectively create a dense distribution of stacking interactions along the fibril axis, making these proteins highly amyloidogenic.
Structural Validation based on biochemical detection of Cys-Ser and Cys-Cys Ladders
Within the hydrophobic core of the RIPK1-RIPK3 hetero-amyloid is an unusual ladder consisting of alternating Ser (RIPK1) and the isosteric Cys (RIPK3) residues (Figure 5A). Cys455 of RIPK3 is not within H-bonding distance of another Cys455 residue in our RIPK1-RIPK3 structure, and disulfide bonds are not observed in the hetero-amyloid by biochemical methods. To test this model we took advantage of the fact that RIPK1 and RIPK3 can form homo-amyloids (Li et al., 2012) that might have structural homology to the hetero-amyloid, and used SDS-PAGE to compare the native RIPK1-RIPK3 hetero-amyloid to RIPK1 or RIPK3 homo-amyloids, and to site-specific mutants (Figure 5B–D). Specifically, in place of a Cys-Ser ladder, a Cys-Cys ladder is predicted for the RIPK3 homo-amyloid, which might be expected exhibit disulfide bond formation at the ladder. When reducing SDS-PAGE was used, RIPK1, RIPK3 and the RIPK1-RIPK3 complex amyloids all displayed monomeric bands (Figure 5B) without evidence for higher molecular weight species. In contrast, when non-reducing SDS-PAGE was used, the RIPK3 fibril sample showed a distinct band consistent with dimer formation in addition to the monomeric band (Figure 5B), supporting the hypothesis of a Cys-Cys ladder and disulfide bond formation. This observation explains why the RIPK3 fibril is difficult to completely denature, as we previously described (Li et al., 2012). These data also support the conclusion that RHIM sequences can form homo-amyloid serpentine structures analogous to the hetero-amyloid structure we report here. To further validate the presence of a Cys-Ser ladder, we tested the hypothesis that RIPK1(S536C)-RIPK3 hetero-complexes would contain Cys-Cys ladders rather than Cys-Ser ladders and so would also form extensive disulfide bonds (Figure 5C and 5D). SDS-PAGE studies of hetero-amyloids formed using this mutation provided support for the presence of a Cys-Cys ladder in this mutant complex (Figure 5D). Furthermore, SDS-PAGE and Western blot analyses of RIPK1 and RIPK1(S537C) homo-amyloids were performed under reducing and non-reducing conditions. Under non-reducing condition, the monomer band of RIPK1(S537C) is greatly reduced with concurrent appearance of higher molecular weight oligomeric bands (Figure 5C). These experiments in aggregate offer strong support for our structures and specifically the presence of a Cys-Ser ladder and a parallel arrangement in the RIPK1-RIPK3 necrosome core complex.
Crystal Structure of the VQVG Motif
To further test whether RIPK3 homo-amyloids and RIPK1-RIPK3 hetero-amyloids have a homologous fold, we crystallized the RHIM VQVG consensus peptide of RIPK3 and determined its structure at 1.27 Å resolution (Figure 6A and 6B, Table S6). The VQVG sequence forms β-strands perpendicular to the long axis of the crystal unit cell. Analogous to many short core structures of amyloid fibrils (Nelson et al., 2005; Sawaya et al., 2007), the VQVG β-strands within each sheet are parallel and in-register in the fibril, and form a cross-β spine with a distance between strands in the sheet of 4.8 Å (Figure S6). Two such β-sheets oppose each other in the observed crystal packing, and they superimpose very well with the NMR derived structure of the RIPK1-RIPK3 larger hetero-amyloid (Figure 6C and S6, Table S6). These data provide strong support for our NMR structure and confirm that homo- and hetero-amyloids formed by RIPK1 and RIPK3 are likely to have the same architecture. In fact RIPK3 homo-amyloids has been reported to capable of inducing necroptosis under some circumstances (Wu et al., 2014). We also attempted to crystallize the RIPK1 consensus sequence IQIG to obtain its structure. Despite of its poor resolution, we found that it adopts a fold similar to that of RIPK1-RIPK3 (data not shown).
Figure 6. RIPK1 and RIPK3 Homo-Amyloids are Analogous to the RIPK1-RIPK3 Hetero-Amyloid.
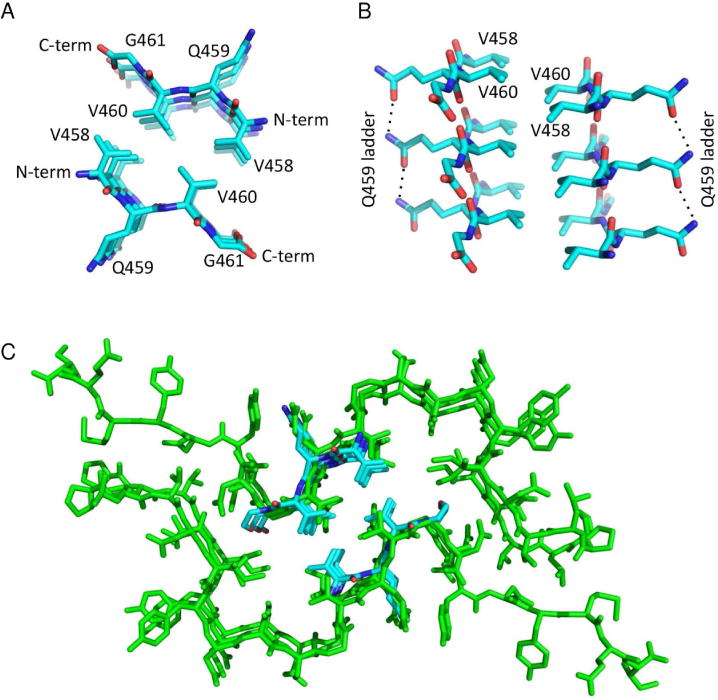
(A) Top view of two antiparallel β-sheets in the RIPK3 homo-amyloid. Each sheet is made from three parallel β-strands of the VQVG sequence.
(B) Side view of the structure. (C) Superposition of the crystal (cyan) and SSNMR (green) structures shows nearly identical topologies for the RIPK3 homo-amyloid and the RIPK1-RIPK3 hetero-amyloid. We recall that crystals from RIPK1 revealed an analogous structure, but were not included in this work due to lower resolution of the data.
See also Figure S6.
Preference for Hetero-Amyloid Formation
Given the similar homo- and hetero-amyloid structures, we wondered why RIPK1 and RIPK3 preferentially formed hetero-amyloids when co-expressed. The additional methyl group in Ile residues in RIPK1 (vs. Val in RIPK3), would be expected to result in stronger interactions for a RIPK1-RIPK1 β-sheet, as compared to RIPK1-RIPK3 or RIPK3-RIPK3 β-sheets. To investigate this question, we used dispersion-corrected (DFT-D) calculations, and found a larger interaction energy when VQVG (RIPK3) creates backbone hydrogen bonds with IQIG (RIPK1) in the strands above and below to form a β-sheet, than we did in the three-stranded homo-RIPK3 β-sheet. The difference in these values is comparable to the accuracy limit of theoretical methods, but was consistently observed in the sense that these calculations were carried out using three different functionals (see Methods for details), obtaining values between −0.38 and −0.55 kcal/mol. As expected, the interaction energy within the corresponding homo-RIPK1 β-sheet is more favorable (by −1 kcal/mol). This supports the idea that the interaction increases from RIPK3-RIPK3 < RIPK1-RIPK3 < RIPK1-RIPK1 within a β-sheet. We then calculated the energetic value of packing of the two β-sheets to create the hydrophobic core; here, the hetero-amyloid formation is substantially preferred over the homo-amyloid formation of IQIG (RIPK1) (ΔEint ~ −7.47 to −10.17 kcal/mol) and is also preferred to the homo-amyloid formation of VQVG (RIPK3). These calculations suggest that the alternation of RIPK1 and RIPK3 within a β-sheet gives efficient packing, maximizing hydrophobic contacts while keeping steric repulsion minimal.
DISCUSSION
A Unique Serpentine Fold with a Dense Hydrophobic Core and Numerous Stabilizing Chains
Solid-state NMR has proven to be a powerful tool for structural studies of amyloids, uncovering details in a number of important and complex amyloid structures (Sawaya et al., 2007). Some examples include β-solenoidal (HET-s) (Wasmer et al., 2008), Greek key (α-synuclein) (Tuttle et al., 2016), or S-shaped (Aβ1-42) (Colvin et al., 2016) topologies. Recently, the finding that an amyloidal peptide from S. aureus can adopt a cross-α spine has expanded our view of the structural versatility of amyloids (Tayeb-Fligelman et al., 2017). Notably, all amyloid structures reported to date build on copies of the same protein.
The topology of the RIPK1-RIPK3 hetero-amyloid bears apparent resemblance to a discrete region of Aβ1-40 E22Δ that spans 19 residues per chain(Schutz et al., 2015) (Figure S7). The pattern of alternating interactions in the RIPK1-RIPK3 hetero-amyloid structure is also somewhat reminiscent of the HET-s fungal prion wherein each protein chain contributes two rings of the β-solenoid. It is remarkable that the small size of the RIPK1-RIPK3 core contains a compact combination of the major previously reported amyloid-stabilizing elements. For instance, the much larger amyloid cores of α-synuclein (~80 residues) or HET-s (~70-residues) only contain one and two Asn ladders, respectively. Strictly defined, the RIPK1-RIPK3 structure is unprecedented, and raises interesting questions about combinatorial amyloids and the source of specificity in their formation.
Cellular activities and biochemical studies provide strong support for this structure. A single mutation in I539, I541, V458 or V460 to D dissociates the hetero-amyloid and causes failure to induce necroptosis (Li et al., 2012). The RIPK1-RIPK3 structure explains these observations, since these residues are the key contributors to the nonpolar interface in the core. Similarly, mutations in either Q540 of RIPK1 or Q459 of RIPK3 to D are detrimental to the formation of the amyloid. Gln residues form strong H-bonded ladders in this and other amyloids (Mompean et al., 2016) and therefore, these changes disrupt the RIPK1-RIPK3 amyloid by electrostatic effects and removal of the H-bonding ladder. Conserved glycine residues found at the edge of the β-sheets (G542 in RIPK1 and G461 in RIPK3) are also crucial. When these residues are mutated, RIPK1 and RIPK3 fail to form an amyloid and to activate necroptosis (Li et al., 2012). This feature is reminiscent of the HET-s amyloid, where conserved glycines were shown to be important for the β-strands during amyloid formation (Daskalov et al., 2014). Our SSNMR structure of the RIPK1-RIPK3 hetero-amyloid indicates that A543 (RIPK1) interacts with (is stacked with) D462 (RIPK3). The A543D mutation prevents the formation of the RIPK1-RIPK3 hetero-amyloid and initiation of necroptosis (Li et al., 2012). These data offer strong support for the presence of a Cys-Ser ladder in the RIPK1-RIPK3 hetero-amyloid necrosome core complex, and a Cys-Cys ladder in the RIPK3 homo-amyloid, providing in aggregate very strong support for the fold reported here.
Specificity of RHIM-RHIM Interactions for the Complex Biological Pathways and Viral Regulation
This is the first reported structure of a hetero-amyloid. Hetero-amyloids are likely to be capable of a range of biological effects not only in necroptosis, but in other biological systems more broadly. For example, putatively noxious amyloid aggregates often contain a mixture of proteins, for example in the context of neurological disorders (Fuentealba et al., 2010). This first example of a hetero-amyloid exhibits highly structurally ordered and specific interactions, with an energetic preference for this hetero-amyloid over the homo-amyloids. Subtle energetic differences due to discrete changes at the amino acid residue level could give rise to interactions that privilege certain complexes above others in the large library of possible hetero-complexes. Preferential formation of specific hetero-amyloids could have implications for a potentially large range of signaling activities. Indeed, this and other RHIM-RHIM amyloids exhibit biological specificity, even despite the high degree of conservation in the consensus sequence element (Figure 7A) and our study helps to elucidate how this is possible. For example, because RIPK1 is upstream of RIPK3 in the TNF signaling pathway, the ability of RIPK1 to preferentially bind RIPK3 may allow RIPK1 to specifically transduce the activation signal to RIPK3. It is likely that the hetero- amyloid formed from RIPK1 and RIPK3 could nucleate the formation of a RIPK3 homo-amyloid (Figure 7B), and therefore only a small amount of RIPK1 would be needed to fully activate RIPK3, which in turn activates MLKL for membrane disruption and lytic cell death in necroptosis. In addition, the murine cytomegalovirus protein M45 with a RIPK1-like consensus sequence of IQIG appears to preferentially target RIPK3 to inhibit necroptosis (Upton et al., 2010), and the second RHIM motif in the cytoplasmic DNA sensor DAI with a sequence of VQLG is less important than its first RHIM motif with a sequence of IQIG for engaging RIPK3 (Rebsamen et al., 2009) (Figure 7A). All of these examples illustrate the potential importance of specificity and structure of mixed amyloids involved in signaling. Mixed amyloids are likely to remain important topics for structural biology.
Figure 7. Alignment of RHIMs and RHIM-Mediated Necroptosis Activation.

(A) Sequence alignment of the RHIM consensus sequences and surrounding regions from human RIPK1 (hsRIPK1, NP_002795), RIPK3 (hsRIPK3, NP_006862), human TRIF (hsTRIF, NP_891549), human DAI (hsDAI RHIMs 1 and 2, NP_110403), and mouse cytomegalovirus M45 (mcM45, Q06A28). Partially conserved residues are highlighted in green outside the consensus I(V)QI(V/L)G, which is shown in red.
(B) Proposed canonical and noncanonical necroptotic pathways in which the RIPK1-RIPK3 and DAI-RIPK3 complexes, once formed, act as a nucleus to induce the polymerization and amyloid formation of additional RIPK3 activated phosphorylated molecular aggregates. Activated RIPK3 can then phosphorylate MLKL, inducing aggregation and leading to MLKL-mediated membrane permeabilization and cell death. In both pathways, specific formation of a hetero-amyloid serves as the crucial first step to activate the pathway.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. Coli DH5α | Thermo Fisher | Cat# 18258012 |
| E. Coli BL21 DE3 (RIPL) | Agilent Technologies | Cat# 230280 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,3-13C Glycerol | CIL company | Cat# CLM-1857-PK |
| 2-13C Glycerol | CIL company | Cat# CLM-1397-PK |
| 13C-Glucose | CIL company | Cat# CLM-420-PK |
| 15N-Ammonium Chloride | CIL company | Cat# NLM-467-PK |
| Peptide: VQVG | Peptide 2.0 Inc | N/A |
| RIPK1 | This paper | N/A |
| RIPK3 | This paper | N/A |
| Deposited Data | ||
| RIPK1-RIPK3 SSNMR amyloid structure | This paper | PDB: 5V7Z |
| RIPK3-RIPK3 RHIM X-Ray structure | This paper | PDB: 5DFO |
| RIPK1-RIPK3 SSNMR Chemical shifts | This paper | BMRB: 30273 |
| Recombinant DNA | ||
| RIPK1 | Human | https://www.ncbi.nlm.nih.gov/nuccore/NM_003804.4 |
| RIPK3 | Human | https://www.ncbi.nlm.nih.gov/nuccore/NM_006871.3 |
| Software and Algorithms | ||
| CYANA | Guntert, 2014 | http://www.cyana.org |
| AMBER | Case et al., 2005 | http://www.ambermd.org |
| PyMOL Molecular Graphics System | Schrödinger | http://www.pymol.org |
| Gaussian 09 | Gaussian Inc. | http://gaussian.com |
| XDS | Kabsh, 2010 | http://xds.mpimf-heidelberg.mpg.de |
| HKL2000 | Otwinowski and Minor, 1997 | http://www.hkl-xray.com |
| CCP4 | Win et al., 2011 | http://www.ccp4.ac.uk |
| PHENIX | Adams et al., 2010 | http://www.phenix-online.org |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead contact Ann McDermott (aem5@columbia.edu).
EXPERIMENTAL METHOD AND SUBJECT DETAILS
Proteins were expressed at 16 °C in BL21(DE3) or DH5α E. coli cells, using M9 medium supplemented with 15NH4Cl, NaH12CO3, and [1,3-13C]glycerol or [2- 13C]glycerol for isotopically labeled proteins.
METHOD DETAILS
Expression and Purification of Isotope Labeled RIPK1 and RIPK3
The RIPK1 (residues 496-583)-RIPK3 (residues 388-518) complex was co-expressed, His-Sumo tagged, with the pET28a (Novagen) vector in E. Coli BL21 DE3 (RIPL) using M9 minimal medium and purified as described previously (Li et al., 2012). More precisely, 0.5 g 15NH4Cl, 2.0 g NaH12CO3 and 2.0 g [1,3-13C]glycerol or 2.0 g [2-13C]glycerol were added into a liter M9 medium. Isopropyl 1-thi-β-D-galactopyranoside (IPTG, 0.5 mM) was added into cells to induce protein expression at 16 °C for 24 h when the optical density at 600 nm (OD600) reached a value of 0.5-0.6. Then the cells were harvested by centrifugation at 6000 rpm for 10 min at 4 °C, and the cell pellets were resuspended in a lysis buffer (50 mM Tris-HCL, 500 mM NaCl, 10 mM imidazole, 5% Glycerol, 2 mM β ME, pH 8.0). After removal of the cell debris, the supernatant was purified by Ni-affinity chromatography. Next, the proteins were precipitated by adding 5% ammonium sulfate at 4 °C, and followed by digestion with the enzyme subtilisin to remove the flanking, unstructured domains which gave no signals, without altering the conformation of the RIPK1-RIPK3 interaction interface as proven previously with chemical shift analysis (Li et al., 2012).
SDS-PAGE Analysis
The constructs RIPK1 (residues 496-583)-RIPK3 (residues 388-518), RIPK1 (residues 496-583), RIPK3 (residues 388-518) were expressed in E. coli and purified via Ni-NTA and size exclusion chromatography as reported earlier (Li et al., 2012). The void peak containing the corresponding proteins were pooled together and concentrated. The samples were subjected to SDS-PAGE either in reducing (sample buffer containing β-mercaptoethanol) or non-reducing (sample buffer without β–mercaptoethanol) conditions.
Crystallization
The tetrapeptide VQVG was crystallized using the hanging drop vapor diffusion method. 1 l of 5 mg/mL VQVG peptide was mixed with 1 l of reservoir solution containing 0.2 M magnesium chloride, 0.1 M sodium cacodylate at pH 6.5 and 20% (v/v) PEG 200. Peptide crystals were mounted directly onto nylon CryoLoops (Hampton Research) without cryo-protectants.
X-ray Diffraction and Data Processing
X-ray diffraction data were collected at the beamline 24-ID-E of the Advanced Photon Source, USA, and the beamline BL19U1 of the National Center for Protein Science Shanghai (NCPSS), China, at a wavelength of 0.979 Å with 5° oscillations. Indexing of diffraction images and scaling of data were performed using the programs XDS (Kabsch, 2010) and HKL2000 (Otwinowski and Minor, 1997). The merged scaled data were imported into CCP4 format with programs under the ‘CCP4i’ interface (Winn et al., 2011). Molecular replacement solutions were found using the program Phenix (Adams et al., 2010) with the search model of VQIV in Tau protein (PDB ID: 2ON9). Crystallographic refinement was performed with the programs Phenix (Adams et al., 2010). The atomic model was built with COOT (Emsley et al., 2010) and illustrated with PyMOL (Schrodinger, 2015). The atomic coordinates and structural factors of the VQVG amyloid core were deposited in the Protein Data Bank with accession code 5ZCK.
Solid State NMR Experiments
MAS SSNMR spectra were recorded on a Bruker Avance I 17.6 T spectrometer (750 MHz 1H frequency) with 3.2 mm E-free probe, on protease-digested samples prepared as previously described (Li et al., 2012). The 2D 13C-13C DARR were recorded on both uniformly-labeled and samples prepared using 1,3–13C and 2–13C glycerol as the carbon source, to ensure sample homogeneity with respect to our previous preparations. These spectra were recorded with different mixing times; namely, 20, 50, 100, 250, 300 and 500 ms. The spectra recorded at 250 ms were optimal for obtaining valuable long-distance restraints. 13C-13C PAR experiments were recorded with long mixing times (12 and 15 ms) on the samples prepared with glycerol as explained in the main text. These samples were also used in the 15N-13C correlation spectra, which consisted of two PAIN spectra (one for each of the samples growth with glycerol), and four TEDOR experiments with 2.7, 4.0, 6.7 and 8.0 ms of mixing. The two latter showed optimal magnetization transfers to obtain distance restraints, as gauged from the comparison with the PAIN spectra. Data were collected at ~10 °C if not mentioned otherwise. Chemical shifts were externally referenced to DSS using adamantane. Further details on the experimental acquisition parameters are given in table S5. All spectra were processed in Topspin 3.1 and analyzed using the program Sparky.
Structure Calculation
The structure of the RIPK1-RIPK3 hetero-amyloid was obtained using CYANA 3.97, following a three-tiered protocol consisting of 1) initial simulated annealing calculation with 21 unambiguous distance restraints with upper limits of 7.5 Å (Table S1), H-bonds and dihedral restraints; 2) inclusion of 21 restraints derived from low-ambiguity peaks (Table S2); and 3) a full automated cross-peak assignment CYANA calculation. In the two first stages, eight non-interacting, extended molecules (four of RIPK1 and four of RIPK3) are submitted to simulated annealing calculations with standard parameters, except for the number of torsion angle dynamics steps, which was set to 20000 (default is 10000).
The final step consisted of a full CYANA calculation with seven cycles of combined automatic cross-peaks assignment and structure calculation, with a tolerance of 0.2 ppm for the matching of input cross-peak positions and chemical shifts of potential assignments, using a cutoff of 7.5 Å. 100 structures were calculated using eight extended, non-interacting molecules and 20000 steps per conformer, from which the 10 lowest-energy structures were selected. This number (100) was chosen to ensure that the set of experimental restraints is strong enough so that convergence to the correct fold does not require calculating larger number of conformers. This prevents over fitting of the resulting structures. Finally, we used the AMBER9 software (Case et al., 2005) to remove clashes and refine this ensemble of structures with 2000 steps of energy minimization with implicit solvation. A cross-check calculation was performed using the full set of experimental restraints without the inclusion of H-bonds (Figure S5).
Dispersion-Corrected DFT (DFT-D) Calculations
To analyze the different interactions in the homo- and hetero-amyloids, quantum mechanical calculations were performed using the Gaussian09 package (Frisch et al., 2009). The interaction energy of the central strand with the strands above and below in a three-stranded RIPK3-RIPK3-RIPK3 β-sheet was compared to that of a RIPK1-RIPK3-RIPK1 β-sheet. This difference provides the gain of interaction energy in the hetero-amyloid (RIPK1-RIPK3-RIPK1) with respect to the homo-amyloid (RIPK3-RIPK3-RIPK3). Similarly, we computed the net gain of interaction energy when the homo-amyloid sheet was RIPK1-RIPK1-RIPK1 with respect to RIPK1-RIPK3-RIPK1.
To study the influence of homo- vs. hetero-amyloids on the energetics of the inter-β-sheet interface, we compared the interaction energies of pairs of RIPK1-RIPK1-RIPK1, RIPK3-RIPK3-RIPK3, and RIPK1-RIPK3-RIPK1. All calculations were performed with DFT including dispersion corrections (DFT-D3) (Grimme et al., 2010). The M06-2X (Zhao and Truhlar, 2006), PBE0 (Adamo and Barone, 1999), and CAM-B3LYP (Yanai et al., 2004) functionals were employed in combination with the D95(d,p) basis set, following literature precedents (Mompean et al., 2016; Plumley and Dannenberg, 2010). All calculations were counterpoise corrected to account for basis set superposition error.
DATA AND SOFTWARE AVAILABILITY
The solid-state NMR bundle and experimental data are deposited in the PDB and BMRB databases under accession codes 5V7Z and 30273, respectively. The crystal structure of the RIPK3 VQVG amyloid is deposited in the PDB under accession code 5ZCK.
Supplementary Material
Figure S1. Chemical Shift Consistency in RIPK1-RIPK3 Samples, Related to Figure 1.
(A) Comparison of DARR spectra from our previous work (black) with the one acquired on new samples used for obtaining the structure of the RIPK1-RIPK3 hetero-amyloid (blue, – this color scheme is used for Figures with assignments in this work). The good match in peak positions supports the assumption that the samples are reproducible and numerous correlations corroborate our previously reported assignments.
(B) DARR spectrum of 13C-, 15N- uniformly labeled (U-[13C-15N]) samples, recorded with a mixing time of 50 ms. A single resonance is detected for each site, which indicates that the conformation and environment of the peptides is homogeneous. The strong peak labeled as ALA with resonances 177.6, 52.40, and 19.35 ppm, which we assigned tentatively to the C’, CA, and CB nuclei of an alanine is not site specifically assigned because it does not appear sufficiently in other data sets to ascertain assignment or tertiary interactions.
Figure S2. Wire Diagrams Illustrate Unambiguous vs. Low-Ambiguity Long-Range Contacts, Related to Figure 2 and Tables S1, S2.
(A) RIPK1–RIPK3 intra β-sheet interactions support a parallel, sequential registration for both proteins (top), with alternating RIPK1–RIPK3 pattern. Inter β-sheet contacts involving hydrophobic side chains support an antiparallel assembly of the two sheets (middle). Other β-sheet correlations indicate that both molecules are bent to enclose a “four-sided” hydrophobic interface (bottom). Red lines represent unambiguous cross-peaks, while black lines are used for resolved low-ambiguity ones.
(B) RIPK1–RIPK1 intramolecular contacts that are in agreement with a bent conformation (top); and intermolecular correlations across the dry interface that support the packing of two β-sheets (bottom).
(C) As in the previous panel B, intramolecular contacts that support a bent conformation are also observed in RIPK1–RIPK1 (top). Similarly, RIPK3–RIPK3 intermolecular correlations across the hydrophobic interface are also observed (bottom).
Figure S3. Ambiguous Restraints and their Structural Information, Related to Figure 3
(A) Expansion of the 15N–13C TEDOR spectrum (mixing time of 8.0 ms). Two ambiguous peaks are shown. If the peak G457/G461 –S536 is interpreted as G457–S536 it is compatible with the RIPK1-RIPK3 structure (black dotted lines), whereas if it is interpreted as G461–S536 it is not (red dotted lines). In contrast, for the peak A543/I452–T532, both possible interpretations are compatible with our structure, yielding either inter- or intra-sheet restraints, respectively.
(B) Expansions of PAR and PAIN spectra recorded with mixing times of 12 and 6 ms, respectively. Blue spectra are recorded on 13C-diluted samples (using 1,3–13C glycerol), and those in red correspond to samples grown with 2–13C glycerol. Black labels illustrate intra-residue correlations, and sequential connectivities are labeled in red. The inter-molecular contacts providing long-distance restraints are represented in blue. An NCA spectrum from prior work (Li et al., 2012) is shown in black for reference. Due to signal-to-noise limitations only residues 457-462 were detected for RIPK3 in that prior work and in this specific NCA spectrum, whereas in the present work we extended the assignments (448-462 for RIPK3, see Figure 2 for visual assignments).
(C) The restraints are displayed on the structural bundle, showing that, all ambiguous cross-peaks from Table S2 (low-ambiguity) can be assigned in such a way that they are compatible with the structure.
Figure S4. DARR Spectrum at 250 ms of Mixing Time, Related to Figure 4.
Cross-peaks providing parallel contacts are indicated in blue. Those in red correspond to long-range interactions between the neighboring sheets, (i.e. across the hydrophobic interface). Peaks in magenta represent restraints around the edges of or enclosing the hydrophobic interface. These restraints contributed significantly to determining the fold illustrated in Figure 4. Underscored peaks are unambiguous (see Table S1). The spectrum with a mixing time of 250 ms is shown in black (to match the color in Figure 1 in the main text). The spectrum shown in blue was recorded at 50 ms (and is that shown in previous Figure S1) and contains many intra-residue correlations.
Figure S5. The fold is determined by experimental distance restraints; the restraints are incompatible with other topologies, Related to Figure 5
(A) Ribbon representation of the 10-lowest energy structures obtained from a simulated annealing calculation without the inclusion of hypothesis-based hydrogen-bond restraints. The experimentally based restraints alone (illustrated in Figure 5) are sufficient to constrain the calculation. RIPK1 is shown in green, and RIPK3 in blue.
(B) Top view of this ensemble showing side chain packing in the hydrophobic core. The average pairwise RMSD of this bundle (backbone atoms) is 1.45 ± 0.4 Å, similar to that of the structure calculated using H-bond restraints (1.20 ± 0.3 Å)
(C) Overlay of the NMR bundle calculated without H-bond restraints (red) and with them (sand color).
(D) Top view of the two bundles showing side chains comprising the hydrophobic core and Asn-and Gln-ladders. The RMSD of these twenty structures (10 with H-bonds and 10 without them) is 1.51 ± 0.5 Å.
(E) Eight possible arrangements were explicitly considered and tested in our calculations. In eight different calculations, we imposed eight different hydrogen bonding patterns to define the spine of different octameric structures. Only the one shown in panel E was compatible with all the spectral restraints. The arrangements shown in panels F–L result in many distance restraints violations and high CYANA target function values when combined with the first set (unambiguous) of restraints (Table S1). The corresponding structure of each (E, F, G, …, L) corresponds to the rows (1, 2, 3, …, 8) of Table S5. We conclude that choosing the specific octamer topology or fold shown in E was essential for the success of these calculations. Both proteins were co-expressed and the fibrils formed in vivo, whereas in vitro assembly based on mixtures of isotopically enriched and isotopically natural abundance samples were not possible in our hands to date. Therefore some of the experimental evidence used in other studies for registration was not possible to obtain, while the interaction surfaces within hetero-amyloid also allowed for a wealth of other highly specific contact information.
Figure S6. Crystal Structure of the VQVG Amyloid Core of RIPK3, Related to Figure 6
(A) The VQVG core forms long needle-shaped crystals.
(B) Electron density map of the VQVG core contoured at 2.0 σ. The water molecule is shown as a red star.
(C) The stick model of the VQVG structure. The distance between two neighboring strands in the β-sheet is around 4.81 Å, which corresponds to that in the RIPK1-RIPK3 amyloid fibril. The amino acid residues are labeled.
(D) The hydrogen bonds formed between the stacked β-strands are shown in dashed lines.
Figure S7. Hydrophobic Packing Similarities Comparing the Aβ1-40 E22Δ Homo-Amyloid, in green, and the RIPK1-RIPK3 Hetero-Amyloid in brown, Related to Figure 7
The structural bundle for both amyloids are shown as ribbon representations, using white spheres to illustrate hydrophobic residues (Ile, Val or Leu).
Highlights.
Solid-state NMR reveals the structure of the human RIPK1-RIPK3 necrosome
The RIPK1 - RIPK3 core is a hetero-amyloid structure
The cross beta spine contains alternating RIPK1 and RIP2 RHIMs with Gln ladders
Two beta sheets envelope a compact hydrophobic interior with a cys-ser ladder
Acknowledgments
This work was supported by grants from National Institute of Health (R01 AI045937) to HW and AEM, the National Science Foundation (MCB 0749381 and MCB 1412253 to AEM), the National Key Research and Development Program of China (2016YFA0500600) and National Natural Science Foundation of China (31670878) to JL. AEM is a member of the New York Structural Biology Center (NYSBC). The NYSBC is a STAR center supported by the New York State Office of Science, Technology and Academic Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and six tables.
AUTHOR CONTRIBUTIONS
M.M. carried out the structure calculations described and wrote the manuscript. W. L., S. L. and A.S. planned and performed the NMR experiments and edited the manuscript. J.L. performed the expression and purification of the isotope labeled RIPK1, RIPK3, and RIPK1-RIPK3 protein complex, and solved the crystal structure of VQVG amyloid core and edited the manuscript. G.B. performed SDS-PAGE analysis of RIPK1, RIPK3 and the RIPK1-RIPK3 complex filament samples under reducing and non-reducing conditions. H.W. and A.M. planned the experiments and cowrote the manuscript.
DECLARATION OF INTEREST
The authors declare no competing financial interest.
References
- Adamo C, Barone V. Toward relilable density functional methods without adjustable parameters: the PBE0 model. The Journal of chemical physics. 1999;110:12. [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, et al. Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils. Journal of the American Chemical Society. 2016 doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Sabate R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2720–2725. doi: 10.1073/pnas.1522361113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe G, Lewandowski JR, Loquet A, Bockmann A, Griffin RG. Proton assisted recoupling and protein structure determination. The Journal of chemical physics. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature chemical biology. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. Gaussian 09. Wallingford, CT, USA: Gaussian, Inc; 2009. [Google Scholar]
- Fuentealba RA, Udan M, Bell S, Wegorzewska I, Shao J, Diamond MI, Weihl CC, Baloh RH. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. The Journal of biological chemistry. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. The Journal of chemical physics. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- Guntert P. Automated NMR structure calculation with CYANA. Methods in molecular biology. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ, Kaiser WJ, Mocarski ES. Herpes simplex virus suppresses necroptosis in human cells. Cell host & microbe. 2015;17:243–251. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Ito Y, Ofengeim D, Najafov A, Das S, Saberi S, Li Y, Hitomi J, Zhu H, Chen H, Mayo L, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–608. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. Journal of the American Chemical Society. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. Journal of immunology. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Lewandowski JR, De Paepe G, Griffin RG. Proton assisted insensitive nuclei cross polarization. Journal of the American Chemical Society. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nature immunology. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Mompean M, Nogales A, Ezquerra TA, Laurents DV. Complex System Assembly Underlies a Two-Tiered Model of Highly Delocalized Electrons. The journal of physical chemistry letters. 2016;7:1859–1864. doi: 10.1021/acs.jpclett.6b00699. [DOI] [PubMed] [Google Scholar]
- Moquin D, Chan FK. The molecular regulation of programmed necrotic cell injury. Trends in biochemical sciences. 2010;35:434–441. doi: 10.1016/j.tibs.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology. 1997;276:20. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Muhlen S, Nachbur U, Pham CL, Zhang Y, Hildebrand JM, Oates CV, Lung TW, Ingle D, et al. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nature microbiology. 2017;2:16258. doi: 10.1038/nmicrobiol.2016.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley JA, Dannenberg JJ. The importance of hydrogen bonding between the glutamine side chains to the formation of amyloid VQIVYK parallel beta-sheets: an ONIOM DFT/AM1 study. Journal of the American Chemical Society. 2010;132:1758–1759. doi: 10.1021/ja909690a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendra BL, Siemer AB, Puthanveettil SV, Hendrickson WA, Kandel ER, McDermott AE. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol. 2013;20:495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K, Vazquez J, Benedict CA, Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Reports. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System. 2015 Version 1.8. [Google Scholar]
- Schutz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Guntert P, Bockmann A, Glockshuber R, Meier BH. Atomic-resolution three-dimensional structure of amyloid beta fibrils bearing the Osaka mutation. Angewandte Chemie. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344:631–637. [Google Scholar]
- Tayeb-Fligelman E, Tabachnikov O, Moshe A, Goldshmidt-Tran O, Sawaya MR, Coquelle N, Colletier JP, Landau M. The cytotoxic Staphylococcus aureus PSMalpha3 reveals a cross-alpha amyloid-like fibril. Science. 2017;355:831–833. doi: 10.1126/science.aaf4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, et al. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nature structural & molecular biology. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell host microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nature reviews Molecular cell biology. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunological reviews. 2011;244:9–28. doi: 10.1111/j.1600-065X.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- Walti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Bockmann A, Guntert P, Meier BH, Riek R. Atomic-resolution structure of a disease-relevant Abeta(1-42) amyloid fibril. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Yang ZH, Wang XK, Zhang Y, Wan H, Song Y, Chen X, Shao J, Han J. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell death and differentiation. 2014;21:1709–1720. doi: 10.1038/cdd.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai T, Tew D, Handy N. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP) Chem Phys Lett. 2004;393:7. [Google Scholar]
- Zhao Y, Truhlar DG. A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions. The Journal of chemical physics. 2006;125:194101. doi: 10.1063/1.2370993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Chemical Shift Consistency in RIPK1-RIPK3 Samples, Related to Figure 1.
(A) Comparison of DARR spectra from our previous work (black) with the one acquired on new samples used for obtaining the structure of the RIPK1-RIPK3 hetero-amyloid (blue, – this color scheme is used for Figures with assignments in this work). The good match in peak positions supports the assumption that the samples are reproducible and numerous correlations corroborate our previously reported assignments.
(B) DARR spectrum of 13C-, 15N- uniformly labeled (U-[13C-15N]) samples, recorded with a mixing time of 50 ms. A single resonance is detected for each site, which indicates that the conformation and environment of the peptides is homogeneous. The strong peak labeled as ALA with resonances 177.6, 52.40, and 19.35 ppm, which we assigned tentatively to the C’, CA, and CB nuclei of an alanine is not site specifically assigned because it does not appear sufficiently in other data sets to ascertain assignment or tertiary interactions.
Figure S2. Wire Diagrams Illustrate Unambiguous vs. Low-Ambiguity Long-Range Contacts, Related to Figure 2 and Tables S1, S2.
(A) RIPK1–RIPK3 intra β-sheet interactions support a parallel, sequential registration for both proteins (top), with alternating RIPK1–RIPK3 pattern. Inter β-sheet contacts involving hydrophobic side chains support an antiparallel assembly of the two sheets (middle). Other β-sheet correlations indicate that both molecules are bent to enclose a “four-sided” hydrophobic interface (bottom). Red lines represent unambiguous cross-peaks, while black lines are used for resolved low-ambiguity ones.
(B) RIPK1–RIPK1 intramolecular contacts that are in agreement with a bent conformation (top); and intermolecular correlations across the dry interface that support the packing of two β-sheets (bottom).
(C) As in the previous panel B, intramolecular contacts that support a bent conformation are also observed in RIPK1–RIPK1 (top). Similarly, RIPK3–RIPK3 intermolecular correlations across the hydrophobic interface are also observed (bottom).
Figure S3. Ambiguous Restraints and their Structural Information, Related to Figure 3
(A) Expansion of the 15N–13C TEDOR spectrum (mixing time of 8.0 ms). Two ambiguous peaks are shown. If the peak G457/G461 –S536 is interpreted as G457–S536 it is compatible with the RIPK1-RIPK3 structure (black dotted lines), whereas if it is interpreted as G461–S536 it is not (red dotted lines). In contrast, for the peak A543/I452–T532, both possible interpretations are compatible with our structure, yielding either inter- or intra-sheet restraints, respectively.
(B) Expansions of PAR and PAIN spectra recorded with mixing times of 12 and 6 ms, respectively. Blue spectra are recorded on 13C-diluted samples (using 1,3–13C glycerol), and those in red correspond to samples grown with 2–13C glycerol. Black labels illustrate intra-residue correlations, and sequential connectivities are labeled in red. The inter-molecular contacts providing long-distance restraints are represented in blue. An NCA spectrum from prior work (Li et al., 2012) is shown in black for reference. Due to signal-to-noise limitations only residues 457-462 were detected for RIPK3 in that prior work and in this specific NCA spectrum, whereas in the present work we extended the assignments (448-462 for RIPK3, see Figure 2 for visual assignments).
(C) The restraints are displayed on the structural bundle, showing that, all ambiguous cross-peaks from Table S2 (low-ambiguity) can be assigned in such a way that they are compatible with the structure.
Figure S4. DARR Spectrum at 250 ms of Mixing Time, Related to Figure 4.
Cross-peaks providing parallel contacts are indicated in blue. Those in red correspond to long-range interactions between the neighboring sheets, (i.e. across the hydrophobic interface). Peaks in magenta represent restraints around the edges of or enclosing the hydrophobic interface. These restraints contributed significantly to determining the fold illustrated in Figure 4. Underscored peaks are unambiguous (see Table S1). The spectrum with a mixing time of 250 ms is shown in black (to match the color in Figure 1 in the main text). The spectrum shown in blue was recorded at 50 ms (and is that shown in previous Figure S1) and contains many intra-residue correlations.
Figure S5. The fold is determined by experimental distance restraints; the restraints are incompatible with other topologies, Related to Figure 5
(A) Ribbon representation of the 10-lowest energy structures obtained from a simulated annealing calculation without the inclusion of hypothesis-based hydrogen-bond restraints. The experimentally based restraints alone (illustrated in Figure 5) are sufficient to constrain the calculation. RIPK1 is shown in green, and RIPK3 in blue.
(B) Top view of this ensemble showing side chain packing in the hydrophobic core. The average pairwise RMSD of this bundle (backbone atoms) is 1.45 ± 0.4 Å, similar to that of the structure calculated using H-bond restraints (1.20 ± 0.3 Å)
(C) Overlay of the NMR bundle calculated without H-bond restraints (red) and with them (sand color).
(D) Top view of the two bundles showing side chains comprising the hydrophobic core and Asn-and Gln-ladders. The RMSD of these twenty structures (10 with H-bonds and 10 without them) is 1.51 ± 0.5 Å.
(E) Eight possible arrangements were explicitly considered and tested in our calculations. In eight different calculations, we imposed eight different hydrogen bonding patterns to define the spine of different octameric structures. Only the one shown in panel E was compatible with all the spectral restraints. The arrangements shown in panels F–L result in many distance restraints violations and high CYANA target function values when combined with the first set (unambiguous) of restraints (Table S1). The corresponding structure of each (E, F, G, …, L) corresponds to the rows (1, 2, 3, …, 8) of Table S5. We conclude that choosing the specific octamer topology or fold shown in E was essential for the success of these calculations. Both proteins were co-expressed and the fibrils formed in vivo, whereas in vitro assembly based on mixtures of isotopically enriched and isotopically natural abundance samples were not possible in our hands to date. Therefore some of the experimental evidence used in other studies for registration was not possible to obtain, while the interaction surfaces within hetero-amyloid also allowed for a wealth of other highly specific contact information.
Figure S6. Crystal Structure of the VQVG Amyloid Core of RIPK3, Related to Figure 6
(A) The VQVG core forms long needle-shaped crystals.
(B) Electron density map of the VQVG core contoured at 2.0 σ. The water molecule is shown as a red star.
(C) The stick model of the VQVG structure. The distance between two neighboring strands in the β-sheet is around 4.81 Å, which corresponds to that in the RIPK1-RIPK3 amyloid fibril. The amino acid residues are labeled.
(D) The hydrogen bonds formed between the stacked β-strands are shown in dashed lines.
Figure S7. Hydrophobic Packing Similarities Comparing the Aβ1-40 E22Δ Homo-Amyloid, in green, and the RIPK1-RIPK3 Hetero-Amyloid in brown, Related to Figure 7
The structural bundle for both amyloids are shown as ribbon representations, using white spheres to illustrate hydrophobic residues (Ile, Val or Leu).


