Abstract
The members of the nitric oxide synthase (NOS) family, eNOS, nNOS and iNOS, are well-characterized enzymes. However, due to the lack of suitable direct NO sensors, little is known about the kinetic properties of cellular NO generation by the different nitric oxide synthase isoenzymes. Very recently, we developed a novel class of fluorescent protein-based NO-probes, the geNOps, which allow real-time measurement of cellular NO generation and fluctuation. By applying these genetic NO biosensors to nNOS-, eNOS- and iNOS-expressing HEK293 cells we were able to characterize the respective NO dynamics in single cells that exhibited identical Ca2+ signaling as comparable activator of nNOS and eNOS. Our data demonstrate that upon Ca2+ mobilization nNOS-derived NO signals occur instantly and strictly follow the Ca2+ elevation while NO release by eNOS occurs gradually and sustained. To detect high NO levels in cells expressing iNOS, a new ratiometric probe based on two fluorescent proteins was developed. This novel geNOp variant allows the measurement of the high NO levels in cells expressing iNOS. Moreover, we used this probe to study the L-arginine-dependency of NO generation by iNOS on the level of single cells. Our experiments highlight that the geNOps technology is suitable to detect obvious differences in the kinetics, amplitude and substrate-dependence of cellular NO signals-derived from all three nitric oxide synthase isoforms.
Keywords: eNOS, geNOps, iNOS, Live-cell fluorescence imaging, Nitric oxide, nNOS
1. Introduction
In higher organisms all three nitric oxide synthase (NOS) isoforms, neuronal, endothelial, and inducible NOS (nNOS, eNOS, iNOS), produce nitric oxide (NO) [1,2] that serves as important signal molecule for many cellular processes and biological functions [3–5]. The nNOS and eNOS isoenzymes are activated by Ca2+/calmodulin [6]. In contrast, iNOS is constitutively active [7] and the amount of iNOS-derived NO generation is predominately controlled on the transcriptional level [8]. Based on their distinct expression profile in particular organs and cell types eNOS-, nNOS- and iNOS-derived NO is involved in either the control of vascular tone [9], neurotransmission [10], or the immune response [11], respectively. While NO rather freely diffuses through bio-membranes [12], the susceptibility of this radical for oxidation clearly restricts its area of action [13]. The rather local intra-, endo- and paracrine actions of NO are mainly mediated by NO-induced nitrosylation of heme-containing proteins and enzymes [14,15] like the soluble guanylate cyclase (sGC) that represents a well-characterized, highly NO-sensitive downstream target [16]. However, due to its chemical reactivity NO may also modify other biomolecules [17] and modulate diverse signaling pathways or, at very high concentrations, initiate cytotoxic damage [18]. In addition to the classical NO-sensitive mechanisms of vasorelaxation, neurotransmission, and immune defense, NO is further discussed to control many other cellular processes in (patho)-physiology [19–21]. Considering the short half-life [22] and limited diffusion [23] of NO, its various physiological activities are mainly determined by its local concentration and spatiotemporal distribution. Most of the current NO detection methods are indirect providing an end-point quantification of NO in cell populations [24] and deliver only limited information about the spatiotemporal patterns of cellular NO kinetics and fluctuation. Due to these technical limitations, the visualization of NO signals from distinct NOS isoforms in real-time and on the level of individual cells has not been comprehensively investigated. In this study we have exploited and refined recently developed genetically-encoded NO probes, the geNOps [25–27], to investigate NO dynamics in single cells. To ensure identical Ca2+ signaling for comparable activation of nNOS and eNOS, we utilized HEK-293 cells expressing either eNOS or nNOS. Our data highlight fast and pulsatile NO signals upon the activation of nNOS in single cells that appear strictly coupled to the cytosolic Ca2+ signal. In contrast, Ca2+-activated eNOS showed a delayed NO generation and slowly increasing NO levels that remained elevated even after Ca2+ levels return to basal values. Cells expressing iNOS (HEK293 cells) showed strong and stable NO production that was merely dependent of extracellular L-arginine supplementation. Our experiments emphasize that the different NOS isoenzymes provoke distinct patterns of cellular NO signals, which most likely shape the activity of tissue-specific downstream signaling pathways and related organ functions.
2. Materials and methods
2.1. Reagents and buffers
Dulbecco's modified Eagle's medium (DMEM) and Nω-nitro-L-arginine (LNNA) were obtained from Sigma-Aldrich (Vienna, Austria). Fura-2-acetoxymethyl ester (fura-2/am), tetramethylrhodamine methyl ester perchlorate (TMRM) was purchased from Invitrogen (San Diego, CA, USA). TransFast™ transfection reagent was obtained from Promega (Mannheim, Germany). Antibodies against eNOS and nNOS were obtained from BD Transduction Laboratories™ (Schwechat, Austria), alpha-tubulin was from Cell Signaling Technology® (Cambridge, UK). Adenosine-5′-triphosphate (ATP) and L-arginine were purchased from Roth (Karlsruhe, Germany). Ionomycin was obtained from Abcam (Cambridge, UK). NOC-7 was from Santa Cruz (San Diego, CA, USA). The geNOps probes and the Iron(II) booster solution were from Next Generation Fluorescence Imaging GmbH - NGFI, Graz, Austria (www.ngfi.eu). Fetal Calf Serum (FCS), 100× Penicilin/Streptomycin, and Amphotericin were purchased form GIBCO (Invitrogen, Austria). Geneticin (G418) was purchased form Sigma Aldrich (Vienna, Austria).
2.2. Molecular cloning
In brief, cloning of the geNOp consisting of seCFP and tagRFP was performed according to standard procedures and all constructs were verified by sequencing. The rigid alpha-helical linker was synthesized by Genscript (Piscataway, NJ, USA) including the restriction sites BamHI at the N-terminus and EcoRI at the C-terminus: GAGGCCGCCGCCCGGGAGGCCGCCGCCAGAGAGGC-CGCCGCC-AGGGAGGCAGCAGCCCGCGAGGCAGCAGCCCGGGAGGCTGCTGCCA-GAGAGGCTGCTGCCAGGGAGGCCGCCGCCCGCGAGGCTGCTGCCCGGGAGGCTGCAGCCAGA. Plasmid DNA encoding for the NO sensitive geNOps was provided from NGFI, Graz. cDNA encoding the double FP based ratiometric geNOp was subcloned into a pcDNA3.1(–) vector via the restriction sites XhoI (N-terminus) and HindIII (C-terminus).
2.3. Cell culture
Human embryonic kidney (HEK293) cells were cultured in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293 cells stably expressing nNOS [28], eNOS or iNOS were cultured in DMEM supplemented with 10% (v/v) heat-inactivated fetal calf serum, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 1.25 μg/ml amphotericin, and 1 mg/ml geneticin (G418) in humidified atmosphere (95% O2/5% CO2) at 37 °C. Culture medium of EA.hy926 cells additionally contained 1% HAT (5 mM hypoxanthin, 20 μM aminopterin and 0.8 mM thymidine). Prior to transfection cells were plated on 30 mm glass cover slips. At 60–80% confluence cells were transfected with 1 ml of serum- and antibiotic-free medium containing 1.5 μg of the appropriate plasmid DNA and 2.5 μg of TransFast transfection reagent (Promega, Madison USA). Cells were maintained in a humidified incubator (37 °C, 5% CO2, 95% air) for 16–20 h before replacing the culture media against DMEM containing antibiotics and FCS. All experiments were performed either 24 or 48 h after transfection.
2.4. Fluorescence imaging
For cytosolic Ca2+ recordings using fura-2, cells were incubated in storage buffer containing 3.3 μM fura-2/AM for 40 min as previously described [29]. For NO imaging cells were loaded with iron(II) fumarate solution for 20 min and subsequently incubated in a storage buffer composed of 138 mM NaCl, 2 mM CaCl2, 5 mM KCl, 1 mM MgCl2, 1 mM HEPES, 2.6 mM NaHCO3, 0.44 mM KH2PO4, 0.34 mM Na2HPO4, 10 mM d-glucose, 0.1% vitamins, 0.2% essential amino acids, and 1% penicillin/streptomycin, pH 7.4 for 1 h prior to experiments. All imaging experiments were performed using a semi-automatic gravity-driven superfusion system. During experiments cells were superfused with a HEPES-buffered solution (HBS) containing 138 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 MgCl2, 10 mM d-glucose, 10 mM HEPES, pH 7.4 (NaOH or HCl adjusted) as previously described [29]. For Ca2+-free conditions HBS containing 1 mM EGTA instead of CaCl2 was used. Fluorescent recordings were taken on an advanced wide-field fluorescent microscope (Till Photonics, Graefling, Germany) equipped with a motorized sample stage, a polychrome V monochromator (Till Photonics), a 40× objective (alpha Plan Fluar 40×, Zeiss, Göttingen, Germany) and a charge-coupled device camera (AVT Stingray F145B, Allied Vision Technologies, Stadtroda, Germany). Fluorescence of C-geNOp and C-geNOpmut transfected cells was recorded at 480 nm upon 430 nm excitation. For detecting NO dynamics of CR-geNOp, expressing cells were alternately excited at 430 and 570 nm and the emissions were collected at 480 and 590 nm, respectively. Multichannel imaging experiments for simultaneous recording of cytosolic Ca2+ and NO, and G-geNOp expressing cells loaded with fura-2/am were alternately excited at 340, 380 and 480 nm and emission was collected at 515 nm (515dcxr). SypHer fluorescence was sequentially excited at 430 and 500 nm. Data acquisition and control was carried out by the Live Acquisition 2.0.0.12 software (Till Photonics).
2.5. Image analysis
The background values recorded by the respective channels were subtracted from the emission of the probe to obtain F and calculate F0 (reflecting the function of fluorescence of the probe over time without stimulation) using an appropriate equation e.g. F0=Finital*exp(-K*Time) + Fplateau in case of a fluorescence decrease reflected by a one exponential decay. To normalize the geNOp signals over time the formula 1-F/F0 was used for calculation, whereby F is defined as the background subtracted raw fluorescence over time.
2.6. Immunoblotting
Wild-type HEK293 cells or HEK293 cells stably expressing either nNOS or eNOS were harvested and homogenized by sonication (3 × 5 s) in ice-cold RIPA lysis buffer (Sigma, Vienna, Austria) containing 2 mM EDTA, protease and phosphatase inhibitors (Complete™, PhosSTOP™, Roche, Vienna, Austria). Protein concentration was determined with the Pierce™ BCA Protein Assay Kit (Fisher Scientific Austria GmbH, Vienna, Austria) using bovine serum albumin as standard. Denatured samples (30 μg) were separated by SDS-PAGE on 10% gels and transferred electrophoretically to nitrocellulose membranes. After blocking with 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) TWEEN-20 for 1 h, membranes were incubated overnight at 4 °C with a primary antibody against eNOS (1:2000; BD Transduction Laboratories), nNOS (1:1000; BD Transduction Laboratories) or β-actin (1:200,000; Sigma-Aldrich). Thereafter, membranes were washed 3 times and incubated for 1 h with a horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (1:5000). Immunoreactive bands were visualized by chemiluminescence using ECL detection reagent (Biozym, Germany) and quantified densitometrically by the Fusion SL system (Peqlab, Erlangen, Germany).
2.7. Structural analysis of geNOps
Structural models of geNOps were generated with the online software Phyre2 (Protein Homology/analogy Recognition Engine V 2.0) which uses a profile-profile alignment algorithm to predict the 3D structure of the protein of interest. The alignment is based on hidden Markov models via HHsearch [30] to significantly improve accuracy of alignment and detection rate. Analysis of the predicted proteins was performed with the software DeepView/Swiss Pdb viewer V4.1.0 obtained from Expasy (Lausanne, Switzerland).
2.8. Statistical analysis
Statistical analysis was performed using the GraphPad Prism software version 5.04 (GraphPad Software, San Diego, CA, USA). Data are presented as mean ± standard error of mean (SEM) of independent experiments (n) throughout the manuscript. For comparison between two groups, two-tailed Student t-test was used for evaluation of statistical significance and a P value of <0.05 was considered significant and indicated by *. For comparison across multiple groups, one-way ANOVA with Barlett's test for equal variances and Benferroni's Multiple Comparison test were used for evaluating statistical significance. At least three independent experiments (n) were performed in at least triplicates for each experimental setup. Data are shown as mean ± standard deviation (SD) as indicated.
3. Results and discussion
3.1. Simultaneous imaging of Ca2+ and NO in single endothelial cells unveils a delayed, slow- but sustained eNOS-mediated NO generation
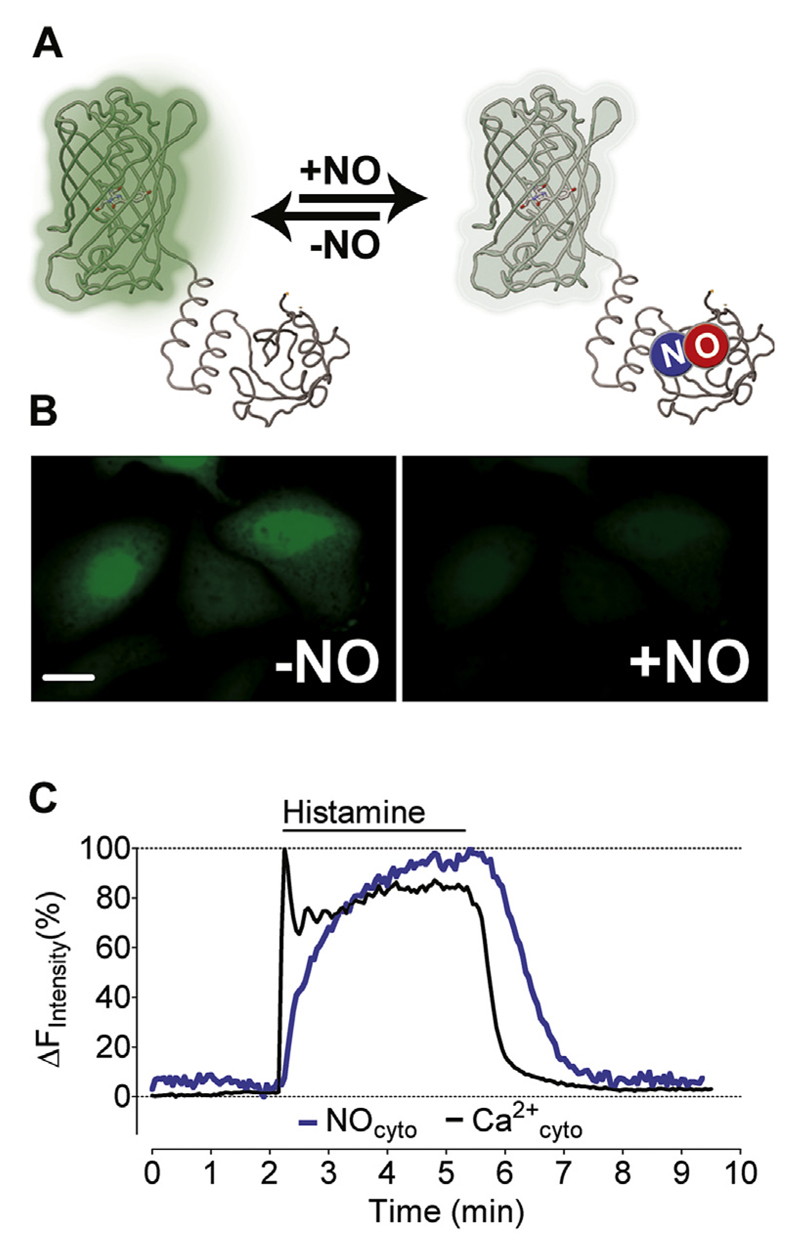
We used fura-2 and the green fluorescent NO-sensitive G-geNOp (Fig. 1A and B) to simultaneously record histamine-induced Ca2+ signal and Ca2+-triggered NO generation by eNOS in the endothelial cell line EAhy.926. Cell stimulation with the inositol-1,4,5-trisphosphat (IP3)-generating agonist histamine (100 μM) yielded an instant rise in cytosolic Ca2+ levels followed by a plateau phase at about 80% of the initial spike (Fig. 1C). In contrast, NO levels slowly increased with a delay of 7.7 ± 3.5 s (n = 8) compared to the Ca2+ signal and maximal NO generation was observed after approximately 4 min of stimulation. Upon washout of the agonist, cytosolic Ca2+ decreased to basal level within 50.8 ± 25.3 s (n = 8) while NO started to decline with a delay of 22.2 ± 13.3 s (n = 8) (Fig. 1C). A similar delay in agonist-induced endothelial cell NO formation (indirectly analyzed by cGMP) compared with cytosolic Ca2+ signal (measured with fura-2) was previously observed in freshly isolated porcine aortic endothelial cells [29]. Our data are also in line with previous studies showing a significant temporal delay between the addition of vasoactive compounds and maximal vessel relaxation [31,32]. Our present findings demonstrate now on the single cellular level that despite fast Ca2+ mobilization, endothelial NO generation through eNOS occurs delayed (with slower kinetics) and that the NO levels remain more sustained than the Ca2+ elevation.
Fig. 1. Simultaneous imaging of Ca2+ and NO in single endothelial cells.
(A) The scheme represents the functional principle of G-geNOp that FP fluorescence is quenched upon NO binding to the probe. (B) Representative wide field images of EA.hy926 cells expressing G-geNOp prior to exposure (left panel) and in the presence of the NO-donor NOC-7 (10 μM, right panel). (C) Representative [Ca2+]cyto (black curve) and [NO]cyto (blue curve) signals shown in percentage of the respective maximal increases in response to 100 μM histamine. Curves represent at least 4 independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. nNOS-derived NO signals occur fast and pulsatile
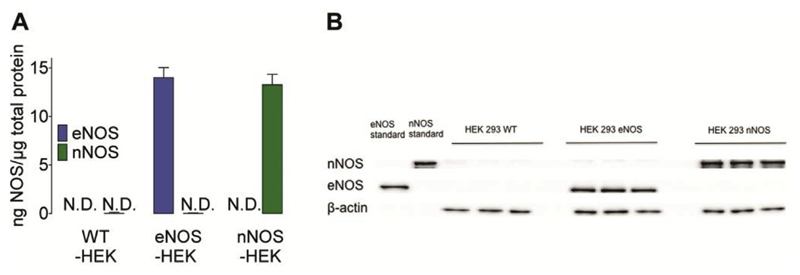
In order to directly compare the NO generation kinetics of eNOS- and nNOS-derived NO signals, we used HEK293 cells stably expressing either eNOS or nNOS as a cell model. This approach allowed us to study NOS kinetics upon identical Ca2+ stimulation in one given cell type. As confirmed by Western blot analyses, neither nNOS nor eNOS were expressed in wild-type HEK293 cells (Fig. 2A and B) but expression levels of the enzymes were comparable in the respective HEK293 cell clones (Fig. 2A and B).
Fig. 2. Western Blot analysis of nNOS and eNOS expression in HEK-cell clones.
(A) Bars represent the expression rate of eNOS (blue bars) and nNOS (green bars) in wild-type (left panel, n = 3), eNOS- (middle bars, n = 3) and nNOS-expressing HEK293 cells (right bars, n = 3). (B) Representative Western blot of total homogenates of wild-type, eNOS- or nNOS-expressing HEK293 cells (10 μg of protein) probed for eNOS (140 kDa), nNOS (155 kDa) and β-actin (43 kDa) expression (n = 3). Mean values are shown ±SD, *P < 0.05 versus control using unpaired t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Several studies have demonstrated that elevated NO levels affect the cellular Ca2+ homeostasis [31,33,34]. Therefore, we studied whether NOS expression affects basal Ca2+ levels or agonist-triggered Ca2+ signals in the NOS-expressing HEK293 cells. Treatment with ATP (100 μM) evoked a sharp and transient cytosolic Ca2+ signal in all HEK293 cells (Fig. 3A, upper panel). In comparison to wild-type HEK293 cells neither the basal Ca2+ levels nor the maximal amplitudes of ATP-induced Ca2+ profiles were altered in HEK293 cells expressing eNOS or nNOS, respectively (Fig. 3A and B). These results indicate that in our experimental conditions, the cellular Ca2+ signaling of HEK293 cells remained unaffected by the stable expression of the NO-generating isoenzymes. Next, we transiently transfected the cell lines with cyan geNOp (C-geNOp) in order to visualize NOS-specific NO dynamics. In wild-type HEK293 cells C-geNOp did not sense NO formation upon IP3-mediated Ca2+ mobilization confirming the absence of any Ca2+/calmodulin dependent NOS isoform (Fig. 3A left lower panel).
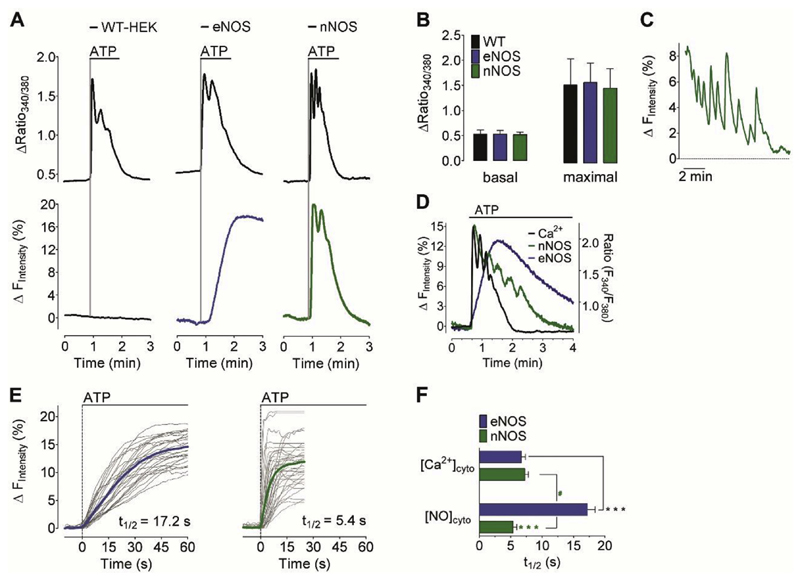
Fig. 3. Ca2+-triggered eNOS- and nNOS-specific NO signals.
(A) Representative time course of cytosolic Ca2+ (upper panel) and NO signals (lower panel) in response to 100 μM ATP in wild-type HEK293 cells (left panel), stably eNOS-expressing HEK293 cells (middle panel), and stably nNOS-expressing HEK293 cells (right panel). NO recordings were performed using C-geNOp. (B) Bars represent mean values in ±SD of basal fura2 ratios (left panel) and maximum cytosolic Ca2+ elevations in response to 100 μM ATP (right panel) of wild-type HEK293 cells (black bars, n = 52/12), eNOS expressing HEK293 cells (blue bars, n = 53/12) and nNOS expressing HEK293 cells (green bars, n = 65/12). (C) Representative pulsatile NO signals in nNOS-expressing HEK cells (D) Overlay of representative cytosolic Ca2+ (black curve) and NO recordings of stably nNOS- (green curve) or eNOS- (blue curve) expressing HEK293. Experiments were at least performed 4 times. (E) Single cell NO responses (light grey curves) and mean curves of eNOS-expressing HEK cells (blue curve, n = 5/32) and nNOS-expressing HEK cells (green curve, n = 5/37). (F) Bars represent the half time (t1/2) of maximum fura-2 ratio (upper blue bar, n = 54/4 for eNOS-expressing cells; upper green bar, n = 66/4 for nNOS expressing cells) and C-geNOp (lower blue bar, n = 32/3 for eNOS-HEK cells and lower green bar, nNOS-expressing cells) signals triggered by 100 μM ATP. Mean values are shown ±SD, *P < 0.05 versus control using unpaired t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
However, HEK293 cells stably expressing eNOS showed prominent NO elevations upon administration of ATP indicating that eNOS expressing HEK293 cells are activated by Ca2+ mobilization. Similar to our experiments with the endothelial cell line EAhy.926. described above and our previous observations in these cells [35], the eNOS-derived NO generation also occurred gradually, slowly and sustained in HEK293 cells compared to the fast arising and declining Ca2+ signal (Fig. 3A, D, and E). This confirms the slower activation kinetics of this isoenzyme. In contrast, triggering the same ATP-induced Ca2+ signal in HEK293 cells expressing nNOS revealed an instant and fast NO signal that nearly instantly followed the Ca2+ signal. The maximal amplitude of the NO signal was higher in eNOS-expressing cells compared to nNOS (Fig. 3E). On a time basis the nNOS-mediated NO formation occurred almost as fast as the respective Ca2+ signals (Fig. 3A, C, and D). Accordingly, upon the identical Ca2+ signal, eNOS exhibits slow activation kinetics but sustained NO activity (Fig. 3E), whereas nNOS derived NO strictly follows the cytosolic Ca2+ signal (Fig. 3D and E). Moreover, our data show that the average Ca2+ -evoked nNOS-derived NO formation occurred even slightly faster than the respective global cytosolic Ca2+-elevation (Fig. 3F). Either this finding might point to a local activation of nNOS or little cytosolic Ca2+ elevations are sufficient to maximally stimulate the enzyme.
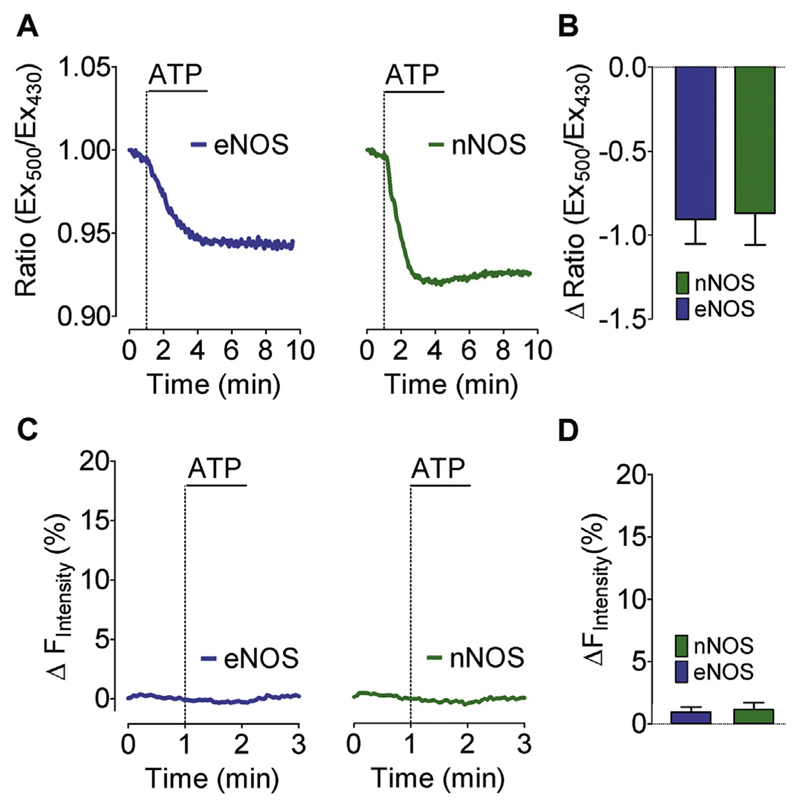
In order to probe whether cytosolic Ca2+ mobilization in eNOS and nNOS expressing cells alters the cytosolic pH, we utilized the pH sensitive genetic biosensor SypHer [65]. In line with another study, cell treatment with an IP3-generating agonist evoked a mild cytosolic acidification in eNOS and nNOS expressing HEK cells (Fig. 4A and B). As geNOps are pH sensitive, this effect might disturb NO measurements. Therefore, we performed control experiments using the NO-insensitive C-geNOpmut, which is still prone to pH fluctuations [25]. This approach revealed that the ATP-induced cellular acidification (Fig. 4A and B) is not strong enough to significantly influence the cyan fluorescence signal of the NO probe in eNOS and nNOS expressing cells (Fig. 4 C and D).
Fig. 4. pH sensitivity of geNOps.
(A) Representative time course of cytosolic pH changes in response to 100 μM ATP in eNOS-HEK cells (left panel) and nNOS-HEK cells (right panel) expressing the cytosolic SypHer. (B) Bars represent mean values in ±SD of maximal sypHer ratio changes in response to 100 μM ATP of eNOS-HEK cells (blue bars, n = 4/41) and nNOS-HEK cells (green bars, n = 4/46). (C) Representative time course of NO signals of eNOS-HEK cells (left panel) and nNOS-HEK cells (right panel) to 100 μM ATP. Cells were transiently transfected with the NO insensitive geNOpmut. (D) Bars represent mean values in ±SD of maximal geNOpmut responses upon administration of 100 μM ATP of eNOS-HEK cells (blue bars,n = 4/21) and nNOS-HEK cells (green bars, n = 4/21). Mean values are shown ±SD, *P < 0.05 versus control using unpaired t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Several studies have shown that the specific enzymatic activity of purified nNOS [36–40] is considerably higher than that of purified eNOS [41–44]. In these studies the enzymatic activity of the different NOS isoforms was controlled by the concentration of several cofactors. Therefore, such experiments are not conclusive in terms of NOS activities in intact cells. Using the geNOps technology, however, allowed us to demonstrate that in HEK cells nNOS-mediated NO formation is indeed significantly faster compared to eNOS-derived NO production. Moreover, our experiments are well in line with mathematical simulations that propose generation of sharp NO pulses by nNOS and a steady-state NO generation on the time scale of seconds to minutes by eNOS [45]. Our data emphasize that the fast and strictly Ca2+-controlled NO signals by nNOS might be essential to locally transmit information for neurotransmission at the short time of neuronal firing while in the vasculature a long-lasting release of NO by eNOS is suitable to ensure long-lasting changes of vessel tone.
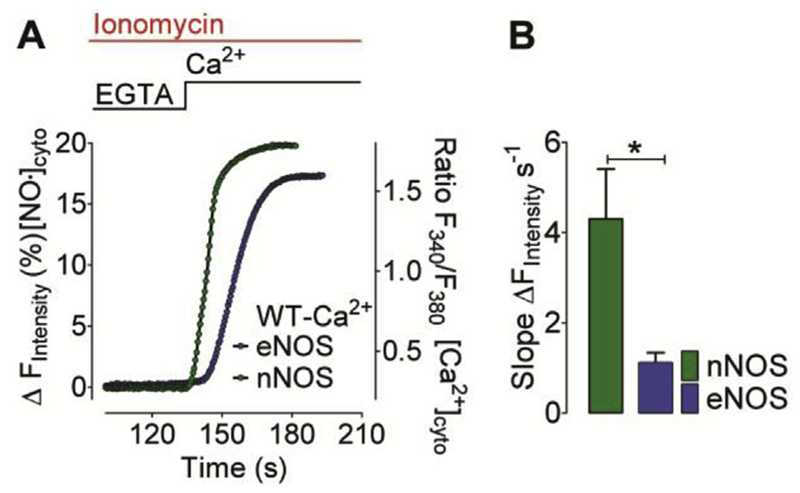
It has been shown that eNOS and nNOS are differentially targeted to distinct subcellular compartments [46,47]. While eNOS is predominately found in the sub-plasmalemma region [48], nNOS is rather located at the ER membrane or in the cytosol [49]. It has been reported that both NOS isoforms translocate upon Ca2+ mobilization [50] and it can be assumed that differences in the subcellular distribution of the enzymes might, at least in part contribute to the isoform-specific differences in the kinetics of NO generation. From this point of view Ca2+-dependent NOS isoforms might be activated much faster in close vicinity of Ca2+ release or Ca2+ entry channels. In order to test whether the kinetic differences of eNOS- versus nNOS-derived NO signals are indeed dependent on cell type-specific intracellular Ca2+ gradients, we treated our model cells with the Ca2+ ionophore in the absence of extracellular Ca2+ and subsequently re-added extracellular Ca2+ to uniformly elevate cytosolic Ca2+ levels. These conditions were chosen to avoid the generation of local Ca2+ gradients that may distinctly impact the kinetics of the various NOS isoenzymes. In HEK293 cells expressing nNOS the production of NO was about 4-times faster compared to eNOS-expressing cells (Fig. 5). These results indicate that the different NO profiles are rather based on the kinetic activities of the enzymes per se than on their subcellular localization. Furthermore, studies have demonstrated that NOS activity is controlled by posttranslational modifications [51–53], so we cannot exclude that the observed differences between NO generation by eNOS and nNOS are also influenced by variations in phosphorylation, acetylation, methylation or palmitoylation of the respective enzymes.
Fig. 5. Ca2+-evoked nNOS- and eNOS-derived NO generation profiles in ionomycin permeabilized cells.
(A) Mean curves represent NO generation profiles derived from eNOS (blue curve) and nNOS (green curve) in response to Ca2+ addition. Experiments were performed in the presence of 5 μM ionomycin. NO recordings were performed using C-geNOp. (B) Bars represent respective statistics of panel A, maximum initial slope of nNOS- (green bar, n = 34/3) and eNOS-derived (blue bar, n = 36/3) NO signals. Mean values are shown ±SD, *P < 0.05 versus control using unpaired t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. The newly developed ratiometric NO probe, CR-geNOp, allows the measurement of high intracellular NO levels in iNOS-expressing cells
The first generation of geNOps bears the drawback that their non-ratiometric characteristic makes them incapable to directly compare NO levels within various cells [25,26]. In order to overcome this limitation we improved the geNOps by engineering a ratiometric NO probe based on two FPs.
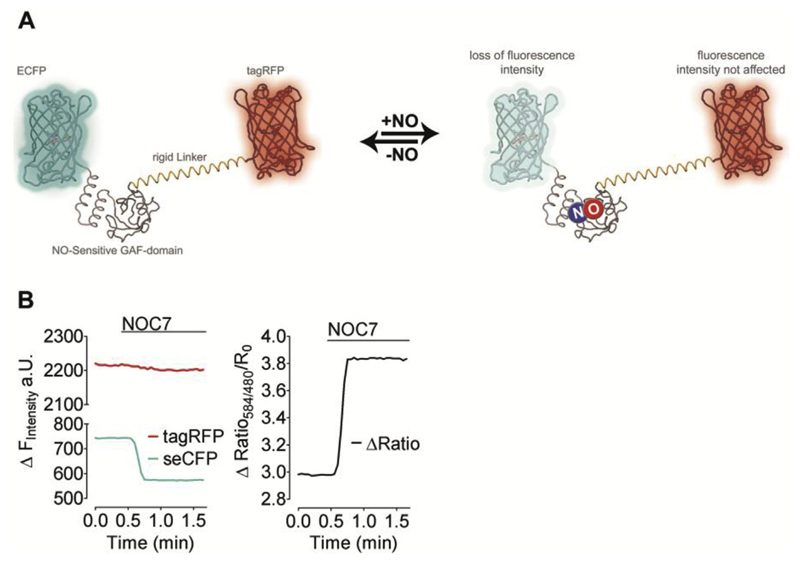
For this purpose we fused tagRFP, a strongly monomeric red FP variant, to the highly NO-sensitive C-geNOp to yield a ratiometric geNOp, referred to as CR-geNOp. To protect the tagRFP from any direct influence of NO, a rigid alpha helical linker based on (EAAAK)n repeats was introduced between the NO-binding GAF domain and the red FP variant (Fig. 6A). For testing this novel ratiometric probe, CR-geNOp expressing HEK293 cells were treated with NOC-7, a potent NO-releasing compound. As shown in Fig. 6B, the red fluorescence of CR-geNOp remained unaffected by the addition of NOC-7 (10 μM) while the cyan signal was strongly quenched in response to this NO donor, demonstrating the ratiometric readout by this new probe.
Fig. 6. CR-geNOp, a ratiometric NO probe based on two FPs.
(A) Schematic structures of CR-geNOp consisting of the cyan FP (CFP), a NO-sensitive GAF domain, a rigid linker (EAAAK)n and the red FP (tagRFP) in the presence and absence of NO, which quenches CFP fluorescence only (B) Left panel: fluorescence intensity CFP (cyan curve) and RFP (red curve) over time in cells expressing CR-geNOp. Cells were treated with 10 μM NOC-7 as indicated. Right panel: respective ratio signal (red/cyan; F584/F480) of CR-geNOp (n = 98/5). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
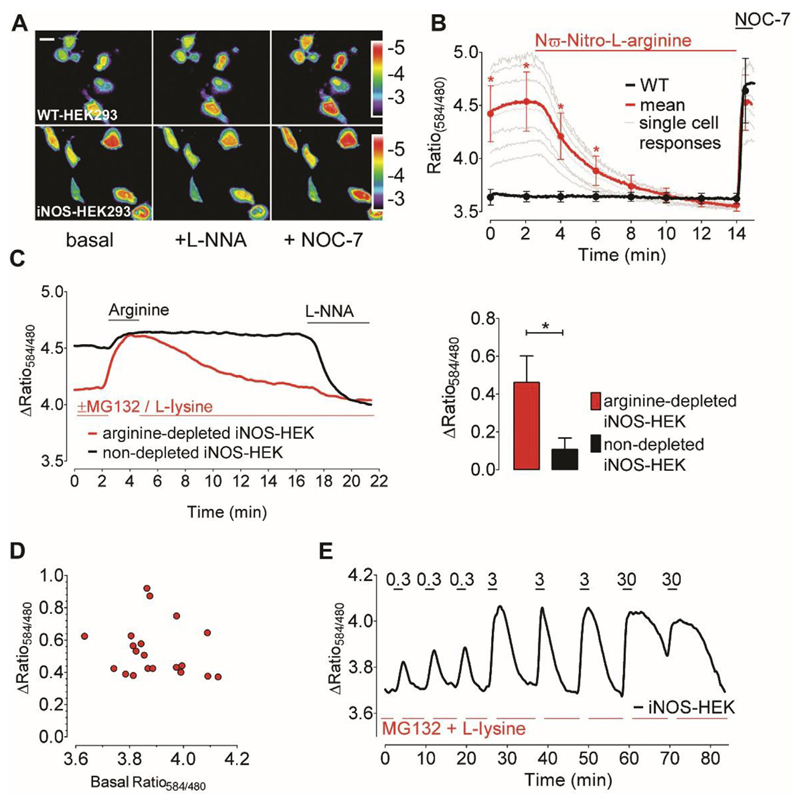
To further test the applicability of the novel double FP-based ratiometric geNOp we utilized HEK293 cells stably expressing the constitutively active iNOS that is known to generate large amounts of NO [54] though, the exact cellular NO concentrations are a matter of debate [55]. Ratiometric imaging of CR-geNOp uncovered strongly elevated NO levels in single iNOS-expressing HEK293 cells (Fig. 7A and B). In contrast to our experiments with nNOS and eNOS we observed clear heterogeneities of the ratio-signals among individual iNOS-expressing cells, possibly indicating differences in expression level and/or enzyme activities in the given cells (Fig. 7B). To confirm that the basal CR-geNOp ratio variances indeed reflect different iNOS-derived intracellular NO levels, cells were treated with NG-nitro-L-arginine (LNNA), a potent, irreversible NOS inhibitor [56,57]. Addition of the inhibitor immediately decreased the CR-geNOp ratio signal in iNOS-expressing HEK cells while that of NOS-negative control cells remained unaffected (Fig. 7B). Subsequent application of high concentrations of the NO-donor NOC-7 increased the fluorescence ratio signal of the NO probe equally in both cell types. Notably, in iNOS-expressing cells the amplitude of the signal induced by the NO-donor was comparable to the increased NO levels based on the iNOS activity in the given cell. These findings demonstrate the high and sustained activity of iNOS in the cell model used. This is in agreement with reports showing that under certain pathological conditions such as sepsis [58,59] high cellular NO concentrations can be persistently formed by iNOS. Eventually, the application of the geNOps technology in iNOS-expressing HEK293 cells proved the benefit of the novel ratiometric NO sensor to dynamically visualize NO fluctuations in single cells at pathophysiological relevant levels.
Fig. 7. Visualizing of NO dynamics in iNOS-expressing cells.
(A) Representative pseudo-colored ratio images (F584/F480) of CR-geNOp-transfected wild-type (upper images) and iNOS-expressing HEK293 cells (lower images) under initial conditions (left panel) and in the presence of 500 μM L-NNA (middle panel) or 10 μM NOC-7 (right panel). Scale bar represents 10 μm. (B) Mean curves (bold line) and single cell traces (thin lines) showing NO signals of stably iNOS-expressing (red bold curve, n = 23/3) and wild-type HEK293 cells (black bold curve, n = 15/3). Cells were treated with 300 μM L-NNA followed by administration of 10 μM NOC-7. (C) Representative time course showing NO signals in arginine-depleted (red curve) versus non-depleted iNOS-expressing HEK293 cells (black curve). Cells were treated with 50 μM L-arginine and 500 μM L-NNA as indicated. Bars represent respective statistics from panel C. Maximal ratio changes of arginine depleted iNOS-expressing (red bar, n = 21/3) and non-depleted iNOS HEK293 cells (black bar, n = 7/3) upon 50 μM L-arginine supplementation. Mean values are shown in ±SD, *P < 0.05 versus control using unpaired t-test. (D) Correlation of maximal arginine-induced ratio changes (y-axis) with respective basal ratio values (x-axis) of arginine-depleted iNOS-expressing HEK293 cells. (E) Representative time course showing cellular NO signals of an arginine-depleted (using MG132 and L-lysine) iNOS-HEK cell in response to increasing concentrations of arginine (0.3 μM, 3 μM and 30 μM). Arginine was added transiently and repetitively as indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. L-Arginine-depletion uncovers the tight substrate dependency of iNOS
The role of L-arginine supplementation to promote NOS activity is controversially discussed [59,60]. Nevertheless, distinct NOS-derived NO signals could be obtained even in the absence of extracellular L-arginine, indicating that L-arginine supplementation is not necessarily required for iNOS activity [61].
This obvious independency of iNOS activity from extracellular L-arginine highlights the high capacity of cells to supply the substrate from adequate internal amino acid pools [62]. To examine whether the internal amino acid pool is sufficient for sustained iNOS activity in our model, we recorded NO levels over time in iNOS-expressing HEK293 cells, upon addition of L-arginine (Fig. 7C). In these experiments, iNOS was found to continuously produce high amounts of NO even in the absence of extracellular L-arginine (Fig. 7C). Supplementation of the amino acid only moderately increased cellular NO levels (Fig. 7C). This is in line with findings that L-arginine washout from the medium of an activated macrophage cell line only slightly affected iNOS-derived NO formation [61]. Next we tested the contribution of endogenous L-arginine to iNOS-derived NO formation. To inhibit the protein degradation machinery of the proteasome as important L-arginine source we applied the proteasome inhibitor MG132 [63]. In our experiments we combined MG132 and the cationic amino acid L-lysine, which is known to replace the remaining freely available L-arginine [64], to deplete cellular L-arginine pools in iNOS-expressing HEK293 cells. As shown in Fig. 7C, the limitation of intracellularly available L-arginine strongly affected iNOS-derived NO generation. Addition of L-arginine to iNOS-expressing HEK293 cells pretreated with MG132 and L-lysine resulted in a prompt increase of NO levels, which gradually decreased upon washout and consumption of the amino acid by the cells (Fig. 7C). The geNOps response induced by L-arginine administration was independent from the initial NO level in arginine depleted iNOS expressing HEK293 cells pretreated with MG132 and L-Lysine (Fig. 7D) supporting our conclusion that the activity of iNOS expressed in HEK293 cells is almost independent from the extracellular L-arginine supply. Repetitive addition of increasing amounts of L-arginine to iNOS-expressing and MG132/L-lysine pretreated cells demonstrated the L-arginine concentration dependency of iNOS-mediated NO formation (Fig. 7E). Interestingly, a concentration as low as 3 μM L-arginine appeared to be sufficient to restore a maximal iNOS activity. This is feasible considering that cells have a high capacity to take up and accumulate extracellularly applied L-arginine. Upon washout of 3 μM L-arginine, elevated NO levels rapidly declined. In contrast, NO levels decreased more slowly upon washout of higher L-arginine concentrations (Fig. 7E), indicating that iNOS-expressing HEK293 cells efficiently take up and store extracellular available L-arginine if necessary. Eventually, these experiments demonstrate that the geNOps technology can be successfully employed to study the consequences of e.g. L-arginine deficiency on subcellular NO dynamics.
Overall by utilizing the new geNOps technology our study reveals different properties of the three NO generating isoenzymes on the single cell level. Hence, the geNOps technology seems to be a promising approach to study NO generation kinetics in different cell types.
Acknowledgement
We are grateful to Professor Thomas Michel (Brigham and Women's Hospital, Harvard Medical School) for his critical comments on this manuscript. The authors acknowledge C.J. Edgell, Pathology Department, University of North Carolina at Chapel Hill, NC, USA for providing the EA.hy926 cells. We acknowledge Prof. Nicolas Demaurex for providing us with SypHer. We also thank Karin Osibow for editing the manuscript. The authors acknowledge the scientific advisory board of Next Generation Fluorescence Imaging (NGFI) GmbH (http://www.ngfi.eu/), a spin-off company of the Medical University of Graz. The researchers are supported by the Doctoral College “Metabolic and Cardiovascular Disease” at the Medical University of Graz that was funded by the FWF (W 1226-B18, DKplus Metabolic and Cardiovascular Disease), the Ph.D. program Molecular Medicine (MOLMED) at the Medical University of Graz and by Nikon Austria within the Nikon-Center of Excellence, Graz. This work was funded by the FWF project P 28529-B27. Microscopic equipment is part of the Nikon-Center of Excellence, Graz that is supported by the Austrian infrastructure program 2013/2014, Nikon Austria Inc., and BioTechMed, Graz.
Abbreviations
- geNOps
genetically encoded fluorescent nitric oxide probes
- eNOS
endothelial nitric oxide synthase
- nNOS
neuronal nitric oxide synthase
- iNOS
inducible nitric oxide synthase
- IP3
inositol 1,4,5-trisphosphate
- NO
nitric oxide
- FP
fluorescent protein
- LNNA
NG-nitro-L-arginine
- [Ca2+]cyto
cytosolic Ca2+ concentration
- [NO]cyto
cytosolic NO concentration
- WT
wild type
References
- [1].Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298(Pt 2):249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lamas S, Marsden PA, Li GK, Tempst P, Michel T. Endothelial nitric oxide synthase: molecular cloning and characterization of a distinct constitutive enzyme isoform. PNAS. 1992;89(14):6348–6352. doi: 10.1073/pnas.89.14.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu Y, Feng Q. NOing the heart: role of nitric oxide synthase-3 in heart development. Differentiation; research in biological diversity. 2012;84(1):54–61. doi: 10.1016/j.diff.2012.04.004. [DOI] [PubMed] [Google Scholar]
- [4].Burger DE, Lu X, Lei M, Xiang F-L, Hammoud L, Jiang M, Wang H, Jones DL, Sims SM, Feng Q. Neuronal nitric oxide synthase protects against myocardial infarction-induced ventricular arrhythmia and mortality in mice. Circulation. 2009;120(14):1345–1354. doi: 10.1161/CIRCULATIONAHA.108.846402. [DOI] [PubMed] [Google Scholar]
- [5].Taylor BS, Kim YM, Wang Q, Shapiro RA, Billiar TR, Geller DA. Nitric oxide down-regulates hepatocyte-inducible nitric oxide synthase gene expression. Archives Surg. 1997;132(11):1177–1183. doi: 10.1001/archsurg.1997.01430350027005. Chicago, Ill. 1960. [DOI] [PubMed] [Google Scholar]
- [6].Schmidt HH, Pollock JS, Nakane M, Forstermann U, Murad F. Ca2+/calmodulin-regulated nitric oxide synthases. Cell calcium. 1992;13(6–7):427–434. doi: 10.1016/0143-4160(92)90055-w. [DOI] [PubMed] [Google Scholar]
- [7].Radomski MW, Palmer RM, Moncada S. Glucocorticoids inhibit the expression of an inducible, but not the constitutive, nitric oxide synthase in vascular endothelial cells. PNAS. 1990;87(24):10043–10047. doi: 10.1073/pnas.87.24.10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Asano K, Chee CB, Gaston B, Lilly CM, Gerard C, Drazen JM, Stamler JS. Constitutive and inducible nitric oxide synthase gene expression, regulation, and activity in human lung epithelial cells. PNAS. 1994;91(21):10089–10093. doi: 10.1073/pnas.91.21.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vallance P, Collier J, Moncada S. Effects of endothelium-derived nitric oxide on peripheral arteriolar tone in man. Lancet. 1989;334(8670):997–1000. doi: 10.1016/s0140-6736(89)91013-1. [DOI] [PubMed] [Google Scholar]
- [10].Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- [11].Nussler AK, Billiar TR. Inflammation, immunoregulation, and inducible nitric oxide synthase. J Leukoc Biol. 1993;54(2):171–178. [PubMed] [Google Scholar]
- [12].Vaughn MW, Kuo L, Liao JC. Effective diffusion distance of nitric oxide in the microcirculation. Am J Physiol - Heart Circulatory Physiol. 1998;274(5):H1705–H1714. doi: 10.1152/ajpheart.1998.274.5.H1705. [DOI] [PubMed] [Google Scholar]
- [13].Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J physiology. 1996;271(5 Pt 1):C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- [14].Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. PNAS. 1992;89(1):444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78(6):931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- [16].Friebe A, Koesling D. Regulation of nitric oxide-sensitive guanylyl cyclase. Circulation Res. 2003;93(2):96–105. doi: 10.1161/01.RES.0000082524.34487.31. [DOI] [PubMed] [Google Scholar]
- [17].Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am J Physiol - Lung Cell Mol Physiol. 2004;287(2):L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- [18].Kroncke KD, Fehsel K, Kolb-Bachofen V. Nitric oxide: cytotoxicity versus cytoprotectionehow, why, when, and where? Nitric oxide Biol Chem. 1997;1(2):107–120. doi: 10.1006/niox.1997.0118. [DOI] [PubMed] [Google Scholar]
- [19].Charles N, Ozawa T, Squatrito M, Bleau A-M, Brennan CW, Hambardzumyan D, Holland EC. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell stem Cell. 2010;6(2):141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG. Moncada, Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72(1):41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Michel T, Feron O. Nitric oxide synthases: which, where, how, and why? J Clin Investigation. 1997;100(9):2146–2152. doi: 10.1172/JCI119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ignarro LJ, Fukuto JM, Griscavage JM, Rogers NE, Byrns RE. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from L-arginine. PNAS. 1993;90(17):8103–8107. doi: 10.1073/pnas.90.17.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lancaster JR. Simulation of the diffusion and reaction of endogenously produced nitric oxide. PNAS. 1994;91(17):8137–8141. doi: 10.1073/pnas.91.17.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bryan NS, Grisham MB. Methods to detect nitric oxide and its metabolites in biological samples. Free Radic Biol Med. 2007;43(5):645–657. doi: 10.1016/j.freeradbiomed.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Eroglu E, Gottschalk B, Charoensin S, Blass S, Bischof H, Rost R, Madreiter-Sokolowski CT, Pelzmann B, Bernhart E, Sattler W, Hallström S, et al. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat Commun. 2016;7:10623. doi: 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eroglu E, Rost R, Bischof H, Blass S, Schreilechner A, Gottschalk B, Depaoli MR, Klec C, Charoensin S, Madreiter-Sokolowski CT, Ramadani J, et al. Application of genetically encoded fluorescent nitric oxide (NO*) probes, the geNOps, for real-time imaging of NO* signals in single cells. J Vis Exp JoVE. 2017;121 doi: 10.3791/55486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Opelt M, Eroglu E, Waldeck-Weiermair M, Russwurm M, Koesling D, Malli R, Graier WF, Fassett JT, Schrammel A, Mayer B. Formation of nitric oxide by aldehyde dehydrogenase-2 is necessary and sufficient for vascular bioactivation of nitroglycerin. J Biol Chem. 2016 Sep; doi: 10.1074/jbc.M116.752071. jbc.M116.752071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schmidt K, Andrew P, Schrammel A, Groschner K, Schmitz V, Kojda G, Mayer B. Comparison of neuronal and endothelial isoforms of nitric oxide synthase in stably transfected HEK 293 cells, American journal of physiology. Heart circulatory physiology. 2001;281(5):H2053–H2061. doi: 10.1152/ajpheart.2001.281.5.H2053. [DOI] [PubMed] [Google Scholar]
- [29].Graier WF, Schmidt K, Kukovetz WR. Bradykinin-induced Ca(2+)-influx into cultured aortic endothelial cells is not regulated by inositol 1,4,5-trisphosphate or inositol 1,3,4,5-tetrakisphosphate. Second messengers phosphoproteins. 1991;13(4):187–197. [PubMed] [Google Scholar]
- [30].Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- [31].Richter C. Reactive Oxygen, Nitrogen Species, Regulate mitochondrial Ca2+ homeostasis and respiration. Biosci Rep. 1997;17(1):53–66. doi: 10.1023/a:1027387301845. [DOI] [PubMed] [Google Scholar]
- [32].Spitaler MM, Hammer A, Malli R, Graier WF. Functional analysis of histamine receptor subtypes involved in endothelium-mediated relaxation of the human uterine artery. Clin Exp Pharmacol physiology. 2002;29(8):711–716. doi: 10.1046/j.1440-1681.2002.03704.x. [DOI] [PubMed] [Google Scholar]
- [33].Brorson JR, Sulit RA, Zhang H. Nitric oxide disrupts Ca2+ homeostasis in hippocampal neurons. J Neurochem. 1997;68(1):95–105. doi: 10.1046/j.1471-4159.1997.68010095.x. [DOI] [PubMed] [Google Scholar]
- [34].Doutheil J, Althausen S, Treiman M, Paschen W. Effect of nitric oxide on endoplasmic reticulum calcium homeostasis, protein synthesis and energy metabolism. Cell calcium. 2000;27(2):107–115. doi: 10.1054/ceca.1999.0099. [DOI] [PubMed] [Google Scholar]
- [35].Charoensin S, Eroglu E, Opelt M, Bischof H, Madreiter-Sokolowski CT, Kirsch A, Depaoli MR, Frank S, Schrammel A, Mayer B, Waldeck-Weiermair M, et al. Intact mitochondrial Ca2+ uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS) Free Radic Biol Med. 2017;102:248–259. doi: 10.1016/j.freeradbiomed.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Riveros-Moreno V, Heffernan B, Torres B, Chubb A, Charles I, Moncada S. Purification to homogeneity and characterisation of rat brain recombinant nitric oxide synthase. Eur J Biochem. 1995;230(1):52–57. doi: 10.1111/j.1432-1033.1995.tb20533.x. [DOI] [PubMed] [Google Scholar]
- [37].Bredt DS, Snyder SH. Isolation of nitric oxide synthetase, a calmodulinrequiring enzyme. Proc Natl Acad Sci U S A. 1990;87(2):682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mayer B, John M, Böhme E. Purification of a Ca2+/calmodulin-dependent nitric oxide synthase from porcine cerebellum. Cofactor-role of tetrahydrobiopterin. FEBS Lett. 1990;277(1–2):215–219. doi: 10.1016/0014-5793(90)80848-d. [DOI] [PubMed] [Google Scholar]
- [39].Harteneck C, Klatt P, Schmidt K, Mayer B. Expression of rat brain nitric oxide synthase in baculovirus-infected insect cells and characterization of the purified enzyme. Biochem J. 1994;304(Pt 3):683–686. doi: 10.1042/bj3040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gerber NC, Ortiz de Montellano PR. Neuronal nitric oxide synthase. Expression in Escherichia coli, irreversible inhibition by phenyldiazene, and active site topology. J Biol Chem. 1995;270(30):17791–17796. doi: 10.1074/jbc.270.30.17791. [DOI] [PubMed] [Google Scholar]
- [41].Presta A, Liu J, Sessa WC, Stuehr DJ. Substrate binding and calmodulin binding to endothelial nitric oxide synthase coregulate its enzymatic activity. Nitric oxide Biol Chem. 1997;1(1):74–87. doi: 10.1006/niox.1996.0110. [DOI] [PubMed] [Google Scholar]
- [42].Martasek P, Liu Q, Liu J, Roman LJ, Gross SS, Sessa WC, Masters BS. Characterization of bovine endothelial nitric oxide synthase expressed in E. coli. Biochem biophysical Res Commun. 1996;219(2):359–365. doi: 10.1006/bbrc.1996.0238. [DOI] [PubMed] [Google Scholar]
- [43].Rodriguez-Crespo I, Gerber NC, Ortiz de Montellano PR. Endothelial nitric-oxide synthase. Expression in Escherichia coli, spectroscopic characterization, and role of tetrahydrobiopterin in dimer formation. J Biol Chem. 1996;271(19):11462–11467. doi: 10.1074/jbc.271.19.11462. [DOI] [PubMed] [Google Scholar]
- [44].Leber A, Hemmens B, Klosch B, Goessler W, Raber G, Mayer B, Schmidt K. Characterization of recombinant human endothelial nitric-oxide synthase purified from the yeast Pichia pastoris. J Biol Chem. 1999;274(53):37658–37664. doi: 10.1074/jbc.274.53.37658. [DOI] [PubMed] [Google Scholar]
- [45].Salerno JC. Neuronal nitric oxide synthase: prototype for pulsed enzymology. FEBS Lett. 2008;582(10):1395–1399. doi: 10.1016/j.febslet.2008.03.051. [DOI] [PubMed] [Google Scholar]
- [46].Busconi L, Michel T. Endothelial nitric oxide synthase. N-terminal myristoylation determines subcellular localization. J Biol Chem. 1993;268(12):8410–8413. [PubMed] [Google Scholar]
- [47].Oess S, Icking A, Fulton D, Govers R, Müller-Esterl W. Subcellular targeting and trafficking of nitric oxide synthases. Biochem J. 2006;396(Pt 3):401–409. doi: 10.1042/BJ20060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Erwin PA, Mitchell DA, Sartoretto J, Marletta MA, Michel T. Subcellular Targeting, Differential S-Nitrosylation, Of endothelial nitric-oxide synthase. J Biol Chem. 2006;281(1):151–157. doi: 10.1074/jbc.M510421200. [DOI] [PubMed] [Google Scholar]
- [49].Zhou L, Zhu D-Y. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric oxide Biol Chem. 2009;20(4):223–230. doi: 10.1016/j.niox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- [50].Michel T. Targeting and translocation of endothelial nitric oxide synthase. Braz J Med Biol Res. 1999;32(11):1361–1366. doi: 10.1590/s0100-879x1999001100006. [DOI] [PubMed] [Google Scholar]
- [51].Du XL, Edelstein D, Dimmeler S, Ju Q, Sui C, Brownlee M. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J Clin Invest. 2001;108(9):1341–1348. doi: 10.1172/JCI11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fulton D, Gratton J-P, Sessa WC. Post-translational control of endothelial nitric oxide synthase: why Isn't calcium/calmodulin enough? J Pharmacol Exp Ther. 2001;299(3):818–824. [PubMed] [Google Scholar]
- [53].Robinson LJ, Busconi L, Michel T. Agonist-modulated palmitoylation of endothelial nitric oxide synthase. J Biol Chem. 1995;270(3):995–998. doi: 10.1074/jbc.270.3.995. [DOI] [PubMed] [Google Scholar]
- [54].Kröncke K-D, Fehsel K, Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clin Exp Immunol. 1998;113(2):147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hall CN, Garthwaite J. What is the real physiological NO concentration in vivo? Nitric Oxide. 2009;21(2):92–103. doi: 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Furfine ES, Harmon MF, Paith JE, Garvey EP. Selective inhibition of constitutive nitric oxide synthase by L-NG-nitroarginine. Biochemistry. 2002;32(33):8512–8517. doi: 10.1021/bi00084a017. [DOI] [PubMed] [Google Scholar]
- [57].Moore PK, Babbedge RC, Wallace P, Gaffen ZA, Hart SL. 7-Nitro indazole, an inhibitor of nitric oxide synthase, exhibits anti-nociceptive activity in the mouse without increasing blood pressure. Br J Pharmacol. 1993;108(2):296–297. doi: 10.1111/j.1476-5381.1993.tb12798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bateman RM, Sharpe MD, Ellis CG. Bench-to-bedside review: microvascular dysfunction in sepsis–hemodynamics, oxygen transport, and nitric oxide. Crit care (London, Engl.) 2003;7(5):359–373. doi: 10.1186/cc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kirkeboen KA, Strand OA. The role of nitric oxide in sepsis–an overview. Acta Anaesthesiol Scand. 1999;43(3):275–288. doi: 10.1034/j.1399-6576.1999.430307.x. [DOI] [PubMed] [Google Scholar]
- [60].Alvares TS, Conte-Junior CA, Silva JT, Paschoalin VMF. Acute L-Arginine supplementation does not increase nitric oxide production in healthy subjects. Nutr Metabolism. 2012;9(1):54. doi: 10.1186/1743-7075-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Closs EI, Scheld JS, Sharafi M, Forstermann U. Substrate supply for nitric-oxide synthase in macrophages and endothelial cells: role of cationic amino acid transporters. Mol Pharmacol. 2000;57(1):68–74. [PubMed] [Google Scholar]
- [62].Escobales N, Rivera-Correa M, Altieri PI, Rodriguez JF. Relationship between NO synthesis, arginine transport, and intracellular arginine levels in vascular smooth muscle cells. Amino Acids. 2000 Sep;19(2):451–468. doi: 10.1007/s007260070023. [DOI] [PubMed] [Google Scholar]
- [63].Simon A, Plies L, Habermeier A, Martiné U, Reining M, Closs EI. Role of neutral amino acid transport and protein breakdown for substrate supply of nitric oxide synthase in human endothelial cells. Circulation Res. 2003;93(9):813–820. doi: 10.1161/01.RES.0000097761.19223.0D. [DOI] [PubMed] [Google Scholar]
- [64].Karbach S, Simon A, Slenzka A, Jaenecke I, Habermeier A, Martiné U, Förstermann U, Closs EI. Relative contribution of different l-arginine sources to the substrate supply of endothelial nitric oxide synthase. J Mol Cell Cardiol. 2011;51(5):855–861. doi: 10.1016/j.yjmcc.2011.07.024. [DOI] [PubMed] [Google Scholar]
- [65].Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem. 2011;286(13):11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]