Abstract
A significant portion of the human genome encodes genes that transcribe long non-protein coding RNAs (lncRNAs). A large number of lncRNAs localize in the nucleus, either enriched on the chromatin or localized to specific sub-nuclear compartments. Nuclear lncRNAs participate in several biological processes, including chromatin organization, transcriptional and post-transcriptional gene expression, and also act as structural scaffolds of nuclear domains. Here, we highlight recent studies demonstrating the role of lncRNAs in regulating gene expression and nuclear organization in mammalian cells. In addition, we update current knowledge about the involvement of the most abundant and conserved lncRNA, MALAT1, in gene expression control.
Keywords: nuclear domain, gene expression, lncRNA, nuclear structure, RNA-binding protein, MALAT1
Overview of nuclear-enriched lncRNAs
It is estimated that ~75% of the human genome is utilized for generating transcripts with no apparent protein-coding potential, and these transcripts are classified as non-coding RNAs (ncRNAs) [1]. Long non-coding RNAs (lncRNAs) are grouped into transcripts that are > 200 nucleotides in length. The human genome is estimated to contain ~16,000 lncRNA genes (Gencode 27). A significant fraction of nuclear localized lncRNAs are transcribed by RNA polymerase II (RNA pol II), and share similar chromatin modifications on their regulatory elements with other RNA pol II-transcribed protein-coding transcripts. Most of the lncRNAs contain normal 5’-caps and 3’ poly-A tails. However, recent studies identified lncRNAs that undergo unusual processing within their 5’ and 3’ends. We would refer to the following reviews that describe this aspect in detail (please also see table 1) [2, 3]. In the present review, we will discuss recent studies on RNA pol II transcribed nuclear lncRNAs, but would refer the reader to the following literature to learn more about non-RNA pol II transcribed nuclear lncRNAs (Please see Box 1) [4, 5].
Table 1.
| Type | Feature | Recommended reviews/articles |
|---|---|---|
| mRNA-like lncRNAs | 5’-capping and 3’ poly-A tails can be spliced | [149, 150] |
| enhancer-derived ncRNAs | Transcribed from enhancer regions, normally non-polyadenylated, that could have sort half-life | [60] |
| Antisense transcripts | Transcribed from divergent promoters, but pol II does not extend on them as efficient as on the corresponding sense genes | [151–153] |
| lncRNAs with noncanonical 3' end | No 3’ poly-A tails have alternative 3’ processing pathways | [154, 155] |
Box1. Introduction of non-Pol II transcribed lncRNAs.
RNA Polymerase III is also known to generate several regulatory lncRNAs. Some of the examples include the repeat containing RNAs such as Alu and B2 lncRNAs and the nuclear speckle-enriched 7SK RNA. Human Alu is a repeat-containing transcript at around 300bp, which is generated from short interspersed elements (SINEs). Alu RNA is known as a transcription repressor, because it competes with RNA pol II thereby negatively regulates the binding of RNA pol II at gene promoters [172]. The corresponding SINE-encoded transcript in mice is B2, which is reported to display similar function to human Alu transcripts [173]. The other pol III transcribed RNA 7SK (~330 nucleotide) is also shown to negatively regulate RNA pol II-mediated transcription by affecting the p-TEFb (positive transcription elongation factor b) activity [174]. We would refer the following literature for a detailed description about RNA pol III-transcribed regulatory lncRNAs [4, 175].
pRNA (Promoter-associated RNA) is a family of RNAs, transcribed by RNA pol I from the rDNA promoter region located upstream of pre-rRNA transcription start site [176]. PRNA interacts with members of the Nucleolar chromatin-remodeling complex (NoRC), and utilizes an RNA-dependent mechanism to activate heterochromatin formation and rDNA silencing [176, 177].
Plants such as Arabidopsis utilize RNA pol V to produce noncoding transcripts from intergenic regions. These transcripts play important roles in regulating heterochromatin formation, defining the boundaries of heterochromatin and gene silencing [178, 179].
Most lncRNAs are expressed at low levels compared to protein-coding mRNAs, and show tissue or cell type specific expression. With the rapid growth of lncRNA research, functions of many lncRNAs have been reported recently. A significant fraction of lncRNA is preferentially localized in the nucleus, and several of the well-studied nuclear-retained lncRNAs are shown to participate in vital molecular processes, including chromatin organization, transcription and RNA processing (please see Table 2) [6–10]. In addition, lncRNAs are shown to play essential roles in the organization of nuclear domains [7–10].
Table 2.
| Category | Nuclear-retained lncRNA names and references |
|---|---|
| Transcription activation | PINCR[49], MANTIS[25], lncKdm2b[12], Linc-RAM[50], LncPRESS1[28], upperhand (UPH)[63], HOTAIRM1[13], RMRP[39], Linc-RoR[29], HoxBlinc[14], DEANR1[156], MEG3[23], lncTCF7[26], myheart(mhrt)[30], BCAR4[32], HERVH[37], THRIL[40], Dali[20], LincRNA-p21[157], alncRNA-EC7[158], PACER[35], Paupar[47], lincRNA-Cox2(also repression)[53], HOXD-AS1[54], RBM5-AS1/LUST[38], Evx1as[159], Khps1[19], TARID[160], TERRA[34], SLERT [56] |
| Transcription repression | HAUNT/Gm15055[22, 44], lincRNA-EPS[51], PAPAS[17, 24], PARTICLE[15], TUG1[161], Oct4P4[18], Evf2/Dlx6as[162], lncRNA-CD244[163], PANDA[52], APTR[145], ANRASSF1[164], H19[21], SChLAP1[31], lincRNA-Cox2(also activation)[53], GNG12-AS1[165], APOA1-AS[45], AS1DHRS4[16] |
| Post-transcriptional regulation | HOTAIR (ubiquitinylation)[166], Linc-RoR (c-myc stability [167]and p53 translation[168]), asFGFR2 (splicing)[27], Pnky (splicing)[64], MALAT1 (pre-mRNA splicing) UCA1 (p16INK stability)[169], lincRNA-p21 (translation)[170] |
| Other regulations: | HAUNT/Gm15055 (DNA locus)[44], Xist (XCI)[9], Neat1 (paraspeckle and pri-miR processing)[9, 80], Firre (Nuclear Organization)[48, 144], DDSR (homologous recombination)[171], TERRA (telomere stability)[34], AluRNA (nucleolar structure) [58] |
Role of Nuclear-retained lncRNAs in chromatin organization
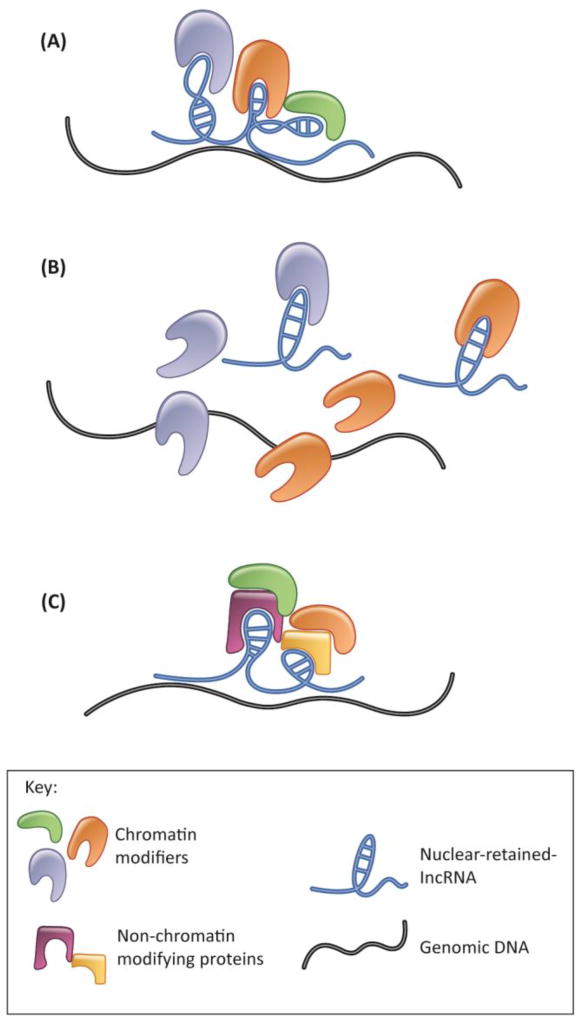
A significant number of nuclear lncRNAs associate with chromatin and thus could be broadly classified as cheRNAs (chromatin-enriched RNAs) [11]. Some nuclear lncRNAs can influence chromatin architecture by interacting with chromatin modulating proteins such as SWI/SNF (see glossary) or PRC subunits (Figure 1A) and promoting their recruitment/association to chromatin, thereby controlling transcriptional activity [12–27]. For example, in hepatocarcinoma cells, lncTCF7 facilitates the transcription of TCF7 by recruiting the SWI/SNF complex to the TCF7 promoter [26]. In differentiating mouse embryonic stem cells, a pseudogene-derived lncRNA called Oct4P4 is transcribed from the X-linked Oct4 pseudogene, which induces transcriptional silencing of the ancestral Oct4 gene by facilitating the deposition of H3K9me3 and HP1a to the Oct4 promoter [18].
Figure 1.
Role of nuclear-retained lncRNAs in chromatin organization. (A) Nuclear-retained lncRNAs modulate chromatin by recruiting chromatin-modulating proteins to chromatin. (B) Nuclear-retained lncRNAs can decoy chromatin-associated proteins away from chromatin. (C) Nuclear-retained lncRNAs can form indirect interaction with chromatin modulators through other kinds of proteins and finally modulate chromatin.
Nuclear lncRNAs can also influence gene expression by preventing the association of specific chromatin factors to specific gene loci. In such a scenario, a lncRNA acts as a ‘decoy’ and prevents proteins such as histone deacetylase [28], methyl transferase [29], or chromatin remodeling complexes [30, 31] from interacting with a certain genomic locus (Figure 1B). In adult mice, Myheart (Mhrt) antisense (AS)-lncRNAs, which are transcribed from the myosin heavy chain 7 gene locus, antagonize the function of the chromatin remodeler Brg1 and thus protect the heart from pathological hypertrophy [30]. Mhrt employs a ‘competitive inhibition mechanism’ by preventing the interaction of Brg1 with chromatin, thereby repressing Brg1 activity.
Several lncRNAs influence chromatin organization and/or activity without directly interacting with chromatin or chromatin modulators; they do so through their association with other proteins (Figure 1C). For example, BCAR4 lncRNA activates the Hedgehog/GLI2 transcriptional program in breast cancer cells by influencing p300 histone acetyl transferase activity [32]. BCAR4, by associating with the RNA-binding protein (RBP) SNIP1, releases SNIP’s inhibitory effect on p300 to enhance the acetylation of H3K18 on GL12 target gene promoters. BCAR4 further recruits another RBP, PNUTS, to acetylated H3K18, ultimately resulting in the activation of GL12 target genes. In this scenario, BCAR4, by interacting with RBPs, influences signal-induced epigenetic regulation of transcription of GL12 target genes.
With the development of new techniques such as CHIRP, CHART, and RAP, all of which determine genome-wide chromatin association of lncRNA [33], one could anticipate that more insights about the chromatin association properties and/or chromatin-modulating functions of lncRNAs will be identified. For instance, a recent study, by performing genome-wide RNA mapping analyses identified that the telomere-associated lncRNA TERRA binds to several other sites on chromatin and modulates the transcription of genes via antagonizing the activity of the RNA helicase ATRX [34].
LncRNAs as Transcriptional regulators
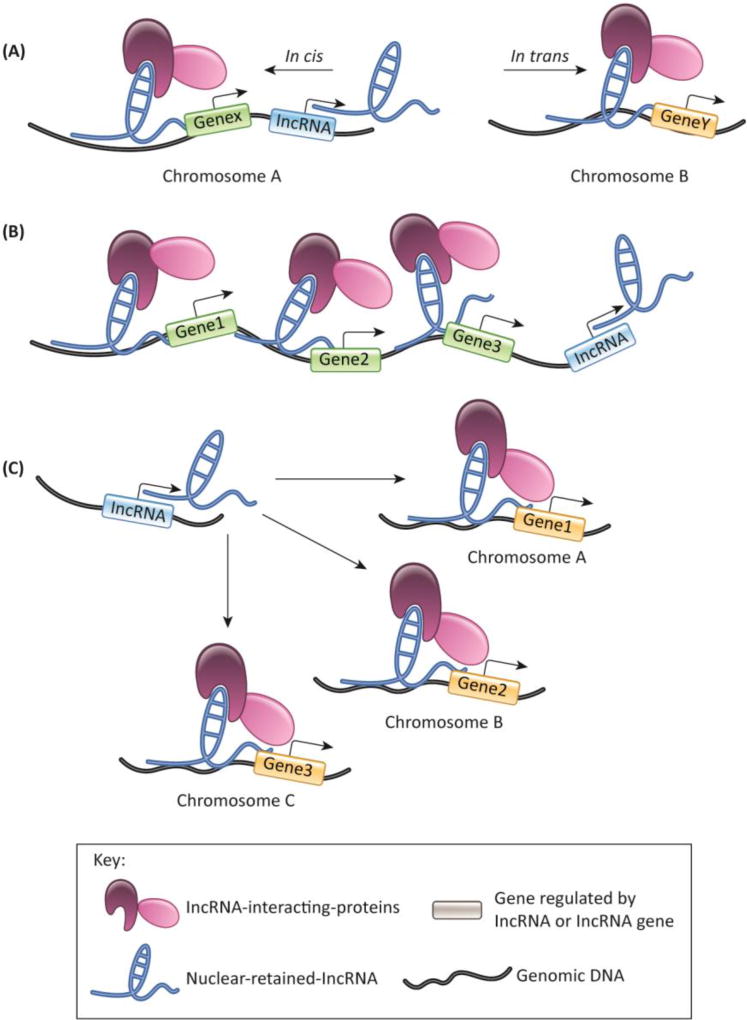
LncRNAs activate or repress transcription (summarized in Table 1) by acting locally (near the sites of their transcription [cis-regulation]) or distally (at sites that are located on other chromosomes [trans-regulation]) (Figure 2A). For instance, lncKdm2b sustains the maintenance of intestinal group 3 innate lymphoid cells (ILCs) by facilitating the transcriptional activation of a transcription factor (TF), zfp292 [12]. LncKdm2b facilitates the recruitment of chromatin organizer protein Satb1 and the nuclear remodeling factor (NURF) complex to the regulatory elements of zfp292. In another example, low-irradiation-induced lncRNA PARTICLE (promoter of MAT2A-antisense radiation-induced circulating lncRNA) represses MAT2A expression by forming a DNA-RNA triplex at the MAT2A locus, and by recruiting transcription-repressive complex proteins G9a and SUZ12 (a subunit of PRC2) to the MAT2A promoter for methylation [15]. LncRNA PACER, which is transcribed from the antisense strand of COX-2 gene, promotes the transcription of COX-2 by sequestering the repressive p50 subunit of NF-KB away from the COX-2 promoter [35]. It is not clear why p50 shows increased affinity for PACER over the COX-2 promoter. In vitro studies may provide more insights into the differential affinity of p50 for PACER over the COX-2 promoter.
Figure 2.
Nuclear-retained lncRNAs act as transcriptional regulators. (A) Nuclear-retained lncRNAs modulate transcription in cis (left) or in trans (right). (B) Nuclear-retained lncRNAs regulate the expression of multiple genes (gene 1,2, and 3 in the model) that are in close proximity. (C) Nuclear-retained lncRNAs regulate transcription of several distally located genes (gene 1, 2, and 3 on different chromosomes in the model).
LncRNAs are also found to regulate transcription by influencing TF activity. A recent study identified a role for a nuclear lncRNA in regulating the basal levels of tumor suppressor protein p53 [36]. In colorectal cancer cells, PURPL lncRNA, by associating with p53 stabilizing protein MYBBP1A, prevents MYBBP1A-p53 complex assembly, and thereby destabilizes the cellular pool of p53. In this case, PURPL modulates transcription indirectly by regulating the levels of a major TF, p53. Recent studies have identified several more examples where lncRNAs are shown to regulate transcription by influencing the localization and/or activity of TFs [37, 38]. It is tempting to speculate that several hundreds of the not-yet-characterized nuclear lncRNAs could contribute to gene regulation by modulating the association of TFs or co-factors to chromatin in a cell- or tissue-specific manner.
RBPs (RNA-binding proteins) constitute a class of proteins that interact with single-stranded or double-stranded RNAs and influence post-transcriptional regulation of gene expression. Interestingly, several RBPs also act as transcriptional regulators, and lncRNAs are found to modulate their activity. In T helper 17 (TH17) lymphocytes, the lncRNA RMRP interacts with the dead-box RNA helicase DDX5 and promotes the interaction between DDX5 and the RORγt transcription factor (RAR-related orphan nuclear receptor) [39]. DDX5 functions as a co-activator to facilitate RORγt-mediated transcription of TH17 genes. RMRP-depleted cells displayed reduced interaction between DDX5 and RORγt and defective transcription of selective TH17 genes, supporting the argument that RMRP acts as a scaffold to facilitate the interaction between an RBP and a TF. In another example, lncRNA THRIL-hnRNP L RNP complex facilitates the transcription of several immune response genes, including TNFα in macrophages [40]. Similarly, lincRNA-p21-hnRNP K complex promotes transcription repression of genes in the p53 pathway and pluripotency [41, 42].
In several instances, lncRNAs modulate the expression of multiple genes. Such lncRNAs could be broadly classified into two groups, primarily based on the genomic location of the genes regulated by the lncRNAs. In the first group, the lncRNA or its loci regulate the expression of multiple genes, which are located in close proximity (Figure 2B). These lncRNAs interact with chromatin, transcription factors, or RBPs, thereby increasing their local concentration near the transcription sites of lncRNAs [43]. Besides the lncRNA, these proteins also interact with genes or RNAs that are present in genomic proximity to potentially regulate their expression. In other instances, lncRNAs actively participate in the recruitment of these proteins to the nearby genomic loci [13, 14, 16, 22, 37, 44, 45]. Finally, transcription from the lncRNA gene locus but not the lncRNA itself could play an important role in regulating the expression of nearby genes. Genetic manipulation of a large number of gene loci, including those of lncRNA and protein-coding genes, revealed that transcription from a particular genomic locus, not necessarily involving the RNA that is synthesized from that locus, could play a crucial role in regulating the expression of nearby genes [46].
In the second group, lncRNAs can control the expression of both locally and distally located genes (Figure 2C). Paupar lncRNA, transcribed from a locus upstream of the PAX6 (paired box 6) gene, regulates the expression of PAX6 as well as several other genes located on distant chromosomes in a transcript-dependent manner [47]. Paupar was found to associate with ~3000 chromatin sites, and a significant fraction of those sites overlapped with functional elements such as promoters. Depletion of Paupar compromised the expression of ~1000 genes, many of which contained Paupar-binding sites within their regulatory elements. Firre is another nuclear-enriched lncRNA that is found to act locally as well as distally [48]. Firre occupies a chromatin area near its transcription site on the X-chromosome and also interacts with additional domains located on four other chromosomes. Firre is thought to act as a scaffold to coordinate the association of these chromosomal loci, as genetic deletion of Firre leads to loss of interaction between these chromosomal loci.
LncRNAs are also known to regulate the expression of genes belonging to specific pathways/biological processes [21, 23, 25, 28, 31, 32, 38, 39, 49–54]. For example, p53-induced lncRNA PINCR promotes the upregulation of a subset of p53 target genes involved in G1 arrest and apoptosis [49]. PINCR, along with Matrin 3 and p53, associates with the enhancer region of the candidate genes and modulates the induction of pro-survival genes upon DNA damage. Another lncRNA, MANTIS, facilitates endothelial angiogenic function by promoting the expression of a subset of endothelial genes [25]. Mechanistically, MANTIS interacts with BRG1 (a subunit of the SWI/SNF chromatin remodeling complex) to collaboratively facilitate the loading of RNA polymerase II onto the endothelial gene promoters. Furthermore, MANTIS is found to improve the ATPase activity of BRG1 by stabilizing its interaction with BAF155. Finally, Firre occupies chromatin regions containing genes involved in energy metabolism and/or adipogenesis, with loss-of-function studies revealing that Firre controls adipogenesis [48]. These studies highlight another important function of lncRNA, which is to bring together unique combinations of chromatin and/or proteins in order to establish gene networks that regulate a particular cellular pathway.
RNA pol II transcribed nuclear lncRNAs have also been shown to regulate the activity of other RNA polymerases within the cell. For instance, lncRNAs such as PAPAS and SLERT are known to regulate the activity of RNA polymerase I (RNA pol I). PAPAS lncRNAs (promoter and pre-rRNA antisense) consist of a group of RNA pol II transcripts that are transcribed in the antisense orientation from a fraction of rDNA repeats [55]. PAPAS recruits suv4-20h2 histone methyl transferase to rDNA gene promoters to mediate rDNA repression by triggering H4K20me3 and chromosome compaction [17, 24]. On the other hand, SLERT (snoRNA-ended lncRNA enhances pre-ribosomal RNA transcription) is synthesized from the intronic region of TBRG4 locus by undergoing unusual processing [56]. SLERT is a member of the recently identified family of sno-lncRNAs [57]. SLERT lacks a canonical 5’ cap or 3’ poly(A) tail, but contains a H/ACA type of sno-RNAs on either end, and is guided to the nucleolus by the snoRNA sequences. In the nucleolus, SLERT interacts with DDX21, and releases the inhibitory interaction between DDX21 and pol I machinery, thereby positively regulating rDNA gene transcription [56]. In addition, a recent study reported that AluRNAs that are processed from intronic Alu elements of RNA pol II-transcribed genes modulate rDNA transcription by influencing nucleolar structure [58].
Some of the enhancer sequences within the genome are transcribed to produce nuclear RNAs such as enhancer RNAs (eRNAs), elncRNAs, and ncRNAa [59] [60]. In general, eRNAs are found to regulate the expression of nearby protein-coding genes in an RNA-dependent manner. Some eRNAs facilitate mediator- and/or cohesion-mediated enhancer-promoter interactions through chromatin looping or activate promoter-mediated transcription. For example, upon estrogen treatment, eRNAs transcribed from ERα-bound enhancers facilitate enhancer-promoter looping of several ERα target genes. Besides such cis regulation, eRNAs also participate in long-range intra-chromosomal interactions to regulate gene expression [61, 62]. Intra-chromosomal interactions between TFF1 and NRIP eRNA gene loci were observed due to chromosomal looping in cells treated with E2 (17β-estradiol), and such interactions appear to be regulated by NRIP eRNA. In the case of eRNA-mediated gene regulation, one needs to be cautious in deciphering whether eRNA transcripts or the transcription from the eRNA-bearing gene loci play vital roles in regulating gene expression. In that sense, some of the eRNAs could turn out to be the by-products of pervasive transcription events simply due to transcriptional activity around enhancer regions. For example, transcription of Upperhand (Uph) lncRNA from a region upstream of the heart-specific TF HAND2 gene is required to maintain the super enhancer signature and the RNA pol II elongation through the Hand2 enhancer locus [63]. However, only blocking of Uph transcription (not the depletion of the mature Uph transcript) inhibited Hand2 transcription, implying that in the case of Uph-Hand2 interaction, only the noncoding transcription is important in establishing a permissive chromatin environment to facilitate Hand2 transcription.
LncRNAs as Post-transcriptional regulators
It is becoming increasingly evident that nuclear-restricted lncRNAs also regulate gene expression by influencing post-transcriptional events. The antisense-FGFR2 lncRNA promotes epithelial-specific alternative splicing of FGFR2 pre-mRNA [27]. AS-FGFR2 facilitates the recruitment of polycomb-group proteins and histone demethylase KDM2a to the FGFR2 regulatory elements, thereby preventing the association and activity of a repressive-splicing adaptor complex that promotes mesenchymal-specific splicing of FGFR2 pre-mRNA. This study uncovers the role of an lncRNA in regulating cell type-specific alternative splicing by modulating the underlying chromatin structure. The neural-specific lncRNA Pnky (Pinky) inhibits neurogenesis by modulating PTBP1-mediated alternative splicing of a set of genes that control neurogenesis [64]. Finally, several studies have identified the involvement of oncogenic nuclear lncRNA MALAT1 in alternative splicing regulation (please see later section on MALAT1 for details). The prostate cancer-specific lncRNA PCA3, synthesized as an ASlncRNA from the PRUNE2 locus, negatively regulates PRUNE2 mRNA levels in an adenosine-to-inosine (A-to-I) editing-dependent manner [65]. Recent studies have documented the role of A-to-I editing in influencing the stability of RNA [66]. However, it needs to be determined how exactly A-to-I editing regulates the stability of PRUNE2 mRNA. Finally, recently identified SPA lncRNAs, which are 5’ snoRNP-ended and 3’ polyadenylated transcripts, are suggested to modulate RNA processing by sequestering several RBPs, including TDP43, RBFOX2 and hnRNP M [67].
Role of Nuclear-retained lncRNAs in the organization of nuclear structure
Xist (X-inactive specific transcript), one of the first functionally annotated nuclear lncRNAs, regulates dosage compensation by promoting X-chromosome inactivation (XCI). Xist is idealized in the scientific community as a perfect example of a nuclear lncRNA, as it coordinates several nuclear processes to achieve XCI. For example, discrete regions within Xist RNA are required for gene silencing, for the association of polycomb repressive complex 2 (PRC2) to the inactive X-chromosome (Xi), and for the localization of Xist to the X-chromosome [68]. Xist interacts with several protein complexes such as SHARP and PRC2, and recruits them to Xi in order to modulate epigenetic changes that favor X-chromosome transcriptional gene silencing [69–71].
Recent studies have also provided compelling data supporting the role of Xist in modulating the 3D architecture of Xi [72]. During the initial stages of XCI, the entire Xi is repositioned near the nuclear lamina (the nuclear compartment that is associated with transcriptionally inactive heterochromatic regions). Interestingly, Xist directly interacts with Lamin B receptor (LBR), an inner nuclear membrane-associated trans-membrane protein that also interacts with the nuclear lamina. Depletion of LBR or disruption of Xist-LBR interaction results in defective recruitment of Xi to the nuclear periphery. In addition, in LBR-depleted cells or cells with disrupted LBR-Xist interaction, Xist failed to associate with active genes across Xi, thus abolishing Xist-mediated gene silencing. These results imply that Xist plays an active role in reshaping the chromatin structure by recruiting Xi to the nuclear lamina, and such tethering of Xi is required for the efficient association of Xist to transcriptionally active genes to exert gene silencing.
Firre, transcribed from the X-chromosome, forms a punctate nuclear compartment containing several chromosomal loci, and facilitates inter-chromosomal interactions and activation of several genes that are transcribed from these gene loci. Firre interacts with nuclear matrix factor hnRNP U and seems to act as a ‘scaffold’ to modulate inter-chromosomal interactions [48].
Finally, lncRNAs are also known to nucleate and/or maintain specific nuclear domains (NDs). NDs are non-membranous structures that are enriched with a unique set of proteins and RNA. Eukaryotic cells contain several NDs, including the nucleolus, nuclear speckles, and paraspeckles. Paraspeckles have been implicated in 1) the nuclear retention of hyper-edited RNAs, 2) the sequestration of certain RBPs in the nucleus, and 3) the immune response [73–75]. Neat1 lncRNA plays a vital role in the nucleation/maintenance of paraspeckles [76–78]. Paraspeckles are initially formed at the site of Neat1 transcription and Neat1 transcription is found to be essential for the formation of paraspeckles. In addition, recent studies also identified potential involvement of Neat1 in oncogene-induced cellular transformation and pri-miRNA processing [79, 80].
Role of MALAT1 in gene regulation
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched abundant transcript 2 (NEAT2), is perhaps the most abundant (~3000 copies/cell) nuclear-retained lncRNA. MALAT1 was initially identified as a prognostic marker for stage I lung adenocarcinoma [81]. MALAT1 is highly conserved among mammalian species [81–84] and its orthologs have been identified in Zebrafish, lizard, and Xenopus [85, 86].
MALAT1 is ubiquitously expressed in all tissues, but its levels are tightly regulated during certain physiological processes, such as the cell cycle [87] and cellular differentiation [88, 89], by various transcriptional and post-transcriptional mechanisms. Transcription factors such as ER/eNOS [90], Sp1 [91], HIF-2a [92–95], and CREB [96] recognize the MALAT1 promoter and induce MALAT1 expression under different stress conditions. At the post-transcriptional level, the long half-life of MALAT1 in many cell lines is attributed to its unique 3’ end RNA processing [83, 97, 98]. The 3’ end of the MALAT1 primary transcript forms a tRNA-like structure and is cleaved by RNase P, yielding a 61 nucleotide RNA called mascRNA [99, 100]. The rest of the 3’ end sequence then folds into a triple-helix structure [101–103], which protects this end against 3’ – 5’ exonucleases [103]. The antisense (AS) lncRNA, TALAM1 was recently shown to form an RNA duplex with the 3’ end of MALAT1, which stabilizes MALAT1 by facilitating its 3’ processing [104].
Molecular Function of MALAT1
MALAT1 is almost strictly retained in the nucleus [105]. A large fraction of MALAT1 is localized within nuclear speckles [82, 83] and interacts with several of the speckle-enriched proteins, including splicing factors. Since MALAT1 preferentially localizes in nuclear speckles, NDs that are suggested to coordinate transcription and pre-mRNA processing, it is proposed that MALAT1 controls gene expression by modulating speckle-ascribed functions, including transcriptional and post-transcriptional gene regulation.
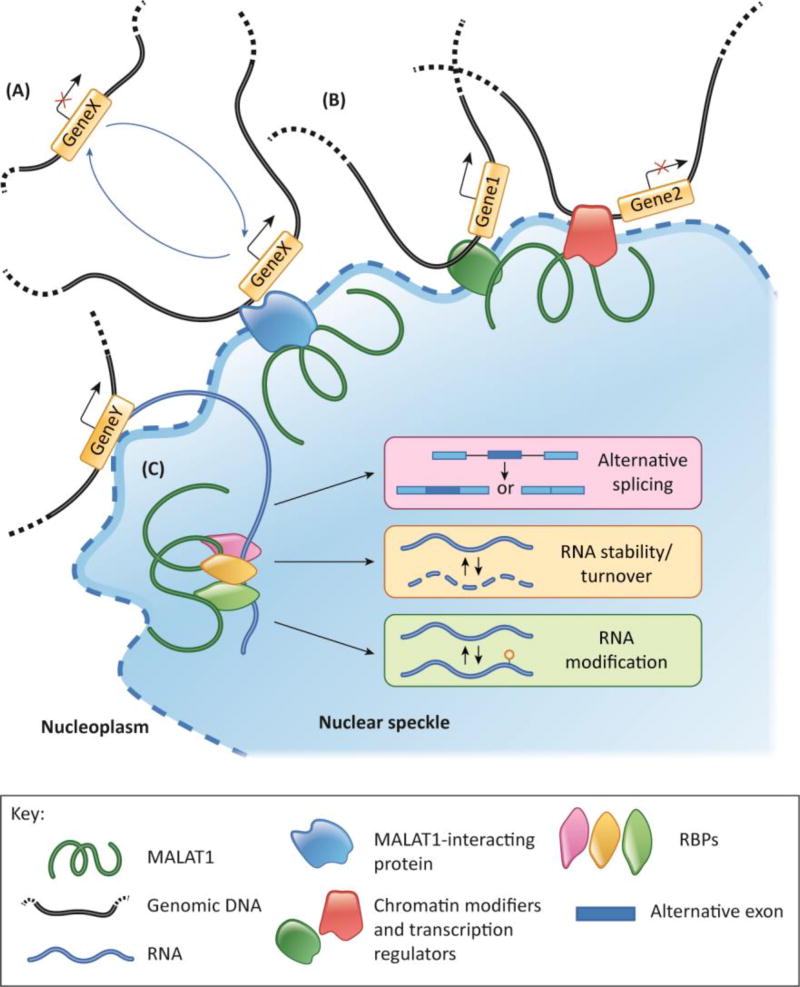
In the nucleus, MALAT1 is found to preferentially associate with transcriptionally active genes [106–108]. MALAT1-depleted cancer cells display reduced expression of cell cycle- or metastasis-associated genes without affecting the processing of their RNAs, implying a direct involvement of MALAT1 in transcription regulation [109, 110]. MALAT1 might act as an “anchorage point’ for the localization of certain genes near nuclear speckles and facilitate their transcriptional activation (Figure 3A). In support of this, MALAT1-depleted cells showed defects in the 1) association of cell cycle genes next to speckles and 2) expression [111]. Such re-localization events of genes partially rely on the interaction between MALAT1 and Pc2 [111]. Pc2 is a member of the PRC1 complex, and its methylation/demethylation plays a vital role in regulating the expression of E2F target genes. Upon serum stimulation, MALAT1 interacted with unmethylated Pc2 and drove the translocation of this protein along with a group of associated cell cycle genes, from polycomb bodies to nuclear speckles. The MALAT1-interacting pool of demethylated Pc2 then promoted the SUMOylation of E2F1, resulting in the activation of the cell cycle genes [111]. In addition to Pc2, several studies showed direct interactions between MALAT1 and PRC2 components, including EZH2 [112–117] and Suz12 [116, 118]. These results imply that MALAT1 could promote the activation or repression of genes by facilitating the recruitment of specific chromatin modulators to target genes in a cell type-specific manner (Figure 3B).
Figure 3.
Model depicting potential mode of action of MALAT1. (A) MALAT1 along with MALAT1-interacting proteins such as unmethylated Pc2 relocates genomic loci from nucleoplasm or other nuclear domains to nuclear speckle periphery and promotes the activation of corresponding genes. (B) MALAT1 interacts with multiple chromatin modifiers and transcription factors to modulate transcription. (C) MALAT1 serves as a scaffold in nuclear speckles for RBPs and influences various co- and post-transcriptional processes.
Recent RAP-seq studies revealed that MALAT1 preferentially interacts with alternatively spliced pre-mRNAs in a protein mediator-dependent manner [107]. In addition, MALAT1 interacts with a large number of RBPs, such as pre-mRNA splicing factors, hnRNPs, and RNA export factors [119–129] (Figure 3C). For example, it was shown that MALAT1 interacts with several members of the SR family of splicing factors [83, 119–121, 130]. SR-proteins are multifunctional proteins that participate in several steps of RNP maturation, including pre-mRNA splicing, RNA export, mRNA decay, and translation [131, 132]. In human cells, MALAT1 modulates the localization of SR proteins to speckles [120] and to transcription sites [83]. Furthermore, MALAT1 expression-altered cells showed changes in the phosphorylation status of SR proteins and displayed defects in the interaction of SR proteins to their target premRNAs [120, 133], implying the involvement of MALAT1 in modulating SR protein activity. This was strengthened by the observation that MALAT1 expression-altered cells showed defects in the alternative splicing of several of the SR target pre-mRNAs [87, 120, 133, 134]. Cancer promoting activity of MALAT1 could also be attributed to its involvement in regulating the alternative splicing of several oncogenic and tumor suppressor gene pre-mRNAs. For example, in breast and liver cancer cells, overexpression or depletion of MALAT1 altered the splicing of several SRSF1-target gene pre-mRNAs [133–135]. In the case of hepatocellular carcinoma, inhibition of SRSF1 abolished the oncogenic properties of cells overexpressing MALAT1 [133]. Given that SR proteins often display collaborative or sometimes redundant functions, MALAT1 may also regulate the binding kinetics of different SR proteins to fine-tune the processing of certain pre-mRNA targets. It is still not clear how exactly MALAT1 controls the activity of speckle-localized proteins.
It was observed that tethering MALAT1 to a chromatin locus facilitates the recruitment of several splicing factors [130]. Further, MALAT1-depleted human cells showed disassembly of proteins from speckles [120]. MALAT1, by associating with several proteins, could act as a nucleation site to facilitate productive interactions between speckle proteins for the assembly of functional complexes. In addition, MALAT1 interacts with nascent transcripts, preferentially alternatively spliced pre-mRNA of several hundreds of transcriptionally active genes through protein intermediates. Based on the observations that MALAT1 localizes to speckles and interacts with splicing factors and alternatively spliced pre-mRNA, we propose that MALAT1 could act as a ‘molecular scaffold’ to enhance protein-protein, protein-RNA, and protein-DNA interactions in or near speckles (Figure 3). By concentrating in speckles, MALAT1 facilitates the interaction between MALAT1-interacting RBPs such as SRSF1 and pre-mRNAs of transcriptionally active genes, thereby influencing their post-transcriptional processing. On the other hand, MALAT1 can help reposition a group of genes to the periphery of speckles via protein mediators and influence their expression.
Most of the data ascribing a role for MALAT1 in transcription and pre-mRNA processing was observed from studies using cell lines. However, MALAT1 knockout mice did not display any abnormality in terms of development, viability, or fertility [84, 136, 137]. There could be several explanations for the absence of phenotype. One possible explanation is that other lncRNAs or alternate cellular pathways compensate for loss of MALAT1. A candidate for this compensation could be the paraspeckle-localized lncRNA NEAT1, which is transcribed from the neighboring genomic locus of MALAT1. NEAT1 has been reported to have overlapping behaviors with MALAT1 in aspects of its gene association and protein interaction [106]. Finally, similar to what has been observed for several microRNAs, MALAT1 could play essential roles under specialized conditions such as pathological stress, and may show significant phenotypes if MALAT1-depleted cells or mice are exposed to such conditions [138]. This idea is supported by the fact that MALAT1 deregulation has been found in increasing numbers of diseases and pathological conditions.
Involvement of MALAT1 in cancer progression and metastasis
Ever since the initial identification of MALAT1 as a marker of metastatic lung cancer, a considerable number of studies have intimately linked MALAT1 to tumor progression and metastasis [139, 140]. Elevated levels of MALAT1 are observed in a broad spectrum of cancers, and are frequently correlated with poor prognoses and chemo- or radiotherapy resistance in patients [114, 134, 141]. Furthermore, alterations in the levels of MALAT1 in multiple cancer cell lines and in animal tumor models significantly affect tumorigenic properties, including cell proliferation, invasion, migration, and metastasis. For example, Malat1 knock out (KO) cells or cells depleted of Malat1 by antisense oligonucleotides in a mouse MMTV-PyMT breast cancer model system showed reduced tumor growth and metastasis, accompanied with enhanced differentiation into cystic tumors [142]. It was recently demonstrated that MALAT1 facilitated cell proliferation, tumor progression, and metastasis of triple-negative breast cancer (TNBC) cells [134]. Finally, increased MALAT1 levels were associated with decreased disease-specific survival in ER negative, lymph node negative patients of the HER2 and TNBC molecular subtypes [134]. These results identify MALAT1 as a metastasis driver and its potential use as a prognostic marker is better suited for ER negative, lymph node negative breast cancer patients, who might otherwise be classified as having low recurrence risk.
Altered expression of MALAT1 is also attributed in hepatocellular carcinoma development. In this case, MALAT1 was found to act as a proto-oncogene by promoting the oncogenic activity of splicing factor SRSF1, ultimately resulting in the induction of Wnt- and mTOR-signaling [133]. In the case of BC and hepatocellular carcinoma, Malat1 depletion altered the expression and pre-mRNA splicing of genes involved in cancer progression and metastasis, indicating that Malat1/MALAT1 regulates processes that are important for cancer pathogenesis [133, 134].
Concluding Remarks
The human genome encodes ~16,000 lncRNAs, of which a significant fraction is retained in the nucleus. Nuclear lncRNAs are involved in almost all physiological/biological and disease-related processes. Most nuclear lncRNAs associate with chromatin and influence gene expression in a cis or trans fashion. Chromatin-associated lncRNAs control the recruitment or stabilization of various chromatin proteins or RNA-binding proteins to regulatory sequences within the gene or RNA, thereby influencing transcriptional or post-transcriptional processes. However, we still do not know how lncRNAs associate to specific chromatin regions in the genome. Some aspects of specificity could be attributed to the ability for lncRNAs to recognize DNA via forming RNA:DNA hybrids. Alternatively, lncRNA could be recruited to a chromatin site by chromatin-associated proteins. Retention of lncRNAs in the nucleus and their localization to specific nuclear compartments could be controlled by active or passive regulatory mechanisms. LncRNAs such as Xist and eRNAs could be retained in the nucleus via their association with chromatin. LncRNAs are also known to contain specific sequence motifs, domains, or repeat sequences within them that potentially act as nuclear retention signals [23, 71, 143–145]. In addition, association of nuclear proteins to lncRNAs could ensure the nuclear retention of lncRNAs [70, 77, 146]. Future studies on a large number of nuclear lncRNAs will help to identify the novel mechanisms utilized by lncRNAs for their nuclear localization and chromatin association (See Outstanding Questions).
Outstanding Questions.
Several nuclear-retained lncRNAs work in trans. How do trans-acting lncRNAs transport from their site of transcription to other gene loci where they act?
Unlike proteins whose nuclear retention signals could be predicted by the existence of NLS (nuclear-localization signal), the nuclear localization of lncRNAs is controlled by several independent mechanisms. What type of sequence or structural elements within lncRNAs dictates their nuclear retention, DNA recognition, and protein association?
Multiple nuclear-retained lncRNAs are reported to interact with polycomb repressive complex 2 (PRC2) or its subunits. However, the field now identifies that at several instances, PRC2 might promiscuously interact with RNA molecules. Hence, one needs to use more stringent measures to confirm functional interactions between lncRNAs and polycomb complex proteins.
Human genome is predicted to contain ~16000 lncRNA (Gencode.org) genes. How many of these lncRNAs play important biological roles and how many of them are by products of spurious transcription?
The known physiological role of lncRNAs is limited due to inadequate organismal studies to date. Some of the recent studies indicate that several of the lncRNAs play a vital role in embryonic and adult tissue development [147, 148]. Future studies aimed at determining the molecular function of the vast number of human nuclear lncRNAs would help us to better appreciate the role of lncRNAs in gene expression and nuclear organization, as well as their potential involvement in diseases.
Trends.
A significant fraction of long non-coding RNAs is retained in the nucleus, several of them are shown to participate in vital nuclear processes, including chromatin organization, transcriptional and post-transcriptional gene expression, and nuclear structure organization.
The advent of new techniques, including RAP, CHIRP, CHART, and MARGI, provides researchers more opportunity to study the chromatin binding features of nuclear-retained lncRNAs, especially at genomic level.
Many nuclear-retained lncRNAs are shown to be biomarkers of diagnosis/prognosis and/or therapeutic targets of diseases, including cancer.
The development of new computational tools/algorithms is largely required to determine the potential correlation between nuclear-retained lncRNA sequences/structures and their functions/localization.
Acknowledgments
We thank Drs. A. Lal and S.G. Prasanth for critical reading and suggestions. We thank J. Roy, S. Sudhakar and S. Adusumilli for proof reading the manuscript. We thank Elsevier’s Illustration service (WebShop) for figure illustration. Work in Prasanth lab is funded by NIH R01 (GM088252) and NSF EAGER (1723008) grants.
Glossary
- SWI/SNF
SWI/SNF (switch/sucrose nonfermentable) is a chromatin-remodeling complex. This complex contains multiple subunits, including ATPase, which allows this complex to remodel nucleosomes from the energy generated through ATP hydrolysis
- PRC
PRC (polycomb repressive complexes) consist of two major types: PRC1 and PRC2. These complexes play important roles in chromatin compaction and transcriptional silencing
- CHIRP, CHART, RAP, and MARGI
Chromatin Isolation by RNA Purification (CHIRP) Capture Hybridization Analysis of RNA Targets (CHART), RNA Antisense Purification (RAP), and MApping RNA Genome Interactions (MARGI) are four techniques developed to map the genomic binding sites of RNA. They are often used to discover the roles and mechanisms of long non-coding RNAs on chromatin
- XCI
X Chromosome Inactivation (XCI) is a phenomenon observed during mammalian female development. Most of the genes are inactivated (transcriptional silencing) on one of the two X-chromosomes in females
- Nuclear Speckle
Speckles are conserved nuclear domains that are present in the form of 10–30 irregularly shaped nuclear structures. Speckles are enriched with RNAs and proteins involved in pre-mRNA processing and mRNP export. Initial studies suggest that speckles act as storage and/or assembly sites of splicing factors from where these proteins are recruited to active genes dispersed throughout the nucleoplasm. Recent studies further indicate that nuclear speckles act as a structural domain that controls the efficiency and integration of distinct steps in gene expression, ranging from transcription, splicing to mRNA export
- Paraspeckle
Paraspeckles are a type of subnuclear domains present in the interchromatin space of mammalian cells. They are nucleated by long non-coding RNA Neat1. The core paraspeckle proteins include PSF/SFPQ, P54NRB/NONO, and PSPC1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, et al. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017;33(8):540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 4.St Laurent G, et al. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasanth KV, et al. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell. 2010;21(23):4184–96. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt AM, Chang HY. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb Perspect Med. 2017;7(7) doi: 10.1101/cshperspect.a026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24(11):651–63. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30(8):348–55. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–8. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41(9):761–72. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Werner MS, Ruthenburg AJ. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015;12(7):1089–98. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 13.Wang XQ, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017;45(3):1091–1104. doi: 10.1093/nar/gkw966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng C, et al. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Rep. 2016;14(1):103–14. doi: 10.1016/j.celrep.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Leary VB, et al. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015;11(3):474–85. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, et al. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci U S A. 2012;109(35):14110–5. doi: 10.1073/pnas.1116597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierhoff H, et al. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell. 2014;54(4):675–82. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 18.Scarola M, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postepska-Igielska A, et al. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60(4):626–36. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Chalei V, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monnier P, et al. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A. 2013;110(51):20693–8. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu GY, et al. The long noncoding RNA Gm15055 represses Hoxa gene expression by recruiting PRC2 to the gene cluster. Nucleic Acids Res. 2016;44(6):2613–27. doi: 10.1093/nar/gkv1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondal T, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z, et al. lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016;14(8):1876–82. doi: 10.1016/j.celrep.2016.01.073. [DOI] [PubMed] [Google Scholar]
- 25.Leisegang MS, et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017;136(1):65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–25. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez I, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22(5):370–6. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain AK, et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol Cell. 2016;64(5):967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan J, et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16:139. doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han P, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102–6. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing Z, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–25. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu C, et al. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 34.Chu HP, et al. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell. 2017;170(1):86–101. e16. doi: 10.1016/j.cell.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XL, et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep. 2017;20(10):2408–2423. doi: 10.1016/j.celrep.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21(4):423–5. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 38.Di Cecilia S, et al. RBM5-AS1 Is Critical for Self-Renewal of Colon Cancer Stem-like Cells. Cancer Res. 2016;76(19):5615–5627. doi: 10.1158/0008-5472.CAN-15-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528(7583):517–22. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Li Z, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002–7. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao X, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25(1):80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joung J, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin Y, et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16(5):504–16. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Halley P, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–30. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vance KW, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33(4):296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhary R, et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife. 2017;6 doi: 10.7554/eLife.23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun. 2017;8:14016. doi: 10.1038/ncomms14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atianand MK, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165(7):1672–85. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puvvula PK, et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat Commun. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu P, et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bierhoff H, et al. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol. 2010;75:357–64. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 56.Xing YH, et al. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell. 2017;169(4):664–678. e16. doi: 10.1016/j.cell.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Yin QF, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48(2):219–30. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 58.Caudron-Herger M, et al. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 2015;34(22):2758–74. doi: 10.15252/embj.201591458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lam MT, et al. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39(4):170–82. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, et al. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17(4):207–23. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 61.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–35. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 63.Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539(7629):433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramos AD, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16(4):439–47. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salameh A, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112(27):8403–8. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anantharaman A, et al. ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res. 2017;45(7):4189–4201. doi: 10.1093/nar/gkw1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H, et al. Unusual Processing Generates SPA LncRNAs that Sequester Multiple RNA Binding Proteins. Mol Cell. 2016;64(3):534–548. doi: 10.1016/j.molcel.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 69.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–6. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.da Rocha ST, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53(2):301–16. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 71.Engreitz JM, et al. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17(12):756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 72.Chen CK, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 73.Morchikh M, et al. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol Cell. 2017;67(3):387–399. e5. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 74.Ma H, et al. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J Virol. 2017;91(9) doi: 10.1128/JVI.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 76.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao YS, et al. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13(1):95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186(5):637–44. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mello SS, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11):1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang L, et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 82.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–93. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eissmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–87. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang B, et al. Identification and Characterization of a Class of MALAT1-like Genomic Loci. Cell Rep. 2017;19(8):1723–1738. doi: 10.1016/j.celrep.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ulitsky I, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tripathi V, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma XY, et al. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics. 2015;16:676. doi: 10.1186/s12864-015-1881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watts R, et al. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013;304(10):C995–1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 90.Aiello A, et al. MALAT1 and HOTAIR Long Non-Coding RNAs Play Opposite Role in Estrogen-Mediated Transcriptional Regulation in Prostate Cancer Cells. Sci Rep. 2016;6:38414. doi: 10.1038/srep38414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li S, et al. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol. 2015;141(11):1909–20. doi: 10.1007/s00432-015-1951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arita T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33(8):3185–93. [PubMed] [Google Scholar]
- 93.Michalik KM, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–97. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 94.Luo F, et al. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016;7(5):5769–87. doi: 10.18632/oncotarget.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lelli A, et al. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia (Auckl) 2015;3:45–52. doi: 10.2147/HP.S90555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koshimizu TA, et al. Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci. 2010;86(11–12):455–60. doi: 10.1016/j.lfs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Tani H, et al. Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov Ther. 2010;4(4):235–9. [PubMed] [Google Scholar]
- 98.Clark MB, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilusz JE, et al. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNAlike cytoplasmic RNA. Cell. 2008;135(5):919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3' end processing mechanisms. RNA. 2010;16(2):259–66. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilusz JE, et al. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown JA, et al. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109(47):19202–7. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown JA, et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21(7):633–40. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zong X, et al. Natural antisense RNA promotes 3' end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res. 2016;44(6):2898–908. doi: 10.1093/nar/gkw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyagawa R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18(4):738–51. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engreitz JM, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–99. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sridhar B, et al. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr Biol. 2017;27(4):602–609. doi: 10.1016/j.cub.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tano K, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–80. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 111.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hirata H, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75(7):1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang D, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6(38):41045–55. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen W, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8(14):22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huo Y, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8(29):46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim SH, et al. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget. 2017;8(19):31305–31317. doi: 10.18632/oncotarget.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qi Y, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016;7(11):12693–703. doi: 10.18632/oncotarget.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan Y, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–41. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 119.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19(3):381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tripathi V, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anko ML, et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13(3):R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Latorre E, et al. The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer. Cancer Res. 2016;76(9):2626–36. doi: 10.1158/0008-5472.CAN-15-2018. [DOI] [PubMed] [Google Scholar]
- 123.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 124.Yoon JH, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ji Q, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111(4):736–48. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li L, et al. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106(31):12956–61. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tripathi V, et al. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell. 2012;23(18):3694–706. doi: 10.1091/mbc.E12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhong XY, et al. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35(1):1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10(10):242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malakar P, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jadaliha M, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7(26):40418–40436. doi: 10.18632/oncotarget.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin R, et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585(4):671–6. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–99. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abbott AL. Uncovering new functions for microRNAs in Caenorhabditis elegans. Curr Biol. 2011;21(17):R668–71. doi: 10.1016/j.cub.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gutschner T, et al. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91(7):791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 140.Qiu MT, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–20. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 141.Lu H, et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37(2):1683–91. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 142.Arun G, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang B, et al. A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol Cell Biol. 2014;34(12):2318–29. doi: 10.1128/MCB.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hacisuleyman E, et al. Function and evolution of local repeats in the Firre locus. Nat Commun. 2016;7:11021. doi: 10.1038/ncomms11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Negishi M, et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014;9(4):e95216. doi: 10.1371/journal.pone.0095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hasegawa Y, et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19(3):469–76. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 147.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Johnsson P, et al. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840(3):1063–71. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 151.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322(5909):1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long noncoding RNAs. Int J Mol Sci. 2015;16(2):3251–66. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Y, et al. Life without A tail: new formats of long noncoding RNAs. Int J Biochem Cell Biol. 2014;54:338–49. doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 155.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859(1):128–38. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang W, et al. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11(1):137–48. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–90. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alvarez-Dominguez JR, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–81. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luo S, et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell. 2016;18(5):637–52. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 160.Arab K, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55(4):604–14. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 161.Huang MD, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cajigas I, et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142(15):2641–52. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Y, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A. 2015;112(29):E3883–92. doi: 10.1073/pnas.1501662112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Beckedorff FC, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9(8):e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stojic L, et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat Commun. 2016;7:10406. doi: 10.1038/ncomms10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yoon JH, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4(2939) doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huang J, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44(7):3059–69. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang A, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23(3):340–50. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kumar PP, et al. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. Elife. 2014;3 doi: 10.7554/eLife.02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sharma V, et al. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 2015;16(11):1520–34. doi: 10.15252/embr.201540437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 173.Allen TA, et al. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11(9):816–21. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 174.Peterlin BM, et al. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3(1):92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dieci G, et al. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 176.Santoro R, et al. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010;11(1):52–8. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Guetg C, et al. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45(6):790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 178.Wierzbicki AT, et al. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135(4):635–48. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bohmdorfer G, et al. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife. 2016;5 doi: 10.7554/eLife.19092. [DOI] [PMC free article] [PubMed] [Google Scholar]