Abstract
Metabolic diseases such as obesity and diabetes are complex diseases resulting from multiple genetic and environmental factors, such as diet and activity levels. These factors are well known contributors to the development of metabolic diseases. One manner by which environmental factors can influence metabolic disease progression is through modifications to chromatin. These modifications can lead to altered gene regulatory programs, which alters disease risk. Furthermore, there is evidence that parents exposed to environmental factors can influence the metabolic health of offspring, especially if exposures are during intrauterine growth periods. In this review, we outline the evidence that chromatin modifications are associated with metabolic diseases, including diabetes and obesity. We also consider evidence that these chromatin modifications can lead to long‐term disease risk and contribute to disease risk for future generations.
This article is categorized under:
Biological Mechanisms > Metabolism
Developmental Biology > Developmental Processes in Health and Disease
Physiology > Organismal Responses to Environment
1. INTRODUCTION
The impact of environmental stimuli on phenotypic traits—and the potential transmission of acquired traits—has been studied since the time of Aristotle. The idea that acquired traits could be inherited was popularized by Jean‐Baptiste Lamarck who, by examining fossils of animals, concluded that species could acquire traits that were advantageous to their survival. His views were later widely dismissed when Darwin's theory of descent with modification gained popularity.
Towards the end of the 1800s, August Weisman performed a series of experiments in which he amputated the tails of several generations of mice to determine what effect this would have on subsequent generations. These experiments led to the concept of the “Weismann barrier” in which germline cells containing genetic information are passed on to future generations and are not affected by environmental conditions as somatic cells.
A half century later, Conrad Waddington demonstrated that by exploiting the developmental plasticity of certain phenotypic traits, environmental factors could give rise to a “sort” inheritance, or the inheritance of an acquired trait. He described this process as “canalization” and represented the developmental process through the classical developmental landscape. Waddington coined the term “epigenetics” as “the branch of biology that studies the causal interactions between genes and their products, which bring the phenotype into being” (Waddington, 1942). Towards the later part of the 1900s, it was realized that changes in gene expression that were not a product of genetic changes could be heritable in mammals (Holliday & Pugh, 1975; Riggs, 1975) and the classical definition of epigenetics as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” was born (Riggs, Martienssen, & Russo, 1996).
It is important to note that the definition of epigenetics has been relaxed in recent times to the point where it is used to refer to any gene regulatory phenomena. There remains debate, however, on the importance of maintaining the strict definition of heritability (see Ptashne, 2007 for a thorough discussion). Herein we have strived to make clear what modifications to chromatin have evidence of adhering to the classical epigenetic definition and which are more likely simply associated with transcriptional or gene regulatory changes.
2. CHROMATIN AND GENE REGULATION
All eukaryotic organisms package their genome into nuclei by wrapping DNA around histone proteins to form chromatin. This chromatin structure is subject to a variety of modifications, such as DNA methylation, histone modifications, histone variants and ATP‐dependent chromatin remodeling (discussed in more detail below). Each individual type of modification to chromatin is generally associated with either “permissive” or “repressive” environments, changing access to the underlying genomic sequence (Figure 1).
Figure 1.
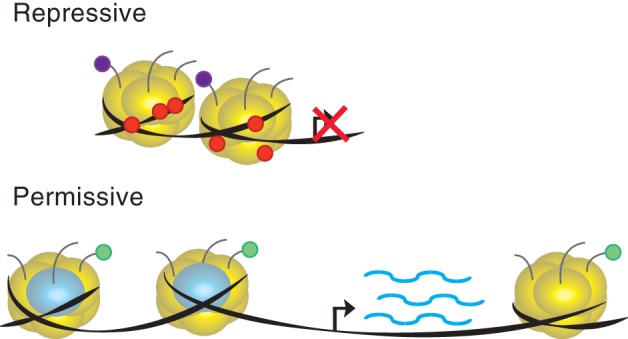
Chromatin modifications associated with “permissive” or “repressive” environments. Modifications to histone tails and DNA methylation are associated with “repressive” states with condensed chromatin or “permissive” state of more accessible chromatin. Red balls, DNA methylation; purple balls, repressive histone modifications; green balls, active histone modifications; yellow globes, histone proteins; blue globes, histone variants
Proper chromatin patterning is essential for normal development and altered chromatin patterning is a hallmark of many human diseases, including obesity, diabetes mellitus, cardiovascular disease, and cancer (Portela & Esteller, 2010). Given that these modifications to chromatin can be mitotically heritable, they have the potential to contribute to long‐term gene expression changes, or even transgenerational effects. Thus, environmental factors that impact chromatin modifications have the potential to lead to long‐term disease risk. We outline the basic mechanisms involved in chromatin modifications and discuss the impact modifications to chromatin can have in the progression of metabolic diseases in the following sections.
2.1. DNA methylation
DNA methylation in mammalian genomes is generally established by the addition of a methyl group to the 5‐carbon of the cytosine in a CpG dinucleotide (5mC). The recent advancement of techniques for whole genome methylation profiling has revolutionized our understanding of the dynamic nature of DNA methylation (see Edwards, Yarychkivska, Boulard, & Bestor, 2017 for a recent discussion). The primary roles for DNA methylation in mammalian genomes are the stable repression of transposon derived sequences (such as long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs).), maintaining mono‐allelic expression of imprinted regions, transcriptional regulation through the regulation of chromatin accessibility, and X chromosome inactivation in female cells.
In mammals, the formation of 5mC is regulated by the DNA methyltransferases (DNMTs) DNMT1, DNMT3A, DNMT3B, and DNMT3L (Bestor, 2000). All DNMT activity requires the metabolite S‐adenosyl methionine (SAM) to methylate cytosine (van der Wijst et al., 2015). In somatic cells, DNMT1 is the most active DNMT. DNMT1 is involved in preserving DNA methylation profiles on newly synthesized DNA (Jin & Robertson, 2013). This is required for daughter cells to maintain the same epigenetic marks as the parent cell. During development, DNMT3a and DNMT3b are responsible for de novo DNA methylation. This de novo methylation is essential for the establishment of different cell lineages (Okano, Bell, Haber, & Li, 1999). These proteins are also responsible for the establishment of methylation in differentiated cells in response to environmental signals. DNMT3L has no DNA methylation activity, but is able to regulate DNMT3a and DNMT3b activity in development and is important for proper targeted methylation (Hata, Okano, Lei, & Li, 2002). In addition to DNMT1, DNMT3A, DNMT3B, and DNMT3L, mammals also have DNMT2, however, it is not required for development. The main function of DNMT2 has been demonstrated to be the methylation of RNA to alter their function and stability (Goll et al., 2006). Recently, DNMT2 has been shown to have a role in DNA methylation in aging macrophages (Khalil et al., 2016). Additionally, DNMT2 has been shown to play a role in hematopoiesis and intergenerational epigenetic inheritance, however, this is not known to a result of its DNA or RNA methylation function (Kiani et al., 2013; Tuorto et al., 2015). In addition to 5mC, N6‐methyladenine has been reported to be present in mice as a novel repressive mark on evolutionarily younger LINE elements (Wu et al., 2016), however, this DNA modification has not yet been found in humans.
The methyl group from 5mC can be removed from DNA by Ten‐eleven translocase (TET) proteins. These proteins are regulated by many metabolites produced by the tricarboxylic acid (TCA) cycle. The process of DNA demethylation requires the 5mC to be converted to 5‐hydroxymethyl cytosine (5hmC). 5hmC has been found in many cell types but has been found at its highest concentrations in neuronal cells (Münzel et al., 2010). 5hmC is then converted to 5‐formylcytosine (5fC), then 5‐carboxylcytosine (5caC), and then ultimately the nucleotide is removed and replaced with unlabeled cytosine (Ito et al., 2011; Shibutani et al., 2014).
There are two waves of demethylation during mammalian development where nearly all 5mC marks are removed. The removal of methylation reverts the cells to a stem‐like state, which allows for proliferation and reprograming. There are a set of evolutionarily younger retrotransposons that “escape” germline reprograming in mice (Hackett et al., 2013; Seisenberger et al., 2012) and humans (Tang et al., 2015; Tang, Kobayashi, Irie, Dietmann, & Surani, 2016) (Figure 2). The persistent DNA methylation at these repeat elements through germline reprograming are attractive candidates for the potential ability to induce altered gene expression in offspring and be responsible for mediating epigenetic inheritance, as discussed in more detail below. Other loci, referred to as imprinted or imprinted differentially methylated regions (DMRs) are methylated in the sperm and oocytes have retained methylation in a sex‐specific manner throughout development.
Figure 2.
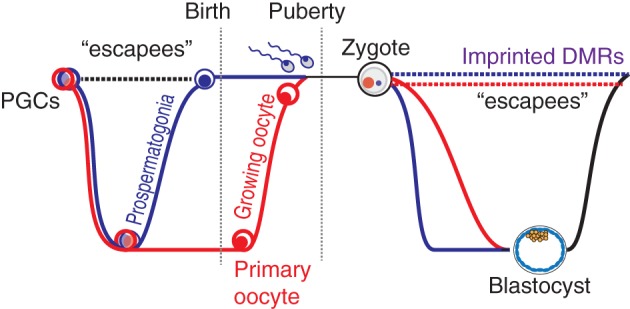
Developmental epigenetic reprograming. There are waves of genome‐wide demethylation followed by the establishment of methylation in primordial germ cells (PGCs) and following fertilization. The vast majority of the genome undergoes this loss of methylation. However, a group of evolutionarily young transposable elements, referred to as “escapees,” avoid the demethylation
2.2. Histone modifications
Histone modifications are involved in the dynamic control of chromatin structure. Histone modifications can positively or negatively influence gene transcription and chromatin availability. The reported forms of histone modifications include acetylation, methylation, ubiquitination, phosphorylation, sumoylation, and biotinylation (Bannister & Kouzarides, 2011), with the most well studied being histone acetylation and methylation. While histone acetylation marks active chromatin, histone methylation can be either activating or repressive depending on its location on the histone tail. For example, H3K4me3 is a mark for transcriptional activation while H3K9me3 is a mark for transcriptional repression. The addition of histone marks has been found to be reliant on the availability of numerous metabolites (Figure 3). These altered histone modifications, in response to metabolic changes, can lead to a dynamic and persistent change in gene expression.
Figure 3.
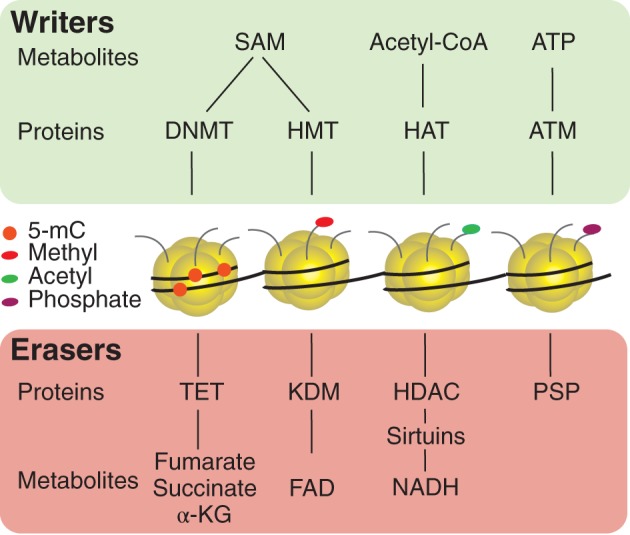
Metabolites and chromatin structure. Metabolites are required by many chromatin modifying proteins. S‐adenosyl methionine (SAM) is required by DNA methyltransferases (DNMTs), as well as histone methyltransferases (HMTs) to add a methyl group to DNA or histone tails (Rea et al., 2000). The metabolites fumartate, succinate and α‐ketogluterate regulate Ten‐eleven translocase (TET) proteins (Klose, Kallin, & Zhang, 2006). TETs are responsible for the removal of methyl groups from DNA. Flavin adenine dinucleotide (FAD) regulates lysine demethylase (KDM) to regulate the removal of methyl groups from histones. Acetyl‐CoA is required for the addition of acetyl groups to histones by histone acetyl transferases (HATs) (Galdieri & Vancura, 2012). Nicotinamide adenine dinucleotide (NADH) interacts with Sirtuins to facilitate acetyl group removal by histone deacetylases (HDACs) (Ions et al., 2013). ATP is a required substrate for serine/threonine kinase (ATM) phosphorylation of histones (Banerjee, Bennion, Goldberg, & Allen, 1991), which is removed by protein serine/threonine phosphatases (PSPs)
2.2.1. Histone variants and nucleosome remodeling
In addition to alterations of histone tails, incorporation of noncanonical histone proteins, known as histones variants (reviewed in Weber & Henikoff, 2014) are associated with altered gene expression. Histone variants such as H2A.Z and H3.3 in humans have been found to mark enhancers and indicate sites of active transcription (Jin & Felsenfeld, 2007; Jin et al., 2009). H3.3 has furthermore been shown to be essential in the regulation of chromatin structure in mammals, with depletion leading to dysregulation of constitutive heterochromatin (Jang, Shibata, Starmer, Yee, & Magnuson, 2015). In addition to modifying the histone complex, translocation of the nucleosomes is another mechanism through which chromatin structure can regulate transcription. This process is regulated by ATP‐dependent nucleosomes remodelers. ATP‐dependent remodelers can “translocate” nucleosomes to allow DNA previously occluded by the nucleosome complex to become accessible (reviewed in Clapier, Iwasa, Cairns, & Peterson, 2017). Briefly, there are subfamilies of ATP‐dependent histone remodeling complexes, including imitation switch proteins (ISWI), chromodomain helicase DNA‐binding proteins (CHD), switch/sucrose nonfermentable proteins (SWI/SNF), and INO80 (Delmas, Stokes, & Perry, 1993; Neigeborn & Carlson, 1984; Shen, Mizuguchi, Hamiche, & Wu, 2000; Tsukiyama, Palmer, Landel, Shiloach, & Wu, 1999). These ATP‐dependent chromatin remodelers can change chromatin structure to allow for altered DNA accessibility. These chromatin remodelers can be guided to specific regions of the chromatin by transcriptional activators and repressors, histone modifications, and long noncoding RNAs. These complexes are reliant upon ATP to disrupt DNA‐histone interactions induce chromatin remodeling (Clapier et al., 2017). Histone modifications and nucleosome remodeling, which are essential for altering gene expression in cell, are all regulated by metabolite levels in the cell.
2.3. RNA‐mediated chromatin regulation
The regulation of epigenetic modifications through noncoding RNAs is an area of growing interest. Noncoding RNAs contribute to gene expression in response to environmental signals. They include long noncoding RNAs (lncRNAs) and small RNAs (sRNAs), which include PIWI‐associated RNAs (piRNAs) and tRNA fragments (tRFs). Each of these RNA species can silence gene expression either pre‐ or posttranscriptionally (Carmell et al., 2007; Hawkins, Santoso, Adams, Anest, & Morris, 2009).
There is growing evidence that environmental factors, such as diet, can impact RNA‐mediated epigenetic modifications. For example, the expression of sRNAs is drastically altered in response to high fat diet and caloric restriction, resulting in an aging phenotype (Green et al., 2017). piRNAs can also be deregulated in response to high‐fat diet, leading to altered DNA methylation in spermatozoa, altered DNA imprinting, and the potential for epigenetic inheritance of metabolic phenotypes (Dai et al., 2017; Kumar, Kuscu, & Dutta, 2016). The tRFs are another type of sRNA with altered expression in response to diet. These tRFs are the cleavage product of tRNAs that are modified my DNMT2 and NSUN2 and circulate in serum in stable nucleoprotein complexes (Tuorto et al., 2012). They have altered production and secretion in response to environmental stressors (Dhahbi et al., 2013). Additionally, tRFs also have been shown to have the potential to mediate transgenerational inheritance of metabolic complications (Chen et al., 2016; Lee, Shibata, Malhotra, & Dutta, 2009; Sharma et al., 2016). Treatment of mouse embryos with tRFs harvested from hyperglycemic and insulin‐resistant mice leads to metabolic complications in offspring as the result of targeted formation and preservation of heterochromatin at transposable elements (Chen et al., 2016; Kumar et al., 2016; Schorn, Gutbrod, LeBlanc, & Martienssen, 2017). lncRNAs can impact chromatin in a variety of ways (reviewed in Sun & Wong, 2016). lncRNAs can guide both histones modifying proteins and DNMTs to targeted sites on the chromosome, which can give these proteins loci specificity (O'Leary et al., 2017, reviewed in Mercer & Mattick, 2013). Additionally, lncRNAs can act as sRNA sponges and stabilize mRNA (Du et al., 2016; Lister et al., 2017). As previously stated, lncRNA can also guide nucleosome remodeling proteins to targeted genomic regions (Tang et al., 2017). Finally, lncRNAs have been demonstrated to be regulators of glucose homeostasis and their dysregulation has been linked to the pathogenesis and progression of diabetes (Gao et al., 2014; Petry et al., 2010).
3. EPIGENETICS AND ENVIRONMENTAL INFLUENCES
Intake of excess sugar and fat leads to the development of obesity, type 2 diabetes and other metabolic diseases (Friedman, 2009). Concurrently, excess sugar and fat consumption has been demonstrated to alter chromatin structure and gene expression. For example, obesogenic diets have been demonstrated to alter DNA methylation (Multhaup et al., 2015) and chromatin accessibility (Leung et al., 2014), allowing for the possibility to impart long‐term gene expression changes and risk for disease, which will be discussed below.
3.1. Epigenetics and metabolic memory
Clinical studies examining blood glucose control for diabetic patients have demonstrated that complications can continue to develop long after blood glucose normalization, a phenomenon that was originally termed “metabolic memory” (Writing Team for the Diabetes, Control, Complications Trial/Epidemiology of Diabetes, Interventions,, & Complications Research Group, 2003). Similarly, many obese people find it difficult to maintain weight loss (Anderson, Konz, Frederich, & Wood, 2001; Kramer, Jeffery, Forster, & Snell, 1989) due to long‐term physiological changes contributing to weight regain (Rosenbaum, Hirsch, Gallagher, & Leibel, 2008; Sumithran et al., 2011). Diet‐induced obesity (DIO) mice develop metabolic dysfunctions, including insulin resistance and impaired glucose tolerance, mimicking dysfunctions observed in many obese patients (Collins, Martin, Surwit, & Robidoux, 2004; Winzell & Ahren, 2004). Studies in mice have demonstrated that mice transitioning from high fat to low fat diet do not completely revert to the same metabolic state as mice only maintained on low fat diet (Guo, Jou, Gavrilova, & Hall, 2009). Given the observation that epigenetic modifications can mediate metabolic signals to influence gene regulation that can persist across cell division, epigenetic modifications represent good candidates for mediating this metabolic memory (Figure 4). However, the evidence for epigenetic modifications mediating metabolic memory remains controversial. It has been demonstrated that chromatin alterations induced by obesogenic diets can persist upon dietary changes (Leung, Trac, Du, Natarajan, & Schones, 2016). However, the genetic background (Leung et al., 2016), dietary composition and feeding time scales (Siersbaek et al., 2017) have been shown to impact the persistence of obesity‐associated chromatin modifications (Siersbaek et al., 2017). More work is needed to decipher the relationship between metabolic phenotypes and epigenetic changes. Given that chromatin modifications can be dynamic and reversible, characterizing persistent versus transient chromatin modifications in response to metabolic signaling is a major challenge for the field (Keating & El‐Osta, 2015).
Figure 4.
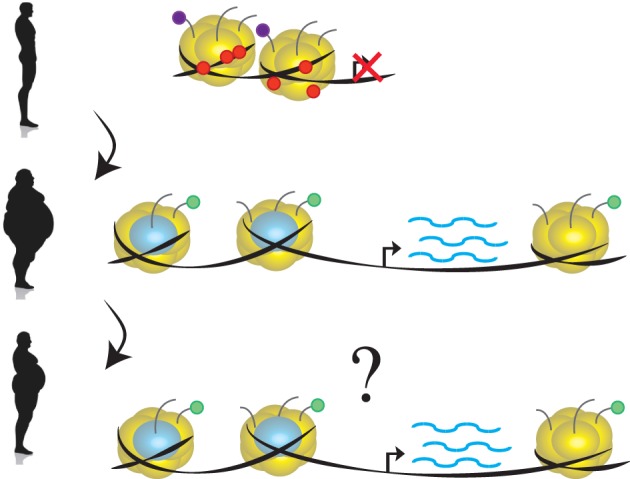
Epigenetics and metabolic memory. The phenomenon of metabolic memory is the observation that complications due to metabolic disease can persist even after metabolic disease is mitigated. Epigenetic modifications are an attractive candidate for mediating this phenomenon
3.1.1. Links between aging and metabolism
There are many links between metabolic diseases and aging (Ahima, 2009). It is now well established that epigenetic alterations are a molecular hallmark of aging. DNA methylation profiles have even been used to define an “epigenetic clock” (Horvath, 2013; Stubbs et al., 2017). Epigenetic alterations associated with aging have been found to be accelerated or slowed as a result of high fat diet and caloric restriction, respectively (Cole et al., 2017; Stubbs et al., 2017; Wang et al., 2017). Recent work profiling transcription in livers of mice found that, while protein coding genes can be activated or repressed with no particular bias in one direction or the other, noncoding RNAs (sRNAs and lncRNAs) and transposable elements were largely activated and this trend could be reversed by lifespan extending interventions such as caloric restriction (Green et al., 2017). Caloric restriction has been shown to promote increased life span for many organisms including, mice, Drosophila and C. elegans (Ja et al., 2009; Lakowski & Hekimi, 1998; Weindruch, Walford, Fligiel, & Guthrie, 1986). However, the evidence for the effectiveness of caloric restriction in primates is not as clear, with there being conflicting reports of its efficacy (Colman et al., 2014; Mattison et al., 2012). In contrast to caloric restriction, high fat diet has been reported to have cells to have aged cellular phenotypes, which both altered noncoding RNA profiles and altered epigenetic marks (Stubbs et al., 2017; Tanaka et al., 2017).
Dietary interventions can affect the transcriptome through altered levels of metabolites. This causes activation of chromatin remodeling pathways, that is, low levels of insulin and low caloric intake lead to activation of FOXO and sirtuins, respectively (Cohen et al., 2004; Qi et al., 2015). These proteins are involved in stem cell maintenance and can slow the aging process. Alternatively, high fat diet has been linked to the induction of disorders strongly associated with increased age (Zhao et al., 2013) through mechanistic target of rapamycin (mTOR) pathway activation that induces stem cell activation and proliferation (Wang et al., 2017). High fat diet has been correlated with an increased risk of cancer development (Khodarahmi & Azadbakht, 2014) as well as inducing inflammation of both the liver and adipose tissue (van der Heijden et al., 2015), which are both disorders related to aging. The impact of diet on aging phenotypes highlights the importance that the environment has on the chromatin structure of cells. These studies provide hope that we can begin to view the deleterious effects associated with aging as treatable and preventable events rather than inevitable.
3.2. Metastable epialleles
Alleles that can be variably expressed through epigenetic modifications in individuals with identical genetic composition are known as metastable epialleles (Rakyan, Blewitt, Druker, Preis, & Whitelaw, 2002). The most well characterized and studied model of a mammalian metastable epiallele is the viable yellow agouti (A vy) mouse (Dolinoy, Das, Weidman, & Jirtle, 2007). This mouse model, named for its coat color, has the paired phenotypes of metabolic dysfunction and coat color.
The A vy allele is regulated by the methylation state of an Intracisternal A‐Particle (IAP) retrotransposon ~100 kb upstream of the gene. A hypomethylated IAP yields a yellow colored obese agouti mouse while, in contrast, a hypermethylated IAP leads to a brown mouse with normal metabolic phenotype. The methylation state is consistent in all somatic tissues, implying that the methylation state is established early in development, before tissue differentiation (Waterland & Jirtle, 2003). As mentioned above, there is an intimate crosstalk between metabolism and chromatin, with many chromatin modifying enzymes relying on the availability of metabolites. Given that metastable epialleles can be impacted by environmental factors, this provides the capability of environmental stimuli to lead to inherited epigenetic changes (or “soft inheritance”). For the A vy mice, feeding A vy dams a diet that is rich in methyl groups decreases the likelihood of having yellow offspring. However, this effect is only transmitted over two generations, indicating that it is intergenerational as opposed to transgenerational (see further discussion below) (Waterland & Jirtle, 2003).
A number of putative metastable epialleles have been found in humans and their regulation has been linked to maternal one‐carbon metabolites. The proopiomelanocortin (POMC) gene has been recently identified as a putative metastable epiallele in humans involved in the regulation of weight (Kühnen et al., 2016). The regulation of the POMC gene is under the control of Alu elements and an LTR (Lam et al., 2015). The methylation state of this epiallele is regulated by both maternal one‐carbon metabolites at conception and paternal transmission of the methylation state. Hypermethylation of the POMC gene leads to increased risk of obesity (Kühnen et al., 2016). Metastable epialleles including CYP2E1 and MGMT have been correlated to the development of Parkinson's and glioblastoma, respectively (Harris, Nagy‐Szakal, & Kellermayer, 2013). As stated above, a number of evolutionary younger TEs have been found to retain their methylation marks even during germline reprograming (“escapees”) (Figure 2), although the mechanisms for this remain unclear. Determining how this process occurs is promising lead to discover how metastable epialleles and phenotypic effects are established through generations.
One mediator of the methylation state of these metastable epialleles is Trim28/Kap1, which is a DNA‐binding protein. Trim28/Kap1 affects genes important in imprinted gene networks, including Nnat and Peg3. In mice, haploinsufficiency of the Trim28 protein leads to a bimodal induction of obesity (polyphenism) (Dalgaard et al., 2016). Furthermore, haploinsufficiency of Trim28/Kap1 and Dnmt3 in mice causes a greater variation in body weight than wild type littermate (Whitelaw et al., 2010). This phenomenon is not limited to mice. In humans, it has been found that the expression of TRIM28/KAP1 is correlated to the body mass index (BMI) (Dalgaard et al., 2016). This work highlights the importance of further research to determine how metastatic epiallele regulation effects human health.
3.2.1. Epigenetic inheritance
A link between metabolic health of parents and the potential for this to impact the metabolic health of offspring has been recognized for some time. For example, a paternal diet (high fat or low protein) or a prior history of intrauterine exposure to maternal caloric restriction can result in increased metabolic risk in offspring (Hales & Barker, 1992). One of the most famous examples of metabolic health affecting subsequent generations is the Dutch Hunger Winter. During the winter of 1944–1945, occupied Netherlands was cut off from supplies and a famine ensued. Pregnant mothers, who experience poor nutrition during the famine, ended up having offspring with increased rates of adiposity, hypertension and glucose tolerance (Lumey et al., 2007), suggesting a link between maternal metabolic health and the health of offspring.
It is also clear that environmental conditions during early development determine susceptibility to disease later in adult organisms, known as the developmental origins of health and disease (DOHaD) hypothesis (Hochberg et al., 2011). Low birth weight is associated with increased risk for metabolic disorders, including obesity and diabetes (Hales & Barker, 1992; McMillen & Robinson, 2005). It has been proposed that this risk can be passed on to future generations (Gluckman, Hanson, & Beedle, 2007). However, it is not clear how this would be mediated through epigenetic mechanisms given germline epigenetic reprograming. These changes in the epigenetic reprograming are differently classified based on how persistent they are in the germline. Environmental pressures on the mother during pregnancy can result in epigenetic changes in the individual, referred to as maternal effects. For example, it has been found in mice that high‐fat or ethinyl‐oestradiol intake during pregnancy increases the risk of the offspring to develop mammary cancer. This effect is then inherited by the offspring of those mice, making this a bona fide case of epigenetic inheritance (de Assis et al., 2012). Epigenetic changes that affect the germ cells of an individual and are thereby passed on one generation are an example of intergenerational epigenetic inheritance. If an epigenetic change persists two or more generations it is known as transgenerational epigenetic inheritance (Figure 5).
Figure 5.
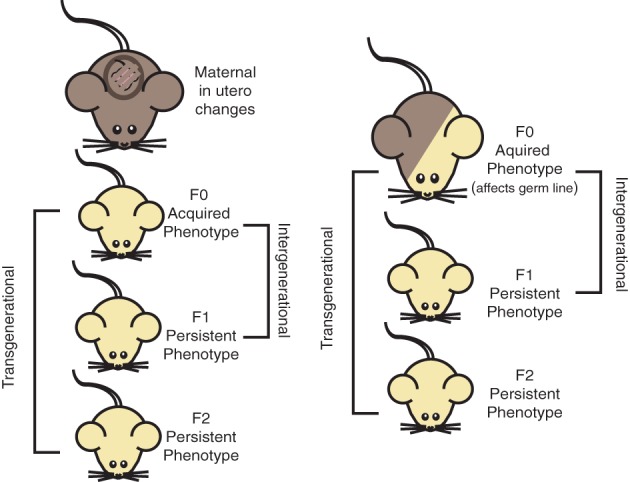
Intergenerational versus transgenerational inheritance. Epigenetic inheritance in mice can be defined by the number of generations the phenotype penetrates. Maternal effects are changes to the offspring caused as a result of changes in the womb, that is, starvation or altered metabolite intake. The children born after these conditions with altered epigenetic structure are the F0 generation. The F0 generation can also be generated if there is some altering effect, that is, obesogenic diet, that alter the germs cells of an individual. If the phenotype is successfully passed on from the F0 generation to their offspring, the F1 generation, the phenotype is viewed as an intergenerational phenotype. If the phenotype is successfully transferred from the F1 generation to the F2 generation and beyond, the phenotype is viewed as a transgenerational phenotype
Transgenerational inheritance of metabolic disease in mice has been well documented. This process could potentially be regulated by the methylation state of metastable epialleles (Rakyan et al., 2002), which can then be passed on through multiple generations (Cropley et al., 2016). One potential method for transmission of metabolic phenotypes to offspring is from epigenetic modification of germ cells. For example, DNA methylation of sperm is extensively remodeled in obesity and can be altered by bariatric surgery (Donkin et al., 2016). RNAs, specifically sRNA, have been shown to have altered levels in sperm in obese males compared to their lean counterparts. Two groups have demonstrated that altered levels of tRFs lead to alterations in gene and transposable element expression (Chen et al., 2016; Sharma et al., 2016). These altered tRFs were shown to be able to induce glucose intolerance independent of the modifications on Sperm DNA. tRFs have been shown to specifically target TEs and metastable epialleles are known to be mediated though TEs hypo‐ or hyper‐methylation, indicating that tRFs are likely candidates for the mediation of epigenetic phenotypes to offspring (Figure 6) (Lane et al., 2003; Schorn et al., 2017). Additionally, other sRNAs have been implicated in the inheritance of phenotypic traits from fathers. Treatment of embryos with miRNA from the Kit−/− mouse, a mouse model with a unique coat coloring that is regulated through small RNA, causes the inheritance of its deregulated pigment phenotype. For both forms of RNA‐mediated inheritance, posttranscriptional modifications are required for intergeneration epigenetic changes(Kiani et al., 2013). This was shown with the miRNA losing its transmissible effect when DNMT2 is knocked out and unmodified tRFs losing their ability to induce metabolic phenotypes. Together this evidence implies that male epigenetic inheritance can be mediated through transmission of small RNA.
Figure 6.
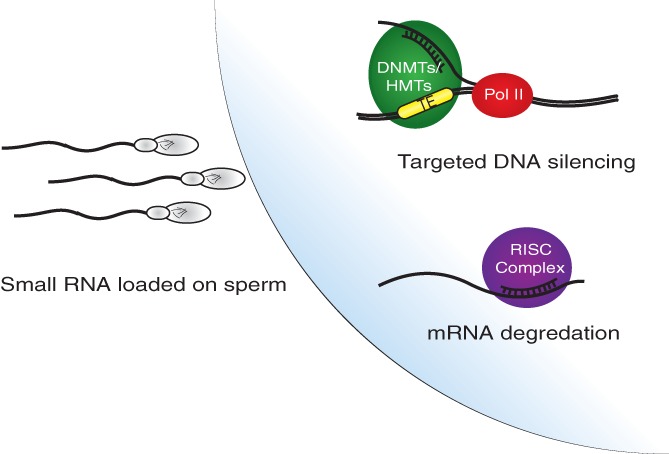
Small RNA‐mediated regulation of gene expression from parental small RNA. Parental small RNAs, in a function independent of the genomic information in sperm, have been shown to transmit phenotypes to offspring. sRNAs can complex to RNA transcripts and induce degradation. Alternatively, sRNAs could bind to actively transcribed RNA in the genome and cause the recruitment of DNA and histone modifying proteins. This could lead to targeted heterochromatin formation in the genome
4. FUTURE DIRECTIONS
The interaction of environmental factors and chromatin modifications—and the impact this interaction can have on metabolic disease—is a rapidly evolving field. Understanding the basic biology underlying how environmental factors can impact chromatin modifications as well as examining how chromatin modifications promote disease progression are important next steps for this field. Much of the advances in these areas are being driven by technological innovation, such as improvements in high‐throughput sequencing.
4.1. Epigenome‐wide association studies
Like genome‐wide association studies (GWAS), epigenome‐wide association studies (EWAS) employ a method of screening populations of individuals to examine differences in methylation profiles. Just as in GWAS, where it was found that variation in the FTO gene correlates with an individual's BMI (Claussnitzer et al., 2015), EWAS has been used to determine the effects that high fat diet and obesity has on the chromatin structure. It has been found that in individuals consuming high fat diet have altered DNA methylation at genes involved in lipid and lipoprotein metabolism, transportation of substrates, and inflammatory response compared to their lean peers (Wahl et al., 2017). EWAS studies have also been used to determine how altered methylation profiles can be used as an indicator for the development of type 2 diabetes (Wahl et al., 2017). While EWAS studies have the potential to improve our understanding of the role of epigenetic variation in human disease, given the dynamic and reversible nature of DNA methylation as well as the great variability between individuals, care must be given to interpretation of results (Lappalainen & Greally, 2017).
4.1.1. Epigenetic‐driven personalized medicine
As described above, tight regulation and maintenance of epigenetic modifications is essential for normal development and alterations to the epigenome are associated with many diseases. Due to changes in the epigenome, patients who suffer from the same disease can have a variable response to the same treatment. With the advent of personalized medicine, patients can be treated based upon their genetic signatures. However, only targeting diseases based on genetic mutations misses the complexity of the epigenome. To truly harness the full potential of personalized medicine, both genetic and epigenetic information will be required. Recent advances in determining the role of metabolism and altered gene transcription in the blood have been made to increase the efficacy of applying personalized medicine to the treatment of immunological disorders (reviewed in Li, Todor, & Luo, 2016). Harnessing epigenetic studies to apply to personalized medicine has the potential to revolutionize standard of care for many diseases (reviewed in Rasool et al., 2015).
5. CONCLUSIONS
In this review, we have considered evidence that environmental factors, such as obesogenic diets, can contribute to modifications to chromatin. We have furthermore examined evidence that the chromatin modifications associated with metabolic diseases can lead to long‐term disease risk. Finally, we discussed the potential impact of inherited chromatin modifications on the health of future generations.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
http://dx.doi.org/10.1002/wsbm.1287
ACKNOWLEDGMENTS
The authors would like to thank Amy Leung and members of the Schones Laboratory for helpful discussion and comments. We gratefully acknowledge funding support from the National Institutes of Health: R01DK112041, R01CA220693 and R56DK071122.
Costello KR, Schones DE. Chromatin modifications in metabolic disease: Potential mediators of long‐term disease risk. WIREs Syst Biol Med. 2018;10:e1416. https://doi.org/10.1002/wsbm.1416
Funding information National Institutes of Health, Grant/Award numbers: R56DK071122, R01CA220693, R01DK112041
References
REFERENCES
- Ahima, R. S. (2009). Connecting obesity, aging and diabetes. Nature Medicine, 15(9), 996–997. https://doi.org/10.1038/nm0909-996 [DOI] [PubMed] [Google Scholar]
- Anderson, J. W. , Konz, E. C. , Frederich, R. C. , & Wood, C. L. (2001). Long‐term weight‐loss maintenance: A meta‐analysis of US studies. American Journal of Clinical Nutrition, 74(5), 579–584. [DOI] [PubMed] [Google Scholar]
- Banerjee, S. , Bennion, G. R. , Goldberg, M. W. , & Allen, T. D. (1991). ATP dependent histone phosphorylation and nucleosome assembly in a human cell free extract. Nucleic Acids Research, 19(21), 5999–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A. J. , & Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21(3), 381–395. https://doi.org/10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor, T. H. (2000). The DNA methyltransferases of mammals. Human Molecular Genetics, 9(16), 2395–2402. [DOI] [PubMed] [Google Scholar]
- Carmell, M. A. , Girard, A. , van de Kant, H. J. , Bourc'his, D. , Bestor, T. H. , de Rooij, R. G. , & Hannon, G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell, 12(4), 503–514. https://doi.org/10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Yan, M. , Cao, Z. , Li, X. , Zhang, Y. , Shi, J. , … Zhou, Q. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science, 351(6271), 397–400. https://doi.org/10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Clapier, C. R. , Iwasa, J. , Cairns, B. R. , & Peterson, C. L. (2017). Mechanisms of action and regulation of ATP‐dependent chromatin‐remodelling complexes. Nature Reviews. Molecular Cell Biology, 18(7), 407–422. https://doi.org/10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer, M. , Dankel, S. N. , Kim, K.‐H. , Quon, G. , Meuleman, W. , Haugen, C. , … Kellis, M. (2015). FTO obesity variant circuitry and adipocyte browning in humans. New England Journal of Medicine, 373(10), 895–907. https://doi.org/10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H. Y. , Miller, C. , Bitterman, K. J. , Wall, N. R. , Hekking, B. , Kessler, B. , … Sinclair, D. A. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science, 305(5682), 390–392. https://doi.org/10.1126/science.1099196 [DOI] [PubMed] [Google Scholar]
- Cole, J. J. , Robertson, N. A. , Rather, M. I. , Thomson, J. P. , McBryan, T. , Sproul, D. , … Adams, P. D. (2017). Diverse interventions that extend mouse lifespan suppress shared age‐associated epigenetic changes at critical gene regulatory regions. Genome Biology, 18, 58 https://doi.org/10.1186/s13059-017-1185-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, S. , Martin, T. L. , Surwit, R. S. , & Robidoux, J. (2004). Genetic vulnerability to diet‐induced obesity in the C57BL/6J mouse: Physiological and molecular characteristics. Physiology & Behavior, 81(2), 243–248. https://doi.org/10.1016/j.physbeh.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Colman, R. J. , Beasley, T. M. , Kemnitz, J. W. , Johnson, S. C. , Weindruch, R. , & Anderson, R. M. (2014). Caloric restriction reduces age‐related and all‐cause mortality in rhesus monkeys. Nature Communications, 5, 3557 https://doi.org/10.1038/ncomms4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropley, J. E. , Eaton, S. A. , Aiken, A. , Young, P. E. , Giannoulatou, E. H. , Joshua, W. K. , … Suter, C. M. (2016). Male‐lineage transmission of an acquired metabolic phenotype induced by grand‐paternal obesity. Molecular Metabolism, 5(8), 699–708. https://doi.org/10.1016/j.molmet.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J. , Wang, Z. , Xu, W. , Zhang, M. , Zhu, Z. , Zhao, X. , … Qiao, Z. (2017). Paternal nicotine exposure defines different behavior in subsequent generation via hyper‐methylation of mmu‐miR‐15b. Scientific Reports, 7(1), 7286 https://doi.org/10.1038/s41598-017-07920-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, K. , Landgraf, K. , Heyne, S. , Lempradl, A. , Longinotto, J. , Gossens, K. , … Pospisilik, J. A. (2016). Trim28 haploinsufficiency triggers bi‐stable epigenetic obesity. Cell, 164(3), 353–364. https://doi.org/10.1016/j.cell.2015.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis, S. , Warri, A. , Cruz, M. I. , Laja, O. , Tian, Y. , Zhang, B. , … Hilakivi‐Clarke, L. (2012). High‐fat or ethinyl‐oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring. Nature Communications, 3, 1053 https://doi.org/10.1038/ncomms2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas, V. , Stokes, D. G. , & Perry, R. P. (1993). A mammalian DNA‐binding protein that contains a chromodomain and an SNF2/SWI2‐like helicase domain. Proceedings of the National Academy of Sciences of the United States of America, 90(6), 2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhahbi, J. M. , Spindler, S. R. , Atamna, H. , Yamakawa, A. , Boffelli, D. , Mote, P. , & Martin, D. I. K. (2013). 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics, 14, 298–298. https://doi.org/10.1186/1471-2164-14-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy, D. C. , Das, R. , Weidman, J. R. , & Jirtle, R. L. (2007). Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatric Research, 61(5 Part 2), 30R–37R. https://doi.org/10.1203/pdr.0b013e31804575f7 [DOI] [PubMed] [Google Scholar]
- Donkin, I. , Versteyhe, S. , Ingerslev, L. R. , Qian, K. , Mechta, M. , Nordkap, L. , … Barres, R. (2016). Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metabolism, 23(2), 369–378. https://doi.org/10.1016/j.cmet.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Du, Z. , Sun, T. , Hacisuleyman, E. , Fei, T. , Wang, X. , Brown, M. , … Liu, X. S. (2016). Integrative analyses reveal a long noncoding RNA‐mediated sponge regulatory network in prostate cancer. Nature Communications, 7, 10982 https://doi.org/10.1038/ncomms10982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J. R. , Yarychkivska, O. , Boulard, M. , & Bestor, T. H. (2017). DNA methylation and DNA methyltransferases. Epigenetics & Chromatin, 10, 23 https://doi.org/10.1186/s13072-017-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. M. (2009). Obesity: Causes and control of excess body fat. Nature, 459(7245), 340–342. https://doi.org/10.1038/459340a [DOI] [PubMed] [Google Scholar]
- Galdieri, L. , & Vancura, A. (2012). Acetyl‐CoA carboxylase regulates global histone acetylation. Journal of Biological Chemistry, 287(28), 23865–23876. https://doi.org/10.1074/jbc.M112.380519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Wu, F. , Zhou, J. , Yan, L. , Jurczak, M. J. , Lee, H. Y. , … Huang, Y. (2014). The H19/let‐7 double‐negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Research, 42(22), 13799–13811. https://doi.org/10.1093/nar/gku1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman, P. D. , Hanson, M. A. , & Beedle, A. S. (2007). Non‐genomic transgenerational inheritance of disease risk. BioEssays, 29(2), 145–154. https://doi.org/10.1002/bies.20522 [DOI] [PubMed] [Google Scholar]
- Goll, M. G. , Kirpekar, F. , Maggert, K. A. , Yoder, J. A. , Hsieh, C. L. , Zhang, X. , … Bestor, T. H. (2006). Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science, 311(5759), 395–398. https://doi.org/10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- Green, C. D. , Huang, Y. , Dou, X. , Yang, L. , Liu, Y. , & Han, J.‐D. J. (2017). Impact of dietary interventions on noncoding RNA networks and mRNAs encoding chromatin‐related factors. Cell Reports, 18(12), 2957–2968. https://doi.org/10.1016/j.celrep.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Guo, J. , Jou, W. , Gavrilova, O. , & Hall, K. D. (2009). Persistent diet‐induced obesity in male C57BL/6 mice resulting from temporary obesigenic diets. PLoS One, 4(4), e5370 https://doi.org/10.1371/journal.pone.0005370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett, J. A. , Sengupta, R. , Zylicz, J. J. , Murakami, K. , Lee, C. , Down, T. A. , & Surani, M. A. (2013). Germline DNA demethylation dynamics and imprint erasure through 5‐hydroxymethylcytosine. Science, 339(6118), 448–452. https://doi.org/10.1126/science.1229277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, C. N. , & Barker, D. J. (1992). Type 2 (non‐insulin‐dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia, 35(7), 595–601. [DOI] [PubMed] [Google Scholar]
- Harris, R. A. , Nagy‐Szakal, D. , & Kellermayer, R. (2013). Human metastable epiallele candidates link to common disorders. Epigenetics, 8(2), 157–163. https://doi.org/10.4161/epi.23438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata, K. , Okano, M. , Lei, H. , & Li, E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development, 129(8), 1983–1993. [DOI] [PubMed] [Google Scholar]
- Hawkins, P. G. , Santoso, S. , Adams, C. , Anest, V. , & Morris, K. V. (2009). Promoter targeted small RNAs induce long‐term transcriptional gene silencing in human cells. Nucleic Acids Research, 37(9), 2984–2995. https://doi.org/10.1093/nar/gkp127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg, Z. , Feil, R. , Constancia, M. , Fraga, M. , Junien, C. , Carel, J. C. , … Albertsson‐Wikland, K. (2011). Child health, developmental plasticity, and epigenetic programming. Endocrine Reviews, 32(2), 159–224. https://doi.org/10.1210/er.2009-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, R. , & Pugh, J. E. (1975). DNA modification mechanisms and gene activity during development. Science, 187(4173), 226–232. [PubMed] [Google Scholar]
- Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10), R115 https://doi.org/10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ions, L. J. , Wakeling, L. A. , Bosomworth, H. J. , Hardyman, J. E. , Escolme, S. M. , Swan, D. C. , … Ford, D. (2013). Effects of Sirt1 on DNA methylation and expression of genes affected by dietary restriction. Age (Dordrecht, Netherlands), 35(5), 1835–1849. https://doi.org/10.1007/s11357-012-9485-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, S. , Shen, L. , Dai, Q. , Wu, S. C. , Collins, L. B. , Swenberg, J. A. , … Zhang, Y. (2011). Tet proteins can convert 5‐methylcytosine to 5‐formylcytosine and 5‐carboxylcytosine. Science, 333(6047), 1300–1303. https://doi.org/10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja, W. W. , Carvalho, G. B. , Zid, B. M. , Mak, E. M. , Brummel, T. , & Benzer, S. (2009). Water‐ and nutrient‐dependent effects of dietary restriction on Drosophila lifespan. Proceedings of the National Academy of Sciences, 106(44), 18633–18637. https://doi.org/10.1073/pnas.0908016106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, C.‐W. , Shibata, Y. , Starmer, J. , Yee, D. , & Magnuson, T. (2015). Histone H3.3 maintains genome integrity during mammalian development. Genes & Development, 29(13), 1377–1392. https://doi.org/10.1101/gad.264150.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, B. , & Robertson, K. D. (2013). DNA methyltransferases (DNMTs), DNA damage repair, and cancer. Advances in Experimental Medicine and Biology, 754, 3–29. https://doi.org/10.1007/978-1-4419-9967-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C. , & Felsenfeld, G. (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes & Development, 21(12), 1519–1529. https://doi.org/10.1101/gad.1547707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C. , Zang, C. , Wei, G. , Cui, K. , Peng, W. , Zhao, K. , & Felsenfeld, G. (2009). H3.3/H2A.Z double variant‐containing nucleosomes mark ‘nucleosome‐free regions’ of active promoters and other regulatory regions. Nature Genetics, 41(8), 941–945. https://doi.org/10.1038/ng.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating, S. T. , & El‐Osta, A. (2015). Epigenetics and metabolism. Circulation Research, 116(4), 715–736. https://doi.org/10.1161/CIRCRESAHA.116.303936 [DOI] [PubMed] [Google Scholar]
- Khalil, H. , Tazi, M. , Caution, K. , Ahmed, A. , Kanneganti, A. , Assani, K. , … Amer, A. O. (2016). Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics, 11(5), 381–388. https://doi.org/10.1080/15592294.2016.1144007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarahmi, M. , & Azadbakht, L. (2014). The association between different kinds of fat intake and breast cancer risk in women. International Journal of Preventive Medicine, 5(1), 6–15. [PMC free article] [PubMed] [Google Scholar]
- Kiani, J. , Grandjean, V. , Liebers, R. , Tuorto, F. , Ghanbarian, H. , Lyko, F. , … Rassoulzadegan, M. (2013). RNA‐mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genetics, 9(5), e1003498 https://doi.org/10.1371/journal.pgen.1003498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, R. J. , Kallin, E. M. , & Zhang, Y. (2006). JmjC‐domain‐containing proteins and histone demethylation. Nature Reviews Genetics, 7(9), 715–727. [DOI] [PubMed] [Google Scholar]
- Kramer, F. M. , Jeffery, R. W. , Forster, J. L. , & Snell, M. K. (1989). Long‐term follow‐up of behavioral treatment for obesity: Patterns of weight regain among men and women. International Journal of Obesity, 13(2), 123–136. [PubMed] [Google Scholar]
- Kühnen, P. , Handke, D. , Waterland, R. A. , Hennig, B. J. , Silver, M. , Fulford, A. J. , … Krude, H. (2016). Interindividual variation in DNA methylation at a putative POMC metastable epiallele is associated with obesity. Cell Metabolism, 24(3), 502–509. https://doi.org/10.1016/j.cmet.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Kumar, P. , Kuscu, C. , & Dutta, A. (2016). Biogenesis and function of transfer RNA‐related fragments (tRFs). Trends in Biochemical Sciences, 41(8), 679–689. https://doi.org/10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski, B. , & Hekimi, S. (1998). The genetics of caloric restriction in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 95(22), 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, D. D. , de Souza, F. S. , Nasif, S. , Yamashita, M. , Lopez‐Leal, R. , Otero‐Corchon, V. , … Low, M. J. (2015). Partially redundant enhancers cooperatively maintain mammalian pomc expression above a critical functional threshold. PLoS Genetics, 11(2), e1004935 https://doi.org/10.1371/journal.pgen.1004935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, N. , Dean, W. , Erhardt, S. , Hajkova, P. , Surani, A. , Walter, J. , & Reik, W. (2003). Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis (New York), 35(2), 88–93. https://doi.org/10.1002/gene.10168 [DOI] [PubMed] [Google Scholar]
- Lappalainen, T. , & Greally, J. M. (2017). Associating cellular epigenetic models with human phenotypes. Nature Reviews Genetics, 18(7), 441–451. https://doi.org/10.1038/nrg.2017.32 [DOI] [PubMed] [Google Scholar]
- Lee, Y. S. , Shibata, Y. , Malhotra, A. , & Dutta, A. (2009). A novel class of small RNAs: tRNA‐derived RNA fragments (tRFs). Genes & Development, 23(22), 2639–2649. https://doi.org/10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A. , Parks, B. W. , Du, J. , Trac, C. , Setten, R. , Chen, Y. , … Schones, D. E. (2014). Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. Journal of Biological Chemistry, 289(34), 23557–23567. https://doi.org/10.1074/jbc.M114.581439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A. , Trac, C. , Du, J. , Natarajan, R. , & Schones, D. E. (2016). Persistent chromatin modifications induced by high fat diet. Journal of Biological Chemistry, 291(20), 10446–10455. https://doi.org/10.1074/jbc.M115.711028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Todor, A. , & Luo, R. (2016). Blood transcriptomics and metabolomics for personalized medicine. Computational and Structural Biotechnology Journal, 14, 1–7. https://doi.org/10.1016/j.csbj.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, N. , Shevchenko, G. , Walshe, J. L. , Groen, J. , Johnsson, P. , Vidarsdóttir, L. , … Morris, K. V. (2017). The molecular dynamics of long noncoding RNA control of transcription in PTEN and its pseudogene. Proceedings of the National Academy of Sciences, 114(37), 9942–9947. https://doi.org/10.1073/pnas.1621490114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey, L. H. , Stein, A. D. , Kahn, H. S. , van der Pal‐de Bruin, K. M. , Blauw, G. J. , Zybert, P. A. , & Susser, E. S. (2007). Cohort profile: The Dutch Hunger Winter Families Study. International Journal of Epidemiology, 36(6), 1196–1204. https://doi.org/10.1093/ije/dym126 [DOI] [PubMed] [Google Scholar]
- Mattison, J. A. , Roth, G. S. , Beasley, T. M. , Tilmont, E. M. , Handy, A. M. , Herbert, R. L. , … de Cabo, R. (2012). Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature, 489(7415), 318–321. https://doi.org/10.1038/nature11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen, I. C. , & Robinson, J. S. (2005). Developmental origins of the metabolic syndrome: Prediction, plasticity, and programming. Physiological Reviews, 85(2), 571–633. https://doi.org/10.1152/physrev.00053.2003 [DOI] [PubMed] [Google Scholar]
- Mercer, T. R. , & Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nature Structural & Molecular Biology, 20, 300–307. https://doi.org/10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- Multhaup, M. L. , Seldin, M. M. , Jaffe, A. E. , Lei, X. , Kirchner, H. , Mondal, P. , … Feinberg, A. P. (2015). Mouse‐human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metabolism, 21(1), 138–149. https://doi.org/10.1016/j.cmet.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel, M. , Globisch, D. , Brückl, T. , Wagner, M. , Welzmiller, V. , Michalakis, S. , … Carell, T. (2010). Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angewandte Chemie International Edition, 49(31), 5375–5377. https://doi.org/10.1002/anie.201002033 [DOI] [PubMed] [Google Scholar]
- Neigeborn, L. , & Carlson, M. (1984). Genes affecting the regulation of gene expression by glucose repression in Saccharomyces cerevisiae . Genetics, 108(4), 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary, V. B. , Hain, S. , Maugg, D. , Smida, J. , Azimzadeh, O. , Tapio, S. , … Atkinson, M. J. (2017). Long non‐coding RNA particle bridges histone and DNA methylation. Scientific Reports, 7(1), 1790 https://doi.org/10.1038/s41598-017-01875-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, M. , Bell, D. W. , Haber, D. A. , & Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99(3), 247–257. [DOI] [PubMed] [Google Scholar]
- Petry, C. J. , Evans, M. L. , Wingate, D. L. , Ong, K. K. , Reik, W. , Constancia, M. , & Dunger, D. B. (2010). Raised late pregnancy glucose concentrations in mice carrying pups with targeted disruption of H19delta13. Diabetes, 59(1), 282–286. https://doi.org/10.2337/db09-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela, A. , & Esteller, M. (2010). Epigenetic modifications and human disease. Nature Biotechnology, 28(10), 1057–1068. https://doi.org/10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- Ptashne, M. (2007). On the use of the word ‘epigenetic’. Current Biology, 17(7), R233–R236. https://doi.org/10.1016/j.cub.2007.02.030 [DOI] [PubMed] [Google Scholar]
- Qi, Y. , Zhu, Q. , Zhang, K. , Thomas, C. , Wu, Y. , Kumar, R. , … Guo, S. (2015). Activation of Foxo1 by insulin resistance promotes cardiac dysfunction and β‐myosin heavy chain gene expression. Circulation: Heart Failure, 8(1), 198–208. https://doi.org/10.1161/circheartfailure.114.001457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan, V. K. , Blewitt, M. E. , Druker, R. , Preis, J. I. , & Whitelaw, E. (2002). Metastable epialleles in mammals. Trends in Genetics, 18(7), 348–351. https://doi.org/10.1016/S0168-9525(02)02709-9 [DOI] [PubMed] [Google Scholar]
- Rasool, M. , Malik, A. , Naseer, M. I. , Manan, A. , Ansari, S. A. , Begum, I. , … Gan, S. H. (2015). The role of epigenetics in personalized medicine: Challenges and opportunities. BMC Medical Genomics, 8(Suppl. 1), S1–S5. https://doi.org/10.1186/1755-8794-8-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S. , Eisenhaber, F. , O'Carroll, D. , Strahl, B. D. , Sun, Z. W. , Schmid, M. , … Jenuwein, T. (2000). Regulation of chromatin structure by site‐specific histone H3 methyltransferases. Nature, 406(6796), 593–599. https://doi.org/10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Riggs, A. D. (1975). X inactivation, differentiation, and DNA methylation. Cytogenetics and Cell Genetics, 14(1), 9–25. [DOI] [PubMed] [Google Scholar]
- Riggs, A. D. , Martienssen, R. A. , & Russo, V. E. A. (1996). Introduction In Epigenetic mechanisms of gene regulation (pp. 1–4). Cold Spring Harbour, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Rosenbaum, M. , Hirsch, J. , Gallagher, D. A. , & Leibel, R. L. (2008). Long‐term persistence of adaptive thermogenesis in subjects who have maintained a reduced body weight. American Journal of Clinical Nutrition, 88(4), 906–912. [DOI] [PubMed] [Google Scholar]
- Schorn, A. J. , Gutbrod, M. J. , LeBlanc, C. , & Martienssen, R. (2017). LTR‐retrotransposon control by tRNA‐derived small RNAs. Cell, 170(1), 61–71.e11. https://doi.org/10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger, S. , Andrews, S. , Krueger, F. , Arand, J. , Walter, J. , Santos, F. , … Reik, W. (2012). The dynamics of genome‐wide DNA methylation reprogramming in mouse primordial germ cells. Molecular Cell, 48(6), 849–862. https://doi.org/10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, U. , Conine, C. C. , Shea, J. M. , Boskovic, A. , Derr, A. G. , Bing, X. Y. , … Rando, O. J. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science, 351(6271), 391–396. https://doi.org/10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. , Mizuguchi, G. , Hamiche, A. , & Wu, C. (2000). A chromatin remodelling complex involved in transcription and DNA processing. Nature, 406(6795), 541–544. https://doi.org/10.1038/35020123 [DOI] [PubMed] [Google Scholar]
- Shibutani, T. , Ito, S. , Toda, M. , Kanao, R. , Collins, L. B. , Shibata, M. , … Kuraoka, I. (2014). Guanine‐ 5‐carboxylcytosine base pairs mimic mismatches during DNA replication. Scientific Reports, 4, 5220 https://doi.org/10.1038/srep05220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek, M. , Varticovski, L. , Yang, S. , Baek, S. , Nielsen, R. , Mandrup, S. , … Grontved, L. (2017). High fat diet‐induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Scientific Reports, 7, 40220 https://doi.org/10.1038/srep40220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs, T. M. , Bonder, M. J. , Stark, A.‐K. , Krueger, F. , von Meyenn, F. , Stegle, O. , & Reik, W. (2017). Multi‐tissue DNA methylation age predictor in mouse. Genome Biology, 18(1), 68 https://doi.org/10.1186/s13059-017-1203-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumithran, P. , Prendergast, L. A. , Delbridge, E. , Purcell, K. , Shulkes, A. , Kriketos, A. , & Proietto, J. (2011). Long‐term persistence of hormonal adaptations to weight loss. The New England Journal of Medicine, 365(17), 1597–1604. https://doi.org/10.1056/NEJMoa1105816 [DOI] [PubMed] [Google Scholar]
- Sun, X. , & Wong, D. (2016). Long non‐coding RNA‐mediated regulation of glucose homeostasis and diabetes. American Journal of Cardiovascular Disease, 6(2), 17–25. [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M. , Yasuoka, A. , Shimizu, M. , Saito, Y. , Kumakura, K. , Asakura, T. , & Nagai, T. (2017). Transcriptomic responses of the liver and adipose tissues to altered carbohydrate‐fat ratio in diet: An isoenergetic study in young rats. Genes & Nutrition, 12(1), 10 https://doi.org/10.1186/s12263-017-0558-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. W. , Dietmann, S. , Irie, N. , Leitch, H. G. , Floros, V. I. , Bradshaw, C. R. , … Surani, M. A. (2015). A unique gene regulatory network resets the human germline epigenome for development. Cell, 161(6), 1453–1467. https://doi.org/10.1016/j.cell.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. W. , Kobayashi, T. , Irie, N. , Dietmann, S. , & Surani, M. A. (2016). Specification and epigenetic programming of the human germ line. Nature Reviews Genetics, 17(10), 585–600. https://doi.org/10.1038/nrg.2016.88 [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Wang, J. , Lian, Y. , Fan, C. , Zhang, P. , Wu, Y. , … Zeng, Z. (2017). Linking long non‐coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Molecular Cancer, 16, 42 https://doi.org/10.1186/s12943-017-0612-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama, T. , Palmer, J. , Landel, C. C. , Shiloach, J. , & Wu, C. (1999). Characterization of the imitation switch subfamily of ATP‐dependent chromatin‐remodeling factors in Saccharomyces cerevisiae . Genes & Development, 13(6), 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto, F. , Herbst, F. , Alerasool, N. , Bender, S. , Popp, O. , Federico, G. , … Lyko, F. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. The EMBO Journal, 34(18), 2350–2362. https://doi.org/10.15252/embj.201591382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto, F. , Liebers, R. , Musch, T. , Schaefer, M. , Hofmann, S. , Kellner, S. , … Lyko, F. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nature Structural & Molecular Biology, 19(9), 900–905. [DOI] [PubMed] [Google Scholar]
- van der Heijden, R. A. , Sheedfar, F. , Morrison, M. C. , Hommelberg, P. P. , Kor, D. , Kloosterhuis, N. J. , … Heeringa, P. (2015). High‐fat diet induced obesity primes inflammation in adipose tissue prior to liver in C57BL/6j mice. Aging (Albany NY), 7(4), 256–268. https://doi.org/10.18632/aging.100738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst, Monique, G. P. , Venkiteswaran, M. , Chen, H. , Xu, G.‐L. , Plösch, T. , & Rots, M. G. (2015). Local chromatin microenvironment determines DNMT activity: From DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase. Epigenetics, 10(8), 671–676. https://doi.org/10.1080/15592294.2015.1062204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington, C. H. (1942). Canalization of development and genetic assimilation of acquired characters. Nature, 150, 563–565. [DOI] [PubMed] [Google Scholar]
- Wahl, S. , Drong, A. , Lehne, B. , Loh, M. , Scott, W. R. , Kunze, S. , … Chambers, J. C. (2017). Epigenome‐wide association study of body mass index, and the adverse outcomes of adiposity. Nature, 541(7635), 81–86. https://doi.org/10.1038/nature20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , Tsui, B. , Kreisberg, J. F. , Robertson, N. A. , Gross, A. M. , Yu, M. K. , … Ideker, T. (2017). Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biology, 18, 57 https://doi.org/10.1186/s13059-017-1186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland, R. A. , & Jirtle, R. L. (2003). Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Molecular and Cellular Biology, 23(15), 5293–5300. https://doi.org/10.1128/MCB.23.15.5293-5300.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, C. M. , & Henikoff, S. (2014). Histone variants: Dynamic punctuation in transcription. Genes & Development, 28(7), 672–682. https://doi.org/10.1101/gad.238873.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch, R. , Walford, R. L. , Fligiel, S. , & Guthrie, D. (1986). The retardation of aging in mice by dietary restriction: Longevity, cancer, immunity and lifetime energy intake. Journal of Nutrition, 116(4), 641–654. [DOI] [PubMed] [Google Scholar]
- Whitelaw, N. C. , Chong, S. , Morgan, D. K. , Nestor, C. , Bruxner, T. J. , Ashe, A. , … Whitelaw, E. (2010). Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biology, 11, R111 https://doi.org/10.1186/gb-2010-11-11-r111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzell, M. S. , & Ahren, B. (2004). The high‐fat diet‐fed mouse: A model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes, 53(Suppl. 3), S215–S219. [DOI] [PubMed] [Google Scholar]
- Writing Team for the Diabetes, Control, Complications Trial/Epidemiology of Diabetes, Interventions, & Complications Research Group (2003). Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: The epidemiology of diabetes interventions and complications (EDIC) study. JAMA, 290(16), 2159–2167. https://doi.org/10.1001/jama.290.16.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T. P. , Wang, T. , Seetin, M. G. , Lai, Y. , Zhu, S. , Lin, K. , … Xiao, A. Z. (2016). DNA methylation on N6‐adenine in mammalian embryonic stem cells. Nature, 532(7599), 329–333. https://doi.org/10.1038/nature17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Tan, Y. S. , Aupperlee, M. D. , Langohr, I. M. , Kirk, E. L. , Troester, M. A. , … Haslam, S. Z. (2013). Pubertal high fat diet: Effects on mammary cancer development. Breast Cancer Research, 15(5), R100 https://doi.org/10.1186/bcr3561 [DOI] [PMC free article] [PubMed] [Google Scholar]


