Abstract
The recognition of RNA functions beyond canonical protein synthesis has challenged the central dogma of molecular biology. Indeed, RNA is now known to directly regulate many important cellular processes, including transcription, splicing, translation, and epigenetic modifications. The misregulation of these processes in disease has led to an appreciation of RNA as a therapeutic target. This potential was first recognized in bacteria and viruses, but discoveries of new RNA classes following the sequencing of the human genome have invigorated exploration of its disease-related functions in mammals. As stable structure formation is evolving as a hallmark of mammalian RNAs, the prospect of utilizing small molecules to specifically probe the function of RNA structural domains and their interactions is gaining increased recognition. To date, researchers have discovered bioactive small molecules that modulate phenotypes by binding to expanded repeats, microRNAs, G-quadruplex structures, and RNA splice sites in neurological disorders, cancers, and other diseases. The lessons learned from achieving these successes both call for additional studies and encourage exploration of the plethora of mammalian RNAs whose precise mechanisms of action remain to be elucidated. Efforts towards understanding fundamental principles of small molecule-RNA recognition combined with advances in methodology development should pave the way towards targeting emerging RNA classes such as long non-coding RNAs. Together, these endeavours can unlock the full potential of small molecule-based probing of RNA-regulated processes and enable us to discover new biology and underexplored avenues for therapeutic intervention in human disease.
Graphical abstract
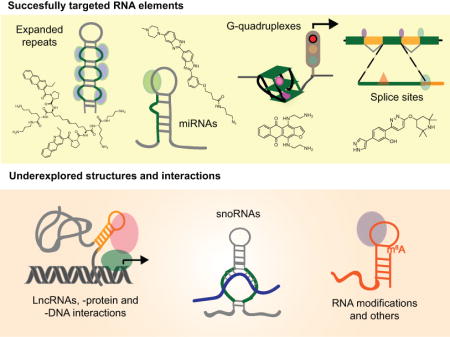
Small molecules have been successfully used to study several mammalian RNA elements in disease. These advances encourage exploration of newly discovered RNA interactions with therapeutic potential.
Introduction
With the advent of next generation sequencing and the characterization of the human genome, the long-standing view of proteins as the only structural and functional building blocks of eukaryotic cells has been challenged.1 Namely, the initiation and findings of the ENCODE and FANTOM projects that aimed to identify the coding elements of the human genome resulted in the unexpected discovery that the majority of human transcripts do not code for proteins and are thus non-coding RNAs.2, 3 It was later found that the number and diversity of these non-coding transcripts increase with organismal complexity, are highest in mammals, and are evolutionarily conserved.4, 5 Highly conserved sequences were also unexpectedly identified in untranslated regions (UTRs) of protein-coding transcripts.6 These discoveries implied a high degree of functionality in noncoding mammalian transcripts and were followed by dedicated efforts to characterize their structure and function.7
Mammalian RNAs as therapeutic targets
Functional studies revealed that mammalian RNAs of varying length and subcellular localization have important roles in nearly all cellular processes (Table 1). RNAs were found to regulate transcription, translation and the epigenetic landscape as well as alter signalling pathways by interacting with individual proteins or protein complexes, lipids, DNA, and other RNAs.5, 8-11 In UTRs, conserved cis-regulatory elements were discovered to serve as binding sites for antisense RNAs and proteins, thus regulating the stability, localization, and translation of mRNAs.12 Importantly, many of these processes are misregulated in a variety of human diseases, ranging from cancers to neurodegenerative and neuromuscular diseases.13-15 The RNAs involved in regulating these processes were found to be differentially expressed in disease states. Together, these discoveries have led to an interest in mammalian RNAs as therapeutic targets.16, 17 The prospect of targeting a new class of biomolecules and thus expanding the “druggable” human genome was met with excitement by the scientific community, resulting in increased efforts towards developing adequate tools to study the druggability of these transcripts.
Table 1.
Classes of mammalian RNAs and their lengths, localization, and function.
| RNA Class | Length | Subcellular Localization | Function |
|---|---|---|---|
| Micro RNA (miRNA) | 20-25 nt | Cytoplasm | Decay of target mRNA |
|
Piwi-interacting RNA (piRNA) |
26-31 nt | Nucleus | Directing chromatin modification to repress transcription |
|
Transfer RNA (tRNA) |
70-90 nt | Cytoplasm | Decoding mRNA sequence during translation |
|
Small nucleolar RNA (snoRNA) |
60-200 nt | Nucleus | Guide for pre-ribosomal RNA processing and modification |
|
Small nuclear RNA (snRNA) |
100-300 nt | Nucleus | Splicing |
|
Long non-coding RNA (lncRNA) |
>200 nt | Nucleus and Cytoplasm | Transcriptional, epigenetic, and translational control |
|
Ribosomal RNA (rRNA) |
2-5 kb | Cytoplasm | Major component of translation machinery |
|
Messenger RNA (mRNA) |
2-5 kb | Nucleus and Cytoplasm | Contains regions that both regulate and code for protein synthesis |
Abbreviations: nt, nucleotide; kb, kilobase.
Oligonucleotides as Sequence-targeted Tools for Studying RNA Function in Mammalian Systems
Most early successes to target mammalian RNAs were achieved with oligonucleotides, which rely on complementary base-pairing with the RNA target of interest in order to degrade or functionally inhibit the transcript.18, 19 Antisense oligonucleotides (ASOs), for example, were indispensable for validating the therapeutic potential of many RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) in cancer and kidney disease, splice sites in Duchenne muscular dystrophy and spinal muscular atrophy, and expanded RNA repeats in Friedrich ataxia.16 RNA-interference- and CRISPR-Cas9-based methods have also become essential tools in studying RNA function, and several recent reviews highlight the properties, utilities, and limitations of these techniques.18, 20, 21 The potential of oligonucleotide-based technologies is increasingly recognized by the pharmaceutical industry, especially as the field begins to find solutions to challenges with stability and tissue permeability in vivo.22, 23 Indeed, several companies have successfully placed ASO-based therapeutics in clinical trials, four of which have obtained FDA approval.24, 25
The sequence-based recognition of RNA by oligonucleotides requires the targeted regions to be accessible to oligonucleotide binding.26, 27 Hence, regions of RNA that form defined structures are difficult to target with these tools.27 For instance, in a recent study of expanded repeat RNAs that cause fragile X-associated tremor ataxia syndrome (FXTAS), the ASO utilized was incapable of reversing FXTAS-associated splicing defects and even displayed some deleterious effects.28 The authors attributed this effect to the significant energetic barrier for ASO binding to the self-complementary structure formed by the expanded repeat RNA. Additionally, ASOs were shown to have limited success in binding RNA regions with low sequence complexity but defined tertiary structure.29 These drawbacks highlight the need for complementary tools that could enable the interrogation of highly structured RNA regions in disease.
Small Molecules as Structure-targeted Tools for Studying RNA Function in Mammalian Systems
For decades, drug-like small molecules have served as invaluable tools for studying structure-function relationships and therapeutic targeting of proteins.30-32 While the field of protein-targeted drug design has flourished for over 50 years, similar efforts towards RNA targeting begun only in the late 1980’s after the realization that several classes of FDA-approved antibacterial drugs exert their function by binding to bacterial ribosomal RNA.33 In the years to follow, the appreciation of RNA as a key regulator and target in both bacterial and viral infections significantly increased.34-37 In 1998, Czarnik and co-workers reported one of the first small molecule inhibitors of the Trans-Activation Response element (TAR) RNA and Trans-activator of transcription (Tat) protein complex that led to a reduction of HIV-1 replication.38 A notable recent example resulted from a phenotypic screening campaign conducted by Merck, which led to a surprising discovery of a small molecule inhibitor of a bacterial riboflavin riboswitch, one of the first riboswitch-targeting ligands with an antibacterial effect in a mouse septicaemia model.39 Successful targeting of viral and bacterial RNAs has served as proof-of-principle to encourage exploration of endogenous structures in mammals.7, 40
As compared to oligonucleotide-based techniques, small molecules offer the unique possibility to target specific RNA structural elements. The prospect of utilizing small molecules to probe the function of distinct structures is particularly exciting given the recent recognition of stable structure formation as a hallmark of mammalian non-coding RNA.41-44 These structures are known to exist in various cellular compartments, interact specifically with other biomolecules, and have major effects on the outcome of various cellular events (Figure 1).45
Figure 1.
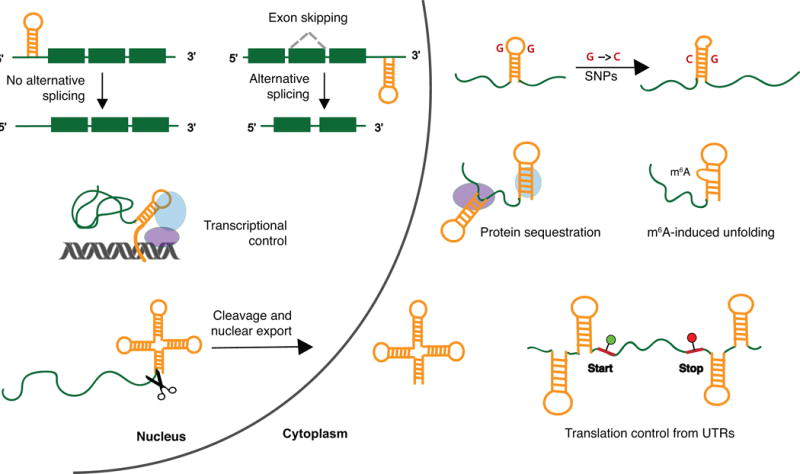
Example cellular processes and interactions regulated by RNA structures in non-coding regions. Left: In the nucleus, structured regions in pre-mRNAs can regulate alternative splicing. Structured non-coding RNAs can recruit transcription factors to genomic loci, interacting with both proteins and DNA. Formation of tRNA-like structures can promote cleavage from a longer transcript and export of the RNA fragment into the cytoplasm. Right: In both the nucleus and the cytoplasm, sequence-based changes such as single nucleotide polymorphisms (SNPs) or modifications such as m6A can alter RNA structure, which can in turn affect RNA function or protein binding. Non-coding RNAs can sequester proteins in specific cytoplasmic regions. RNA structures in UTRs can limit translation rates by impeding the initiation step. Despite the simple hairpin structures shown for clarity, cellular RNAs are known to adopt various complex structures. Adapted from Bevilacqua et al. 2016.45 Abbreviations: SNP, single nucleotide polymorphism; m6A, 6-methyladenosine; UTR, untranslated region.
From a pharmaceutical industry perspective, RNA-targeted chemical probes can assist in establishing tractability as well as clinical translatability of a specific RNA target.46 The tunability of small molecules, in addition to their generally better cellular and tissue distribution in vivo poses them as an attractive alternative strategy to oligonucleotide-based techniques.47 For certain diseases such as neuromuscular disorders, small molecule-based therapies are advantageous over ASO-based therapies because ASO delivery to muscle or the central nervous system remains challenging with a few notable exceptions.48-50 As a result, the small molecule-based targeting of RNA in disease is an emerging avenue of exploration in industry, and several companies aiming to expand the scope of small molecule drug targets with RNA were recently established.51
In this review, we will survey disease-associated mammalian RNA classes that have successfully been targeted with drug-like small molecules in a biological setting, including cell culture and animal models. Aminoglycosides and peptides will be excluded as they have different physicochemical properties from the particular classes of small molecules reviewed herein and have been extensively surveyed elsewhere.35, 52-57 Additionally, we note that several excellent and recent reviews highlight a subset of the small molecules described here, as well as those targeting bacterial and viral RNA, while surveying the drug-like properties and discovery methods in various in vitro and biological systems.35, 52, 53, 58-61 This review will thus place a unique emphasis on the specific RNA structural elements or RNA-mediated interactions that enable disease-related functions in mammalian systems and the phenotypic changes observed upon treatment with targeted ligands. Further, we will draw attention to underexplored mammalian RNA targets in which chemical probe development can aid our understanding of their precise mechanism of action and therapeutic potential. Finally, we will discuss challenges as well as solutions that can advance the field and realize the full potential of the RNA revolution.
MAMMALIAN RNAs TARGETED WITH BIOACTIVE SMALL MOLECULES
Selectivity is undoubtedly the largest challenge in RNA targeting. The majority of the RNA mass in mammalian cells is composed of ribosomal and transfer RNA (80-90 and 10-15 %, respectively).62 Their high abundance, defined structure, and cytoplasmic localization have rendered rRNA and tRNA as major obstacles for achieving selectivity when targeting other, less abundant RNA.63 Nevertheless, reports of selective recognition of less abundant RNA with phenotypic outcomes in cells and in vivo have risen over the recent decade, and efforts to understand the basis of this selectivity are ongoing.58, 64-66 To date, small molecules have been found to regulate ~30 unique disease-associated mammalian RNAs apart from the ribosome. These RNAs are transcribed from various regions of the genome and can be categorized into four general structural elements: expanded repeats, miRNAs, G-quadruplexes, and splice sites (Table 2). The following sections will briefly discuss the roles that these RNA structures and their interactions play in disease networks, followed by select small molecules that were shown to specifically modulate those networks, and finally important considerations for future work in these areas.
Table 2.
Mammalian RNA structural elements in disease that have been successfully targeted with small molecules.
| RNA Structural Element | Location | Disease Association |
|---|---|---|
| Expanded repeats | Introns, 5′ and 3′ UTRs | Huntington’s disease, Fragile X mental retardation 1, Myotonic dystrophy, Spinocerebellar ataxia 1 |
| miRNAs | N/A | Various cancers |
| G-quadruplexes | 5′-UTRs, splice sites | Alzheimer’s disease, breast cancer, pancreatic cancer, prostate cancer |
| Splice sites | Exon-intron junctions | Frontotemporal dementia, Parkinson’s disease, obesity, spinal muscular atrophy |
Abbreviations: UTR, Untranslated region; N/A, not applicable.
Expanded Repeats
While repeat RNA segments of 20-30 nucleotides are found in healthy cells, expansions of several hundred to thousands are hallmarks of some incurable neuronal diseases.67 These expansions are thought to promote pathogenesis through several distinct mechanisms, including but not limited to: (1) toxic RNA gain-of-function, (2) toxic protein gain-of-function; and (3) aberrant loss-of-transcript and loss-of-protein function.68, 69 In the first, RNA-dominant mechanism, the expanded repeats sequester essential proteins such as splicing and transcription factors by localizing them to nuclear foci, thereby causing an excess of alternatively spliced isoforms (Figure 2(a)).70 While the nucleotide composition of the expanded regions is gene-specific, the majority of these expansions are characterized by repeating tri- or hexa-nucleotide motifs that form internal loops with terminal hairpins (Figure 2(b)).71
Figure 2.
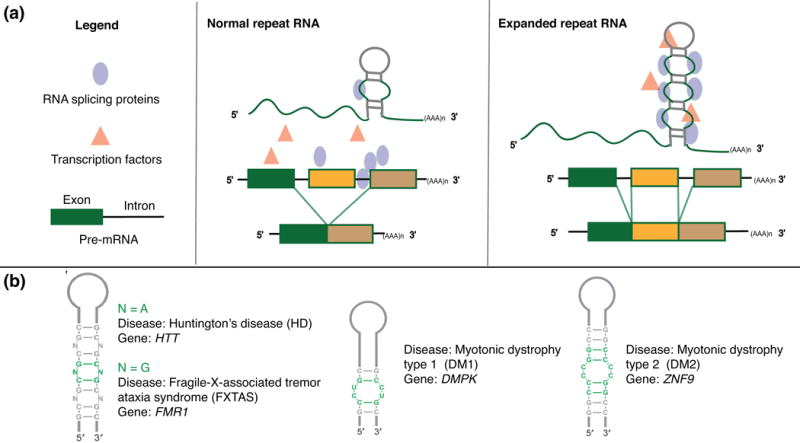
The RNA processing effect induced by expanded repeats and representative structures of single expansions in select diseases. (a) In normal repeat RNA, splicing proteins and transcription factors are available for proper processing to mature mRNA isoforms. When repeats are expanded in disease, proteins needed for efficient splicing are sequestered, leading to excess of mis-spliced mRNA isoforms. Adapted from Todd and Paulson, 2010.70 (b) Secondary structures of repeat RNA and their associated diseases. Adapted from Blaszczyk et al, 2017.71 Abbreviations: N, nucleotide; HTT, Huntington gene; FMR1, Fragile X mental retardation 1; DMPK, DM1 protein kinase; ZNF9, Zinc finger protein 9.
Expanded repeats are the most successfully targeted mammalian RNA class in a biological setting to date. The triumphs in small molecule-based targeting of these structures are attributed to several factors. These factors include intrinsic properties of the RNA, such as: (1) well-defined secondary structure, (2) repeated binding motifs that provide multiple binding sites and aid in cellular specificity, and (3) specific localization and molecular crowding in nuclear foci.72 In addition, a variety of design and screening methodologies for these targets have been established. In silico screening has enabled rational ligand design due the availability of solved structures, and in vitro methods such as time-resolved fluorescent resonance energy transfer (TR-FRET) and electrophoretic mobility shift assays (EMSA) provide measurements of protein displacement from the RNA.73-75 Finally, the availability of in vivo mouse and Drosophila models significantly aid in thorough assessment of ligand-induced phenotypic changes.73 Indeed, 34 bioactive small molecule ligands were discovered only in the last few years.
A current paradigm for small molecule-expanded repeat RNA targeting is the r(CUG)exp in type 1 myotonic dystrophy (DM1). One of the characteristic phenotypes of this incurable disease is manifested when toxic repeats from the DM1 Protein Kinase (DMPK) locus sequester Muscleblind-like protein 1 (MBNL1) away from the canonical splice site, leading to splicing defects (Figure 3(a)).13, 76, 77 In 2016, a clinical trial of an ASO-based treatment for DM1 was halted due to insufficient target engagement, presumably due to low accumulation in muscle,78-80 emphasizing the importance of complementary small molecule approaches such as those pursued by the Disney, Miller, and Zimmerman laboratories.66, 74, 81-85 Although a variety of small molecule design strategies have been utilized, these molecules share the common property of being multivalent and therefore targeting several repeating units at a time. This approach is considered particularly advantageous in the case of r(CUG)exp because the mean repeat expansion is 4,400, suggesting that even if the RNA is not highly expressed, each transcript has the capacity to bind many small molecules.85
Figure 3.
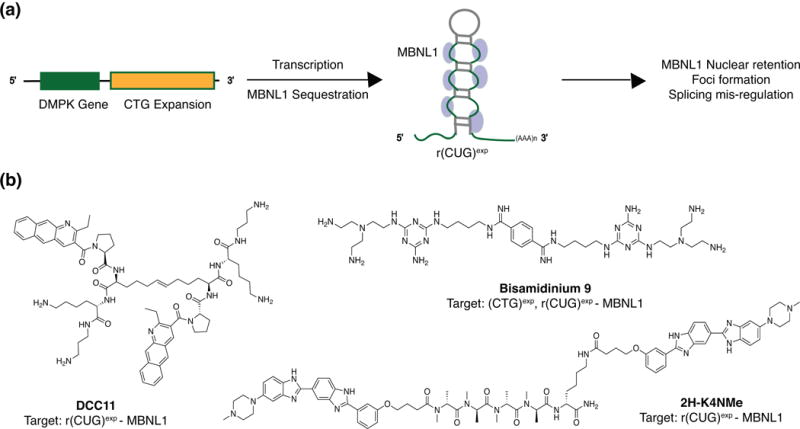
Small molecule-based targeting of r(CUG)exp repeats in DM1. (a) CTG expansion in the DMPK locus results in the transcription of r(CUG)exp repeats that then sequester MBNL proteins, leading to some of the characteristic phenotypes in DM1. Abbreviations: DMPK, DM1 protein kinase; MBNL, Muscle-blind protein 1. (b) Representative small molecule inhibitors of steps in DM1 repeat pathogenesis.
To exploit this opportunity, Miller and co-workers utilized a disulfide-based, resin-bound dynamic combinatorial chemistry (RBDCC) approach for constructing multivalent ligands.84 Specifically, a benzo(g)quinolone scaffold-based multivalent ligand was first discovered through RBDCC to bind r(CUG)exp in vitro.86 In their follow-up work, further scaffold diversification and disulfide replacement with an olefin bioisostere led to the development of two bioactive multivalent ligands that were successful in restoring splicing in a DM1 mouse model, including DCC11 (Figure 3(b)).84 Similarly, Disney and co-workers conjugated two units of a known small molecule binder with an optimized peptide linker, yielding a modularly assembled ligand 2H-4KNMe that led to improvement of DM1-associated defects in a mouse model (Figure 3(b)).87 Biophysical characterization of ligand-RNA interactions in this study revealed that the multivalent ligands have increased residence times and faster on rates than their individual binding units, ultimately resulting in bioactivity and lending further support for multivalent ligand design to target repeat RNAs.
The successes achieved with multivalent ligands have encouraged novel and creative strategies in targeting DM1 repeats. For instance, a synergistic effect in DM1 targeting was achieved by Zimmerman and co-workers with a rational design strategy to target both the DNA (CTG)exp and RNA (CUG)exp repeats in this disease (Figure 3(b)).81 The resulting bioactive ligand, Bisamidinium 9, was able to reverse repeat-induced phenotypes in DM1 Drosophila models. Additionally, inspired by a recent report of oxidized guanosine residues causing translation stalling and its link to neurological disease, Disney and co-workers are pursuing small molecule-targeted photochemically induced oxidation of RNA in order to directly chemically alter the DM1 repeats.88, 89
Small molecule-based interrogation of expanded repeats has provided valuable insights into the RNA gain-of-function pathways that contribute to neurological disease. As mentioned previously, however, the pathology of these disorders is complex and involves multiple pathways. Emerging evidence of the roles of bi-directional transcription, antisense RNA, and repeat-associated non-ATG (RAN) translation further complicate our understanding of disease pathogenesis while offering unique opportunities for small molecules that probe individual pathways.68 For example, Disney and co-workers recently reported two small molecule probes that separately inhibit r(CGG)exp-induced protein sequestration in nuclear foci and RAN translation in FXTAS.90 These probes enabled the elucidation of how those individual pathways contribute to r(CGG)exp toxicity in this disease. In order to independently probe the sequestration of splicing proteins and RAN translation pathways in other disorders, it is necessary to conduct phenotype assessments that extend beyond measuring nuclear foci dispersion or splicing restoration alone. Together, these efforts may lead to the identification of additional RNA-associated drug targets in neurological disease and enable combination therapy to achieve synergistic effects.
Micro-RNAs (miRNAs)
MiRNAs are short (20-25 nt) transcripts that have important cellular functions and are misregulated in a variety of human diseases, including cancers, cardiovascular, neurodegenerative and autoimmune diseases, as well as diabetes and obesity.91-93 It is proposed that their disease-related roles are achieved through two distinct modes of gene silencing: mRNA decay or direct translational repression.91, 93-95 The biogenesis of these transcripts begins with nucleus-localized primary miRNA (pri-miRNA) hairpins that are digested by Drosha to form precursor miRNA (pre-miRNA). The pre-miRNA is then transported to the cytoplasm for further processing by Dicer to form a dsRNA molecule that can be loaded into the RNA-induced silencing complex (RISC) to bind mRNAs through complementary base-pairing and mediate gene silencing (Figure 4(a)).96 While miRNAs are involved in multiple diseases, most of the small molecule targeting has been focused on miRNAs in cancer. This interest is in part due to the upregulation of select miRNAs in cancer, where they act as oncomirs and inhibit expression of tumor suppressors.97 Additionally, one miRNA can regulate several tumor suppressor mRNAs. As a result, inhibition of the processing pathway of one miRNA can lead to a more significant outcome as multiple pathways can be affected simultaneously.
Figure 4.
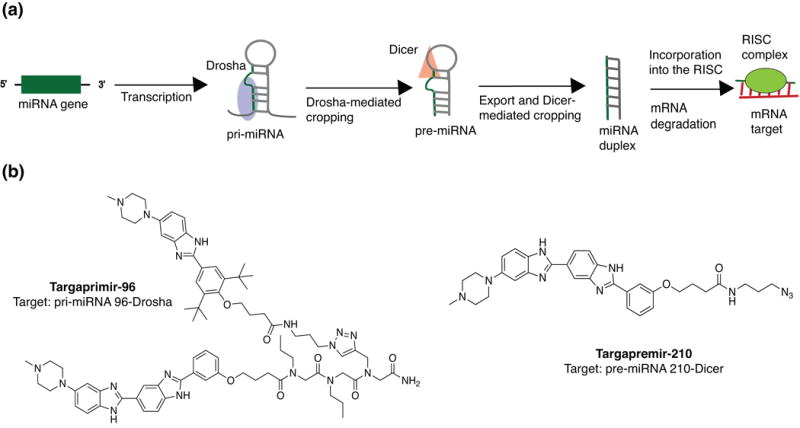
Small molecule-based targeting of the miRNA biogenesis pathway. (a) Mechanism of action of miRNA-mediated gene silencing. (b) Example small molecule inhibitors of miRNA biogenesis. Abbreviations: pri-miRNA, primary miRNA; pre-miRNA, precursor miRNA; RISC, RNA-induced silencing complex.
Small molecule inhibitors of both Drosha- and Dicer-miRNA interactions have been reported by several laboratories, including a number of important studies by Disney and co-workers. In their laboratory, an RNA motif-small molecule database named Inforna was generated using data obtained through 2-dimensional combinatorial screening (2DCS).98, 99 Inforna was applied to parse secondary structures in miRNAs that can be matched with RNA motif-small molecule pairs.99 Precise linking of modules that bind near and in the Drosha processing site yielded the multivalent molecule Targaprimir-96. This ligand enabled selective targeting of pri-miRNA 96 and subsequent inhibition of Drosha processing in triple negative breast cancer (TNBC) cell lines (Figure 4(b)). Notably, similar small molecule-induced phenotypes were observed in a TNBC xenograft mouse model. In a separate report, Disney and co-workers found that ligand Targapremir-210 inhibited Dicer processing of pre-miRNA-210 under hypoxic conditions (Figure 4(b)).65 The results of this study led the authors to propose general guidelines for specific miRNA targeting to alleviate oncogenic phenotypes. Namely, they suggest that (1) binding must occur in a functional site, e.g. Dicer or Drosha processing region, (2) the ligand must avidly bind to that site, and (3) both RNA abundance and molecule affinity can affect the target occupancy necessary to elicit a biological response. It will be interesting to see if these guidelines hold true when applied to other biological systems in which different miRNAs play crucial roles.
Despite the reported successes in small molecule targeting of different steps in miRNA biogenesis and the emergence of general guidelines for miRNA targeting, no miRNA-targeted small molecules are reported to be undergoing pre-clinical testing.59 Researchers have attributed this issue to similar secondary structures among different miRNAs, as well as other cellular RNAs, and a lack of tertiary structure. The impact of the sparse structural diversity is supported by the continuous identification of the same small molecule scaffolds to target these motifs. For instance, an Inforna-based search conducted to target miRNA-544 yielded a bis-benzimidazole ligand previously used by Disney and co-workers to target r(CUG)exp.101 Nonetheless, successful miRNA inhibition was achieved by taking advantage of high miRNA-544 expression levels and the targeting of a functional miRNA processing site. These findings can be further exploited through increased understanding of the fundamental basis of differential miRNA molecular recognition by proteins that interact with these functional sites.102 Structural insights into these interactions as well as their changes in disease are therefore in high demand.97 Indeed, a recent comparative study of mammalian RNA hairpins reported novel structure- and sequence-based requirements for efficient miRNA processing that change with single nucleotide polymorphisms (SNPs) in human disease.103
Lastly, we propose that the development of reliable, function-based in vitro and cellular assays that directly reflect miRNA engagement is of high significance. Currently, reporter-based assays are most commonly used to identify small molecule inhibitors of miRNA-mediated gene silencing.97 This method typically relies on luciferase activity that is suppressed upon miRNA binding to its target sequence in the 3′-UTR of the luciferase gene. The luciferase readout can increase if the small molecule leads to decreased miRNA binding of the labelled mRNA (Figure 5(a)). While conceptually attractive, this assay fails to provide information about direct miRNA engagement or the specific miRNA-protein interaction that the molecule may be inhibiting. On the other hand, several methods that measure direct miRNA binding are available, including small molecule microarrays developed by Schneekloth and co-workers and the aforementioned 2DCS-Inforna approaches developed by Disney and co-workers, among others.104-107 These approaches have led to the discovery of bioactive ligands, though the mechanisms of action were determined during follow-up experiments rather than during the screen. To compensate for these drawbacks, an alternative and promising high-throughput screening methodology was recently reported by Garner and co-workers.108 This approach enables detection of direct and allosteric inhibitors of Dicer-mediated miRNA cleavage and may thus be a promising new approach for discovering small molecules that target and inhibit specific pre-miRNA processing steps (Figure 5(b)).
Figure 5.
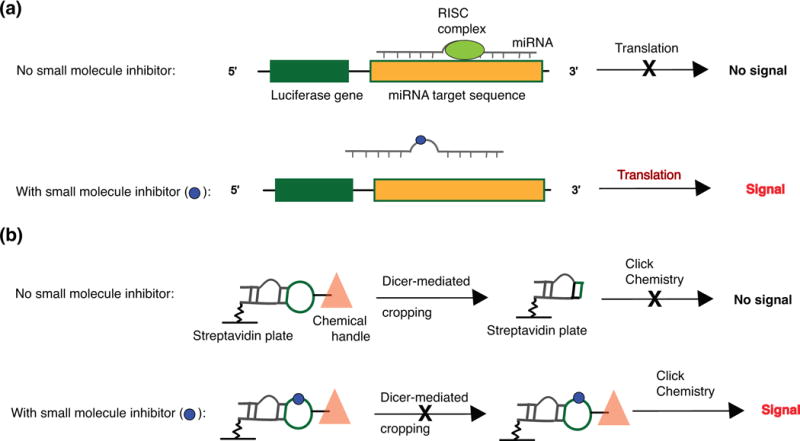
Traditional and emerging screens to identify small molecule miRNA inhibitors. (a) A standard luciferase-based reporter system used to identify small molecule miRNA inhibitors. Increase in luciferase signal indicative of translation is assumed to be caused by small molecule binding to miRNA or one if its precursors, thereby reducing binding of the mature miRNA to its target sequence. Adapted from Mahato et al.97 (b) A click-chemistry-based assay to identify small molecule inhibitors of Dicer-mediated processing. Adapted from Garner et al.108 Abbreviations: RISC, RNA-induced silencing complex.
G-quadruplexes in 5′-UTRs and Splice Sites
Formation of secondary and tertiary structures in untranslated regions of mRNAs are known to influence gene expression, including that of oncogenes.109 The relatively recent discovery of G-quadruplex structures in regulatory mRNA regions, particularly UTRs and splice sites, has led to increased efforts toward targeting RNA G-quadruplexes.110 G-quadruplex structures form upon stacking of cation-stabilized guanine tetrads in close proximity (Figure 6(a)).111 G-quadruplexes are perhaps best known for acting as translation regulators, controlling cap-dependent and independent translation at 5′-UTRs (Figure 6(b)). Additionally, the presence of these structures at splice sites or 3′-UTRs can impact the production of alternative splice variants or polyadenylation, respectively.112, 113 While the lifetime of RNA G-quadruplex structures in cells has recently been questioned by Bartel and co-workers,114 the selective activity of G-quadruplex binding ligands in cell culture suggests that small molecules may be able to perturb the equilibrium between stem-loop structures and G-quadruplexes.115-117 This section will first discuss examples of small molecules that have been shown to target G-quadruplexes in 5′-UTRs, followed by the first example of targeting a functional G-quadruplex in a splice site.
Figure 6.
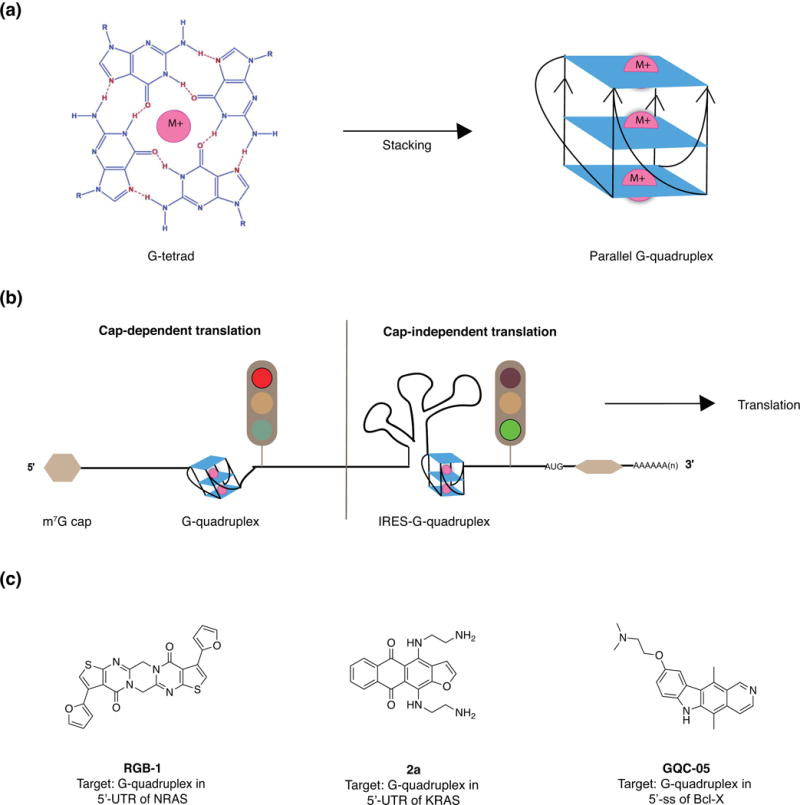
Structure, function, and small molecule binders of G-quadruplexes. (a) Nucleotide composition and base-pairing interactions in an RNA G-quadruplex and stacking of multiple G-quadruplexes. Adapted from Maiti et al.111 (b) Regulation of cap-dependent and independent translation by G-quadruplexes. Adapted from Balasubramanian et al.109 (c) Examples of small molecule stabilizers of G-quadruplexes in 5′-UTRs. Abbreviations: M+, monovalent metal cation; m7G, 7-methylguanylate; IRES, internal ribosomal entry site; NRAS, Neuroblastoma RAS viral oncogene homolog; KRAS, Kirsten rat sarcoma viral oncogene homolog; ss, splice site; Bcl-X, B-cell lymphoma-extra.
As G-quadruplexes are predicted to form in the 5′-UTRs of several known oncogenes, many small molecule-targeting endeavours have focused on oncogenes whose protein products are considered undruggable. For example, the RAS oncogene is overexpressed in many human cancers, and no anti-RAS drugs have reached the clinic.115 In a study by Uesugi and co-workers, the authors identified a small molecule (RGB-1) that targets a 5′-UTR G-quadruplex in the NRAS protooncogene (Fig 6(c)).117 In breast cancer cell lines, treatment with RGB-1 led to a significant decrease in NRAS protein expression. Additionally, the KRAS oncogene was explored for potential G-quadruplex structures by Xodo and co-workers.115 Using G-quadruplex-prediction software, the authors identified a putative G-quadruplex motif in the 5′-UTR region, which was then validated in vitro with several experimental techniques, including circular dichroism, UV-melting, and EMSAs. This motif was found to exist in equilibrium with a stem-loop structure, and the discovered molecule (2a) that stabilized the G-quadruplex form was thought to shift this equilibrium to inhibit KRAS translation (Fig 6(c)). When administered to pancreatic cancer cells, the molecule decreased KRAS protein levels, activated apoptosis, and inhibited colony formation.
While G-quadruplex structures are continuously identified in vitro, verifying the existence and function of these structures in biological systems has been difficult.118 For example, the equilibrium between competing secondary structures in these regions may be perturbed when a G-quadraplex targeted antibody or small molecule is used to test putative G-quadruplexes. In addition, mutagenesis studies can affect both secondary structure formation and protein binding.119 To overcome these challenges, Eperon, Dominguez and co-workers developed a strategy in which guanines are substituted by 7-deaza-guanines (7-deaza-G), preventing Hoogsteen base-pairing and subsequent G-quadruplex formation while allowing the formation of other secondary structures.120 Footprinting of 7-deaza-G-modified RNA sequences enabled the identification of several G-quadruplexes in a splice site of the Bcl-X gene, whose alternative splice variants are associated with prostate and breast cancer, among others.120-122 Eperon and Dominguez soon followed up with a report of a small molecule-stabilizer (GQC-05) of the G-quadruplex conformation that inhibited the disease-dominant splicing pathway of this gene in HeLa cells (Figure 6(c)).118 While the precise mechanism by which GQC-05 induces structural and therefore functional changes remains to be elucidated, this exciting work suggests that discrete tertiary structures concealed in long, unspliced RNA transcripts can indeed be targeted with small molecules in a bioactive setting.
As new G-quadruplex structures and their respective small molecule binders are being discovered, special care should be taken in designing in vitro experiments to identify functional binding events in biologically relevant constructs. As evidenced by the work of Balasubramanian and co-workers, positioning of the G-quadruplex-forming segment in a sequence that closely resembles its natural context is crucial for drawing conclusions about its cellular function.123 Lastly, the continued success in identifying functional G-quadruplexes in long RNAs and splice sites could be greatly accelerated by the development of high-throughput methods to enable fast discovery of these structures.119
Pre-mRNA splice sites
As mentioned briefly in the previous section, pre-mRNA splice sites present an attractive and seemingly tractable target for RNA-targeting small molecules. Even in healthy cells, alternative splicing events occur in over 95% of the human genome and generate a diverse array of RNA transcripts.124 These events involve skipping or the use of different 5′- and 3′-splice sites, which rely on complex RNA-protein interactions mediated through small nuclear ribonucleoproteins (snRNPs).125 Additionally, splicing can be developmental-, cell-, and tissue-type specific. In humans, aberrant splicing is involved in a variety of diseases including cancer, hemophilia, cystic fibrosis, and several genetic disorders.126-128 For genetic disorders specifically, alternative splicing is considered a favorable point for therapeutic intervention because it offers the possibility to target an early step of gene expression without altering the genome.127 Finally, targeting the specific disease-associated RNAs involved in this process is considered more favorable than targeting protein-centric processes, as altering the protein splicing machinery may induce widespread splicing changes that in turn generate aberrant and dysfunctional proteins.129 The recognition of secondary structure formation in pre-mRNA splice sites has consequently led to increased interest in targeting these sites and their associated interactions with small molecules.130
The most promising example of small molecules targeting RNA splice sites is the discovery of modulators of the survival motor neuron (SMN2) mRNA in spinal muscular atrophy (SMA). The SMN2 gene is a duplicate copy of the SMN1 gene whose expression is lost in this disease.127 The SMN2 variant contains an SNP that causes skipping of exon 7 and produces an unstable, truncated protein.131-133 Therefore, the goal of therapeutic targeting of this gene, as first demonstrated by Krainer and co-workers, is to enhance the production of full-length SMN mRNA and protein by promoting exon 7 inclusion (Figure 7(a)).134, 135 This strategy was tested in the clinic by Ionis and Biogen, and resulted in accelerated FDA approval of an ASO, with clinical studies ongoing.136 In 2015, Sivasankaran and co-workers at Novartis discovered two small molecule enhancers of exon 7 inclusion via a luciferase-based SMN reporter screen.137 NVS-SM1 was found to increase the levels of the full- length SMN mRNA and protein in human fibroblasts, as well as extend survival in a severe SMA mouse model. Detailed mechanistic studies revealed that these molecules act by stabilizing the transient double-stranded RNA complex formed by the SMN2 pre-mRNA and the U1 snRNP complex in the 5′ splice site (ss) of exon 7 (Figure 7(b)). NVS-SM1 is in Phase II clinical trials for the treatment of SMA.138 Concurrently, Metzger and co-workers from Roche reported a different class of molecules that demonstrated a dose-dependent correction of SMN2 splicing to include exon 7.139 In their follow-up mechanistic studies, the group utilized a variety of techniques ranging from NMR, SPR, transcriptome-wide RNA-seq, and protein chemistry methods to elucidate the rationale behind the surprising specificity of these compounds for the SMN2 gene.140 The most potent molecule, SMN-C5, was found to bind to two distinct sites. One site was the 5′ ss on exon 7 in SMN2, which is identical to the binding site of NVS-SM1. The other site is at a purine-rich exonic splicing enhancer 2 (ESE2) motif in exon 7 that binds to the hnRNP G protein. The authors hypothesized that this additional binding site promotes the specificity of SMN-C5 by dislocating hnRNP G and allowing the U1 snRNP complex to bind to ESE2 (Figure 7(b)). Additionally, it was proposed that the specific recognition of the RNA-protein interaction at the ESE is enabled by the quaternary structure of this complex. Given the emerging importance of alternative splicing in a wide variety of genetic diseases, these findings and their implications offer exciting avenues to achieve selective enhancement or alteration of splicing by small molecules.
Figure 7.
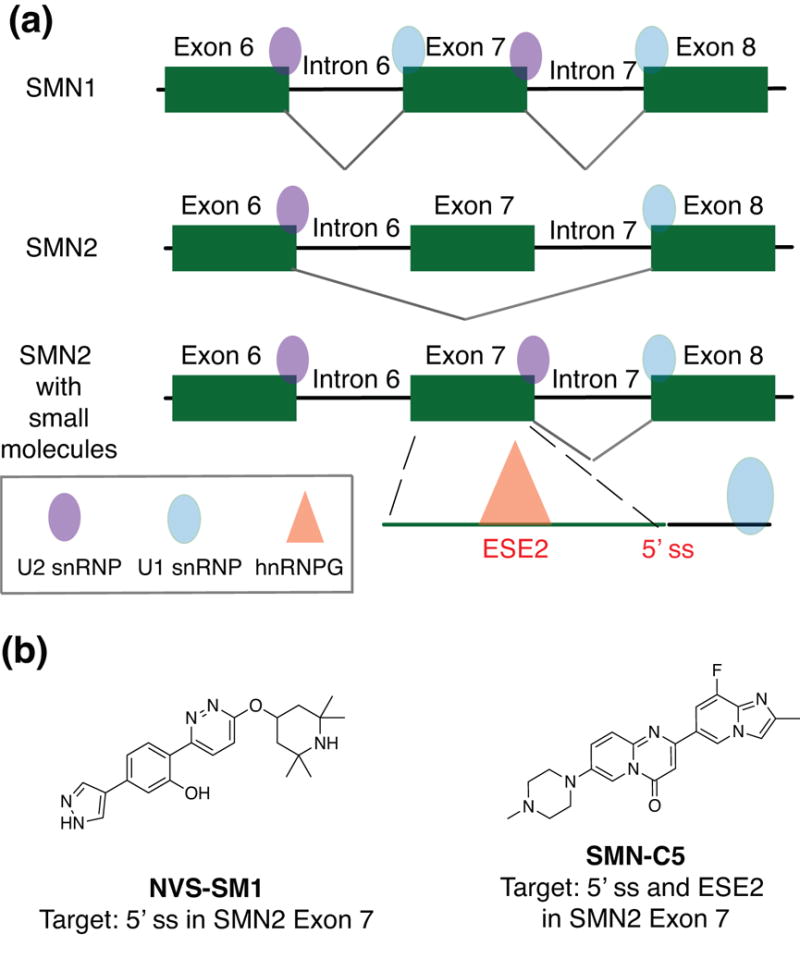
Small molecule-induced regulation of the SMN2 splicing mechanism. (a) The splicing mechanism of SMN1 (healthy protein) and SMN2 (disease protein) in which Exon 7 is included due to a SNP. (b) Small molecules shown to induce Exon 7 stabilization in SMN2 gene. Abbreviations: SNP, single nucleotide polymorphism; SMN, survival motor neuron; ESE, exonic splicing enhancer; hnRNP G, heterogeneous nuclear ribonucleoprotein G; snRNP, small nuclear ribonucleoprotein; ss, splice site.
UNDEREXPLORED MAMMALIAN RNA TARGETS FOR SMALL MOLECULE INTERVENTION
The four classes of RNA that have been successfully targeted with small molecules inspire exploration of more recently discovered mammalian RNA classes or RNA structural domains whose exact functions, mechanisms of action, and effects on disease-related phenotypes are less characterized. In this section, we will draw attention to underexplored RNA classes that are particularly attractive for small molecule-based targeting approaches. These highlights involve well-characterized RNA structures with emerging roles in disease, as well as RNA-protein or RNA-DNA interactions whose in vitro inhibition by small molecules was previously demonstrated and hence offers substantial promise. We note that some disease-relevant mammalian RNAs such as tRNA-derived fragments (tRFs) and circular RNAs are not highlighted herein as current research suggests that these RNAs lack stable structure, which would render oligonucleotide-based approaches more feasible than small molecule-based approaches.141, 142
Mammalian Ribosomal RNA
While the ribosome is often considered an obstacle for selective small molecule RNA-targeting, recent determination of a 3.6 Å X-ray diffraction structure of the human 80S ribosome has inspired creative targeting efforts, particularly toward the elevated protein synthesis that is a hallmark of cancer.143 After their report of the crystal structure, Peyron, Klaholz, and co-workers leveraged its availability to conduct computational modeling studies and repurpose a eukaryote-specific antibiotic, cycloheximide, as an anticancer drug.144 The molecule induced anti-proliferative effects in several cancer cell lines. In addition, the misregulation of specific post-transcriptional modifications in the human ribosome in cancer was shown to impact tRNA and mRNA binding, impairing translation of mRNAs important for cellular transformation.145 Many of these modifications have recently been mapped onto the reported 80S ribosome structure, offering new opportunities for structure-based ligand design to specifically target differentially modified ribosomes in cancer.146, 147
Long non-coding RNAs (lncRNAs)
LncRNAs are classified as transcripts greater than 200 nucleotides, although many are several kilobases in length.148 Their diverse and disease-related functions, along with tissue- or temporally-restricted expression, make them attractive therapeutic targets.148, 149 While low sequence conservation throughout evolution has been interpreted by some as indicative of a lack of function, an alternative interpretation is the conservation of structure.150, 151 Indeed, structural domains of disease-relevant lncRNA that interact with specific proteins or protein complexes are continuously identified.42, 152-157 Further, some lncRNA structural domains or interactions have evolutionarily conserved functions.151, 158 Given the ability of small molecules to detect RNA structural elements and target specific biomolecular interactions, these tools seem perfectly poised to provide new insights into the exciting field of lncRNA biology.
With the variety of lncRNA binding partners and complex folds these molecules undergo, several types of unique lncRNA targeting avenues emerge: 1) complex structures of lncRNA domains, 2) lncRNA-protein interactions, and 3) lncRNA-DNA interactions. Examples of potentially targetable lncRNA are discussed below.
LncRNA structural domains
X-inactive specific transcript (XIST)
Xist is one of the first discovered and most well characterized lncRNAs.159, 160 Xist functions as the major effector of the X chromosome inactivation (XCI) process in mammals and has been implicated in oncogenic processes.161-163 The scaffolding and 3D-organization of Xist necessary for XCI are achieved through a complex interactome that consists of hundreds of potential interactions.160 Importantly, mutational studies revealed that specific structural domains, namely six tandem hairpin repeats (A-F) are crucial for its function in XCI.164 The mechanisms through which individual structural domains and protein interactions mediate the functions of Xist are currently under investigation.160 Structural insights into the A- and F-repeat region (RepA) of Xist by Pyle and co-workers revealed an intricate tertiary architecture within specific functional modules.152 Moreover, Weeks and co-workers conducted an in cellulo chemical probing study of the entire 18 k.b. transcript and discovered an additional domain in the 3′ end that may have a role in Xist localization.165 These valuable findings should accelerate the discovery of small molecules that can recognize specific Xist structural elements to illuminate their roles in development and disease.
Metastasis-associated lung-adenocarcinoma transcript 1 (MALAT1)
MALAT1 is a highly abundant nuclear lncRNA that is overexpressed in several human cancers.166 A range of functions have been proposed, including regulation of expression and alternative splicing of oncogenes.167 Structurally, the recent determination of a 3.1 Å structure solved through X-ray diffraction by Steitz and co-workers revealed a bipartite triple helix at the 3′ end.168 Single-point mutations assumed to disrupt the triple helix stability led to reduced MALAT1 levels in cells, suggesting a pivotal role of this structure in enabling MALAT1 accumulation.168-170 Lastly, evidence of selective recognition of the triple helix by a methyltransferase protein further supports a functional role of this domain.171 Small molecule-induced destabilization of the triple helix could offer significant insights into the role of this structure in driving oncogenic phenotypes and encourage its pursuit as a therapeutic target in cancers.
LncRNA-protein interactions
HOX-transcript antisense RNA (HOTAIR)
Chang and co-workers first demonstrated that HOTAIR is involved in promoting breast cancer metastasis and can be up to 125-times overexpressed in patient-derived metastatic breast cancer samples.172 The role of HOTAIR in transcription is exerted through its scaffolding of chromatin-modifying complexes Polycomb-repressive complex 2 (PRC2) and Lysine-specific demethylase 1 (LSD1), presumably localizing these complexes to specific loci. The binding of PRC2 and LSD1 complexes, which are themselves anticancer targets, have been localized to distinct structural domains at the 5′- and 3′-ends of HOTAIR, respectively.173 HOTAIR was also recently reported to function post-translationally by scaffolding ubiquitin proteins.174 Spatiotemporal, small molecule-based interrogation of individual complex interactions with HOTAIR would enable the elucidation of discrete roles and their possible co-dependence in driving HOTAIR-associated oncogenic processes.
From a therapeutic standpoint, the possibility of targeting the HOTAIR:PRC2 interaction is an attractive anticancer strategy as compared to targeting PRC2 alone because this protein complex has been shown to have both oncogenic and tumor-suppresive functions.175 Indeed, a recent study by Nephew and co-workers demonstrated the ability of a peptide-nucleic acid conjugate to specifically inhibit the HOTAIR:PRC2 interaction, resulting in decreased cancer-related phenotypes in ovarian and breast cancer cell lines as well as mice with ovarian cancer.176 Further, Faghihi and co-workers conducted an Alpha screen in search of small molecule inhibitors of the HOTAIR:PRC2 complex in 2015.177 Though a promiscuous intercalator elipticine was identified as a hit, this in vitro study validated the ability of small molecules to inhibit lncRNA-protein complex formation and motivates follow-up studies with diverse small molecule libraries to fully realize the potential of this strategy.
Second chromosome locus associated with prostate 1 (SChLAP1)
SChLAP1 lncRNA was initially found to be exclusively expressed in aggressive prostate cancer and was recently reported in bladder cancer.178, 179 The oncogenic effects of SChLAP1 on gene expression are at least in part exerted through its binding to the switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex.178 This binding event impairs the ability of the SWI/SNF subunit SNF5 to bind to the genome, thereby antagonizing its tumor-suppressive functions in prostate cancer. Specific targeting of the SChLAP1:SWI/SNF complex would elucidate whether this interaction is sufficient for SChLAP1-associated phenotypes and also represents an attractive therapeutic strategy to target prostate cancer without affecting healthy cells.
LncRNA-DNA interactions
Many lncRNAs that regulate transcription hybridize with a DNA duplex, resulting in the unique topological structure of a lncRNA-DNA triple helix hybrid.180 For example, five triple helix target sites in mesenchymal stem cells were recently predicted for HOTAIR, and these binding events were proposed to aid in guiding epigenetic modifiers such as PRC2 to relevant loci.181 Similarly, triple helix-mediated changes in chromatin structure were reported for lncRNA Khps1.182 This interaction is proposed to regulate the expression of the proto-oncogene SPHK1. Likewise, lncRNA Fendrr is known to bind to the mouse Foxf1 locus, forming a triple helix hybrid that is predicted to recruit PRC2 and in turn inhibit transcription of Foxf1.183 Importantly, Foxf1 is critical for embryonic development in mice. Targeting these triple helices with small molecules could thus provide valuable insights into whether these structures promote recruitment of epigenetic modifiers to target loci or if these processes occur independently, ultimately deciphering the mechanisms that enable the roles of these lncRNAs in both oncogenic and developmental processes. Promisingly, many examples of ligands that bind this structural class in vitro have been reported.184
Underexplored G-quadruplexes, including lncRNA
As alluded to previously, while the majority of G-quadruplex structures have so far been found in 5′- or 3′-UTRs, their existence in splicing regions and non-coding RNA transcripts is becoming increasingly recognized.185 The paradigm of a G-quadruplex-containing lncRNA is the telomeric repeat-containing RNA (TERRA). This lncRNA has long been recognized as an attractive anticancer target and has drawn more attention since the discovery that these molecules form G-quadruplexes in vivo.186 This fold was shown to be required for telomere heterochromatin formation in cancer cells.187, 188 In 2014, Campos-Olivas, Gonzalez and co-workers reported selective in vitro ligands for TERRA G-quadruplexes using a fluorine-NMR screen.189 In 2017, the first bioactive TERRA-targeting small molecule was reported by Li and co-workers.190 This quindoline derivative was shown to induce DNA damage response and apoptosis, inhibit proliferation and cause G2/M phase arrest in osteosarcoma cancer cells. The authors propose that this effect occurred due to the molecules ability to stabilize the TERRA G-quadruplex, promoting its interaction with the Telomeric Repeat Binding Factor 2 protein, causing it to dissociate from telomeric duplex DNA and ultimately inducing a DNA damage response.
This finding offers exciting avenues for designing new or repurposing existing G-quadruplex-targeted small molecule libraries to further explore the roles of these structures in the transcriptome. For instance, recent reports by Cech and co-workers suggest that G-quadruplexes are the main structural element recognized by the catalytic subunit of PRC2, the aforementioned protein complex known to interact with HOTAIR as well as other lncRNAs.191 Moreover, functional G-quadruplexes were recently found in other disease-associated lncRNAs such as GSEC and H19.192, 193 Lastly, therapeutically relevant G-quadruplexes have been identified in the CD44 gene isoform important for the epithelial-mesenchymal transition in breast cancer, as well as different non-coding RNAs involved in neurodegenerative diseases.194, 195
Underexplored repeats
Expanded repeats are associated with more than 30 neurological disorders and no treatment options are currently available.68 While expanded RNA repeats are among the most commonly targeted mammalian RNAs with small molecules, several repeat sequences remain unexplored and/or have limited examples of successful targeting. With the plethora of repeat-focused screening methods available, it could be possible to rapidly identify small molecules for the many unexplored repeat RNAs, both to discover novel biology and explore additional therapeutic potential.73 For example, small molecules may enable the identification and study of higher-order alternative structures that exist in equilibrium with hairpin-terminated repeats. Petrucelli, Disney and co-workers recently found that r(GGGGCC)exp in frontotemporal dementia/amyotrophic lateral sclerosis (FTD/ALS) exist in an equilibrium between hairpin and G-quadruplex structures.196 They identified several small molecules that selectively bound to the hairpin structure, leading to the dispersion of nuclear foci and inhibition of RNA translation in patient fibroblasts. To continue exploring higher-order structure formation in other expanded repeats, structural information on full-length repeat constructs is in high demand.69
Unexplored splice variants
As demonstrated by the successes in targeting SMN splice variants, small molecule-based modulation of RNA splicing pathways opens up the possibility of myriad interventions to understand disease processes. For example, recent work by Lee and co-workers resulted in the discovery of novel and race-specific alternative splicing events in aggressive prostate cancer.197 Specifically, the group found that a PIK3CD-S variant enriched in African-American patient samples was associated with increased proliferation, invasion, and resistance. Importantly, exon-level variations were the only differentiable factors explaining the five-fold higher mortality rate in African-American men with prostate cancer. This discovery further challenges the dogma of DNA-level mutations/alterations as primary drivers of cancer aggressiveness and further emphasizes the importance of studying aberrant RNA splicing events in disease.198, 199
Promoter-associated RNAs (paRNAs)
PaRNAs are a class of non-coding RNA molecules that are thought to function as transcriptional regulators of downstream genes in cis.200 Catapano and co-workers recently showed that a paRNA in the promoter region of the tumor suppressor E-cadherin has a defined secondary structure domain that forms an alternate structure as a result of an SNP, which is linked to increased cancer risk.201 This structural rearrangement affects paRNA’s interactions with miRNA and epigenetic regulators, downregulating the transcription of E-cadherin in epithelial cancers. This discovery suggests an underexplored mechanism by which SNPs in non-coding regions can impact the epigenetic landscape and therefore human disease. Given that the knockdown of paRNA affects proliferation in prostate cancer, the authors propose that development of inhibitors for this structured target can enable gene-selective transcriptional reprogramming.
Small nucleolar RNAs (snoRNAs)
SnoRNAs primarily function by promoting post-transcriptional modifications in ribosomal RNA to enable maturation of the ribosome.202 In addition, snoRNAs have recently been implied in regulating alternative splicing, mRNA targeting and miRNA production as well as tumorigenic processes.203-205 SnoRNAs form well-defined secondary structures, a characteristic that poses them as interesting targets for small molecule intervention, although their disease-related roles are still emerging and remain to be fully validated.206 A potential model system to test their function in tumorigenesis is snoRNA 42 (SNORA42), a commonly overexpressed RNA in lung cancer.207 Its knockdown in non-small cell lung cancer (NSCLC) inhibited tumorigenicity and apoptosis in a p53-dependent manner. As the exact mechanism by which SNORA42 regulates p53 remains unknown, there is an opportunity to obtain new insights and establish a paradigm of snoRNA function in cancer.
RNA modifications
RNA modifications are beginning to emerge as important new modulators of RNA structures involved in human diseases.208 For instance, 2′-O-methylation, 5-methylcytidine, and adenosine-to-inosine modifications modulate disease-related properties in small RNAs such as miRNAs, PIWI-interacting RNAs, and tRNA-derived small RNAs. Researchers in this field have expressed the need for chemical probes that can specifically recognize these modified structures, which would eliminate time- and resource-intensive antibody development.209 Additionally, N6-methyladenosine (m6A) has been implicated in many diseases, including cancer and metabolic disease.210 A recent discovery shows that m6A RNA methylation on ADAM10 mRNA is critical for tumorigenesis and self-renewal of glioblastoma stem cells, implicating this modified RNA as an attractive target for glioblastoma treatment.211
Conclusion and Outlook
The field of RNA-small molecule targeting in mammals has significantly advanced over the past five years. Continued exploration of both fundamental principles and biological applications will further the exponential growth in the discovery of selective RNA-targeted ligands with biological activity. From the small molecule perspective, it is important to continually identify and validate guiding principles for efficient design of RNA-targeted ligands.64 The discovery of these principles may lead to the design of large screening libraries enriched in RNA-binding chemotypes that can yield higher hit rates and avoid the continuous identification of promiscuous RNA-binding ligands. These efforts can be complemented by the simultaneous development of activity- or function-based in vitro assays that are efficient, reliable, and RNA-centric. Conducting rigorous assays for selectivity and target-engagement, as well as defining specificity landscapes, will also be crucial for establishing guidelines for small molecule targeting of RNA moving forward.26, 212, 213
From the RNA perspective, it is urgent that the community elucidates how to select the RNAs best suited for targeting. This work would significantly aid our understanding of the limited successes of targeting RNA with small molecules in a biological setting despite the many RNAs that have been targeted in vitro. While it is possible that these rules may be RNA class-specific, identifying the importance of factors such as: 1) RNA abundance; 2) RNA localization; 3) the availability of functional sites for binding; 4) structural rigidity; and 5) the complexity of the RNA 3D and/or quaternary structures will be crucial as researchers pursue small molecule development for the underexplored RNA classes described above. Advances in methods that enable robust characterization of RNA 3D structure, dynamics and in-cell interactions, as well as characterization of RNA-ligand interfaces, could accelerate our understanding of these principles.214-219
While oligonucleotide-based approaches have been a starting point in validating the biological and disease relevance of many mammalian RNAs, many questions remain with regards to the role of RNA structural domains and RNA:biomolecule interactions, and these questions could be uniquely addressed by utilizing small molecules. The exciting prospect of small molecule-based targeting of RNA in disease is increasingly recognized by the pharmaceutical industry, as evident in new efforts by Merck, Novartis, and Pfizer as well as the establishment of new companies aiming to expand the scope of current RNA drug targets.51 In this review, we have surveyed four different mammalian RNA classes that have been successfully targeted with small molecules to elicit a disease-relevant biological response, proposed emerging targets for small molecule modulation, and identified several areas and future directions that will advance RNA-small molecule discovery. Like many others, we believe that progress can be further accelerated by collaborative efforts using multi-disciplinary approaches from experts in many areas of science. Hence, we hope that this collection of promising results combined with highlights of underexplored RNA targets draws the attention and excitement of medicinal chemists as well as computational and structural biologists to join the next phase of the RNA revolution.
Acknowledgments
We thank the Hargrove lab members for stimulating discussion and proofreading of the manuscript. We specifically thank Jordan Forte for help with the TOC graphic. AEH wishes to acknowledge Duke University, the Prostate Cancer Foundation Young Investigator Award, and the National Institute of Health (NIH) Maximizing Investigator’s Research Award (MIRA) (R35GM124785). AD was supported through the NIH MIRA award.
Footnotes
No conflicts of interest.
Contributor Information
Anita Donlic, 0000-0002-0423-618X, Department of Chemistry, Duke University.
Amanda E. Hargrove, 0000-0003-1536-6753, Department of Chemistry, Duke University; Department of Biochemistry, Duke University Medical Center
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 5.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 6.Duret L, Dorkeld F, Gautier C. Strong Conservation of Noncoding Sequences during Vertebrates Evolution – Potential Involvement in Posttranscriptional Regulation of Gene-Expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glinsky GV. Phenotype-defining functions of multiple non-coding RNA pathways. Cell Cycle. 2008;7:1630–1639. doi: 10.4161/cc.7.11.5976. [DOI] [PubMed] [Google Scholar]
- 10.Fu XD. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederichs S. Non-coding RNA and disease. RNA Biol. 2012;9:701–702. doi: 10.4161/rna.20972. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 16.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muntoni F, Wood MJ. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov. 2011;10:621–637. doi: 10.1038/nrd3459. [DOI] [PubMed] [Google Scholar]
- 18.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 20.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 21.Watts JK, Corey DR. Gene silencing by siRNAs and antisense oligonucleotides in the laboratory and the clinic. J Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barata P, Sood AK, Hong DS. RNA-targeted therapeutics in cancer clinical trials: Current status and future directions. Cancer Treat Rev. 2016;50:35–47. doi: 10.1016/j.ctrv.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Donner A. A platform for RNA. 2013;6 [Google Scholar]
- 25.Donner A. It is an RNA world. 2014;7 [Google Scholar]
- 26.Childs-Disney JL, Disney MD. Approaches to Validate and Manipulate RNA Targets with Small Molecules in Cells. Annu Rev Pharmacol Toxicol. 2016;56:123–140. doi: 10.1146/annurev-pharmtox-010715-103910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers TA, Wyatt JR, Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran T, Childs-Disney JL, Liu B, Guan L, Rzuczek S, Disney MD. Targeting the r(CGG) repeats that cause FXTAS with modularly assembled small molecules and oligonucleotides. ACS Chem Biol. 2014;9:904–912. doi: 10.1021/cb400875u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mruk K, Chen JK. Thinking big with small molecules. J Cell Biol. 2015;209:7. doi: 10.1083/jcb.201501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blagg J, Workman P. Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell. 2017;32:9–25. doi: 10.1016/j.ccell.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cundliffe E. On the nature of antibiotic binding sites in ribosomes. Biochimie. 1987;69:863–869. doi: 10.1016/0300-9084(87)90213-6. [DOI] [PubMed] [Google Scholar]
- 34.Hermann T. Small molecules targeting viral RNA. Wiley Interdiscip Rev RNA. 2016;7:726–743. doi: 10.1002/wrna.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas JR, Hergenrother PJ. Targeting RNA with Small Molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 36.Hermann T, Tor Y. RNA as a target for small-molecule therapeutics. Expert Opin Ter Pat. 2005;15:49–62. [Google Scholar]
- 37.Deigan KE, Ferre-D’Amare AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mei HY, Cui M, Heldsinger A, Lemrow SM, Loo JA, Sannes-Lowery KA, Sharmeen L, Czarnik AW. Inhibitors of protein-RNA complexation that target the RNA: Specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 39.Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 40.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Gen. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 41.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci U S A. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 2013;41:8220–8236. doi: 10.1093/nar/gkt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhartiya D, Scaria V. Genomic variations in non-coding RNAs: Structure, function and regulation. Genomics. 2016;107:59–68. doi: 10.1016/j.ygeno.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Bevilacqua PC, Ritchey LE, Su Z, Assmann SM. Genome-Wide Analysis of RNA Secondary Structure. Annu Rev Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 46.Garbaccio RM, Parmee ER. The Impact of Chemical Probes in Drug Discovery: A Pharmaceutical Industry Perspective. Cell Chem Biol. 2016;23:10–17. doi: 10.1016/j.chembiol.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Angelbello AJ, Chen JL, Childs-Disney JL, Zhang P, Wang Z-F, Disney MD. Using Genome Sequence to Enable the Design of Medicines and Chemical Probes. Chemical Reviews. 2018 doi: 10.1021/acs.chemrev.7b00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornton CA, Wang E, Carrell EM. Myotonic dystrophy: approach to therapy. Curr Opin Genet Dev. 2017;44:135–140. doi: 10.1016/j.gde.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. The Lancet. 378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. The Lancet. 388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 51.Mullard A. Small molecules against RNA targets attract big backers. Nat Rev Drug Discov. 2017;16:813–815. doi: 10.1038/nrd.2017.239. [DOI] [PubMed] [Google Scholar]
- 52.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 53.Shortridge MD, Varani G. Structure based approaches for targeting non-coding RNAs with small molecules. Curr Opin Struct Biol. 2015;30:79–88. doi: 10.1016/j.sbi.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilch DS, Kaul M, Barbieri CM, Kerrigan JE. Thermodynamics of aminoglycoside-rRNA recognition. Biopolymers. 2003;70:58–79. doi: 10.1002/bip.10411. [DOI] [PubMed] [Google Scholar]
- 55.Turner JJ, Jones S, Fabani MM, Ivanova G, Arzumanov AA, Gait MJ. RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol Dis. 2007;38:1–7. doi: 10.1016/j.bcmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol Life Sci. 2007;64:1841–1852. doi: 10.1007/s00018-007-7034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arya DP, editor. Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. Hoboken: John Wiley & Sons, Inc; 2007. [Google Scholar]
- 58.Connelly CM, Moon MH, Schneekloth JS., Jr The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem Biol. 2016;23:1077–1090. doi: 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizvi NF, Smith GF. RNA as a small molecule druggable target. Bioorg Med Chem Lett. 2017;27:5083–5088. doi: 10.1016/j.bmcl.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 60.Disney MD, Yildirim I, Childs-Disney JL. Methods to enable the design of bioactive small molecules targeting RNA. Org Biomol Chem. 2014;12:1029–1039. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Angelbello AJ, Chen JL, Childs-Disney JL, Zhang P, Wang ZF, Disney MD. Using Genome Sequence to Enable the Design of Medicines and Chemical Probes. Chem Rev. 2018 doi: 10.1021/acs.chemrev.7b00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6 doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Disney MD. Rational design of chemical genetic probes of RNA function and lead therapeutics targeting repeating transcripts. Drug Discov Today. 2013;18:1228–1236. doi: 10.1016/j.drudis.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan BS, Forte JE, Culver RN, Zhang Y, Hargrove AE. Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-Targeted Bioactive Ligands. Angew Chem Int Ed Engl. 2017;56:13498–13502. doi: 10.1002/anie.201707641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, Disney MD. Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit. J Am Chem Soc. 2017;139:3446–3455. doi: 10.1021/jacs.6b11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rzuczek SG, Southern MR, Disney MD. Studying a Drug-like, RNA-Focused Small Molecule Library Identifies Compounds That Inhibit RNA Toxicity in Myotonic Dystrophy. ACS Chem Biol. 2015;10:2706–2715. doi: 10.1021/acschembio.5b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson DL, Orr HT, Warren ST. The unstable repeats–three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang N, Ashizawa T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr Opin Genet Dev. 2017;44:17–29. doi: 10.1016/j.gde.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ciesiolka A, Jazurek M, Drazkowska K, Krzyzosiak WJ. Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front Cell Neurosci. 2017;11:97. doi: 10.3389/fncel.2017.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blaszczyk L, Rypniewski W, Kiliszek A. Structures of RNA repeats associated with neurological diseases. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1412. [DOI] [PubMed] [Google Scholar]
- 72.Rzuczek SG, Colgan LA, Nakai Y, Cameron MD, Furling D, Yasuda R, Disney MD. Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat Chem Biol. 2017;13:188–193. doi: 10.1038/nchembio.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konieczny P, Selma-Soriano E, Rapisarda AS, Fernandez-Costa JM, Perez-Alonso M, Artero R. Myotonic dystrophy: candidate small molecule therapeutics. Drug Discov Today. 2017;22:1740–1748. doi: 10.1016/j.drudis.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 74.Kumar A, Parkesh R, Sznajder LJ, Childs-Disney JL, Sobczak K, Disney MD. Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem Biol. 2012;7:496–505. doi: 10.1021/cb200413a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen L, Lee J, Wong CH, Zimmerman SC. Small molecules that target the toxic RNA in myotonic dystrophy type 2. ChemMedChem. 2014;9:2455–2462. doi: 10.1002/cmdc.201402095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic Dystrophy in Transgenic Mice Expressing an Expanded CUG Repeat. Science. 2000;289:1769. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 77.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.ClinicalTrials.gov Bethesda (MD):National Library of Medicine (US) Identifier NCT0231 2011. A Safety and Tolerability Study of Multiple Doses of ISIS-DMPKRx in Adults With Myotonic Dystrophy Type 1. 2014 Dec 9; 2016 Nov 3. Available at: https://clinicaltrials.gov/ct2/show/NCT02312011. (Accessed January 18, 2018)
- 79.Preclinical data behind the Ionis trial published. Available at: http://www.myotonic.org/preclinical-data-behind-ionis-trial-published. (Accessed January 24, 2018)
- 80.Madsen A. Ionis Reports Setback on DMPKRx Program for Myotonic Dystrophy. Available at: https://strongly.mda.org/ionis-reports-setback-dmpkrx-program-myotonic-dystrophy/. (Accessed January 24, 2018)
- 81.Nguyen L, Luu LM, Peng S, Serrano JF, Chan HY, Zimmerman SC. Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J Am Chem Soc. 2015;137:14180–14189. doi: 10.1021/jacs.5b09266. [DOI] [PubMed] [Google Scholar]
- 82.Jahromi AH, Fu Y, Miller KA, Nguyen L, Luu LM, Baranger AM, Zimmerman SC. Developing bivalent ligands to target CUG triplet repeats, the causative agent of myotonic dystrophy type 1. J Med Chem. 2013;56:9471–9481. doi: 10.1021/jm400794z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ofori LO, Hoskins J, Nakamori M, Thornton CA, Miller BL. From dynamic combinatorial ‘hit’ to lead: in vitro and in vivo activity of compounds targeting the pathogenic RNAs that cause myotonic dystrophy. Nucleic Acids Res. 2012;40:6380–6390. doi: 10.1093/nar/gks298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoskins JW, Ofori LO, Chen CZ, Kumar A, Sobczak K, Nakamori M, Southall N, Patnaik S, Marugan JJ, Zheng W, et al. Lomofungin and dilomofungin: inhibitors of MBNL1-CUG RNA binding with distinct cellular effects. Nucleic Acids Res. 2014;42:6591–6602. doi: 10.1093/nar/gku275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rzuczek SG, Gao Y, Tang ZZ, Thornton CA, Kodadek T, Disney MD. Features of modularly assembled compounds that impart bioactivity against an RNA target. ACS Chem Biol. 2013;8:2312–2321. doi: 10.1021/cb400265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guan L, Luo Y, Ja WW, Disney MD. Small molecule alteration of RNA sequence in cells and animals. Bioorg Med Chem Lett. 2017 doi: 10.1016/j.bmcl.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simms Carrie L, Hudson Benjamin H, Mosior John W, Rangwala Ali S, Zaher Hani S. An Active Role for the Ribosome in Determining the Fate of Oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang WY, He F, Strack RL, Oh SY, Frazer M, Jaffrey SR, Todd PK, Disney MD. Small Molecule Recognition and Tools to Study Modulation of r(CGG)(exp) in Fragile X-Associated Tremor Ataxia Syndrome. ACS Chem Biol. 2016;11:2456–2465. doi: 10.1021/acschembio.6b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 92.Garofalo M, Condorelli G, Croce CM. MicroRNAs in diseases and drug response. Curr Opin Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 93.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 94.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 96.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wen D, Danquah M, Chaudhary AK, Mahato RI. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J Control Release. 2015;219:237–247. doi: 10.1016/j.jconrel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tran T, Disney MD. Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat Commun. 2012;3:1125. doi: 10.1038/ncomms2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M, Childs-Disney JL. Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs. ACS Chem Biol. 2016;11:1720–1728. doi: 10.1021/acschembio.6b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR, Phinney DG, Disney MD. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:5898–5903. doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haga CL, Velagapudi SP, Strivelli JR, Yang WY, Disney MD, Phinney DG. Small Molecule Inhibition of miR-544 Biogenesis Disrupts Adaptive Responses to Hypoxia by Modulating ATM-mTOR Signaling. ACS Chem Biol. 2015;10:2267–2276. doi: 10.1021/acschembio.5b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu B, Childs-Disney JL, Znosko BM, Wang D, Fallahi M, Gallo SM, Disney MD. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinformatics. 2016;17:112. doi: 10.1186/s12859-016-0960-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roden C, Gaillard J, Kanoria S, Rennie W, Barish S, Cheng J, Pan W, Liu J, Cotsapas C, Ding Y, et al. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017;27:374–384. doi: 10.1101/gr.208900.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chirayil S, Chirayil R, Luebke KJ. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res. 2009;37:5486–5497. doi: 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Connelly CM, Boer RE, Moon MH, Gareiss P, Schneekloth JS. Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. Acs Chem Biol. 2017;12:435–443. doi: 10.1021/acschembio.6b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shortridge MD, Walker MJ, Pavelitz T, Chen Y, Yang W, Varani G. A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chem Biol. 2017;12:1611–1620. doi: 10.1021/acschembio.7b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Velagapudi SP, Disney MD. Two-dimensional combinatorial screening enables the bottom-up design of a microRNA-10b inhibitor. Chem Commun (Camb) 2014;50:3027–3029. doi: 10.1039/c3cc00173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lorenz DA, Garner AL. A click chemistry-based microRNA maturation assay optimized for high-throughput screening. Chem Commun (Camb) 2016;52:8267–8270. doi: 10.1039/c6cc02894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Agarwala P, Pandey S, Maiti S. The tale of RNA G-quadruplex. Org Biomol Chem. 2015;13:5570–5585. doi: 10.1039/c4ob02681k. [DOI] [PubMed] [Google Scholar]
- 112.Fay MM, Lyons SM, Ivanov P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J Mol Biol. 2017;429:2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 114.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353 doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miglietta G, Cogoi S, Marinello J, Capranico G, Tikhomirov AS, Shchekotikhin A, Xodo LE. RNA G-Quadruplexes in Kirsten Ras (KRAS) Oncogene as Targets for Small Molecules Inhibiting Translation. J Med Chem. 2017 doi: 10.1021/acs.jmedchem.7b00622. [DOI] [PubMed] [Google Scholar]
- 116.Pandey S, Agarwala P, Jayaraj GG, Gargallo R, Maiti S. The RNA Stem-Loop to G-Quadruplex Equilibrium Controls Mature MicroRNA Production inside the Cell. Biochemistry. 2015;54:7067–7078. doi: 10.1021/acs.biochem.5b00574. [DOI] [PubMed] [Google Scholar]
- 117.Katsuda Y, Sato S, Asano L, Morimura Y, Furuta T, Sugiyama H, Hagihara M, Uesugi M. A Small Molecule That Represses Translation of G-Quadruplex-Containing mRNA. J Am Chem Soc. 2016;138:9037–9040. doi: 10.1021/jacs.6b04506. [DOI] [PubMed] [Google Scholar]
- 118.Weldon C, Dacanay JG, Gokhale V, Boddupally PVL, Behm-Ansmant I, Burley GA, Branlant C, Hurley LH, Dominguez C, Eperon IC. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weldon C, Eperon IC, Dominguez C. Do we know whether potential G-quadruplexes actually form in long functional RNA molecules? Biochem Soc Trans. 2016;44:1761–1768. doi: 10.1042/BST20160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weldon C, Behm-Ansmant I, Hurley LH, Burley GA, Branlant C, Eperon IC, Dominguez C. Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat Chem Biol. 2017;13:18–20. doi: 10.1038/nchembio.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 122.Scherr AL, Gdynia G, Salou M, Radhakrishnan P, Duglova K, Heller A, Keim S, Kautz N, Jassowicz A, Elssner C, et al. Bcl-xL is an oncogenic driver in colorectal cancer. Cell Death Dis. 2016;7:e2342. doi: 10.1038/cddis.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Havens MA, Duelli DM, Hastings ML. Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Douglas AGL, Wood MJA. RNA splicing: disease and therapy. Brief Funct Genomics. 2011;10:151–164. doi: 10.1093/bfgp/elr020. [DOI] [PubMed] [Google Scholar]
- 130.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 134.Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 135.Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.FDA approves first drug for spinal muscular atrophy. Available at: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm534611.htm. (Accessed January 19, 2018)
- 137.Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 138.ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US) Identifier NCT02268552. An Open Label Study of LMI070 (Branaplam) in Type 1 Spinal Muscular Atrophy (SMA) 2014 Oct 20; 2017 Oct 30. Available at: https://clinicaltrials.gov/ct2/show/NCT02268552. (Accessed January 18, 2018)
- 139.Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KK, Karp GM, Qi H, Woll MG, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 140.Sivaramakrishnan M, McCarthy KD, Campagne S, Huber S, Meier S, Augustin A, Heckel T, Meistermann H, Hug MN, Birrer P, et al. Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers. Nat Commun. 2017;8:1476. doi: 10.1038/s41467-017-01559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life (Basel) 2015;5:1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 144.Myasnikov AG, Kundhavai Natchiar S, Nebout M, Hazemann I, Imbert V, Khatter H, Peyron JF, Klaholz BP. Structure-function insights reveal the human ribosome as a cancer target for antibiotics. Nat Commun. 2016;7:12856. doi: 10.1038/ncomms12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 146.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017;551:472–477. doi: 10.1038/nature24482. [DOI] [PubMed] [Google Scholar]
- 147.Marcel V, Catez F, Diaz J-J. Ribosomes: the future of targeted therapies? Oncotarget. 2013;4:1554–1555. doi: 10.18632/oncotarget.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 150.Jandura A, Krause HM. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 151.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu F, Somarowthu S, Pyle AM. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol. 2017;13:282–289. doi: 10.1038/nchembio.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xi L, Cech TR. Protein-RNA interaction restricts telomerase from running through the stop sign. Nat Struct Mol Biol. 2015;22:835–836. doi: 10.1038/nsmb.3118. [DOI] [PubMed] [Google Scholar]
- 155.Sunwoo H, Wu JY, Lee JT. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc Natl Acad Sci U S A. 2015;112:E4216–4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McFadden EJ, Hargrove AE. Biochemical Methods To Investigate lncRNA and the Influence of lncRNA:Protein Complexes on Chromatin. Biochemistry. 2016;55:1615–1630. doi: 10.1021/acs.biochem.5b01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 159.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pintacuda G, Young AN, Cerase A. Function by Structure: Spotlights on Xist Long Non-coding RNA. Front Mol Biosci. 2017;4:90. doi: 10.3389/fmolb.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 162.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 163.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 164.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 165.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A. 2016;113:10322–10327. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 167.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta Gene Regul Mech. 2016;1859:192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 168.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Hoogsteen-position pyrimidines promote the stability and function of the MALAT1 RNA triple helix. RNA. 2016;22:743–749. doi: 10.1261/rna.055707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 176.Ozes AR, Wang Y, Zong X, Fang F, Pilrose J, Nephew KP. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci Rep. 2017;7:894. doi: 10.1038/s41598-017-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pedram Fatemi R, Salah-Uddin S, Modarresi F, Khoury N, Wahlestedt C, Faghihi MA. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. J Biomol Screen. 2015;20:1132–1141. doi: 10.1177/1087057115594187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang J, Shi Z, Nan Y, Li M. Inhibiting malignant phenotypes of the bladder cancer cells by silencing long noncoding RNA SChLAP1. Int Urol Nephrol. 2016;48:711–716. doi: 10.1007/s11255-016-1230-2. [DOI] [PubMed] [Google Scholar]
- 180.Li Y, Syed J, Sugiyama H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem Biol. 23:1325–1333. doi: 10.1016/j.chembiol.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 181.Kalwa M, Hanzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631–10643. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 183.Bacolla A, Wang G, Vasquez KM. New Perspectives on DNA and RNA Triplexes As Effectors of Biological Activity. PLoS Genet. 2015;11:e1005696. doi: 10.1371/journal.pgen.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shaw NN, Arya DP. Recognition of the unique structure of DNA:RNA hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 185.Rouleau S, Jodoin R, Garant JM, Perreault JP. RNA G-Quadruplexes as Key Motifs of the Transcriptome. Adv Biochem Eng Biotechnol. 2017 doi: 10.1007/10_2017_8. [DOI] [PubMed] [Google Scholar]
- 186.Hirashima K, Seimiya H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015;43:2022–2032. doi: 10.1093/nar/gkv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Martadinata H, Heddi B, Lim KW, Phan AT. Structure of long human telomeric RNA (TERRA): G-quadruplexes formed by four and eight UUAGGG repeats are stable building blocks. Biochemistry. 2011;50:6455–6461. doi: 10.1021/bi200569f. [DOI] [PubMed] [Google Scholar]
- 188.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Garavis M, Lopez-Mendez B, Somoza A, Oyarzabal J, Dalvit C, Villasante A, Campos-Olivas R, Gonzalez C. Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through 19F-NMR based fragment screening. ACS Chem Biol. 2014;9:1559–1566. doi: 10.1021/cb500100z. [DOI] [PubMed] [Google Scholar]
- 190.Zhang Y, Zeng D, Cao J, Wang M, Shu B, Kuang G, Ou TM, Tan JH, Gu LQ, Huang ZS, et al. Interaction of Quindoline derivative with telomeric repeat-containing RNA induces telomeric DNA-damage response in cancer cells through inhibition of telomeric repeat factor 2. Biochim Biophys Acta. 2017;1861:3246–3256. doi: 10.1016/j.bbagen.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 191.Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, Youmans DT, Cech TR, Davidovich C. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell. 2017;65:1056–1067 e1055. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 192.Matsumura K, Kawasaki Y, Miyamoto M, Kamoshida Y, Nakamura J, Negishi L, Suda S, Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36:1191–1199. doi: 10.1038/onc.2016.282. [DOI] [PubMed] [Google Scholar]
- 193.Fukuhara M, Ma Y, Nagasawa K, Toyoshima F. A G-quadruplex structure at the 5′ end of the H19 coding region regulates H19 transcription. Sci Rep. 2017;8:45815. doi: 10.1038/srep45815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huang H, Zhang J, Harvey SE, Hu X, Cheng C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017 doi: 10.1101/gad.305862.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015;589:1653–1668. doi: 10.1016/j.febslet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 196.Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS. Neuron. 2014;84:239. doi: 10.1016/j.neuron.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 197.Wang BD, Ceniccola K, Hwang S, Andrawis R, Horvath A, Freedman JA, Olender J, Knapp S, Ching T, Garmire L, et al. Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat Commun. 2017;8:15921. doi: 10.1038/ncomms15921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 201.Pisignano G, Napoli S, Magistri M, Mapelli SN, Pastori C, Di Marco S, Civenni G, Albino D, Enriquez C, Allegrini S, et al. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat Commun. 2017;8:15622. doi: 10.1038/ncomms15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 203.Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. 2012;1826:121–128. doi: 10.1016/j.bbcan.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Thorenoor N, Slaby O. Small nucleolar RNAs functioning and potential roles in cancer. Tumour Biol. 2015;36:41–53. doi: 10.1007/s13277-014-2818-8. [DOI] [PubMed] [Google Scholar]
- 205.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 206.Yoshihama M, Nakao A, Kenmochi N. snOPY: a small nucleolar RNA orthological gene database. BMC Res Notes. 2013;6:426. doi: 10.1186/1756-0500-6-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang X, Cozen AE, Liu Y, Chen Q, Lowe TM. Small RNA Modifications: Integral to Function and Disease. Trends Mol Med. 2016;22:1025–1034. doi: 10.1016/j.molmed.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 210.Batista PJ. The RNA Modification N(6)-methyladenosine and Its Implications in Human Disease. Genomics Proteomics Bioinformatics. 2017;15:154–163. doi: 10.1016/j.gpb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Carlson CD, Warren CL, Hauschild KE, Ozers MS, Qadir N, Bhimsaria D, Lee Y, Cerrina F, Ansari AZ. Specificity landscapes of DNA binding molecules elucidate biological function. Proc Natl Acad Sci U S A. 2010;107:4544–4549. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Eubanks CS, Forte JE, Kapral GJ, Hargrove AE. Small Molecule-Based Pattern Recognition To Classify RNA Structure. J Am Chem Soc. 2017;139:409–416. doi: 10.1021/jacs.6b11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lu Z, Gong J, Zhang QC. PARIS: Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution. Methods Mol Biol. 2018;1649:59–84. doi: 10.1007/978-1-4939-7213-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Baird NJ, Inglese J, Ferre-D’Amare AR. Rapid RNA-ligand interaction analysis through high-information content conformational and stability landscapes. Nat Commun. 2015;6:8898. doi: 10.1038/ncomms9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Feng C, Chan D, Joseph J, Muuronen M, Coldren WH, Dai N, Corrêa IR, Jr, Furche F, Hadad CM, Spitale RC. Light-activated chemical probing of nucleobase solvent accessibility inside cells. Nat Chem Biol. 2018 doi: 10.1038/nchembio0318-325. [DOI] [PubMed] [Google Scholar]
- 219.Eubanks CS, Hargrove AE. Sensing the impact of environment on small molecule differentiation of RNA sequences. Chem Commun (Camb) 2017;53:13363–13366. doi: 10.1039/c7cc07157d. [DOI] [PubMed] [Google Scholar]


