DSC-MR imaging data were collected after a preload and during a bolus injection of gadolinium contrast agent using a gradient recalled-echo-EPI sequence. Forty-nine low-grade and high-grade glioma datasets were uploaded to The Cancer Imaging Archive. Datasets included a predetermined arterial input function, enhancing tumor ROIs, and ROIs necessary to create normalized relative CBV and CBF maps. Seven sites computed 20 different perfusion metrics. For normalized relative CBV and normalized CBF, 93% and 94% of entries showed good or excellent cross-site agreement. All metrics could distinguish low- from high-grade tumors.
Abstract
BACKGROUND AND PURPOSE:
Standard assessment criteria for brain tumors that only include anatomic imaging continue to be insufficient. While numerous studies have demonstrated the value of DSC-MR imaging perfusion metrics for this purpose, they have not been incorporated due to a lack of confidence in the consistency of DSC-MR imaging metrics across sites and platforms. This study addresses this limitation with a comparison of multisite/multiplatform analyses of shared DSC-MR imaging datasets of patients with brain tumors.
MATERIALS AND METHODS:
DSC-MR imaging data were collected after a preload and during a bolus injection of gadolinium contrast agent using a gradient recalled-echo–EPI sequence (TE/TR = 30/1200 ms; flip angle = 72°). Forty-nine low-grade (n = 13) and high-grade (n = 36) glioma datasets were uploaded to The Cancer Imaging Archive. Datasets included a predetermined arterial input function, enhancing tumor ROIs, and ROIs necessary to create normalized relative CBV and CBF maps. Seven sites computed 20 different perfusion metrics. Pair-wise agreement among sites was assessed with the Lin concordance correlation coefficient. Distinction of low- from high-grade tumors was evaluated with the Wilcoxon rank sum test followed by receiver operating characteristic analysis to identify the optimal thresholds based on sensitivity and specificity.
RESULTS:
For normalized relative CBV and normalized CBF, 93% and 94% of entries showed good or excellent cross-site agreement (0.8 ≤ Lin concordance correlation coefficient ≤ 1.0). All metrics could distinguish low- from high-grade tumors. Optimum thresholds were determined for pooled data (normalized relative CBV = 1.4, sensitivity/specificity = 90%:77%; normalized CBF = 1.58, sensitivity/specificity = 86%:77%).
CONCLUSIONS:
By means of DSC-MR imaging data obtained after a preload of contrast agent, substantial consistency resulted across sites for brain tumor perfusion metrics with a common threshold discoverable for distinguishing low- from high-grade tumors.
Current Response Assessment in Neuro-Oncology criteria,1 which include anatomic imaging only, are often insufficient for determining tumor response and progression after therapy. Frequently, imaging changes on postcontrast MR imaging or FLAIR are not the result of increased tumor activity but rather a consequence of the treatment itself. These changes, which can include edema, inflammation, or increased vascular or blood-brain barrier permeability, make it difficult to distinguish true tumor progression from treatment response.2–6 Because of these difficulties, patients are often precluded from switching to potentially more effective therapies within treatment windows of 3–5 months.1 Clearly, better indications of tumor response that are not confounded by these treatment adverse effects are needed.
Perfusion MR imaging methods, which have repeatedly demonstrated the ability to provide biologically relevant information for treatment management, have the potential to overcome these limitations. For brain perfusion, the DSC-MR imaging methods have been most commonly used. With DSC-MR imaging, T2- or T2*-weighted images are acquired with high temporal resolution during a bolus administration of a gadolinium contrast agent.7 The derived relative CBV (rCBV) maps have demonstrated the ability to predict tumor grade8,9 and survival,10 distinguish treatment effects from recurrent tumor,11,12 and predict response to antiangiogenic therapy more reliably than standard MR imaging.13–15
Despite this promise, the translation of DSC-MR imaging for routine clinical use has been hindered by a lack of consistency in the methods used and the rCBV values reported to make the noted distinctions. However, often a threshold determined for one purpose, such as distinguishing low- from high-grade tumor,16 is used for another purpose such as predicting outcomes.17 Consequently, the present confusion may be due to the lack of well-defined studies performed under carefully controlled conditions that test a specific outcome. This study addresses these limitations by providing carefully curated DSC-MR imaging datasets of low-grade glioma (LGG) and high-grade glioma (HGG) to multiple sites that participate in the National Cancer Institute Quantitative Imaging Network. With this approach, variations in image acquisition and preprocessing are eliminated and postprocessing methods can be directly compared in their ability to distinguish LGGs from HGGs. In addition, the threshold for this distinction can be identified both for each individual site and as a consensus recommendation.
Materials and Methods
Patients
All subjects recruited from a single site provided informed written consent according to institutional review board policy. Subjects with histologically confirmed, newly diagnosed, and treatment-naïve glial tumors who had preoperative DSC-MR imaging were included in this study. Subjects with purely oligodendroglial lesions were not included due to demonstrated differences in rCBV compared with astrocytic tumors.18 Due to the disparity in the number of subjects histologically diagnosed with low- and high-grade tumors, consecutive subjects with low-grade tumors between 2008 and 2014 and high-grade tumors from 2010 to 2014 were identified. Subjects were excluded if anatomic images were not available for lesion delineation or when DSC-MR imaging data were of poor quality.
Imaging
Studies were performed on 1.5T or 3T MR imaging systems. Standard precontrast MRIs were acquired including FLAIR (TE/TR/NEX/matrix = 126/9000 ms/2200–2500 ms/2/252 × 215) and T1WI spin-echo (TE/TR/NEX/matrix = 11 ms/650 ms/2/256), after which gadolinium contrast agent was administered. Either 0.05 mmol/kg of gadobenate dimeglumine (MultiHance; Bracco Diagnostics, Princeton, New Jersey) (n = 48) or 0.1 mmol/kg of gadodiamide (Omniscan; GE Healthcare, Piscataway, New Jersey) (n = 1) was administered before obtaining the postcontrast T1-weighted images. This initial gadolinium administration serves as a preload for the subsequent DSC-MR imaging acquisition. A preload results in more accurate rCBV because it reduces the T1 leakage effects that can confound the DSC-MR imaging data analysis.9,19–21 After collecting the postcontrast T1WI (T1+C), using parameters equivalent to the precontrast settings, we acquired DSC-MR imaging data (gradient recalled-echo–EPI: TR = 1100–1250 ms, TE = 30 ms, flip angle = 70°–80°, 12–17 slices, slice thickness = 4–5 mm, interslice gap = 0–1.5 mm, matrix = 96 × 96/128 × 128, FOV = 165–240 × 165–240 mm2). The gradient recalled-echo–EPI data were collected for 120 seconds with 0.1 mmol/kg of gadolinium injected at 60 seconds. For ease of coregistration of the DSC-MR images to anatomic images, a spin-echo “reference” scan (TE/TR/NEX/matrix = 14 ms/500 ms/1/256 × 192) was obtained using a slice prescription matching the more limited slice prescription of the DSC-MR imaging examination.
Central Preprocessing
The preprocessing workflow is schematized in Fig 1. All preprocessing was performed in OsiriX Imaging Software (http://www.osirix-viewer.com) using the IB Delta Suite (Imaging Biometrics, Elm Grove, Wisconsin). Six datasets provided for each case included the following: 1) T1+C images; 2) DSC-MR imaging time-series co-registered to the T1+C images; 3) an arterial input function (AIF), which included 3 AIF locations in each file; 4) a whole-brain mask and ROIs of 5) normal-appearing white matter (NAWM) and 6) tumor. The NAWM was used to compute normalized rCBV (nRCBV) and normalized CBF (nCBF) maps.
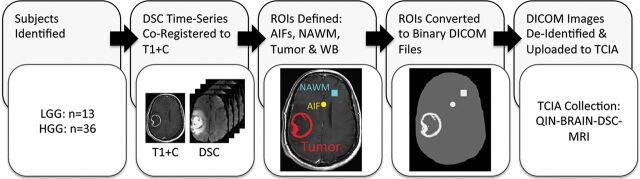
Fig 1.
Preprocessing workflow. Forty-nine subjects were identified, 13 of whom had a diagnosis of low-grade glioma and 36 with a diagnosis of high-grade glioma. The DSC-MR image volume was co-registered to the T1+C images via the reference scan obtained with the same slice prescription as the DSC-MR imaging. Six datasets were provided for each case including the following: 1) T1+C images; 2) DSC-MR imaging time-series registered to the T1+C images; 3) an AIF, which included 3 AIF locations in each file; 4) a whole brain (WB) mask and ROIs of 5) normal-appearing white matter (NAWM), and 6) tumor. Each ROI was created as a 16-bit binary DICOM file that included only whole voxels rather than contoured points. Anonymized datasets were uploaded to The Cancer Imaging Archive. QIN indicates Quantitative Imaging Network.
The DSC-MR imaging volume was co-registered to the T1+C images via the reference scan obtained with the same slice prescription as the DSC-MR imaging. The AIF locations were determined semiautomatically using IB Neuro (Imaging Biometrics) with manual adjustments when necessary. An average signal generated from 3 pixels constituted the AIF. The whole-brain mask was made available to prevent disparity in values that could result from threshold variations that each software platform might use. Using the IB Delta Suite, we determined tumor ROIs from deltaT1 maps, which are standardized difference maps22 that enable clear visualization of enhancing lesions free of bright signal from blood products or proteinaceous material. Nonenhancing lesions, apparent as areas of dark signal on T1WIs, were delineated by a neuroradiologist with >20 years of experience. Each ROI was created as a 16-bit binary DICOM file that included only whole voxels rather than contoured points. This approach prevents differences in the applied ROIs because each software platform applies different rules regarding whether a voxel is considered inside or outside an ROI. Anonymized datasets were uploaded to The Cancer Imaging Archive,23,24 where they were further vetted to ensure the compatibility of complete datasets for the analysis platform of each site. All sites were blinded to tumor grade.
Image and Statistical Analyses
Seven sites (1–7) using 7 different software platforms provided 20 different rCBV measurements (Table 1) and 12 different CBF measurements (Table 2). Details for each software platform are listed in the tables. Several sites used >1 platform or analysis method. When applicable, the analysis for measurements of standardized rCBV was grouped separately from nRCBV due to image-scale differences. Agreement between each pair of values was assessed by computing the Lin concordance correlation coefficient (LCCC). An LCCC > 0.8 indicates good agreement, and LCCC > 0.9 indicates excellent agreement.
Table 1:
Teams and rCBV analysis methods
| Team/Entry No. | Software | Scaling | Leakage Correction | Description |
|---|---|---|---|---|
| A1 | IB Neuro | NAWM | Yes | Modified BSW21 |
| B1 | IB Neuro | NAWM | Yes | Modified BSW21 |
| C1 | IB Neuro | NAWM | Yes | Modified BSW21 |
| B2 | Matlaba | NAWM | Yes | 120-point TI (BSW21) |
| B3 | Matlab | AIF | Yes | 120-point TI (BSW21) |
| D1 | nordicICE | NAWM | Yes | 120-point TI (BSW21) |
| D2 | nordicICE | Population AIF | Yes | 120-point TI (BSW21) |
| E1 | PGUI (LC)b | NAWM | Yes | 120-point TI (BSW21) |
| E2 | PGUI (C) | NAWM | Yes | AUC of C(t) ratio for ROI/AIF |
| E3 | PGUI (PM LC) | NAWM | Yes | Bayesian parametric FT44 |
| D3 | Slicerc | NAWM | Yes | 120-point TI (BSW21) |
| F1 | FIATd | NAWM | Yes | Modified Weisskoff45,46 |
| G1 | MCcbve | NA | Yes | Modified Weisskoff45,46 |
| A2 | IB Neuro | NA (standardized)47 | Yes | Modified BSW21 2006 |
| A3 | IB Neuro | NAWM | No | Leakage correction not used |
| B4 | IB Neuro | NAWM | No | Leakage correction not used |
| E4 | PGUI | NAWM | No | Leakage correction not used |
| E5 | PGUI | NAWM | No | Deconvolution of residue function (sSVD) |
| E6 | PGUI | NAWM | No | Deconvolution of residue function (oSVD) |
| A4 | IB Neuro | N/A (standardized)47 | No | Leakage correction not used |
Note:—TI indicates trapezoidal integration; BSW, Boxerman-Schmainda-Weisskoff; NA, not applicable; FT, Fourier Transform; sSVD, standard singular value decomposition; oSVD, block circulant singular value decomposition; C(t) ratio, concentration time course.
MathWorks, Natick, Massachusetts.
Perfusion Graphical User Interface; MINDlab, Center of Functionally Integrative Neuroscience, Aarhus University Hospital Norrebrogade, Denmark.44
Functional Image Analysis Tool (University of Michigan).
Mayo Clinic cerebral blood volume (Mayo Clinic, Rochester, Minnesota).37
Table 2:
Teams and nCBF analysis methods
| Team/Entry No. | Software | Scaling | Description |
|---|---|---|---|
| A5 | IB Neuro | NAWM | SVD: deconvolution of ΔR2* with AIF |
| B5 | IB Neuro | NAWM | SVD: deconvolution of ΔR2* with AIF |
| C2 | IB Neuro | NAWM | SVD: circular deconvolution of ΔR2* with AIF |
| B6 | Matlab | NAWM | SVD applied to leakage-corrected tissue ΔR2* and AIF ΔR2* curves |
| D4 | nordicICE | NAWM | No AIF |
| D5 | nordicICE | Population AIF | Population AIF |
| E7 | PGUI | NAWM | nCBF |
| E8 | PGUI | NAWM | sSVD |
| E9 | PGUI | NAWM | oSVD |
| E10 | PGUI | NAWM | Bayesian FT deconvolution44 |
| D6 | Slicer | NAWM | No AIF |
| F2 | FIAT | NAWM | nCBF |
Note:—SVD indicates singular value decomposition; sSVD, standard singular value decomposition; oSVD, block circulant singular value decomposition; FT, Fourier Transform..
The ability of each metric to distinguish LGG from HGG was determined using the Wilcoxon rank sum test, with P < .05 indicating significance. A receiver operating characteristic analysis was performed to identify the threshold that provides the optimal sensitivity (SN) and specificity (SP) to distinguish LGG from HGG. The DeLong test for comparing ≥2 receiver operating characteristic curves was used to determine whether there were differences in the ability of each measurement to classify tumors.
To determine “consensus” cutoff points, we created boxplots of the sum of SN and SP. Optimal thresholds were identified as those with maximum SN + SP mean, according to the Youden selection criteria with small variance. The random effects model was used to assess the reliability of measurements across sites and platforms. The reliability is quantified by the following:
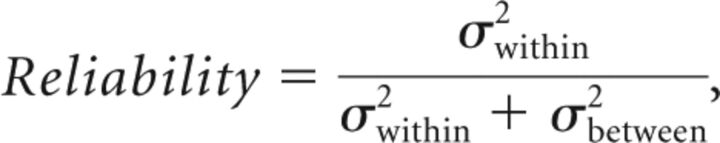 |
where σ is the SD within or between software platforms.
Finally, to assess the clinical relevance of the study observations, we determined the false-positive rate from T1+C images in comparison with each of the perfusion parameters (nRCBV, standardized rCBV, nCBF). The false-positive rate is defined as the proportion of low-grade tumors thought to be aggressive and/or high-grade, as indicated by the decision for tumor resection, relative to all tumors resected. The false-negative rate was not determined because data from all patients, including those who did not undergo an operation, were not available. By means of the T1+C images, the false-positive rate was defined as the proportion of tumors that are low-grade and had contrast agent–enhancing lesions. For the perfusion parameters, a false-positive rate is defined as a low-grade tumor with a value above the threshold determined for distinguishing low- from high-grade tumors.
Results
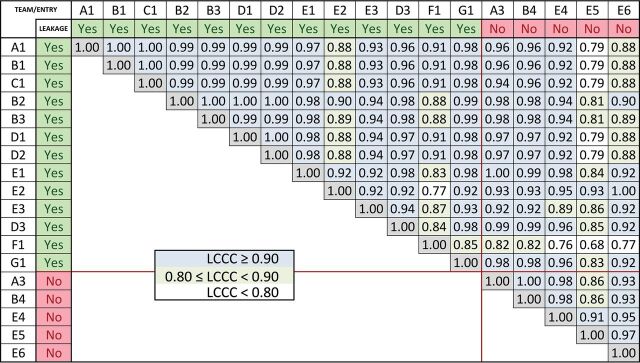
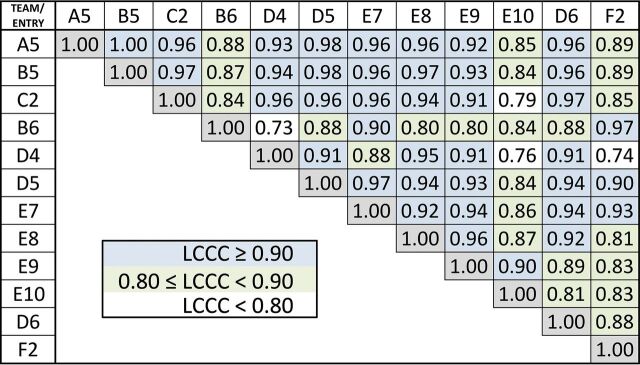
Sixty-three subjects met inclusion criteria for this study, with 14 excluded for the following reasons: The contrast agent bolus was delayed during DSC acquisition, preventing capture of the postbolus steady-state signal (n = 4); the contrast agent bolus was injected too slowly and in an irregular pattern (n = 4); the contrast agent bolus was not present during acquisition of images (n = 2); there were severe ghosting and motion artifacts of DSC images (n = 2); or anatomic images were not available for lesion delineation (n = 2). Forty-nine co-registered LGG (n = 13) and HGG (n = 36) DSC-MR imaging datasets were preprocessed, anonymized, and uploaded to The Cancer Imaging Archive (Fig 1). Tumor grade was confirmed with histopathology a median of 3 days (range, 0–4 days) following MR imaging. Examples of postprocessed datasets are shown in Fig 2. LCCC results are displayed in a matrix listing each nRCBV (Fig 3) or nCBF (Fig 4) entry on both the x- and y-axes. For tumor nRCBV, 75% of the entries showed excellent agreement with LCCC ≥ 0.9 and 19% with good agreement (0.80 ≤ LCCC < 0.90), leaving only 6% with poor concordance (LCCC < 0.80). The concordance was best for nRCBV values determined with leakage correction. For nCBF, only 59% had 0.90 ≤ LCCC ≤ 1.0 and 34% had 0.80 ≤ LCCC ≤ 0.89.
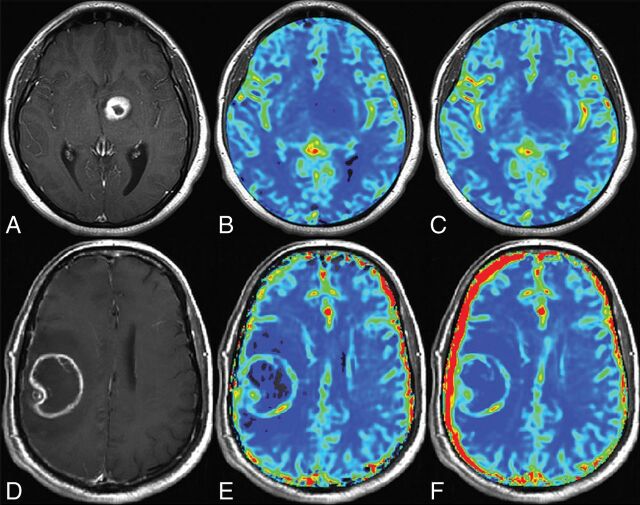
Fig 2.
Sample parameter maps. Shown are postcontrast T1-weighted images (A and D) and corresponding rCBV (B and E) and CBF (C and F) maps for one patient with a low-grade glioma (A–C) and another patient with a high-grade glioma (D–F).
Fig 3.
Lin concordance results for nRCBV. Listed are the Lin concordance correlation coefficients for the nRCBV values for each pair of sites. Whether leakage correction was applied is indicated by green labels, with red labels indicating no leakage correction. Pairs with excellent concordance (LCCC > 0.90) are highlighted with blue-filled squares; very good concordance (0.80 < LCCC < 0.90), with green squares; and weaker concordance (LCCC < 0.80), with white squares.
Fig 4.
Lin concordance results for nCBF. Listed are the Lin concordance correlation coefficients for the normalized CBF values for each pair of sites. Pairs with excellent concordance (LCCC > 0.90) are highlighted with blue-filled squares; very good concordance (0.80 < LCCC <0.90), with green squares; and weaker concordance (LCCC < 0.80), with white squares.
For all software platforms, both nRCBV and nCBF showed statistically significant differences between LGG and HGG (Tables 3 and 4), with a mean nRCBV = 1.4 ± 0.13 and mean nCBF = 1.57 ± 0.24. The SN/SP for nRCBV ranged from 81%–97%/77%–85% and was slightly worse for nCBF with SN/SP = 64%–97%/69%–85%. By means of the DeLong test, no significant differences were found among the 18 nRCBV (P = .72) metrics to distinguish LGG from HGG. While differences among the nCBF metrics were borderline significant (P = .05), if the entry with the lowest area under the curve (0.658) was excluded, there was no significant difference between the remaining measures (P = .49). The DeLong test for the standardized rCBV showed no significant distinction between the 2 submissions for this measure (P = .23).
Table 3:
nRCBV low- vs high-grade thresholds and ROC analysis results
| Team/ Entry No. | Software Platform | Leakage Correction Used | LG vs HG (P Value) | Threshold | SN | SP | AUC |
|---|---|---|---|---|---|---|---|
| A1 | IB Neuro | Yes | 5.34E-06 | 1.605 | 0.861 | 0.846 | 0.889 |
| B1 | IB Neuro | Yes | 5.36E-06 | 1.605 | 0.861 | 0.846 | 0.889 |
| C1 | IB Neuro | Yes | 5.39E-06 | 1.605 | 0.861 | 0.846 | 0.889 |
| B2 | Matlab | Yes | 2.02E-06 | 1.315 | 0.972 | 0.769 | 0.892 |
| B3 | Matlab | Yes | 1.75E-06 | 1.330 | 0.972 | 0.769 | 0.896 |
| D1 | nordicICE | Yes | 3.30E-06 | 1.305 | 0.972 | 0.769 | 0.895 |
| D2 | nordicICE | Yes | 3.30E-06 | 1.305 | 0.972 | 0.769 | 0.895 |
| E1 | PGUI (LC) | Yes | 7.32E-06 | 1.615 | 0.861 | 0.846 | 0.892 |
| E2 | PGUI (C) | Yes | 1.14E-05 | 1.400 | 0.861 | 0.769 | 0.871 |
| E3 | PGUI (PM LC) | Yes | 1.18E-05 | 1.400 | 0.889 | 0.769 | 0.856 |
| D3 | Slicer | Yes | 1.08E-06 | 1.415 | 0.944 | 0.769 | 0.891 |
| F1 | FIAT | Yes | 2.50E-05 | 1.235 | 0.944 | 0.769 | 0.853 |
| G1 | McCBV | Yes | 1.26E-06 | 1.425 | 0.944 | 0.769 | 0.891 |
| A3 | IB Neuro | No | 1.55E-06 | 1.330 | 0.917 | 0.769 | 0.895 |
| B4 | IB Neuro | No | 1.56E-06 | 1.330 | 0.917 | 0.769 | 0.895 |
| E4 | PGUI | No | 6.94E-06 | 1.450 | 0.861 | 0.769 | 0.887 |
| E5 | PGUI | No | 5.67E-05 | 1.585 | 0.806 | 0.846 | 0.872 |
| E6 | PGUI | No | 1.19E-05 | 1.395 | 0.861 | 0.769 | 0.873 |
Note:—AUC indicates area under the curve; LG, low-grade; HG, high-grade.
Table 4:
nCBF low- vs high-grade thresholds and ROC analysis results
| Team/Entry No. | Software | LG vs HG (P Value) | Threshold | SN | SP | AUC |
|---|---|---|---|---|---|---|
| A5 | IB Neuro | 2.90E-05 | 1.815 | 0.806 | 0.846 | 0.861 |
| B5 | IB Neuro | 2.38E-05 | 1.555 | 0.917 | 0.769 | 0.864 |
| C2 | IB Neuro | 3.82E-06 | 1.415 | 0.944 | 0.769 | 0.880 |
| B6 | Matlab | 2.61E-04 | 2.045 | 0.722 | 0.846 | 0.814 |
| D4 | nordicICE | 9.81E-06 | 1.255 | 0.972 | 0.769 | 0.868 |
| D5 | nordicICE | 3.03E-05 | 1.580 | 0.861 | 0.846 | 0.866 |
| E7 | PGUI | 1.30E-04 | 1.575 | 0.806 | 0.846 | 0.846 |
| E8 | PGUI | 3.85E-05 | 1.375 | 0.917 | 0.692 | 0.845 |
| E9 | PGUI | 1.67E-04 | 1.460 | 0.833 | 0.769 | 0.812 |
| E10 | PGUI | 7.86E-03 | 1.920 | 0.639 | 0.692 | 0.658 |
| D6 | Slicer | 1.04E-06 | 1.405 | 0.944 | 0.769 | 0.887 |
| F2 | FIAT | 2.65E-04 | 1.460 | 0.861 | 0.769 | 0.816 |
Note:—AUC indicates area under the curve; LG, low-grade; HG, high-grade.
Alternatively, the data can be pooled, as shown by the boxplots of SN + SP (Fig 5), for which median and quartile values are indicated. The maximum sums were the following: nRCBV = 1.4 (SN/SP = 90%/77%) and nCBF = 1.58 (SN/SP = 86%/77%). For these consensus thresholds, the minimum individual SN/SP was 83%/77% for nRCBV and 80%/70% for nCBF.
Fig 5.
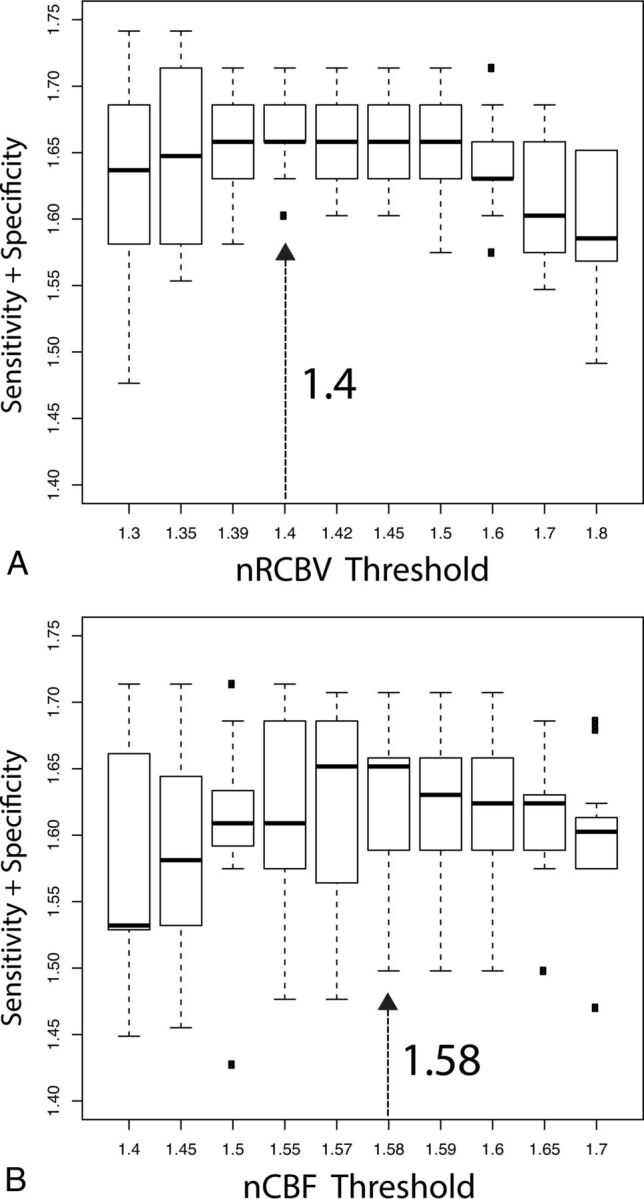
Boxplots showing the sum of sensitivity and specificity values for nRCBV (A) and nCBF (B) for all sites and metrics. From these data, values that gave a maximum sum of specificity plus sensitivity and minimal variance were chosen as the consensus threshold values.
For the 18 nRCBV measurements, the reliability was determined to be 0.93, indicating that 93% of the variation can be attributed to differences in tumors, and 7%, to differences in analysis methods. The reliability was 95% for nRCBV determined with leakage correction and 93% for the group without leakage correction. For the nRCBV computed with one of the most common leakage-correction algorithms (Boxerman-Schmainda-Weisskoff21), the reliability improved to 98%. The reliability of standardized rCBV was 96%. For the 12 nCBF measurements, the reliability was 61%.
Finally, the false-positive rate using T1+C was found to be 31%. In comparison, the false-positive rate across all software platforms was 15%–23% for nRCBV, 15%–31% for nCBF, and 8%–15% for standardized rCBV.
Discussion
By means of carefully curated DSC-MR imaging datasets, obtained with a single acquisition approach, all nRCBV and nCBF metrics, processed by 7 different sites, could distinguish LGG from HGG. The optimal nRCBV and nCBF thresholds varied by only 9% and 15%, respectively. Unique to this study, consensus thresholds of nRCBV = 1.4 and nCBF = 1.58 were determined, indicating good accuracy overall and for each individual site. These results should bolster confidence in the ability of DSC-MR imaging to provide reliable and consistent cross-platform perfusion metrics for the evaluation of brain tumors and, specifically, for distinguishing low- from high-grade gliomas.
The range of nRCBV threshold values determined in this study is much tighter than the 0.7–3.0 range previously reported for distinguishing tumor grade,25,26 predicting differences in survival,10,17,27–29 and distinguishing true progression from pseudoprogression30 and tumor from treatment effect.11,12,31 While this large range of threshold values has been attributed to different acquisition and postprocessing schemes,20 differences in patient populations and the clinical questions addressed also contribute to the variabilities. While it is unlikely that a single threshold can be universally applied for all clinical questions, these studies suggest that with well-defined studies to address a specific outcome under carefully controlled conditions, it is possible to reach consensus.
The present study also demonstrates a greater cross-platform concordance than that previously reported. For example, in one study,32 2 commercial software packages (nordicICE; NordicNeuroLab, Bergen, Norway and BrainSTAT; GE Healthcare) were compared. Like the present study, 1 dataset of 24 patients with de novo glioblastoma was used and ROIs of tumor and reference brain were predetermined. However, unlike the present study, vastly different mathematic algorithms were applied, resulting in very disparate definitions for nRCBV and CBF; thus, a wide range of values was reported. In the present study, most algorithms involved the integration of the concentration-time course and the application of Boxerman-Schmainda-Weisskoff leakage correction,21 which, in a subanalysis, also showed better reliability. In the previous study, 5 of 10 algorithms relied on the determination of the AIF.32 Using AIF to compute nRCBV resulted in coefficients of variation of 15%, but only 2% when AIF was not used. The challenges of reliably determining the AIF are well-known and may largely explain the poor repeatability.33,34 Most software platforms in this study did not incorporate AIF for nRCBV calculation and may therefore also explain the excellent concordance across sites. Yet, the computation of CBF requires the determination of AIF and is likely a primary reason for the greater variance in comparison with nRCBV (Figs 3 and 4). Also, the individual nCBF thresholds calculated using IB Neuro varied across sites because some sites chose to use circular deconvolution of the AIF for processing while others did not.
Five of 10 analysis methods in the previous software comparison study often used γ-variate fitting.32 Several studies reported a lower SNR35 as well as greater inaccuracy when γ-variate fitting was used for brain tumor DSC-MR imaging data, especially in the presence of contrast agent leakage.19,20 Although γ-variate fitting suppresses the postbolus baseline, making it appear that leakage has been corrected, there is no physiologic basis for this correction and it does not appropriately consider leakage that can occur during the bolus.36 Gamma-variate fitting was not used by any of the software platforms evaluated in the present study.
In another study,37 nRCBV values were generated from 3 FDA-approved software packages including IB Neuro 1.1, FuncTool software 4.5.3 (GE Healthcare) and nordicICE 2.3.13 and 1 in-house software platform. While effort was made to use the tools in a similar way, more user interaction was required of some (FuncTool, nordicICE), and FuncTool did not have the option for leakage correction. The largest differences between the in-house and commercial software occurred with the tool that required the most user interaction (nordicICE), further motivating the development of more automated workflows with less need for user interaction. Yet another study comparing these same 3 packages also found significant differences, with the outlying package depending heavily on the type of rCBV metric used.38 This finding again suggests that it is imperative that the same output metric be used when making such comparisons.
Of relevance to the current study, rCBV maps generated with IB Neuro showed superior leakage correction and stronger correlation with image-guided microvessel quantification as well as higher accuracy in distinguishing tumor recurrence from pseudoprogression/radiation necrosis compared with other software platforms.39 These results are relevant, given the number of sites in the present study that chose to use IB Neuro for their processing.
A limitation of the current study is the use of a DSC-MR imaging dataset that was obtained at a single center using a single approach. Use of a range of acquisition methods would likely result in greater variation in the DSC-MR imaging perfusion results. A previous study confirmed this by comparing a range of acquisition and analysis methods, which also influenced the ability to distinguish high-grade tumor from reference brain.20 However, a consensus regarding best practices for DSC-MR imaging data acquisition is being reached, as described in a recent review,40 and includes the approach used for this study. Specifically, use of a preload of contrast agent and a flip angle <90° is proving to be one of the most accurate approaches, further confirmed by 2 recent studies,19,41 both incorporating sophisticated simulations of DSC-MR imaging data representative of brain tumor. Use of a preload might also be an important reason for greater consistency across postprocessing methods in this study compared with previous studies (eg, Orsingher et al32). Collecting DSC-MR imaging data after preload was shown to decrease the dependence of tumor rCBV on the chosen method of analysis.20
An additional limitation of this study is the use of laboratory or proprietary commercial packages for which many of the details of the algorithmic implementation are not available and thus cannot be further evaluated as potential sources of differences. Also, the software platforms used for this study were dictated entirely by platforms being used at each participating site. Consequently, this is not a comprehensive comparison of all available software platforms with DSC-MR imaging postprocessing capabilities.
The general application of the results of this study is somewhat limited because the preprocessing steps were carefully controlled so that consistent input data were provided to all sites and software platforms. In practice, subjective manipulation of the preprocessing steps is common; therefore, consistency is less likely, as the discussion of the previous studies reveals. Yet the identification of preprocessing as a key confound should not inhibit use of DSC-MR imaging but rather motivate improving automation of the preprocessing steps. In fact, several efforts to automate tumor segmentation are well underway,42,43 which remove this source of discrepancy entirely.
Conclusions
This study demonstrates that nRCBV and nCBF can be used to distinguish LGG from HGG in a consistent fashion and using a single consensus threshold. This result should increase confidence in using nRCBV primarily, but also nCBF, on a routine basis, potentially motivating its incorporation into the updated Response Assessment in Neuro-Oncology criteria. Finally, these results provide strong motivation for the development of more automated preprocessing workflows that are less dependent on subjective user interaction.
ABBREVIATIONS:
- AIF
arterial input function
- HGG
high-grade glioma
- LCCC
Lin concordance correlation coefficient
- LGG
low-grade glioma
- NAWM
normal-appearing white matter
- nCBF
normalized cerebral blood flow
- nRCBV
normalized relative cerebral blood volume
- rCBV
relative cerebral blood volume
- SN
sensitivity (in the context of receiver operating characteristic analysis)
- SP
specificity (in the context of receiver operating characteristic analysis)
- T1+C
postcontrast T1WI
Footnotes
Disclosures: Kathleen M. Schmainda—RELATED: Grant: National Institutes of Health/National Cancer Institute, Comments: U01 176110, R01 CA082500*; UNRELATED: Grants/Grants Pending: National Institutes of Health, Comments: National Institutes of Health grants submitted on various topics*; Other: Imaging Biometrics, Comments: ownership interest. Ying Liu—UNRELATED: Employment: Medical College of Wisconsin*; Grants/Grants Pending: National Institutes of Health.* Jayashree Kalpathy-Cramer—RELATED: Grant: National Institutes of Health, Comments: National Institutes of Health/National Cancer Institute grants U24CA180927, U24CA180918, U01CA154601*; UNRELATED: Consultancy: INFOTECHsoft; Grants/Grants Pending: National Institutes of Health/National Science Foundation, Comments: R01EY019474-05 National Science Foundation.* Thomas L. Chenevert—RELATED: Grant: National Institutes of Health*; UNRELATED: Grants/Grants Pending: National Institutes of Health*; Patents (Planned, Pending or Issued): University of Michigan*; Royalties: University of Michigan.* Benjamin Hoff—RELATED: Grant: National Institutes of Health, Comments: U01 CA085878.* Brian Ross—RELATED: Grant: National Institutes of Health, Comments: This was part of an National Institutes of Health grant to the University of Michigan.* Yue Cao—RELATED: Grant: National Institutes of Health.* Panagiotis Korfiatis—RELATED: Grant: National Cancer Institute, Comments: CA160045.* Bradley Erickson—RELATED: Grant: National Cancer Institute, Comments: CA160045.* C.C. Quarles—RELATED: Grant: National Cancer Institute.* Laura Bell—RELATED: Grant: National Institutes of Health/National Cancer Institute R01 CA158079.* Timothy Dondlinger—RELATED: Grant: National Institutes of Health*; UNRELATED: Employment: Imaging Biometrics LLC. P.E. Kinahan—RELATED: Grant: GE Healthcare, Comments: research grant*; Other: co-founder of PET/X LLX.* *Money paid to the Institution.
This work was supported by the National Institutes of Health/National Cancer Institute: U01 CA176110 (K.M.S., M.A.P., S.D.R., T.D.), U01 CA148131 (M.M., P.E.K.), U01 CA166104 (T.L.C., B.H.), U01 CA154601 (X.D., Y.-F.Y., J.K.-C.), U01 CA160045 (B.E., P.K.), U01 CA 183848 (Y.C., M.P.A.), R01 CA158079 (L.B., C.C.Q.), R01 CA082500 (K.M.S., M.A.P., S.D.R.), R50 CA211270 (M.M.), and P01 CA085878, U24CA180927, U24CA180918 (J.K.-C.).
Paper previously presented, in part, at: Annual Meeting and Exhibition of the International Society of Magnetic Resonance in Medicine, April 22–27, 2017; Honolulu, Hawaii.
REFERENCES
- 1. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol 2010;28:1963–72 10.1200/JCO.2009.26.3541 [DOI] [PubMed] [Google Scholar]
- 2. Brandsma D, van den Bent MJ. Pseudoprogression and pseudoresponse in the treatment of gliomas. Curr Opin Neurol 2009;22:633–38 10.1097/WCO.0b013e328332363e [DOI] [PubMed] [Google Scholar]
- 3. Wen PY, Norden AD, Drappatz J, et al. Response assessment challenges in clinical trials of gliomas. Curr Oncol Rep 2010;12:68–75 10.1007/s11912-009-0078-3 [DOI] [PubMed] [Google Scholar]
- 4. Macdonald DR, Cascino TL, Schold SC Jr, et al. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol 1990;8:1277–80 10.1200/JCO.1990.8.7.1277 [DOI] [PubMed] [Google Scholar]
- 5. Vymazal J, Wong EY. Response patterns of recurrent glioblastomas treated with tumor-treating fields. Semin Oncol 2014;41(Suppl 6):S14–24 10.1053/j.seminoncol.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 6. Aquino D, Gioppo A, Finocchiaro G, et al. , MRI in glioma immunotherapy: evidence, pitfalls, and perspectives. J Immunol Res 2017;2017:5813951 10.1155/2017/5813951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosen BR, Belliveau JW, Vevea JM, et al. Perfusion imaging with NMR contrast agents. Magn Reson Med 1990;14:249–65 10.1002/mrm.1910140211 [DOI] [PubMed] [Google Scholar]
- 8. Lee SJ, Kim JH, Kim YM, et al. Perfusion MR imaging in gliomas: comparison with histologic tumor grade. Korean J Radiol 2001;2:1–7 10.3348/kjr.2001.2.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmainda KM, Rand SD, Joseph AM, et al. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. AJNR Am J Neuroradiol 2004;25:1524–32 [PMC free article] [PubMed] [Google Scholar]
- 10. Law M, Oh S, Babb JS, et al. Low-grade gliomas: dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging: prediction of patient clinical response. Radiology 2006;238:658–67 10.1148/radiol.2382042180 [DOI] [PubMed] [Google Scholar]
- 11. Hu LS, Eschbacher JM, Heiserman JE, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol 2012;14:919–30 10.1093/neuonc/nos112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prah MA, Al-Gizawiy MM, Mueller WM, et al. Spatial discrimination of glioblastoma and treatment effect with histologically-validated perfusion and diffusion magnetic resonance imaging metrics. J Neurooncol 2018;136:13–21 10.1007/s11060-017-2617-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmainda KM, Prah M, Connelly J, et al. Dynamic-susceptibility contrast agent MRI measures of relative cerebral blood volume predict response to bevacizumab in recurrent high-grade glioma. Neuro Oncol 2014;16:880–88 10.1093/neuonc/not216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kickingereder P, Wiestler B, Burth S, et al. Relative cerebral blood volume is a potential predictive imaging biomarker of bevacizumab efficacy in recurrent glioblastoma. Neuro Oncol 2015;17:1139–47 10.1093/neuonc/nov028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris RJ, Cloughesy TF, Hardy AJ, et al. MRI perfusion measurements calculated using advanced deconvolution techniques predict survival in recurrent glioblastoma treated with bevacizumab. J Neurooncol 2015;122:497–505 10.1007/s11060-015-1755-8 [DOI] [PubMed] [Google Scholar]
- 16. Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared to conventional imaging. AJNR Am J Neuroradiol 2003;24:1989–98 [PMC free article] [PubMed] [Google Scholar]
- 17. Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247:490–98 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito T, Yamasaki F, Kajiwara Y, et al. Role of perfusion-weighted imaging at 3T in the histopathological differentiation between astrocytic and oligodendroglial tumors. Eur J Radiol 2012;81:1863–69 10.1016/j.ejrad.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 19. Semmineh NB, Stokes AM, Bell LC, et al. A population-based Digital Reference Object (DRO) for optimizing dynamic susceptibility contrast (DSC)-MRI methods for clinical trials. Tomography 2017;3:41–49 10.18383/j.tom.2016.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology 2008;249:601–13 10.1148/radiol.2492071659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 2006;27:859–67 [PMC free article] [PubMed] [Google Scholar]
- 22. Bedekar D, Jensen T, Rand S, et al. Delta T1 method: an automatic post-contrast ROI selection technique for brain tumors. In: Proceedings of the Annual Meeting and Exhibition of the International Society for Magnetic Resonance in Medicine, Stockholm, Sweden May 1–7, 2010 [Google Scholar]
- 23. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26:1045–57 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmainda KM, Prah MA, Connelly JM, et al. Glioma DSC-MRI perfusion data with standard imaging and ROIs. The Cancer Imaging Archive. 10.7937/K9/TCIA.2016.5DI84Js8. Accessed July 27, 2016. [DOI]
- 25. Sugahara T, Korogi Y, Tomiguchi S, et al. Posttherapeutic intraaxial brain tumor: the value of perfusion-sensitive contrast-enhanced MR imaging for differentiating tumor recurrence from nonneoplastic contrast-enhancing tissue. AJNR Am J Neuroradiol 2000;21:901–09 [PMC free article] [PubMed] [Google Scholar]
- 26. Lev MH, Ozsunar Y, Henson JW, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol 2004;25:214–21 [PMC free article] [PubMed] [Google Scholar]
- 27. Hirai T, Murakami R, Nakamura H, et al. Prognostic value of perfusion MR imaging of high-grade astrocytomas: long-term follow-up study. AJNR Am J Neuroradiol 2008;29:1505–10 10.3174/ajnr.A1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Z, Le Bas JF, Grand S, et al. Prognostic value of perfusion MR imaging in patients with oligodendroglioma: a survival study. J Neuroradiol 2011;38:53–61 10.1016/j.neurad.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 29. Saraswathy S, Crawford FW, Lamborn KR, et al. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J Neurooncol 2009;91:69–81 10.1007/s11060-008-9685-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong DS, Kim ST, Kim EH, et al. Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: the role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. AJNR Am J Neuroradiol 2011;32:382–87 10.3174/ajnr.A2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am J Neuroradiol 2009;30:552–58 10.3174/ajnr.A1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orsingher L, Piccinini S, Crisi G. Differences in dynamic susceptibility contrast MR perfusion maps generated by different methods implemented in commercial software. J Comput Assist Tomogr 2014;38:647–54 10.1097/RCT.0000000000000115 [DOI] [PubMed] [Google Scholar]
- 33. Knutsson L, Larsson EM, Thilmann O, et al. Calculation of cerebral perfusion parameters using regional arterial input functions identified by factor analysis. J Magn Reson Imaging 2006;23:444–53 10.1002/jmri.20535 [DOI] [PubMed] [Google Scholar]
- 34. Jafari-Khouzani K, Emblem KE, Kalpathy-Cramer J, et al. Repeatability of cerebral perfusion using dynamic susceptibility contrast MRI in glioblastoma patients. Transl Oncol 2015;8:137–46 10.1016/j.tranon.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perkio J, Aronen HJ, Kangasmäki A, et al. Evaluation of four postprocessing methods for determination of cerebral blood volume and mean transit time by dynamic susceptibility contrast imaging. Magn Reson Med 2002;47:973–81 10.1002/mrm.10126 [DOI] [PubMed] [Google Scholar]
- 36. Paulson ES, Prah DE, Schmainda KM. Spiral Perfusion Imaging With Consecutive Echoes (SPICE) for the simultaneous mapping of DSC- and DCE-MRI parameters in brain tumor patients: theory and initial feasibility. Tomography 2016;2:295–307 10.18383/j.tom.2016.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korfiatis P, Kline TL, Kelm ZS, et al. Dynamic susceptibility contrast-MRI quantification software tool: development and evaluation. Tomography 2016;2:448–56 10.18383/j.tom.2016.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelm ZS, Korfiatis PD, Lingineni RK, et al. Variability and accuracy of different software packages for dynamic susceptibility contrast magnetic resonance imaging for distinguishing glioblastoma progression from pseudoprogression. J Med Imaging (Bellingham) 2015;2:026001 10.1117/1.JMI.2.2.026001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu LS, Kelm Z, Korfiatis P, Dueck, et al. Impact of software modeling on the accuracy of perfusion MRI in glioma. AJNR Am J Neuroradiol 2015;36:2242–49 10.3174/ajnr.A4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boxerman JL, Shiroishi MS, Ellingson BM, et al. Dynamic susceptibility contrast MR imaging in glioma: review of current clinical practice. Magn Reson Imaging Clin N Am 2016;24:649–70 10.1016/j.mric.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 41. Leu K, Boxerman JL, Ellingson BM. Effects of MRI protocol parameters, preload injection dose, fractionation strategies, and leakage correction algorithms on the fidelity of dynamic-susceptibility contrast MRI estimates of relative cerebral blood volume in gliomas. AJNR Am J Neuroradiol 2017;38:478–84 10.3174/ajnr.A5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal 2017;35:18–31 10.1016/j.media.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 43. Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993–2024 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bjornerud A, Sorensen AG, Mouridsen K, et al. T1- and T2*-dominant extravasation correction in DSC-MRI, Part I, theoretical considerations and implications for assessment of tumor hemodynamic properties. J Cereb Blood Flow Metab 2011;31:2041–53 10.1038/jcbfm.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weisskoff RM, Zuo CS, Boxerman JL, et al. Simultaneous blood volume and permeability mapping using a single Gd-based contrast injection. In: Proceedings of the Annual Meeting and Exhibition of the Society of Magnetic Resonance in Medicine, San Francisco, California August 6–12, 1994 [Google Scholar]
- 46. Liu HL, Wu YY, Yang WS, et al. Is Weisskoff model valid for the correction of contrast agent extravasation with combined T1 and T2* effects in dynamic susceptibility contrast MRI? Med Phys 2011;38:802–09 10.1118/1.3534197 [DOI] [PubMed] [Google Scholar]
- 47. Bedekar D, Jensen T, Schmainda KM. Standardization of relative cerebral blood volume (rCBV) image maps for ease of both inter- and intrapatient comparisons. Magn Reson Med 2010;64:907–13 10.1002/mrm.22445 [DOI] [PMC free article] [PubMed] [Google Scholar]