Abstract
The first circular RNA (circRNA) was identified more than 40 years ago, but it was only recently appreciated that circular RNAs are common outputs of many eukaryotic protein-coding genes. Some circular RNAs accumulate to higher levels than their associated linear mRNAs, especially in the nervous system, and have clear regulatory functions that result in organismal phenotypes. The pre-mRNA splicing machinery generates circular RNAs via backsplicing reactions, which are often facilitated by intronic repeat sequences that base pair to one another and bring the intervening splice sites into close proximity. When spliceosomal components are limiting, circular RNAs can become the preferred gene output, and backsplicing reactions are further controlled by exon skipping events and the combinatorial action of RNA binding proteins. This allows circular RNAs to be expressed in a tissue- and stage-specific manner. Once generated, circular RNAs are highly stable transcripts that often accumulate in the cytoplasm. The functions of most circular RNAs remain unknown, but some can regulate the activities of microRNAs or be translated to produce proteins. Circular RNAs can further interface with the immune system as well as control gene expression events in the nucleus, including alternative splicing decisions. Circular RNAs thus represent a large class of RNA molecules that are tightly regulated, and it is becoming increasingly clear that they likely impact many biological processes.
Keywords: circRNA, pre-mRNA splicing, backsplicing, spliceosome, translation, alternative splicing, noncoding RNA, microRNA, immune surveillance, CDR1as, Laccase2, scrambled exons, R-loop, transposable elements, exon skipping
Graphical Abstract
Besides generating a linear mRNA, many protein-coding genes produce circular RNAs that are tightly regulated and have biological functions.
INTRODUCTION
As most eukaryotic genes are arranged in a split manner, nascent transcripts must be spliced so that each of the noncoding intronic segments are removed and the exonic segments are joined to one another (e.g., exon 1 is joined to exon 2, which is joined to exon 3, etc.). It has long been recognized that this step in gene expression is highly regulated and can be exploited to generate a diverse set of mRNAs from a given gene, with each mature isoform potentially having a unique function, localization pattern, or regulatory role (reviewed in 1, 2, 3). An extreme case is the D. melanogaster Dscam (Down syndrome cell adhesion molecule) gene that expresses 38,016 distinct linear mRNAs, and the ability to generate a diverse set of Dscam isoforms is critical for proper neural circuit assembly.4, 5 Broadly speaking, >95% of human genes are alternatively spliced,6 and these splicing decisions are thought to be regulated in a combinatorial manner by both cis-regulatory elements and trans-acting factors (reviewed in 7, 8). Besides generating a variety of linear mRNAs, it is now clear that many eukaryotic genes also surprisingly produce circular RNAs that have covalently linked ends.9–11
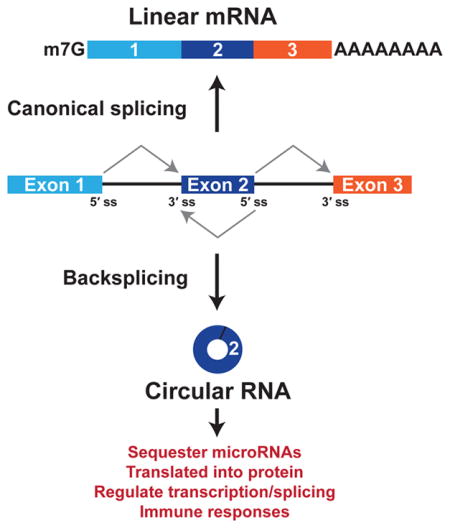
Circular RNAs are generated when the pre-mRNA splicing machinery “backsplices” to join a splice donor to an upstream splice acceptor (e.g., the end of exon 2 is joined to the beginning of exon 2) (Figure 1). Despite the first circular RNA being identified more than 40 years ago,12 these transcripts had largely been assumed to be very rare in cells, dismissed as noise, and/or forgotten about. This is, in part, because the prevailing dogma has been that most splicing events occur co-transcriptionally as soon as an intron has been fully transcribed.13–15 Thus, the intron along with its associated splice acceptor (3′ splice site) should be removed before any downstream splice donors (5′ splice sites) have been transcribed, making most backsplicing reactions impossible. It is now recognized that some introns are slowly or post-transcriptionally spliced,16–18 thereby allowing the opportunity for backsplicing reactions to occur. Most circular RNAs are rarely generated and accumulate to low levels,19 but some are expressed at levels 10-fold higher than their associated linear mRNA.9, 10 This suggests that the main function of some protein-coding genes may be to generate circular RNAs rather than linear mRNAs or proteins.
Figure 1. Pre-mRNAs can be alternatively spliced to generate a linear mRNA or a circular RNA.
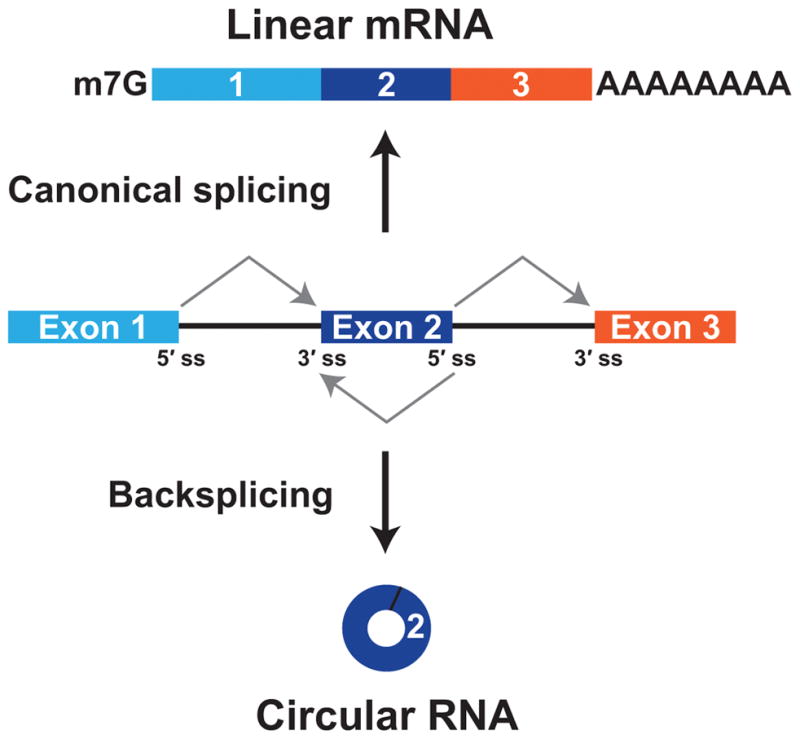
If the pre-mRNA splice sites (ss) are joined in a linear order, a mature linear mRNA is generated that is also capped and polyadenylated (top). Alternatively, the pre-mRNA splicing machinery can backsplice and join a 5′ ss to an upstream splice acceptor (3′ ss), resulting in production of a circular RNA whose ends are covalently linked by a 3′–5′ phosphodiester bond (bottom).
Here, I will highlight the tremendous progress that has been made over the last few years to reveal the diversity of circular RNAs present in cells as well as discuss key insights into how these transcripts are made, regulated, and function. High-throughput RNA sequencing (RNA-seq) has identified thousands of circular RNAs across eukaryotes, some of which have clear biological functions that result in organismal phenotypes.20, 21 Production of circular RNAs is often facilitated by intronic repeat sequences, which base pair to one another to bring the intervening splice sites into close proximity. The efficiency of backsplicing reactions is further regulated by RNA binding proteins, exon skipping events, as well as by the levels of core spliceosomal components, thereby allowing circular RNAs to be expressed in a tissue- and stage-specific manner. Once produced, circular RNAs are naturally resistant to degradation by exonucleases, and thus have long half-lives and some accumulate to high levels. Functions for the vast majority of circular RNAs remain unknown, but some of these transcripts regulate microRNAs, alternative splicing patterns, the immune system, or are bound by ribosomes to produce proteins.
Early insights into circular RNAs
Sänger and colleagues serendipitously identified the first circular RNA in 1976 as part of an effort to understand how viroids function as plant pathogens.12 It was previously known that viroids were similar to viruses in that they were infectious and pathogenic, but it was unclear how viroids function as they appeared to be rather small (<400 nt) RNAs that were not protected by protein coats. By characterizing highly purified viroid RNA molecules, Sänger and colleagues demonstrated that viroid RNA was unable to be enzymatically labeled at its 5′ or 3′ ends nor able to be degraded by snake venom phosphodiesterase.12 Electron microscopy revealed single-stranded circles of viroid RNA, and subsequent sequencing of the viroid nucleotide sequence unequivocally demonstrated that viroids are indeed true circular RNAs.22 Hepatitis delta (δ) virus (HDV), a satellite virus of hepatitis B virus, was subsequently shown to be a ~1700 nt circular RNA molecule23 and, unlike plant viroids, to encode a single open reading frame that is translated to regulate viral replication.24, 25
Circular RNAs were further found to be generated from self-splicing introns in unicellular eukaryotes26 and from ribosomal RNA introns in archaea,27 but it was not until the 1990s that the first endogenous circular RNAs expressed in higher eukaryotes were identified. In an effort to characterize the DCC (Deleted in Colorectal Carcinoma) tumor suppressor gene, Nigro and colleagues performed a number of PCRs to map the exact order of exons in the mature DCC mRNA.28 Exons were always joined accurately at consensus splice sites, but sometimes in an order different from that present in the nascent transcript (e.g., exon 3 appeared to be upstream of exon 2 in some mature mRNAs). These scrambled transcripts did not appear to be polyadenylated and were highly enriched in the cytoplasm, but were expressed at ~1/1000th the level of the normally spliced DCC transcript.28 It was thus proposed that RNAs containing scrambled exons may simply represent errors in the normal splicing process. By studying the human ETS-1 gene, Cocquerelle and colleagues similarly identified processed ETS-1 transcripts containing exons in a scrambled order that were expressed at ~1/100th the level of the normally spliced transcript.29 Further characterization of the scrambled ETS-1 transcripts revealed that they were highly stable (half-life >48 h) circular RNAs (with a 3′-5′ phosphodiester bond at the junction site) that contained only a subset of the exons that are encoded in the full length ETS-1 gene.30 Whereas the DCC and ETS-1 circular RNAs were expressed at very low levels, Capel and colleagues found that the mouse Sry gene, which determines sex in mammals, predominately generates a circular RNA in mouse testis.31 More than 90% of the Sry transcripts in the adult testis corresponded to the single-exon Sry circular RNA, which accumulated in the cytoplasm, but was not associated with polysomes.
Beyond demonstrating that circular RNAs can be the predominant output of some protein-coding genes, the mouse Sry gene provided some of the first mechanistic insights into how specific exons could be selected for circularization. ~50-kb of near perfectly complementary sequences (>99.7%) are present in the introns flanking the Sry exon that forms a circular RNA.31, 32 Capel and colleagues suggested that these flanking sequences may be able to base pair to one another, thereby positioning the splice donor close to its upstream splice acceptor and facilitating backsplicing. Consistent with this model, it was shown using plasmids that Sry exon circularization in cells indeed requires the presence of both inverted repeats and that ~400 base pairs were sufficient.33 Upon examining backsplicing in vitro using nuclear extracts, it was likewise shown that intronic inverted repeats promote circular RNA production, especially as exon size increased.34–36 These in vitro experiments further strongly suggested that circular RNAs were generated by the spliceosome, but that backsplicing may be >100-fold less efficient that canonical splicing reactions.
A handful of additional endogenous circular RNAs were identified over the ensuing years.37–44 For the rat cytochrome P450 2C24, human cytochrome P450 2C18, and human dystrophin genes, alternatively spliced mRNAs lacking the exons present in the mature circular RNA were identified, suggesting that exon skipping and backsplicing could sometimes (but not always) be intimately linked.37, 38, 40 In addition, the promise of circular RNAs for biomedical applications was recognized (e.g., Ref 45). However, it was not until the rise of high-throughput RNA-seq methods (See Sidebar) and some more serendipity that the widespread nature of circular RNAs was revealed.
Sidebar. (Boxed Information).
High-throughput identification of circular RNAs
To identify circular RNAs by RNA-seq, it has become standard to deplete ribosomal RNAs (rRNAs) and use random priming. Nevertheless, many circular RNAs are expressed at low levels and rRNA depletion is not sufficient to ensure annotation of all circular RNAs, unless very high depth RNA-seq is used. Therefore, in many protocols, either exonucleases, such as RNase R, or poly(A) selection steps are further employed to deplete linear mRNAs, thereby allowing greater sequencing coverage of circular RNAs.10, 46–48 Alternatively, commercially available microarrays that include probes to backsplicing junctions are available. On the computational side, multiple pipelines have been developed to identify circular RNAs from RNA-seq datasets, including find_circ,11 MapSplice,49 CIRCexplorer,50 circRNA_finder,51 CIRI,52 KNIFE,53 NCLScan,54 Segemehl,55 DCC,56 UROBORUS,57 and PTESFinder.58 Although all of these algorithms support the idea that circular RNAs are widely expressed, they unfortunately differ significantly in the sets of circular RNAs that they predict.59–61 When the same RNA-seq dataset was inputted independently into five of the algorithms, many (~40%) putative circular RNAs were predicted by only a single algorithm.60 The overall number of circular RNAs predicted ranged from 1532 to 4067, and only 854 were identified by all five algorithms.60 There are thus striking differences in sensitivity and precision among the algorithms, and the field currently lacks a clear gold-standard method to identify circular RNAs from RNA-seq datasets. It is thus strongly recommended that multiple independent pipelines be used to identify circular RNA candidates, and that candidates are subsequently verified using independent approaches, including RT-PCR across backspliced junctions and Northern blotting.62 With further improvements to the RNA-seq analysis algorithms (reviewed in 63), it should become easier to computationally eliminate artifacts and identify only true circular RNAs.
High-throughput sequencing approaches sparked a renaissance for circular RNAs
Beginning ten years ago, many groups started to use RNA-seq to characterize “full” transcriptomes and were able to identify a variety of previously unannotated splice variants and noncoding RNAs.6, 64–68 These initial analyses missed circular RNAs, however. This is largely due to (i) the widespread use of poly(A) selection steps in library preparation protocols, which depleted circular RNAs and other transcripts that lack poly(A) tails, and (ii) the use of computational algorithms that required RNA-seq reads to align in a linear manner to the genome, thereby causing all reads corresponding to backspliced junctions to be discarded (reviewed in 63, 69). It was not until Salzman and colleagues used RNA-seq to try to identify cancer-specific transcripts caused by chromosomal rearrangements that it was serendipitously discovered that thousands of circular RNAs are expressed in cells.9 In that study, hundreds of transcripts with a scrambled exon order were identified in leukemia cells, but the vast majority of these transcripts were surprisingly also observed in normal cells. This indicated that these unusual RNAs were not due to structural rearrangements of the genomic DNA, but from splicing processes that were active in all cells.9 Consistent with these scrambled transcripts being circular RNAs, they were enriched in poly(A) depleted fractions48 and were resistant to RNase R, which digests nearly all linear RNAs.70
Subsequent RNA-seq studies confirmed these initial observations and identified thousands of circular RNAs that are generated from protein-coding genes across various eukaryotes, including metazoans, plants, fungi, and protists.10, 11, 50, 51, 71–77 Many of these putative circular RNAs have been assembled into several searchable online databases, including circBase,78 CIRCpedia,79 and CircInteractome.80 In general, circular RNAs can be detected from over 10% of expressed genes in a given cell type, and there is often no clear correlation between levels of circular RNAs and their corresponding mRNAs.10, 72 Most circular transcripts are tissue-specific and expressed at low levels, but some are expressed in multiple species,10, 11, 72, 73, 81 and there are hundreds of genes that express circular RNAs at higher levels than the canonical linear mRNA.9, 73, 82 This is especially true in the nervous system, which is a hotspot for circular RNA expression. For example, in Drosophila, >90% of annotated circular RNAs can be detected in heads, and half of these transcripts are not detected in any other tissue.51 As circular RNAs are often up-regulated during neurogenesis and can be enriched in synapses, it has been suggested that many may respond to and/or regulate neuronal functions.19, 73, 83, 84
This review will only focus on the biogenesis and functions of circular RNAs derived from backsplicing of eukaryotic exons, but it should be noted that circular RNAs have been found in archaea47, 85 and that some circular RNAs in metazoans are generated from tRNA splicing reactions86 or a failure to debranch certain intron lariats87, 88 (reviewed in 89–91).
Base pairing between intronic repeats facilitates production of many circular RNAs
All internal exons (excluding the first and last exons of genes) have splicing signals at their 5′ and 3′ ends and theoretically can circularize. However, only a small subset of possible backsplicing events is ever observed in cells, in part because these reactions often occur at an extremely low efficiency.92 When backsplicing does occur, it can generate a circular RNA that contains a single exon, which is typically longer than the average expressed exon, or one that contains multiple exons. The most commonly observed circular RNAs have 2 to 3 exons with the internal intron(s) removed. As the introns flanking exons that circularize are typically longer than average,10, 50, 51 Jeck and colleagues searched for intronic motifs that are enriched around human circular RNAs and found that pairs of Alu repetitive elements (~300-nt in length) were commonly observed, especially in an inverted orientation.10 This arrangement is analogous to the complementary intronic repeats that flank the mouse Sry circular RNA,31 and suggested that intronic repeats may be a common driver of backsplicing (reviewed in 93). Indeed, many highly expressed circular RNAs in humans, mice, pigs, C. elegans, and Drosophila are flanked by complementary repeats [but not always,51, 94–96 discussed further below], and one can accurately predict many circular RNAs by simply searching for complementary sequences in the flanking introns.19, 50, 77, 84, 97 Besides Alu elements (which are specific to primates), a variety of complementary sequences can flank circular RNAs, including non-repetitive sequences.50 It has thus been proposed that most backsplicing events do not require particular sequence motifs (beyond the splice sites), but are instead promoted by base pairing interactions between flanking intronic elements.
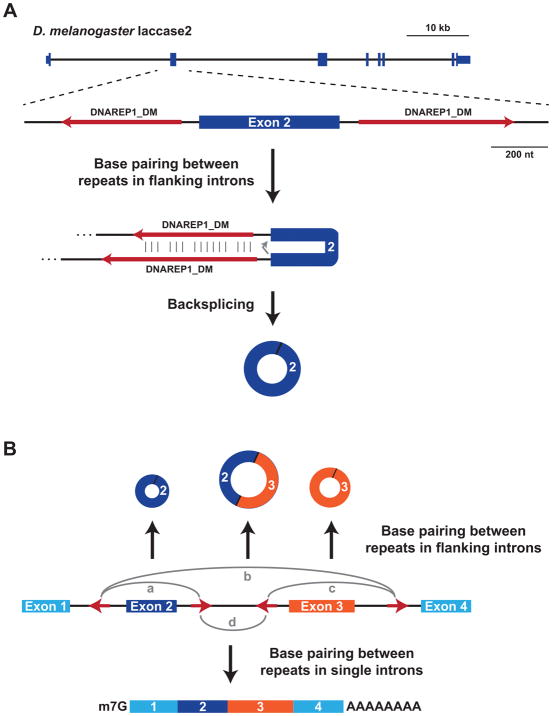
Using expression plasmids or by removing intronic repeats from endogenous gene loci using CRISPR-Cas9, we and others have proven that base pairing between intronic repeats can indeed promote circular RNA biogenesis.19, 50, 97–101 As an example, exon 2 of the D. melanogaster laccase2 gene generates a 490-nt circular RNA and is flanked by a pair of inverted DNAREP1_DM family transposons (Figure 2A).97 Mutational analysis revealed that ~100-nt of each repeat (which are highly complementary over a ~55-nt region) is sufficient for exon circularization, even when there is competition between canonical splicing and backsplicing.97, 102 As expected, disrupting base pairing between the repeats eliminated circular RNA production, while the introduction of compensatory mutations was sufficient to restore backsplicing. Interestingly, the DNAREP1_DM repeats are not present at the laccase2 gene in other Drosophila species, and thus only D. melanogaster generates the Laccase2 circular RNA.97 It is thus important to keep in mind that the genomic repeat landscape varies significantly among species103 and that distinct circular RNAs can be expressed in different eukaryotes. Even if the protein-coding exons are evolutionarily conserved, the ultimate output and function of a gene may differ across organisms.93
Figure 2. Base pairing between intronic repeats facilitates many backsplicing events.
(A) Exon/intron structure of the D. melanogaster laccase2 gene, highlighting exon 2 that can generate a 490-nt circular RNA. A pair of DNAREP1_DM transposons (red arrows) flanks exon 2. Base pairing between these intronic sequences brings the intervening splice sites into close proximity, facilitating backsplicing.97 (B) When multiple intronic repeat elements (red arrows) are present in a pre-mRNA, distinct mature RNAs can be generated depending on which repeats base pair to another (denoted by gray arcs).50 (a–c) Base pairing between repeats in different introns results in backsplicing and production of a circular RNA that contains a single exon (a and c) or multiple exons (b). (d) In contrast, a linear mRNA is produced when repeats in a single intron base pair to one another.
At some gene loci, base pairing between short (~30-nt) intronic repeats (including simple repeats) is able to promote circular RNA production.97, 98 Nevertheless, the presence of intronic repeats is not always sufficient to trigger backsplicing (and, hence, circular RNAs can be expressed in a tissue-specific manner). This is due to the co-transcriptional nature of splicing, thermodynamics, regulation by RNA binding proteins, and the fact that most introns contain multiple repetitive elements that compete for base pairing. Depending on how the repetitive elements base pair to one another, very different spliced isoforms can be generated (Figure 2B).50 When base pairing occurs across different introns (like at the laccase2 gene), backsplicing is induced to generate a circular RNA that is composed of the intervening exon(s). In contrast, when base pairing occurs locally within a single intron, canonical splicing occurs to produce a linear mRNA. The number of repetitive elements, the distance between them, and their degree of complementarity all affect which sequences base pair to one another and, hence, the splicing outcome.50 This competition additionally allows a single protein-coding gene to generate multiple distinct circular RNAs, a process known as alternative circularization.71, 72, 79, 103, 104
Exon skipping can sometimes (but not always) be coupled to circular RNA production
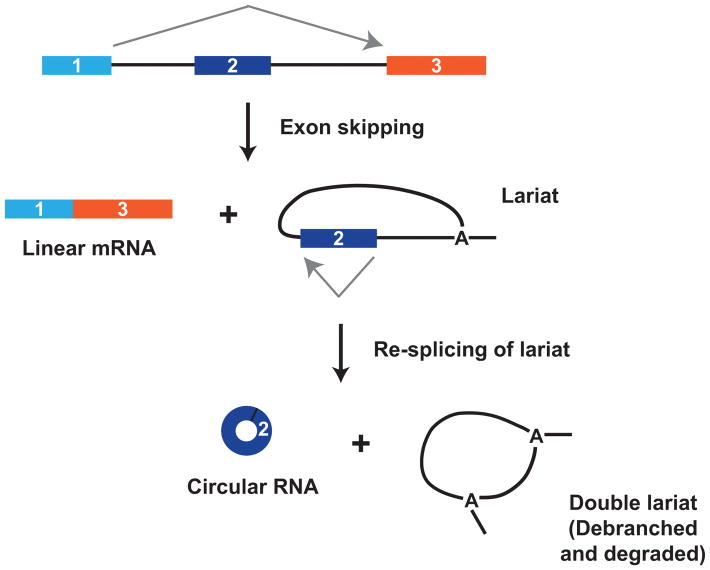
Consistent with earlier studies,37, 38, 40 a recent global transcriptome analysis suggests that the biogenesis of some circular RNAs can be coupled to exon skipping events, thereby allowing a linear mRNA and a circular RNA to be generated from a single pre-mRNA (Figure 3).105 In some cases, the exon-skipped mRNA variant is expressed at very low levels and only detectable by RT-PCR, not by RNA-seq.21 At the S. pombe mrps16 gene, Barrett and colleagues have beautifully demonstrated a mandatory coupling between exon skipping and circular RNA production.95 Splicing exon 1 to exon 3 of mrps16 releases an intron lariat containing exon 2 that is subsequently spliced again to join the beginning and end of exon 2 together. This re-splicing event thereby generates a mature circular RNA that accumulates in cells, whereas the double lariat is rapidly degraded.106 It remains unclear why the mrps16 intron lariat is subjected to a second round of splicing when other lariats are not, but it may be due to topological effects or differences in the speed of lariat debranching. Regardless of the underlying details, coupling exon skipping to circular RNA production provides a mechanism for generating circular RNAs in the absence of intronic repeats.
Figure 3. Exon skipping can be directly coupled to circular RNA production.
At the S. pombe mrps16 gene, exon skipping results in production of a mature linear mRNA as well as an intron lariat containing exon 2. This lariat is re-spliced to generate a mature circular RNA as well as a double lariat structure that is subsequently debranched and degraded.95
Combinatorial control of circular RNA levels by RNA binding proteins
Besides cis-acting intronic repeats and exon skipping events, it is now clear that RNA binding proteins are critical regulators of circular RNAs, allowing these transcripts to be expressed in tissue-specific patterns or induced/repressed as cells change their state.51, 53, 71, 73, 74, 82–84, 96, 107 For example, the intronic repeats themselves can be directly bound by trans-acting factors, including NF90/NF110108 (discussed below) or the RNA helicase DHX9 (DExH-Box Helicase 9) that binds to double-stranded RNAs.109 DHX9 binding inhibits backsplicing reactions, perhaps by directly unwinding the double-stranded intronic regions or by recruiting ADAR (adenosine deaminase acting on RNA) enzymes that convert adenosines to inosines.73, 77, 109 As DHX9 is up-regulated during differentiation processes, it has been proposed that these DHX9-driven mechanisms may allow cells to actively suppress production of a wide variety of circular RNAs.96 On the other hand, the alternative splicing factor Quaking (QKI) promotes the production of hundreds of circular RNAs during human epithelial-mesenchymal transition (EMT).74 This appears to be because QKI binds to flanking introns and forms dimers, thereby bringing the intervening splice sites into close proximity (analogous to how inverted repeats promote backsplicing). The Drosophila Muscleblind (Mbl) protein can likewise bind to intronic elements in its own pre-mRNA to promote production of circular RNAs, thereby auto-regulating host gene expression and ensuring that Mbl protein levels are maintained in a tight range.94
Circular RNA production can be further controlled by FUS110 as well as by multiple hnRNP (heterogeneous nuclear ribonucleoprotein) and SR (serine-arginine) proteins.97, 102, 111 As discussed above, production of the Drosophila Laccase2 circular RNA is promoted by base pairing between intronic repeat elements, but its expression is also inhibited by multiple hnRNPs and SR proteins. Interestingly, simultaneous depletion of these splicing factors resulted in additive increases in Laccase2 circular RNA levels, indicating that each factor plays a non-redundant role to impact splicing decisions.97 Circular RNAs are thus controlled in a combinatorial manner with intronic repeats generally providing the opportunity for backsplicing to occur, and a variety of trans-acting factors acting to fine-tune the efficiency. Considering that each exon or intron has its own unique set of protein-binding sites, one can easily imagine how this code can be used to generate a variety of circular RNA expression patterns.
The output of genes shifts to circular RNAs when the splicing machinery is limiting
Although mutating the splice sites completely eliminates circular RNA production,94, 98, 100, 112 we recently showed in Drosophila that circular RNAs can become the preferred gene output when core spliceosome or transcription termination factors are depleted from cells.102 The spliceosome is formed by a stepwise and dynamic assembly of five snRNAs (small nuclear RNAs) and ~170 proteins (reviewed in 2). As expected, depletion of core spliceosomal components resulted in decreased expression of spliced linear mRNAs. In stark contrast, we found that the expression of many single-exon circular RNAs, including the Laccase2 circular RNA, increased when factors with very different functions in the spliceosome were depleted from cells. For example, individually depleting snRNP-U1-70K or snRNP-U1-C (which are involved in 5′ splice site recognition), Prp8 (the largest spliceosomal protein component, which crosslinks with all the critical sites of chemistry), or Slu7 (which acts in the second catalytic step of splicing) each resulted in significant increases in the expression of many endogenous circular RNAs.102 Backsplicing was further found to be significantly less sensitive than canonical splicing to pharmacological inhibition of the SF3b complex, a component of the U2 snRNP that is involved in pre-spliceosome assembly.
We propose that the early steps in spliceosome assembly, specifically the mechanism by which exons are initially identified, explain these differences in sensitivity between canonical splicing and backsplicing (Figure 4).102 During exon definition,113 U1 and U2 snRNPs bind at opposite ends of each exon and are stabilized by a network of protein-protein interactions that includes additional factors such as SR proteins. These cross-exon interactions then must be converted to cross-intron interactions (to pair splice sites from separate introns) to yield a canonically spliced linear mRNA. However, to generate a circular RNA, these initial exon definition complexes likely do not need to be disrupted and instead may be directly converted into catalytically competent backsplicing complexes. When core spliceosomal components are depleted from cells, we propose that cross-exon interactions are not easily replaced with cross-intron interactions, thereby causing single exon circular RNAs to be the preferred splicing outcome.102 It remains unclear if backsplicing uses the exact same set of core spliceosomal components as canonical splicing reactions, but this work provides a framework for thinking about how global changes in circular RNA levels (at least those generated from single exons) can be obtained. If a tissue has limiting amounts of spliceosomal components, it would be expected that higher levels of circular RNAs should be produced, especially from highly transcribed genes. It will thus be very informative to determine if this model holds true in neuronal or aging tissues, which produce the highest amounts of circular RNAs.9, 73, 82
Figure 4. Model for how early steps in spliceosome assembly determine whether canonical splicing or backsplicing occurs.
In pre-mRNAs with long introns, spliceosomal components first assemble across each exon. U1 snRNP (red) and U2 snRNP (green) recognize the 5′ and 3′ splice sites, respectively, with additional factors, such as SR proteins, serving to stabilize the exon definition complex. These cross-exon interactions must then be replaced with cross-intron interactions in order generate a mature linear mRNA (left). However, when spliceosome activity is limiting (e.g., due to depletion of core spliceosomal components), recent work suggests that cross-exon interactions may not be easily disrupted and the full spliceosome instead assembles across an exon, resulting in backsplicing.102
Circular RNAs can regulate microRNA activity and levels
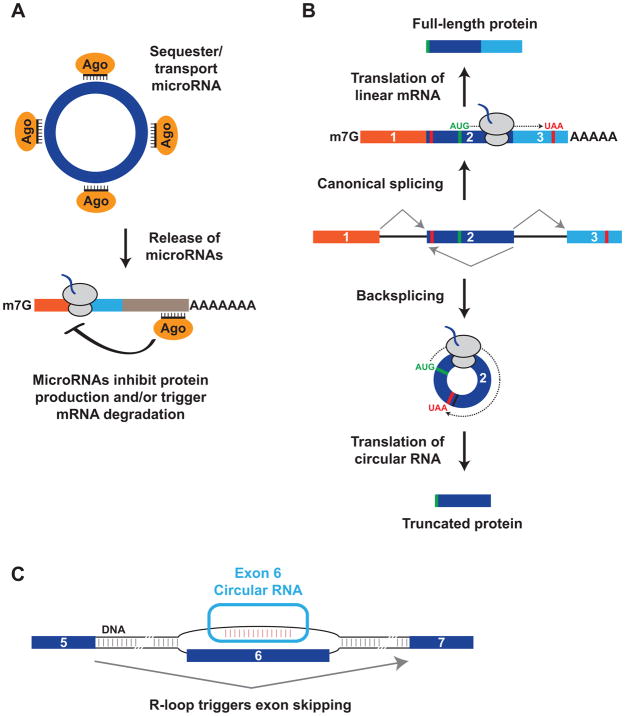
Once generated, circular RNAs are naturally resistant to degradation by exonucleases and accumulate as stable transcripts (e.g., the median half-life of 60 human circular RNAs was calculated to be ~24 hours114). What then are the molecular functions of circular RNAs? For >99.9% of identified circular RNAs, the answer remains unknown. Nevertheless, some regulate microRNAs, which are ~21-nt small RNAs that function post-transcriptionally by base pairing to mRNAs and repressing protein production and/or triggering mRNA degradation (reviewed in 115) (Figure 5A). By searching for conserved microRNA target sites within circular RNAs, it was revealed that the mouse Sry and human CDR1as (also known as ciRS-7) circular RNAs represent extreme and notable cases.11, 99 The Sry circular RNA contains 16 binding sites for the microRNA miR-138, whereas CDR1as contains more than 70 evolutionarily conserved binding sites for miR-7. It was thus proposed that these circular RNAs may function as decoys or sponges116, 117 that reduce the number of freely available miR-138 or miR-7 transcripts, respectively.11, 99
Figure 5. Circular RNAs can have a variety of molecular functions.
(A) Some circular RNAs, including CDR1as and Sry, are able to bind many copies of particular microRNAs (in complex with an Argonaute (AGO) protein). In response to a specific stimulus (e.g. cleavage of CDR1as by miR-671), the circular RNA can potentially release these microRNA transcripts, which then bind to mRNAs to down-regulate their expression. (B) Recent work suggests that linear mRNAs and circular RNAs from the same gene locus can be translated to generate distinct protein products. In the example shown, the mature linear mRNA (top) and the circular RNA (bottom) use the same AUG start codon (green), but the circular RNA generates a truncated protein that terminates at a stop codon encountered after the backsplicing junction. (C) Some copies of the Arabidopsis SEP3 circular RNA are retained in the nucleus, where they form R-loops (an RNA:DNA hybrid) at the endogenous SEP3 gene loci to impact alternative splicing of its host gene.
CDR1as is derived from an antisense long noncoding RNA44, 106 and is expressed several orders of magnitude higher than the CDR1 (Cerebellar Degeneration-Related protein 1) gene from the opposite strand.20, 44 None of the >70 miR-7 binding sites present in CRD1as are perfectly complementary to miR-7, which allows the circular RNA to be densely bound but not sliced by miR-7. Consistent with this idea, depleting CDR1as in cell lines resulted in down-regulation of miR-7 target mRNAs, presumably because miR-7 was no longer sequestered.11, 99 CDR1as additionally contains a region near perfectly complementary to miR-671, which enables the circular RNA to be endonucleolytically cleaved by Argonaute 2 (AGO2) in a miR-671-dependent manner.44 This has led to a proposed model in which CDR1as helps store and/or transport miR-7 (bound to an Argonaute protein) until miR-671 slices the circular RNA, releasing the miR-7 cargo so that it can regulate the expression of mRNAs in a specific subcellular location (Figure 5A).
In mice, the CDR1as circular RNA is highly expressed in excitatory neurons, with minimal expression in inhibitory neurons, glial cells, or non-neuronal tissues.20 Upon deleting the CDR1as genomic region, knockout mice were viable, fertile, and displayed no gross abnormalities in adult brain anatomy. Nevertheless, defects in excitatory synaptic transmission were observed and the knockout mice exhibited impaired sensorimotor gating, a behavioral phenotype associated with neuropsychiatric disorders.20 These effects are presumably due to loss of the CDR1as circular RNA, but it remains possible that the phenotypes are caused, at least in part, by the long noncoding RNA from which the circular RNA is derived106 or the overlapping CDR1 gene. Interestingly, at least in the tissues examined, CDR1 does not appear to be expressed.20 On the molecular level, the expression of the vast majority of microRNAs was unchanged in the brains of CDR1as knockout mice.20 However, miR-7 was markedly down-regulated in the cerebellum, cortex, hippocampus, and olfactory bulb (but unaffected in tissues where CDR1as is not expressed). This resulted in increased expression in the brain of miR-7 target genes, including immediate early genes, such as Fos, that serve as markers of neuronal activity. In contrast, miR-671 was consistently up-regulated in the cerebellum, cortex, and olfactory bulb of CDR1as knockout mice.
Considering that expression of the miR-7 and miR-671 passenger (miR*) strands were unperturbed in CDR1as knockout mice, these specific changes in microRNA expression likely occur post-transcriptionally.20 Binding to near-perfect target sites (similar to the miR-671 site in CDR1as) can cause tailing and degradation of microRNAs,118 and thus the observed increase in miR-671 levels is likely directly due to the lack of CDR1as expression in the knockout mice.20 Likewise, when miR-7 is unable to bind CDR1as, it binds other RNAs in cells, including the long noncoding RNA Cyrano,119 which has a near perfect miR-7 target site that may promote miR-7 trimming and degradation.20
The CDR1as locus thus beautifully demonstrates how circular RNAs and microRNAs can have a dynamic interplay with important biological effects, but there is still debate as to whether other circular RNAs function in a similar manner. This is because most annotated circular RNAs contain few microRNA binding sites.72, 114 Nevertheless, there is emerging evidence that a subset of circular RNAs, including human circHIPK3, human circZNF91, human circHECTD1, and many circular RNAs in Drosophila, may function to sponge specific microRNAs.51, 96, 101, 120 For example, circZNF91, which is induced as human epidermal stem cells differentiate, contains 24 binding sites for miR23b-3p, a microRNA that is known to regulate keratinocyte differentiation.96 As other circular RNAs up-regulated during this differentiation process also have significant numbers of microRNA binding sites,96 it has been suggested that a subset of circular RNAs may sponge key microRNAs as cells change their state. Given that microRNAs can confer robustness during development,121, 122 it will be interesting to determine if circular RNAs also help buffer regulatory networks against perturbation.
Translation of circular RNAs
Initial experiments suggested that eukaryotic ribosomes can not be loaded onto circular RNAs due to their lack of a 5′ cap structure,123 but it was subsequently demonstrated that the presence of an internal ribosome entry site (IRES) allowed synthetic circular RNAs to be translated in vitro124 and in cells.97, 108, 112, 125 IRES elements are present in a variety of viruses and directly bind translation initiation factors or the ribosome itself, thereby allowing translation to occur in a cap-independent manner (reviewed in 126). As initial reports failed to find circular RNAs co-sedimenting with ribosomes10, 127 or in ribosome profiling libraries,72, 83 it appeared that endogenous circular RNAs may not be able to recruit ribosomes. However, several recent reports suggest that a subset of endogenous circular RNAs may, in fact, be translated (reviewed in 128, 129).
Human circ-ZNF609 contains a 753-nt open reading frame that starts at the same AUG codon used to initiate linear mRNA translation and terminates at a stop codon that is encountered three nucleotides past the backsplicing junction (Figure 5B).130 A small, but significant portion of this circular RNA co-sediments with polysomes, and expression plasmids revealed that circ-ZNF609 could be weakly translated (two orders of magnitude less efficiently than a linear mRNA). Interestingly, knock-down of circ-ZNF609 results in proliferation defects in human and mouse myoblasts,130 but it remains unknown whether the protein derived from circ-ZNF609 contributes to this phenotype.
Using mass spectrometry, peptides spanning endogenous backsplicing junctions were identified for 19 additional human circular RNAs,131 and deeply sequenced ribosome profiling datasets revealed 122 Drosophila circular RNAs associated with ribosomes.132 For 40% of these fly circular RNAs, translation is predicted to use the same start codon as the host linear mRNA and to terminate after the backsplicing junction (like circ-ZNF609) (Figure 5B). Many of the putative ORFs have at least one identifiable protein domain, but it must be noted that only a single sequencing read currently supports the vast majority of these translation events. It is near impossible to use ribosome profiling to look for ribosome phasing or start/stop codons on circular RNAs, so a combination of minigene expression plasmids, sucrose gradients, mutational analysis, and mass spectrometry were used to further support the idea that some of these circular RNAs may indeed be translated.132 Notably, this analysis suggested that circular RNA translation may often occur on membrane-associated ribosomes or in subcellular compartments rich in membranes (e.g. synapses).
Consistent with expectations, the regions upstream of the circular RNA start codons have detectable IRES activity when inserted into bicistronic reporter constructs.130, 132 For circ-ZNF609, splicing was required for IRES activity, suggesting that deposition of the exon junction complex133 may enhance translation of some circular RNAs.130 Yang et al. further demonstrated that N6-methyladenosine (m6A) residues are enriched in circular RNAs (~13% of circular RNAs contain m6A) and that these modified nucleotides can act to promote translation.131 In fact, a single m6A site may be sufficient to initiate translation of synthetic circular RNAs. Upon binding m6A, it was proposed that the YTHDF3 reader protein recruits translation initiation factors, including eIF4G2 and eIF3A to the circular RNA.131
It remains possible that a number of these recently described translation events represent technical or biological noise as other genome-wide studies failed to identify any evidence for circular RNA translation.72, 82, 83 Further work is now required to determine how many circular RNAs are truly translated to significant levels. It is tempting to speculate that significant amounts of protein could be made over the long lifetimes of circular RNAs (which could be useful for biotechnology purposes), and that these endogenous proteins may serve as dominant negatives and/or regulate the same processes as proteins generated from their associated host linear mRNAs. Besides being translated themselves, there is emerging evidence that some circular RNAs, including circPABPN1, can regulate translation of their associated linear mRNAs.134 Mature circular RNAs have also been found to bind to a variety of other proteins,94, 127, 135, 136 which suggests that these transcripts may impact a variety of other protein activities or functions.
Circular RNAs in the nucleus regulate transcription and alternative splicing of their host genes
Most characterized circular RNAs predominately localize in the cytoplasm,9, 10 but there are now emerging examples of circular RNAs that function in the nucleus.21, 84, 137 Exon-intron circular RNAs are incompletely spliced and have a retained intron that allows them to interact with U1 snRNP and promote transcription of their host gene.137 In Arabidopsis, a circular RNA is generated from exon 6 of SEPALLATA3 (SEP3), a key developmental regulatory gene, and ~15% of these circular transcripts are retained in the nucleus (Figure 5C).21 Remarkably, plants over-expressing the SEP3 circular RNA (but not a linear version of exon 6) produced flowers that had fewer stamen and more petals than normal. This appears to be because the ectopically expressed circular RNA forms an R-loop (an RNA:DNA hybrid) at the endogenous SEP3 genomic loci, which causes increased production of a linear SEP3 splice variant that has exon 6 skipped (Figure 5C). As over-expression of this SEP3 splice variant was sufficient to cause the same homeotic phenotype as over-expression of the SEP3 circular RNA, this circular RNA appears to function solely by modulating alternative splicing of its host gene.21 It remains to be determined how exactly the circular RNA does this, but it was proposed that the R-loop causes a pause in SEP3 transcription that allows alternative splicing regulators to be recruited that promote exon skipping. Of note, ~5% of R-loops in Arabidopsis are resistant to RNase R treatment,21 suggesting that other circular RNAs may likewise bind genomic DNA to modulate transcription or alternative splicing patterns.
Interplay between the immune system and circular RNAs
Considering that some circular RNAs, including viroids and HDV, are pathogens, one would expect that eukaryotic immune systems should somehow be able to sense and destroy these foreign invaders. Indeed, when HeLa cells were transfected with circular RNAs that had been made in vitro using self-splicing group I introns138, 139 or split ligation,140 a number of well-known pattern recognition receptors, including RIG-I and MDA5, as well as other innate immunity regulators and cytokines were strongly induced.135 Cells are thus able to mount a significant immune response to foreign circular RNAs, one that was, in fact, stronger than when a linear RNA encoding the same sequence was transfected into cells.
It remains incompletely understood how a foreign circular RNA is sensed, but some important aspects have been revealed (reviewed in 141). First, RIG-I is required for the immune response and co-localizes with foreign circular RNAs,135 even though the transcripts lack the 5′ triphosphate moiety that RIG-I is thought to require.142 Second, the mechanism by which the circular RNA is made is the key determinant of whether the transcript is recognized as self or non-self.135 Whenever a circular RNA was made using self-splicing group I introns (even when it was made in cells from expression plasmids), a robust immune response was observed. In stark contrast, circular RNAs made using the spliceosome were always recognized as self (regardless of the sequence of the mature circular RNA or its flanking introns). This is presumably because the spliceosome deposits a number of RNA binding proteins, such as the exon junction complex or the nuclear export machinery, onto the circular RNA that somehow mark the circular RNA as self. In this manner, cells are able to sense and respond to a diverse set of foreign circular RNAs, irrespective of their primary sequence, while not responding to their many endogenous circular RNAs.135 Further work is now required to understand how exactly RIG-I recognizes foreign circular RNAs as well as how backsplicing by the spliceosome prevents autoimmune reactions.
Beyond ensuring that foreign circular RNAs are recognized and neutralized, multiple immune regulators, including RIG-I and the dsRNA binding proteins NF90/NF110, appear to regulate the expression of endogenous circular RNAs.108 In normally growing cells, NF90/NF110 bind to A/U-rich elements (including base paired Alu elements) in the introns flanking many (~30%) exons that yield circular RNAs and act to promote these backsplicing events. However, upon viral infection, NF90/NF110 shuttle to the cytoplasm to bind viral transcripts and suppress viral replication.143 This causes NF90/NF110 to bind fewer nascent pre-mRNAs in the nucleus, thereby resulting in reduced production of circular RNAs.108 Interestingly, NF90/NF110 are also able to bind to some mature circular RNAs in the cytoplasm and, in fact, show higher affinity to circular RNAs than linear RNAs. It has thus been proposed that the normal production of circular RNAs allows the establishment of a molecular reservoir of NF90/NF110 in the cytoplasm, which can be released when needed to enable prompt responses to viral infection.108
Circular RNAs in aging and disease
Alternative splicing patterns have been shown to change with aging,144, 145 and there is a genome-wide trend for increased circular RNA expression in the nervous system as Drosophila, C. elegans, and mice age.51, 146 For example, out of 2,513 detected Drosophila circular RNAs, 262 were significantly up-regulated >2-fold in the heads of 20 day old animals compared to 1 day old animals.51 Notably, these increases in expression were largely independent of changes in expression of linear mRNAs from their host genes and were not observed in aging mouse hearts146 or in rhesus monkey muscles.147 This suggests that circular RNAs may be preferentially stabilized or generated in the aging nervous system, but it remains unclear whether this accumulation of circular RNAs is protective, detrimental, or an innocuous biomarker of aging. At least in mice, it appears that a circular RNA derived from the Pwwp2a gene can help protect from pathological hypertrophy and heart failure.120
Compared to normal tissues, cancer cells generally have reduced expression of circular RNAs,107 which may in part be due to dilution via cell division (reviewed in 148, 149). Consistent with this idea, it was recently shown that circular RNA levels often increase as cells differentiate and stop proliferating.96 Down-regulation of some circular RNAs, including circITCH150–152 and circFOXO3,136 has been suggested to promote cell growth, whereas over-expression of others appears to promote proliferation.153 Of particular note, cancer-associated chromosomal translocations can result in production of aberrant fusion circular RNAs when intronic sequences that flank the breakpoint base pair to one another.154 These fusion circular RNAs can promote transformation, cell viability, and resistance upon therapy, and thus represent promising new therapeutic targets. Furthermore, because some circular RNAs are present in exosomes/extracellular vesicles and bodily fluids,92, 155–158 circular RNAs may represent promising cancer biomarkers. Packaging of circular RNAs into vesicles may be a way to eliminate circular RNAs from cells and/or a mechanism that enables cell-to-cell communication.
Perspectives
Thousands of putative circular RNAs have now been identified by RNA-seq, but it remains important to keep in mind that the vast majority have never been verified by orthogonal techniques or studied in any detail. There is thus still much work to do to eliminate sequencing artifacts from these lists, to distinguish backsplicing events from trans-splicing events, and to fully characterize circular RNA expression patterns. Nevertheless, a growing number of circular RNAs have now been characterized in sufficient detail such that we have important insights into how they can be generated, regulated, and function. In particular, insights into the biogenesis mechanisms have been translated into efficient methods for expressing circular RNAs in cells,62, 79, 97, 99, 108, 112 and this has already enabled the functions of some circular RNAs to be revealed.11, 21, 99 By combining over-expression studies with circular RNA knockouts generated by CRISPR-Cas9,20 the functions for many more circular RNAs will likely be identified over the coming years. It should be possible to use CRISPR-Cas9 to remove the flanking intronic repeat elements, thereby knocking out the circular RNA of interest while not affecting linear RNA production from the host gene.
Intronic repeats are critical for the biogenesis of many circular RNAs (Figure 2), but it remains incompletely understood how genes can generate circular RNAs in the absence of repeats.51, 76, 95 For example, most of the circular RNAs up-regulated during human epidermal stem cell differentiation96 or in Drosophila51 are not flanked by repeats, and it is unclear how these exons (and not others) are selected for backsplicing. Besides exon skipping events (Figure 3), there are likely a number of cis- and trans-acting regulatory mechanisms that await discovery which enable specific circular RNA expression patterns. Further work is additionally needed to understand the detailed mechanisms of backsplicing reactions and to determine whether all of the canonical core spliceosomal components are involved.2
Once a circular RNA is generated, little is known about how they accumulate in the cytoplasm, especially in non-dividing cells. Specific structural features in other classes of RNA are recognized by receptor proteins, enabling those transcripts to be brought to the nuclear pore complex for export to the cytoplasm (reviewed in 159). It is likely that an analogous mechanism exists for circular RNAs and a better understanding of the proteins that interact with circular RNAs, especially during their biogenesis, may help reveal how circular RNA localization is controlled and can change during development.84 For those circular RNAs that associate with ribosomes (Figure 5B), how exactly is the ribosome assembled on the transcript and do the generated proteins accumulate to high enough levels to have biological effects? Abundant circular RNAs that are not translated may help form large RNA-protein complexes or otherwise impact the same pathway as the protein produced from its parental gene. Alternatively, production of the circular RNA may itself be the key functional event, as backsplicing reactions can be used to turn down expression of the linear mRNA94 or terminate readthrough transcription events.102
How are endogenous circular RNAs recognized as self, and is it possible to generate circular RNAs in vitro that do not invoke an immune response? This avenue could open up many biotechnology applications where treating cells or patients with long-lasting circular RNAs could be ideal. Many circular RNAs have long half-lives, but the mechanisms by which circular RNAs are ultimately degraded remain largely unknown. The CDR1as circular RNA can be cleaved by AGO2 (in complex with miR-671),44 but this appears to be an isolated mechanism. Other RNA endonucleases may generally facilitate decay of circular RNAs by providing access points for exonucleases (reviewed in 160), and identification of circular RNAs that are rapidly degraded may provide a starting point for defining such mechanisms. In summary, the last few years have revealed that circular RNAs are not simply rare oddities, but common outputs of eukaryotic genes. Many surprises have been revealed, and the field is well poised to reveal more insights into how these transcripts are regulated and function to impact normal and disease states.
Acknowledgments
I thank members of my laboratory for discussions and suggestions. Supported by NIH grants R35-GM119735 and R01-NS099371. J.E.W is a Rita Allen Foundation Scholar.
Footnotes
The author has declared no conflicts of interest for this article.
References
- 1.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 5.Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 8.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 9.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 16.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 20.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017:357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 21.Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 22.Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sanger HL. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978;273:203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 23.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 24.Weiner AJ, Choo QL, Wang KS, Govindarajan S, Redeker AG, Gerin JL, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J Virol. 1988;62:594–599. doi: 10.1128/jvi.62.2.594-599.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sureau C, Negro F. The hepatitis delta virus: Replication and pathogenesis. J Hepatol. 2016;64:S102–S116. doi: 10.1016/j.jhep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Grabowski PJ, Zaug AJ, Cech TR. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell. 1981;23:467–476. doi: 10.1016/0092-8674(81)90142-2. [DOI] [PubMed] [Google Scholar]
- 27.Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54:693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- 28.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 29.Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 31.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 32.Gubbay J, Vivian N, Economou A, Jackson D, Goodfellow P, Lovell-Badge R. Inverted repeat structure of the Sry locus in mice. Proc Natl Acad Sci U S A. 1992;89:7953–7957. doi: 10.1073/pnas.89.17.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245–248. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 34.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 35.Schindewolf C, Braun S, Domdey H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic Acids Res. 1996;24:1260–1266. doi: 10.1093/nar/24.7.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun S, Domdey H, Wiebauer K. Inverse splicing of a discontinuous pre-mRNA intron generates a circular exon in a HeLa cell nuclear extract. Nucleic Acids Res. 1996;24:4152–4157. doi: 10.1093/nar/24.21.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997;17:2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 40.Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet. 1999;8:493–500. doi: 10.1093/hmg/8.3.493. [DOI] [PubMed] [Google Scholar]
- 41.Li XF, Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J Biol Chem. 1999;274:8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 42.Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O’Dell KM, Monckton DG, Artero RD. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered. 2006;97:253–260. doi: 10.1093/jhered/esj037. [DOI] [PubMed] [Google Scholar]
- 43.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohjanen PR, Colvin RA, Puttaraju M, Been MD, Garcia-Blanco MA. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 1996;24:3733–3738. doi: 10.1093/nar/24.19.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, Dudekula DB, Abdelmohsen K, Gorospe M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44:e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermuller J, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 57.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Izuogu OG, Alhasan AA, Alafghani HM, Santibanez-Koref M, Elliott DJ, Jackson MS. PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. BMC Bioinformatics. 2016;17:31. doi: 10.1186/s12859-016-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatomer DC, Liang D, Wilusz JE. Inducible Expression of Eukaryotic Circular RNAs from Plasmids. Methods Mol Biol. 2017;1648:143–154. doi: 10.1007/978-1-4939-7204-3_11. [DOI] [PubMed] [Google Scholar]
- 63.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 66.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 71.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 74.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 75.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 78.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong R, Ma XK, Chen LL, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–1074. doi: 10.1080/15476286.2016.1269999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garrett RA, Dalgaard J, Larsen N, Kjems J, Mankin AS. Archaeal rRNA operons. Trends Biochem Sci. 1991;16:22–26. doi: 10.1016/0968-0004(91)90011-j. [DOI] [PubMed] [Google Scholar]
- 86.Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 88.Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20:1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017;33:540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 90.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14:978–984. doi: 10.1080/15476286.2017.1317911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilusz JE. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements. 2015;5:1–7. doi: 10.1080/2159256X.2015.1045682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 95.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kristensen LS, Okholm TLH, Veno MT, Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2017:1–12. doi: 10.1080/15476286.2017.1409931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 100.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 101.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, Cherry S, Wilusz JE. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol Cell. 2017;68:940–954. e943. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 104.Ye CY, Zhang X, Chu Q, Liu C, Yu Y, Jiang W, Zhu QH, Fan L, Guo L. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 2017;14:1055–1063. doi: 10.1080/15476286.2016.1245268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 106.Barrett SP, Parker KR, Horn C, Mata M, Salzman J. ciRS-7 exonic sequence is embedded in a long non-coding RNA locus. PLoS Genet. 2017;13:e1007114. doi: 10.1371/journal.pgen.1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation - exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214–227. e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 109.Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 110.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 114.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 121.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ren Z, Ambros VR. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci U S A. 2015;112:E2366–2375. doi: 10.1073/pnas.1422858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 124.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 125.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, Bindereif A. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. doi: 10.1038/srep31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tatomer DC, Wilusz JE. An Unchartered Journey for Ribosomes: Circumnavigating Circular RNAs to Produce Proteins. Mol Cell. 2017;66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 129.Schneider T, Bindereif A. Circular RNAs: Coding or noncoding? Cell Res. 2017;27:724–725. doi: 10.1038/cr.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22–37. e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L, Wang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017 doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell. 2017;67:228–238. e225. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 138.Ford E, Ares M., Jr Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci U S A. 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Puttaraju M, Been MD. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20:5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 143.Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24:2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rodriguez SA, Grochova D, McKenna T, Borate B, Trivedi NS, Erdos MR, Eriksson M. Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging Cell. 2016;15:267–278. doi: 10.1111/acel.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tollervey JR, Wang Z, Hortobagyi T, Witten JT, Zarnack K, Kayikci M, Clark TA, Schweitzer AC, Rot G, Curk T, et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011;21:1572–1582. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, de Cabo R, et al. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015;7:903–910. doi: 10.18632/aging.100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2017;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2017;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/beta-Catenin Pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, Wang Z, Wen P, Yang H, Shi X, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169–48177. doi: 10.18632/oncotarget.18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 155.Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Preusser C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, Santoso S, Bindereif A. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7:1424473. doi: 10.1080/20013078.2018.1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 159.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 160.Schoenberg DR. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:582–600. doi: 10.1002/wrna.78. [DOI] [PMC free article] [PubMed] [Google Scholar]