Abstract
Introduction
Pregnane X receptor (PXR) and the constitutive androstane receptor (CAR) are two members of the nuclear receptor superfamily that play major roles in the expression of various drug metabolism enzymes and are known for their ligand promiscuity. As with other nuclear receptors, PXR and CAR are each composed of a ligand-binding domain (LBD) and a DNA-binding domain (DBD) connected by a hinge region.
Areas covered
This review focuses on the information obtained over the last 15+ years from X-ray crystallography studies of the structure of PXR and CAR. Areas of focus include the mobility of each structure, based on temperature factors (B factors); multimeric interactions; the binding of coregulators and ligands; and how the crystal structures were obtained. The first use of hydrogen-deuterium exchange coupled with mass spectroscopy (HDX-MS) to study compound-protein interactions in the PXR-LBD is also addressed.
Expert opinion
X-ray crystallography studies have provided us with an excellent understanding of how the LBDs of each receptor function; however, many questions remain concerning the structure of these receptors. Future research should focus on determining the co-crystal structure of an antagonist bound to PXR and on studying the structural aspects of the full-length CAR and PXR proteins.
Keywords: Constitutive androstane receptor, HDX-MS, pregnane X receptor, structural biology, X-ray crystallography
1. Introduction
Nuclear receptors (NRs) are a large superfamily of ligand-activated transcription factors that are believed to have evolved from a common ancestor [1]. Among these receptors, pregnane X receptor (PXR, NR1I2) and the constitutive androstane receptor (CAR, NR1I3) have been characterized as the major actors in the modulation of enzymes involved in the biotransformation, metabolism, and elimination of xenobiotics and endobiotics [2, 3]. These two receptors play diverse roles in physiologic processes such as the homeostasis of bile acids, steroids, and thyroid hormone; gluconeogenesis; and lipid metabolism [4]. However, their most prominent function is in regulating the expression of an array of genes encoding phase I and II drug-metabolizing enzymes (DMEs), as well as phase III ATP-binding cassette drug transporters [5, 6]. The target genes of PXR and CAR include those encoding oxidative enzymes such as cytochrome p450 (CYPs), conjugative enzymes such as UDP-glycosyltransferases (UGTs), glutathione S-transferases (GSTs), sulfotransferases (SULTs), and transporter proteins such as multidrug resistance proteins (MDRs) and multidrug resistance-associated proteins (MRPs) [7]. PXR and CAR upregulate overlapping sets of genes, including CYP3A4 and CYP2B6 [8], but they also control the expression of distinct groups of genes with diverse signaling pathways [9]. Because of their importance in regulating the expression of these DMEs and transporters, PXR and CAR have a significant impact on drugs in clinical use in terms of adverse drug-drug interactions and reduced efficacy [10].
PXR and CAR are recognized as orphan NRs, which are characterized as having no identifiable endogenous ligands and do not conform to the classical steroid hormone receptor model. Initially, there were some obstacles to the characterization and establishment of PXR and CAR as xenobiotic sensors; these difficulties were largely attributed to the ability of PXR and CAR to be modulated by a broad range of chemicals and to the overlapping regulation of gene expression by the two NRs in response to the same ligands. The mouse PXR (mPXR) was discovered based on the homology of its ligand-binding domain (LBD) sequence to that of other known NRs, and it was so named because it was activated by pregnane steroids. mPXR was subsequently found to be activated by both naturally occurring and synthetic steroids [11]. The human PXR (hPXR) was initially designated SXR, which stood for steroid and xenobiotics receptor [12]. The protein was subsequently ascertained to be the human homolog of the Xenopus benzoate X receptor [13] and was later identified as a homolog of mPXR [14]. Human CAR (hCAR) was initially identified as MB67 [15], but it was referred to as hCAR after the discovery of the mouse CAR (mCAR) [16]. Subsequently, the role of CAR in xenobiotic sensing was recognized [17].
PXR controls gene expression through the PXR responsive element module (PXRRE) that is present in the promoter region of target genes, whereas CAR mediates gene expression through the phenobarbital response enhancer module (PBREM) [18]. Nuclear receptors interact with these promoter regions through the zinc fingers, which are conserved in PXR and CAR, located in the N-terminal DNA-binding domain (DBD) [10]. In both receptors, the DBD is connected to the C-terminal ligand binding domain (LBD) through a flexible hinge, although there is very little amino acid similarity between the hinges of PXR and CAR [4]. PXR and CAR are modular proteins that share these domains with other NRs; however, both receptors lack the long N-terminal A/B domain and the C-terminal F domain often seen in other NRs [19].
The hallmark of NRs is the LBD, where ligands are situated in a mostly hydrophobic pocket. PXR and CAR are notable for having a large, flexible, and dynamic LBD that can accommodate a wide range of ligands with diverse structural and physicochemical properties; this is the basis of their ligand promiscuity. Some ligands, including both xenobiotics and endobiotics, are common to both PXR and CAR [3]. Ligand specificity is observed across species, possibly reflecting an evolutionary adaptation to environmental differences. For instance, pregnenolon-16α-carbonitrile (PCN) interacts potently with mPXR but not with hPXR, whereas SR12813 and rifampicin are selective for hPXR [20]. CITCO has a preference for hCAR over mCAR, whereas TCPOBOP is more selective for mCAR as compared to hCAR [3, 21]. PXR and CAR heterodimerize with retinoic acid X receptor (RXR) to form heterodimers, and ligand binding triggers conformational changes to recruit or dissociate from partner proteins such as coregulators. Discrimination between coactivators (e.g., steroid receptor coactivator 1 [SRC-1] or transcriptional mediator/intermediary factor 2 [TIF2]) or corepressors (e.g., nuclear receptor corepressor [NCoR] or silencing mediator for retinoid or thyroid hormone receptors [SMRT]) is mainly attributable to the positioning of the C-terminus activation function 2 (AF-2) helix (helix 12), which is believed to act as a switch, directing coregulators to engage the NRs through the AF-2 domain [22].
In the past two decades, considerable efforts have been made to obtain crystal structures of PXR and CAR with various ligands bound to them. These studies have provided a wealth of information and have increased our understanding of the nature of the ligand promiscuity and other notable features exclusive to these xenosensors. In this review, we compare the reported crystal structures and discuss the most recent discoveries in this field. In addition, we describe the first structural studies of PXR to use hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS), which have shed light on the dynamic consequences of ligand and coactivator/corepressor peptide binding.
2. Crystal structures of PXR and CAR
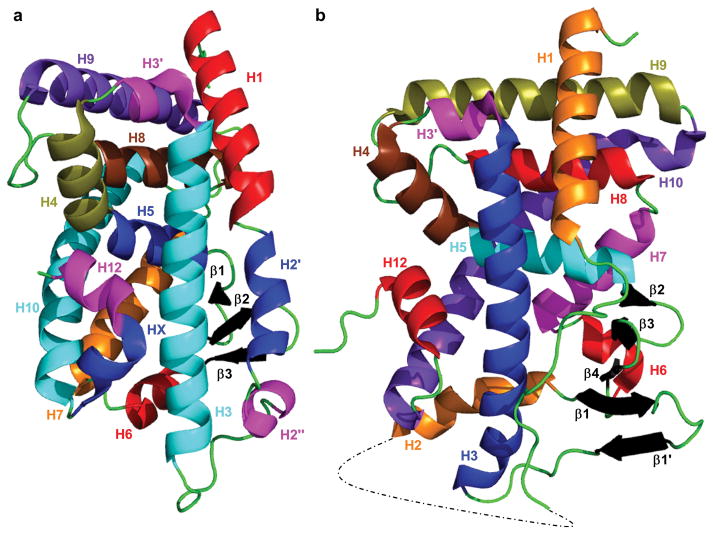
The crystal structure of the LBD of hPXR was first solved in 2001 [23], with the crystal structure of the LBD of hCAR following in 2004 [24]. The structure of the mCAR-LBD was also published in 2004 [25, 26]. These initial crystal structures and subsequent ones (Table 1) have shown that the LBDs of CAR and PXR share similarities and differences with other NRs, as well as with each other. The LBDs of both CAR and PXR, like those of other NRs, contain a three-layered helical sandwich (H1/H3, H4/H5/H8, and H7/H10). CAR contains a fourth helical layer consisting of two 310 helices (H2′ and H2″) (Figure 1). The LBD structures have a total of 11 α-helices and a binding pocket filled primarily by hydrophobic residues that is formed in part by an antiparallel β-sheet; however, in many PXR-LBD structures, H6 is disordered and is modeled as an extended flexible loop. The size of the ligand-binding pocket (LBP) differs between the two receptors (~600 Å3 for CAR vs. 1200–1600 Å3 for PXR), in part because PXR has a large insert between H1 and H3 that effectively replaces the two 310 helices seen in CAR with two β-strands (β1 and β1′) and an additional α-helix (H2) (Figure 1) [23, 25, 27]. The two β-strands help to form the unique five-strand antiparallel β-sheet of PXR that constitutes half of the LBP. H2, which is not modeled in every PXR-LBD crystal structure, is believed to help provide a channel for ligand entry and exit from the ligand-binding site [28].
Table 1.
Summary of published structures of PXR-LBD and CAR-LBD.
| CAR-LBD Crystal Structures | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PDB ID | Species | Space Group | Co-peptide | Other Proteins | Oligomer in Asymmetric Unit | Ligand(s) | Resolution (Å) | Year | Ref. |
| 1XNX | mouse | C2221 | None | None | homodimer | 16,17-Androstene-3-ol | 2.90 | 2004 | 25 |
| 1XVP | human | P3221 | SRC-1 | RXRα | 2 heterodimers | CITCO | 2.60 | 2004 | 24 |
| 1XV9 | human | P3221 | SRC-1 | RXRα | 2 heterodimers | 5b-Pregnane-3,20-dione | 2.70 | 2004 | 24 |
| 1XLS | mouse | P1 | TIF2 | RXRα | 4 heterodimers | TCPOBOP | 2.96 | 2004 | 26 |
| PXR-LBD Crystal Structures | |||||||||
| PDB ID | Species | Space Group | Co-peptide | Other Proteins | Oligomer in Asymmetric Unit | Ligand(s) | Resolution (Å) | Year | Ref. |
| 1ILG | human | P43212 | None | None | monomer | None | 2.52 | 2001 | 23 |
| 1ILH | human | P43212 | None | None | monomer | SR12813 | 2.76 | 2001 | 23 |
| 1M13 | human | P43212 | None | None | monomer | Hyperforin | 2.15 | 2003 | 27 |
| 1NRL | human | P212121 | SRC-1 | None | homodimer | SR12813 | 2.00 | 2003 | 28 |
| 1SKX | human | P43212 | None | None | monomer | Rifampicin | 2.80 | 2005 | 41 |
| 2O9I | human | P212121 | SRC-1 | None | homodimer | T0901317 | 2.80 | 2007 | 49 |
| 2QNV | human | P43212 | None | None | monomer | Colupulone | 2.80 | 2008 | 43 |
| 3CTB | human | P212121 | Tethered SRC-1 | None | homodimer | None | 2.00 | 2008 | 47 |
| 3HVL | human | P212121 | Tethered SRC-1 | None | homodimer | SR12813 | 2.10 | 2008 | 47 |
| 3R8D | human | P43212 | None | None | monomer | PNU-142721 | 2.80 | 2011 | 44 |
| 4J5W | human | P212121 | Tethered SRC-1 | RXRα | heterotetramer | None | 2.80 | 2013 | 33 |
| 4J5X | human | P212121 | Tethered SRC-1 | RXRα | heterotetramer | SR12813 | 2.80 | 2013 | 33 |
| 4NY9 | human | P43212 | None | None | monomer | CCR1 Antagonist 1a | 2.80 | 2014 | 50 |
| 4XHD | human | P43212 | None | None | monomer | CCR1 Antagonist 2b | 2.40 | 2015 | 30 |
| 4S0T | human | P4 | Adnectin-1 | None | homodimer | CCR1 Antagonist 2b | 3.14 | 2015 | 30 |
| 4S0S | human | P4 | Adnectin-1 | None | homodimer | None | 2.80 | 2015 | 30 |
| 4X1F | human | P43212 | None | None | monomer | 17-α-Ethinylestradiol | 2.00 | 2015 | 46 |
| 4X1G | human | P43212 | None | None | monomer | 17-α-Ethinylestradiol, trans-Nonachlor | 2.25 | 2015 | 46 |
| 4XAO | human | P43212 | None | None | monomer | None | 2.58 | 2015 | 46 |
| 5A86 | human | P212121 | Tethered SRC-1 | None | homodimer | S1P1 Antagonistc | 2.25 | 2015 | 48 |
| 5X0R | human | P212121 | SRC-1 | None | homodimer | SJB7 | 2.67 | 2017 | 45 |
| 6BNS | human | P212121 | Tethered SRC-1 | None | homodimer | RORγt Inverse Agonistd | 2.56 | 2018 | 51 |
N-((R)-1-((S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl)-3-methyl-1-oxobutan-2-yl)-3-hydroxy-3-methylbutanamide
N-((2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl)-2-cyclopropylacetamide
4-chloro-N-[(1R)-1-[1-ethyl-6-(trifluoromethyl)benzimidazol-2-yl]ethyl]benzenesulfonamide
2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide
CCR1=CC chemokine receptor-1 S1P1= sphingosine 1-phosphate receptor 1
RORγt=retinoic acid receptor-related orphan receptor gamma truncated
Figure 1.
Crystal structure of (a) hCAR-LBD (PDB 1XV9) and (b) hPXR-LBD (PDB 1M13). The dotted line in the hPXR-LBD structure represents residues 178 to 197, which are not modeled. Secondary structure elements are color coded for easier recognition.
The structures of PXR-LBD and CAR-LBD are similar to their NR subfamily member vitamin D receptor (VDR, NR1I1). VDR is not a xenobiotic sensing NR but has a role in vitamin D hormone signaling which plays a primary role in calcium homeostasis [29]. The first VDR-LBD crystal structure was published in 2000 [30], and unlike for PXR and CAR, there are also crystal structures of the VDR-DBD, first published in 2002 [31], and structures for the full length receptor in complex with RXR solved by either small angle X-ray scattering (SAXS) [32] or cryo-electron microscopy (cryo-EM) [33]. VDR-LBD shares the classical three-layered helical sandwich with PXR and CAR and has a three-strand antiparallel β-sheet like CAR, but has an insert between H1 and H3 like PXR, though the VDR-LBD insert seems to lack much secondary structure and is usually deleted before crystallization [30]. Further structural aspects of VDR, including mutations that affect its structure and function, and other proteins involved in vitamin D signaling are further covered in a recent review by Rochel and Molnár [34].
3. B factors of PXR and CAR crystal structures
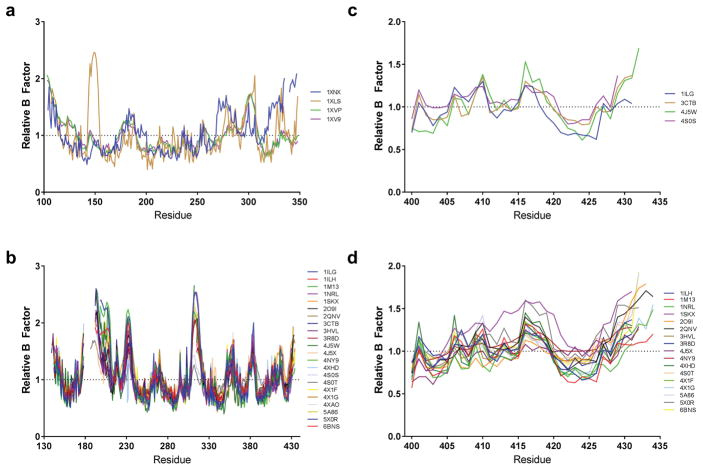
Many of the differences between the LBDs of CAR and PXR relate to the thermal (or dynamic) motion in different regions of the protein. As crystal structures represent the average position of each atom in the entire crystal, there is some uncertainty about the exact position of a given atom. The uncertainty of the overall structure is quantified by the resolution, whereas the uncertainty for each individual atom is quantified by the temperature factor (B factor). Examining the B factors in a protein structure can reveal areas of dynamic mobility within the structure, assuming the model is correct—errors in model building will also cause high B factors. Comparing the average B factor of each residue, as calculated by Baverage in CCP4 [35], to the average B factor of that residue’s protein chain revealed some consistent regions of dynamic motion (Figure 2). In the hCAR-LBD (Figure 2a), these regions include H1 (residues 103–130), H3′ (residues 174–190), the loop between H7 and H8 (residues 251–259), and the loop between H9 and H10 (residues 293–309). The mCAR-LBD shows dynamic motion in the loop between H2″ and H3 (residues 153–162), H3′ (residues 184–200), H9 and the loop between H9 and H10 (residues 279–309), and H10-HX-H12 (residues 328–358). The disorder in H3′ and in the loop between H9 and H10 is consistent in CAR across species and structures. The disorder in the H10-HX-H12 region of the mCAR-LBD structure (protein data bank [PDB] code 1XNX) is caused by the binding of the inverse agonist androstenol, which disrupts the conformation of this region [25]. The source of the highest relative B factors in mCAR is the loop between H2″ and H3 reported in PDB 1XLS [26]. The extensive disorder in this area, as compared to the other CAR structures, might at first raise the suspicion that there had been an error in the model building. However, it is probable that a difference in the space group and, therefore, different contacts within the crystal lattice cause the massive differences in the relative B factors when comparisons are made to the other mCAR structure (1XNX). The loop between H2″ and H3 in 1XNX is more than 14 Å away from the nearest protein chain, whereas in the 1XLS model this loop (specifically, P157 of chain E) comes within 2.5 Å of R342 of chain G. Similar distances are seen with the same amino acid pairs in other monomers. This close contact helps cause P157 to adopt unfavorable backbone torsion angles. The close contact with the loop containing R342 might also explain the higher B factor seen in that region (residues 337–345) in 1XLS.
Figure 2.
(a, b) Comparisons of temperature factors (B factors) between different CAR (a) and PXR (b) structures. The average B factor of each residue was normalized to the average B factor throughout that particular protein chain to look for regions of thermal mobility. If a structure had multiple chains of either CAR or PXR, the average value was taken. mCAR structures were plotted using the equivalent residue number in the human sequence. (c, d) The N-termini of ligand-free PXR (c) and agonist-bound PXR (d) were examined to see what effect the binding of agonist had on the mobility of H12 (residues 422–430).
Examination of the B factors of the 22 published PXR-LBD crystal structures reveals a fairly consistent profile across all structures (Figure 2b). The three regions with the highest degree of thermal motion are the loop between H1 and β1, which includes H2 (residues 172–211); the loop between β1′ and H3 (residues 227–238); and the loop between β4 and H7 that contains the sometimes-modeled H6 (residues 307–323). Other disordered regions include the loop between H8 and H9 (residues 351–364), the loop between H9 and H10 (residues 378–386), and the end of the AF-2 region (residues 425–434). Many of these flexible regions, especially the loop between H1 and β1 and that between β4 and H7, are defined by the absence of residues in the model, indicating that in multiple structures the regions were greatly disordered to the point of being impossible to model. As with the CAR structures, the hPXR-LBD was crystallized in multiple space groups, and this can pose an additional challenge to comparing B factors. In this case, there are no apparent changes in the B factor due to the PXR-LBD structures being in the P212121 or P43212 space groups, as P212121 is a subgroup of P43212. The PXR-LBD structures reported in PDB 4S0T and 4S0S, both of which are P4 structures, show major differences from the other 20 structures, particularly in the areas of the model that often have unmodeled residues [36]. 4S0T has the largest proportion of residues 172 to 211 modeled, and both 4S0T and 4S0S have substantially lower B factors for the sequence between residues 307 and 323 (which contains H6). 4S0S and 4S0T also have one region of increased B factor relative to every other structure; this region is located between residues 386 and 398, which is the beginning of H10. Both 4S0T and 4S0T were modeled using a unique cofactor, Adnectin-1, so it is unclear whether the changes in B factor are due to the unique cofactor, the unique space group, or a combination of both, with the latter being most likely. The other major oddity revealed by the PXR B-factor comparison concerns residues 231 and 232 of 4XHD [36], where a region of normally higher B values exhibits a sudden and drastic dip to a low B value. Closer examination of the structure reveals a torsional strain between these residues that is not present in other structures, highlighting the usefulness of examining B factors in comparisons of crystal structures.
The B factor traces of the human PXR-LBD and CAR-LBD (Figure 2a and 2b) show that the PXR-LBD has a much greater variance in B factor across the whole structure. Also, the complete LBD of CAR has been modeled in every structure, whereas no complete model is available for the LBD of PXR. Together, these findings suggest that the PXR-LBD is more disordered, i.e., more flexible, than the CAR-LBD. Relative B factors also show that the AF-2 region is more ordered in hCAR (residues 341–348) than in hPXR (residues 422–430); this is believed to relate to the constitutive activity of CAR. Interestingly, bound agonist appears to have little to no effect on the mobility of H12 in the PXR LBD (Figure 2c and 2d), even though one would intuitively expect binding of agonist to stabilize the conformation and reduce the relative B factor. One possible explanation for this observation is that the crystallization process itself forces H12 of the PXR-LBD into the active conformation regardless of the ligand, whereas in solution, the presence of ligand increases the stability of the PXR-LBD and prolongs its time in the active conformation by comparison to ligand-free PXR. More effects of ligand binding are discussed below.
4. CAR/PXR and multimeric interactions
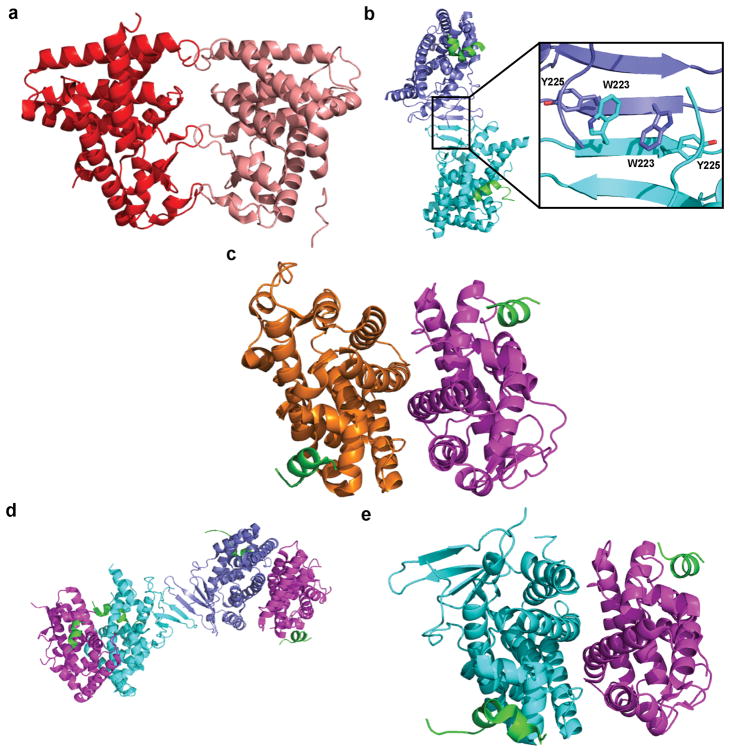
mCAR-LBD has been seen to form a homodimer with an interface consisting of the three loops located between H9 and H10, β1 and β2, and H2″ and H3 (Figure 3a) [25]. The same dimer orientation has not been seen in any hCAR-LBD structure; however, the hCAR-LBD has always been crystallized with the hRXRα-LBD, which might prevent homodimer formation. Other experiments have suggested that dimer formation with this particular interface also occurs with the hCAR-LBD [37]. The PXR-LBD homodimer interface consists of the two β1′ sheets interacting via a tryptophan zipper to form a 10-strand antiparallel β-sheet that spreads across both monomers (Figure 3b) [28]. Half of the structures (11 in total) in the PDB show the complete homodimer in the asymmetric unit; in the other 11 structures, the dimer exists via crystallographic symmetry. Analytical ultracentrifugation has indicated that a homodimer species of PXR-LBD exists in solution, but the homodimer does not form when W223 and Y225 of the tryptophan zipper are mutated to alanine [38]. The PXR homodimer appears to be necessary for transcriptional activation via the recruitment of SRC-1 in transient transfection experiments, but it does not affect the subcellular localization of PXR or the binding of ligand/DNA to the receptor [38].
Figure 3.
(a) Crystal structure of the mCAR-LBD homodimer (red/salmon) (PDB 1XNX). (b) Crystal structure of the hPXR-LBD homodimer (blue/cyan) with bound SRC-1 (green), with a close-up of the tryptophan zipper that composes the dimer interface (PDB 209I). (c) Crystal structure of the hCAR-LBD (orange) and RXRα-LBD (magenta) heterodimer interface with bound SRC-1 (PDB 1XVP). (d, e) Crystal structure of the PXR-LBD and RXRα-LBD heterotetramer (d) and the heterodimer interface (e) with bound SRC-1 (PDB 4J5W).
The CAR-LBD and PXR-LBD have both been co-crystallized with the RXRα-LBD [24, 26, 39]. RXR heterodimers typically form when the N-terminal halves of H10 form a coiled-coil structure through ϕKϕϕ, where ϕ is a hydrophobic residue, and CAR/RXRα LBD and PXR/RXRα LBD heterodimers form in a similar fashion [40]. When heterodimerized with the hRXR-LBD, the hCAR-LBD is rotated through 10° compared to the peroxisome proliferator-activated receptor γ (PPARγ) LBD, and the mCAR-LBD shows a similar rotation (16°) [24, 26]. The full dimer interface (Figure 3c) consists of H7, H9, H10, and the loop between H8 and H9, with an interface size of 995 Å2, which is slightly larger than that of the PPARγ/RXRα heterodimer interface (905 Å2) [26]. H7 in hCAR is relatively straight when in complex with RXRα, unlike PPAR H7, which enables areas of direct contact between the LBDs [24]. Heterodimerization appears to stabilize the RXRα AF-2 helix in its active conformation; however, a functionally active AF-2 helix of RXRα does not appear to be necessary for the constitutive activity of CAR [22, 24]. RXRα may function to further stabilize CAR in the active conformation as RXRα increases the ligand-binding affinity of both TCPOBOP and androstanol and the binding of the coactivator TIF2 to CAR [26].
hPXR and hRXRα form a heterotetramer in the crystal structure (Figure 3d), maintaining the β1′ interface seen in the PXR-LBD homodimer [39]. Binding of an agonist to PXR compresses the length of the complex by 4 Å and stabilizes a portion of the RXRα structure, supporting the general belief that activating ligands serve to stabilize NR complexes in a similar manner to coactivators [39]. The PXR/RXRα interface consists of several electrostatic and polar interactions between H5, H9, and H10 of PXR and H7, H9, and H10 of RXR (Figure 3e). The PXR/RXRα LBD interface (approximately 1200 Å2) is much larger than the CAR/RXRα LBD interface, although they do have similar contacts [39]. As seen with the hCAR/hRXRα heterodimer, both PXR and RXRα have significantly higher affinity for the coactivator SRC-1 when existing as a part of the heterotetramer, indicating that the higher-order structure affects coactivator binding and thereby gene transcription [39].
5. The AF-2 region of CAR
The most distinctive structural feature of CAR is the rigidity of helix 12 in the absence of ligand, which is believed to be the basis of the constitutive activity of CAR. Helix X (HX), located between H10 and H12 (i.e., the AF-2 helix), consists of one helical turn in humans and two turns in mice [24, 26]. This region is normally an extended loop in other NRs, including PXR, but HX in CAR can pack tightly against H3 and H10 as a result of H10 being one turn shorter in CAR than in other NR-LBDs (Figure 1). The shorter distance between H10 and H12 is crucial for the constitutive activity of CAR, as inserting three amino acids between H10 and HX disrupted constitutive activity in mCAR [22]. There is one residue between HX and H12, which reduces the dynamic motion of H12 seen in other NRs and causes CAR to remain in a more favorable conformation for coactivator recruitment and RXR interaction. Although other structures possess helical elements similar to those of HX, the linker between HX and H12 is slightly longer (2+ residues) in those cases [24]. Also aiding in maintaining the rigidity of H12 is a charge-charge interaction between H4 and the C-terminal carboxyl group of H12, which is enabled by the short length of the CAR H12 as compared to that of other NRs. H12 and HX sit atop four amino acids (F161, N165, F234, and Y326) that shield the LBP [24]. These four amino acids are not conserved in PXR, thus allowing ligand easier access to H12. In CAR, receptor activation appears to require interaction between H12 and these four barrier residues. In other NRs that lack this barrier, the H12 appears to act like a “cap” on the LBD [41].
6. Coregulator binding to the AF-2 region
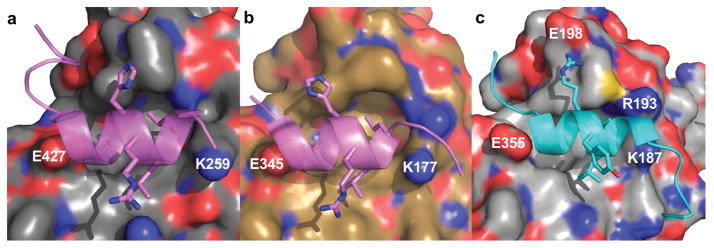
The orientation of H12 (AF-2) has a direct effect on the ability of coactivators or corepressors to bind to the receptor. The active conformation consists of H12 folded against the body of the protein, where it can form part of the LBP, sometimes directly interacting with bound agonist. In the absence of ligand, H12 of CAR is already in the active conformation, whereas H12 of PXR is believed to be much more mobile. When in the active conformation, coactivators bind with the motif LXXLL, which is a two-turn α-helix docked into a charge clamp pocket formed by H12 and H3 [42, 43, 44]. The three leucine residues orient into the coactivator-binding site (Figure 4). The charge clamp of PXR (Figure 4a) consists of E427 of H12 and K259 of H3, whereas that of CAR (Figure 4b) consists of E345 of H12 and K177 of H3. A second charge clamp has been observed in CAR between R193 of H3′ and E198 of H4 in the structure of mCAR when bound to TIF2 (Figure 4c) [26]. According to molecular dynamics simulations, the binding of coactivator to the LBD of PXR or other NRs restricts the motion of the entire LBD [45]. There has been some evidence using total internal reflection fluorescence microscopy that ligand binding to PXR-LBD will not increase coactivator binding, unlike with many other NRs [46]; however, another study using a mammalian two-hybrid assay in addition to TR-FRET showed that ligand binding does indeed increase association with coactivator [47].
Figure 4.
(a, b) The charge clamp pocket in which SRC-1 (lavender) binds is located between K259 and E427 in hPXR (gray) (PDB 1NRL) (a) and between K177 and E355 in hCAR (brown) (PDB 1XVP) (b). (c) The structure of TCBOPOP-bound mCAR (silver) (PDB 1XLS) reveals a second charge clamp between R193 and E198 when the complex is bound to TIF2 (cyan).
Corepressors will bind to the AF-2 region when H12 is not in the active conformation. There appear to be multiple inactive conformations, as H12 has been observed in many different positions in structural studies of other NRs [41]. The temperature factor (B factor) for H12 in many of these structures is greater than 100 Å, indicating that the helix is highly mobile. When H12 is displaced from the active position, corepressors such as SMRT can bind with the motif LXXXIXXL [48]. In other NRs, this motif adopts a three-turn α-helix that docks in the same site as the coactivator. The additional turn extends into the AF-2 region and prevents the active conformation. So far, no crystal structure has been solved for CAR or PXR with a bound corepressor.
Classic coactivators and corepressors are not the only binders to the AF-2 region of PXR or CAR; Adnectins, targeted biologics derived from the tenth type III domain of human fibronectin by Adnexus Therapeutics, also can bind to the AF-2 region of PXR [36]. Although the specific amino acid interactions of Adnectins differ from those of a coactivator, the size of the interface is quite similar, and the residues involved on the PXR side are also similar. Adnectin-1 did not significantly alter ligand orientation, pocket size, or overall secondary structure of PXR when bound, and it apparently displaced already-bound SRC-1 [36].
7. Effect of ligand binding
The binding of ligand to the ligand-binding site of CAR or PXR affects the overall structure of the LBD. We refer readers to a recent review by Chai et al. [49] for depictions of binding interactions which we discuss here. Currently, there is only one structure showing an inverse agonist bound to CAR (i.e., that of androstenol bound to mCAR [PDB 1XNX]), and no structure of an antagonist bound to PXR has been reported. Binding of the inverse agonist androstenol disrupts the rigidity of H12 (AF-2) in CAR, leaving the orientation more random [25]. The only polar interactions in this binding interaction are between the 3α-hydroxyl moiety of androstenol and residues N175 (directly) and H213 (via a water molecule); without these interactions, CAR does not respond to the compound [25]. Androstenol binding does not directly displace H12 but results in the formation of a kink between H10 and H11, which is anchored by a hydrogen bond between E339 and the backbone of Q245. This disrupts the charge-charge interaction between H4 and the C-terminal carboxyl group of H12 that helps lock H12 in the transcriptionally active conformation [25]. Agonists that directly bind to CAR function by further stabilizing HX and H12. TCPOBOP, CITCO, and 5β-pregnanedione bind to CAR mainly through hydrophobic interactions [24, 26]. TCBOBOP forms two indirect hydrogen bonds to mCAR via a water molecule, and 5β-pregnanedione forms one hydrogen bond between the C21 ketone and H203 of hCAR [24, 26]. CITCO has multiple modes of binding to hCAR, with no specific hydrogen bonds being required [24].
Agonists bound to PXR also function by stabilizing the AF-2 region of the receptor. In this case, however, it is believed that H12 is not already in an active confirmation before ligand binding while in solution. The PXR-LBD will undergo induced fit in order to bind to a variety of ligands, a characteristic not yet seen in CAR-LBD structures. H2 and the loops between β4 and H7 and between β1′ and H3 provide a flexible side to the LBP opposite the antiparallel β-sheet (Figure 1) [50]. Six residues within the LBP of PXR commonly interact with ligands (M243, S247, Q285, W299, H407, and F420) [51]. The PXR agonist SR12813 has been used in several structural studies of the PXR-LBD. SR12813 has three different binding orientations to PXR in the absence of coactivator [23], but it has a further, unique binding orientation in the presence of an SRC-1 peptide, which traps SR12813 in an orientation that directly contacts H12 [28]. Binding of hyperforin to PXR enables H6 to appear, which is not seen in the apo- or SR12813-bound structures, and shifts the position of H2, indicative of the ability of PXR to undergo induced fit [27]. Neither does hyperforin directly contact any residue in the AF-2 helix. Colupulone, which is structurally similar to hyperforin, adopts a vastly different orientation in the LBP, highlighting the challenge of predicting how similar compounds will interact with the PXR-LBD [52]. The hydrophobic contacts generated by colupulone more closely resemble those of rifampicin, which may be the best example of the ability of PXR to undergo induced fit. When rifampicin binds to PXR, the entire flexible side of the LBP becomes disordered, and the 4-methyl-1-piperazinyl ring of rifampicin remains disordered [50]. In fact, rifampicin would be unable to bind in the LBP of the apo-structure unless this region expanded. This region of the LBP also remains disordered in crystals with smaller ligands such as PNU-142721 or SJB7 bound [47, 53]. In the case of SJB7, the interaction between L428 and F429 in H12 and the para-methoxy group is believed to drive agonist action, and the lack of this interaction with other SJB7 analogs, such as SPA70, is believed to lead to antagonism [47]. The LBP of PXR is large enough to allow multiple compounds to bind simultaneously, as seen recently in the crystal structure of trans-nonachlor (TNC) and 17α-ethinylestradiol (EE2) bound to PXR (PDB 4X1G) [54]. Although TNC and EE2 are weak PXR agonists by themselves, they synergistically agonize PXR to a much higher degree [54]. This suggests that some of the in vivo activity of PXR may be driven by the combination of several ligands acting together rather than by single ligands acting independently. This result also provides a new strategy for developing potent PXR agonist/antagonists by first screening combinations of smaller chemicals, including fragments, with the aim of ultimately combining them to form a larger compound.
8. Crystallization techniques
8.1 Purification strategies
Many of the PXR-LBD and CAR-LBD crystal structures were derived from proteins expressed through similar processes (Table 2). The first structure of the PXR-LBD to be solved used protein co-expressed with residues 623 to 710 of the coactivator SRC-1 in a separate vector [23]. The SRC-1 sequence was subsequently inserted in the same vector as the PXR-LBD [47]. Another expression strategy has been to tether a shorter SRC-1 sequence (residues 678–700) to the PXR-LBD with a glycine-serine pentapeptide linker [39, 55, 56]. Both the PXR-LBD and the RXRα-LBD were purified with this tethered SRC-1 to solve the heterotetramer structure [39]. The tethered SRC-1 did not alter the binding interactions of SR12813, as compared to those of the previous structure containing un-tethered SRC-1 peptide [55]. The presence of SRC-1 peptide, whether tethered, co-expressed, or added to the purified protein, appears to be essential for the solubility of the PXR-LBD at the higher concentrations needed for crystallization. Due to the possibility of dithiothreitol forming covalent complexes with PXR, some structures were generated by first mutating C284 in the LBP to serine [52, 57].
Table 2.
Summary of published crystallization conditions for PXR-LBD and CAR-LBD.
| CAR-LBD Crystallization Conditions | |||||||
|---|---|---|---|---|---|---|---|
| PDB ID | Co-peptide | [Protein] (mg/mL) | Crystallization Buffer | Diffusion Method | Temp (°C) | Ref. | |
| In Purification | Added Pre-Crystallization | ||||||
| 1XNX | SRC-1 (153-mer) | None | 6–8 | 0.1 M HEPES, pH 7.2, 18% PEG400, 0.2 M CaCl2, 0.01 M L-cysteine | H | 14 | 25 |
| 1XVP | None | SRC-1 (13-mer) | 10 | 0.1 M TRIS, pH 8.0, 12–14% glycerol, 0.8–0.9 M LiSO4 | H | 4 | 24 |
| 1XV9 | None | SRC-1 (13-mer) | 10 | 0.1 M TRIS, pH 8.0, 12–14% glycerol, 0.8–0.9 M LiSO4 | H | 4 | 24 |
| 1XLS | None | TIF2 (18-mer) | 8 | 0.1M (NH4)2C4H4O6, pH 6.5, 10–15% PEG2000, 3% 1,6-hexanediol | H | 22 | 26 |
| PXR-LBD Crystallization Conditions | |||||||
| PDB ID | Co-peptide | [Protein] (mg/mL) | Crystallization Buffer | Diffusion Method | Temp (°C) | Ref. | |
| In Purification | Added Pre-Crystallization | ||||||
| 1ILG | SRC-1 (88-mer) | None | 5 | 50 mM imidazole, pH 7.2, 10% 2-propanol | H | 22 | 23 |
| 1ILH | SRC-1 (88-mer) | None | 4 | 50 mM imidazole, pH 7.2, 10% 2-propanol | H | 22 | 23 |
| 1M13 | SRC-1 (88-mer) | None | 5 | 50 mM imidazole, pH 7.6, 12% 2-propanol | H | 22 | 27 |
| 1NRL | SRC-1 (88-mer) | SRC-1 (25-mer) | 11 | 50 mM imidazole, pH 7.2, 10% 2-propanol | H | 22 | 28 |
| 1SKX | SRC-1 (88-mer) | None | 3 | 50 mM imidazole, pH 7.2, 9% 2-propanol | H | 25 | 41 |
| 2O9I | SRC-1 (88-mer) | None | 5 | 50 mM imidazole, pH 7.1, 10% 2-propanol | H | 22 | 49 |
| 2QNV | SRC-1 (88-mer) | None | 10 | 50 mM imidazole, pH 8.0, 10% 2-propanol, 50 mM dithiothreitol | H | 25 | 43 |
| 3CTB | Tethered SRC-1 | None | 10 | 100 mM imidazole, pH 8.0, 10–30% 2-methyl-2,4-pentanediol | H | 4 | 47 |
| 3HVL | Tethered SRC-1 | None | 10 | 100 mM imidazole, pH 8.0, 10–30% 2-methyl-2,4-pentanediol | H | 4 | 47 |
| 3R8D | SRC-1 (88-mer) | None | 3 | 50 mM imidazole, pH 7.8, 16% 2-propanol | H | 22 | 44 |
| 4J5W | Tethered SRC-1 | None | NL | 100 mM BIS-TRIS propane, pH 7.0, 16–20% PEG8000, 0.1–0.2 M MgCl2, 0.02% sodium azide | H | 4 | 33 |
| 4J5X | Tethered SRC-1 | None | NL | 101 mM BIS-TRIS propane, pH 7.0, 16–20% PEG8000, 0.1–0.2 M MgCl2, 0.02% sodium azide | H | 4 | 33 |
| 4NY9 | SRC-1 (88-mer) | None | NL | 100 mM imidazole, pH 7.8, 8–10% 2-propanol | H | 23 | 50 |
| 4XHD | SRC-1 (88-mer) | None | 3 | 100 mM imidazole, pH 7.8, 10% 2-propanol | H | 20 | 30 |
| 4S0T | SRC-1 (88-mer) | Adnectin-1 | 3 | 100 mM TRIS, pH 7.5, 10% 2-propanol, 3.5% 2-methyl-2,4-pentanediol | S | 20 | 30 |
| 4S0S | SRC-1 (88-mer) | Adnectin-1 | 3 | 100 mM HEPES, pH 7.0, 18.3% 1,6-hexanediol, 3% 2-methyl-2.4-pentanediol | H | 12 | 30 |
| 4X1F | SRC-1 (88-mer) | None | 2.4 | 100 mM imidazole, pH 7.1, 10% 2-propanol, 100 mM NaCl | H | 18 | 46 |
| 4X1G | SRC-1 (88-mer) | None | 2.4 | 100 mM imidazole, pH 7.1, 10% 2-propanol, 50 mM NaCl, 50 mM LiCl | H | 18 | 46 |
| 4XAO | SRC-1 (88-mer) | None | 2.4 | 100 mM imidazole, pH 7.1, 10% 2-propanol | H | 18 | 46 |
| 5A86 | Tethered SRC-1 | None | 15 | 50 mM HEPES, pH 7.0, 10–16% 2-propanol | H | NL | 48 |
| 5X0R | SRC-1 (88-mer) | SRC-1 (25-mer) | 9.1 | 50 mM imidazole, pH 7.8, 10% 2-propanol | H | 20 | 45 |
| 6BNS | Tethered SRC-1 | None | NL | 100 mM HEPES, pH 7.5, 10–15% PEG10000, 150 mM NaCl | NL | 23 | 51 |
(H)anging and (s)itting drop vapor diffusion
NL= Not Listed
PEG= polyethylene glycol
The CAR-LBD was expressed in two different manners for crystallization, depending on whether the RXRα-LBD was used. The mCAR-LBD homodimer was co-expressed with a SRC-1 peptide in a separate vector, an approach that is similar to the initial strategy employed for the PXR-LBD [25]. For both the mouse and human CAR/RXRα structures, protein was produced by co-expressing each LBD in a separate plasmid with no coactivator present [24, 26]. The presence of the other dimer partner appears to keep each receptor soluble. However, coactivator was added to the protein mixture before the crystallization trials were completed.
8.2 Crystallization conditions
The PXR-LBD and CAR-LBD have some similarities in their crystallization conditions. Both have been stored in similar buffers for crystallography, which is an important component of the crystallization condition that is often overlooked. The storage buffer usually consists of 20 mM Tris, pH 7.8, 250 mM NaCl, 5 mM DTT, and 2.5 mM EDTA. PXR-LBD and CAR-LBD crystals were formed primarily through hanging drop vapor diffusion, with sitting drop vapor diffusion being listed for only one crystal (PDB 4S0T) [36]. Almost all structures with ligand were formed via co-crystallization, as the LBP is in the interior of the crystal structure with limited solvent access. Only two ligands have been soaked into the LBD, both with PXR: N-((R)-1-((S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl)-3-methyl-1-oxobutan-2-yl)-3-hydroxy-3-methylbutanamide (PDB 4NY9) [58] and N-((2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl)-2-cyclopropylacetamide (PDB 4XHD) [36].
The differences between PXR-LBD and CAR-LBD crystallization lay primarily in the crystallization buffer, as is the case with many proteins (Table 2). Three different conditions for CAR-LBD crystallization have been published, resulting in three different space groups; this is not surprising, as the conditions are from three different groups published simultaneously and contain three vastly different protein mixtures (mCAR, mCAR + hRXR, and hCAR + hRXR). There are no apparent similarities between the three conditions beyond those already mentioned, as each crystallization was completed at a different protein concentration, pH, and temperature.
The conditions for PXR-LBD crystallization are fairly consistent across multiple laboratories (Table 2). The basic conditions include 50–100 mM imidazole, pH 7–8, and 10% 2-propanol. Occasionally, a different alcohol is added to or replaces the 2-propanol. Two conditions do not fit this pattern: the condition for the PXR/RXRα-LBD heterotetramer (PDB 4J5W and 4J5X) [39] and the recently published structure of 2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide bound to the PXR-LBD (PDB 6BNS) [59]. The former exception is not surprising, as this is a different protein mixture. The crystallization conditions for the PXR-LBD have generally produced crystals in one of two space groups, P43212 or P212121. The P43212 crystals contain a monomer in the asymmetric unit, whereas the P212121 crystals contain the full PXR-LBD homodimer. As mentioned earlier, the dimer exists in the P43212 crystals by symmetry. When the PXR-LBD was co-expressed with only an 88–amino acid SRC-1 peptide, the peptide was not present, let alone seen, in the final crystal except in one case (PDB 2O9I) [23, 57]. In all other cases, SRC-1 was seen in the crystal structure only when a smaller 25–amino acid SRC-1 peptide was added to the purified protein or when the tethered SRC-1 construct was used. In the P43212 crystal lattice, the peptide-binding site of PXR is involved in a crystal packing conformation that would appear to leave insufficient space for the 88–amino acid peptide to bind to the receptor. The P212121 space group allows enough room for the tethered SRC-1 sequence or the smaller 25–amino acid peptide. Two PXR-LBD crystal structures in the P4 space group (PDB 4S0T and 4S0S) were solved by using similar conditions as for other PXR-LBD structures [36]. These structures were solved using Adnectin-1, which binds in the coactivator site replacing the 88–amino acid SRC-1 peptide, whose purpose is to act as a crystallization chaperone. Adnectins have been used in other crystal complexes (EGFR, IL23, and PCSK9) before [60, 61].
9. Hydrogen-deuterium mass spectrometry of PXR
HDX-MS has emerged as a powerful tool in the investigation of protein-ligand interactions in solution, particularly for examining the structural dynamics that result from ligand binding. This technique can impart information that cannot be obtained from static crystal structures. It has been successfully applied to several NRs for ligand-binding mapping, to rationalize structure-activity relationships (SARs), to uncover graded agonism, and to elucidate the mechanism of ligand activation. Nuclear receptors studied with HDX-MS include farnesoid X receptor [62], glucocorticoid receptor [63], PPARγ [64], RXRα [65], and vitamin D receptor (VDR) [66], among others. The investigation of a growing list of NRs with this technique has yielded important information that complements the data obtained by other structural and biophysical methods.
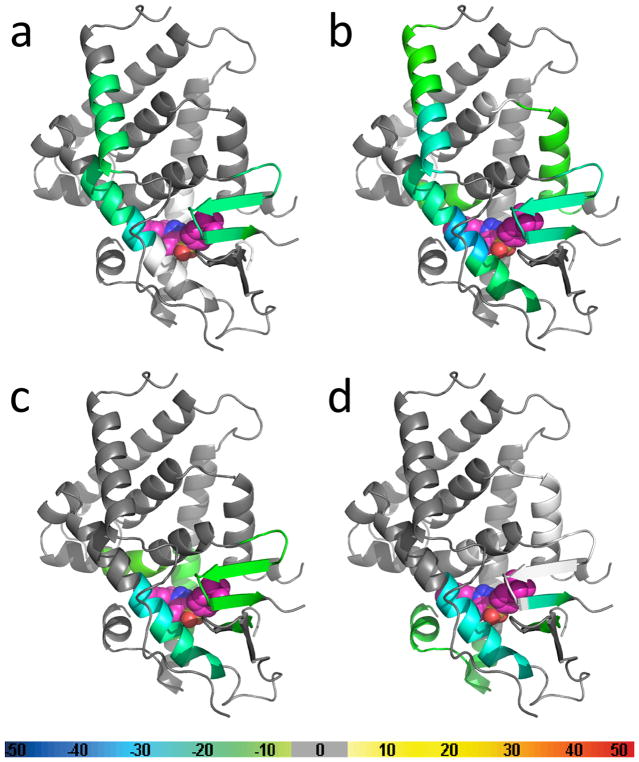
The results of the first HDX-MS analysis of PXR were recently reported (Figure 5). That study explored solution conformational changes due to the complexation of PXR with the agonist SJB7 or the antagonist SPA70 [47]. The differential HDX signature of PXR in the presence and absence of SJB7 indicated that the agonist induced PXR structural stability, with several regions being protected from deuterium exchange (Figure 5a). This is consistent with the notion that ligand binding induces a transition to a less fluid state and that the ligand is an integral component of protein compactness and stability. Areas of local stabilization distant from the site where SJB7 resides have been observed, and these would not have been inferred from the crystal structure. Interestingly, no protection in the AF-2 helix has been detected. This is not without precedent: RXRα likewise showed no HDX difference in the AF-2 helix in the presence or absence of agonists and inhibitors [65]. Unlike full agonists, partial agonists failed to stabilize the AF-2 helix in VDR and PPARγ [64, 66].
Figure 5.
Results of hydrogen-deuterium exchange mass spectrometry (HDX-MS) studies of the PXR LBD in the presence of SJB7 and SPA70. The crystal structure of the PXR LBD (PDB 5X0R) was used as the template for overlays of the differential HDX data for (a) SJB7, (b) SPA70, (c) SJB7 with the subsequent addition of SRC-1 peptide, and (d) SPA70 with the subsequent addition of NCoR peptide. The coloring of the PXR structures is based on the color bar at the bottom of the figure. Regions that were not covered are represented in white. SJB7 and SPA70 are represented as spheres, with carbon atoms depicted in pink. The SJB7 binding location is taken from the crystal structure, whereas that for SPA70 was determined by docking studies.
Differences in protein conformation compared to apo PXR have also been detected in the presence of the antagonist SPA70 (Figure 5b), where the HDX mapping region is similar to that of SJB7 (Figure 5a). This indicates that SPA70 resides in a site in the LBP analogous to that of SJB7, suggesting similar PXR-ligand contacts. Because there is currently no crystal structure available for the PXR-SPA70 complex, the evidence provided by the comparative HDX-MS analysis of SJB7 and SPA70 is significant. Given that the scaffolds of SPA70 and SJB7 are almost identical, with only minor functional group differences, these HDX-MS studies, together with docking modeling, provided a rationale for describing PXR antagonism by SPA70. In short, it was proposed that the absence of a para-methoxy group in SPA70 (which is present in SJB7 and interacts with residues in the AF-2 helix, according to the crystallographic model) leads to a decreased stabilization of the AF-2 helix for coactivator binding.
Global analyses of solvent exchange due to the presence of the coactivator peptide SRC-1 (CPSSHSSLTERHKILHRLLQEGSPS) or corepressor peptide NCoR (ASNLGLEDIIRKALMGSFD) have also been conducted (Figures 5c and 5d, respectively). The addition of the corepressor peptide NCoR to the PXR-SPA70 complex leads to the stabilization of additional regions (Figure 5d). In particular, the AF-2 helix is seen to have reduced flexibility, although HDX-MS is not capable of establishing the spatial position of the AF-2 helix. Curiously, the coactivator peptide SRC-1 failed to restrict its flexibility (Figure 5c). A peptide longer than the 25-residue SRC-1 used in this study might provide further stabilization.
The same HDX-MS approach has recently been used to study CAR-ligand interaction [67], where this technique demonstrated direct binding of the small-molecule inhibitor CINPA1 to hCAR-LBD. Ligand-binding was shown to stabilize certain regions of the protein, while the addition of the corepressor peptide NCoR1-3 further changed the solvent-protection profile.
10. Conclusion
X-ray crystallographic studies of the CAR-LBD and PXR-LBD over the past 15+ years have revealed unique structural features that enable CAR and PXR to respond to a variety of ligands. The LBPs of CAR and PXR are largely hydrophobic and do not selectively target one particular ligand. This enables them to respond to a wide range of exogenous and endogenous ligands and thereby affect drug metabolism. Comparisons of the structural models and the respective B factors help to explain some of the observed differences in the function of CAR and PXR. The LBD of PXR is much more flexible than that of CAR as several regions of the PXR-LBD remain unmodeled and more regions have higher relative B factors. This increased flexibility enables the PXR-LBD to undergo induced fit to bind larger ligands. Meanwhile, the rigidity of the CAR-LBD, manifested by the smaller fluctuations in its relative B factor, contributes to its constitutive activity, as H12 is fixed in place by the unique HX. The promiscuity of ligand binding makes it challenging to predict how a chemical scaffold will interact with PXR or CAR, though stabilization of H12, either directly or indirectly, seems to be a key interaction for agonism. Crystal studies of receptor-ligand-coactivator complexes have explored the mechanisms of agonism, how agonists interact with the receptor to enhance coactivator binding, and how coactivator binding can affect the binding of agonists, whereas other studies have demonstrated how both CAR and PXR interact with RXRα in the heterodimer, showing that PXR, uniquely, remains dimerized when bound to RXRα, thus forming a heterotetramer.
HDX-MS has proved to be an effective tool for probing the conformational dynamics of the PXR-LBD in complex with the agonist SJB7 and the antagonist SPA70, which could not be addressed by X-ray crystallography. Changes in the HDX protection profile of PXR-SPA70 complexes resulted in the mapping of the SPA70 interactions within the canonical LBP, which, alongside docking studies, aided in the generation of a hypothesis for PXR antagonism by SPA70. These findings provide a foundation for the improvement of PXR antagonists by providing opportunities to explore the chemical scaffold of SPA70.
11. Expert opinion
Crystallographic studies on PXR and CAR have provided ample evidence of the general structure of the LBD of both receptors, shown how structural differences account for differences between the two receptors in their ligand response and overall activity, explored the binding orientations of numerous ligands, and mapped the interactions between the two receptors and RXR in their respective complexes. However, we still lack information on certain aspects, such as the exact mechanism(s) of antagonism of PXR. Although HDX-MS techniques can provide information on the general binding area of PXR antagonists such as SPA70, a crystal structure of PXR with bound antagonist and/or corepressor would provide more direct evidence of which interactions with PXR are key to antagonism. A major obstacle to obtaining this type of crystal structure is the reliance on a coactivator in the purification and crystallization of the PXR-LBD. It is most likely that an antagonist will never successfully bind in the LBP of PXR in a pertinent orientation if the LBD is already saturated by a coactivator; likewise, it would be extremely difficult to replace the entire coactivator population that is already bound to the LBD with corepressor to achieve the homogenous mixture needed for crystal formation. Using a different expression system, such as one in which the commonly used SRC-1 88-mer is replaced by an equivalent section of corepressor, might alleviate this challenge and enable the successful crystallization of an antagonist bound in the PXR-LBP. Although multiple laboratories have crystallized the PXR-LBD by using essentially the same conditions (a level of reproducibility that we find to be rare), obtaining the structure of antagonist-bound PXR-LBD with corepressor will probably require screening for a new condition. An ideal condition would be one that forms PXR-LBD crystals both with and without coactivator/corepressor, enabling easier comparisons of the effects of agonists, antagonists, and those ligands that appear to lie somewhere between the two. For binding sites on the surface of PXR-LBD, such as those proposed for the antagonist ketoconazole, the binding pocket may be too shallow for efficient binding to achieve a crystal structure without the presence of other cofactors [68]. In the absence of such crystal structures (and as a supplement to them), HDX-MS profiles with diverse sets of ligands would provide a better understanding of the relation between gradual agonism and protein stabilization. HDX signatures could also help to predict the pharmacology of these ligands. Taking as a basis the first reported PXR antagonist, this technique would be invaluable for the characterization and development of more potent and selective antagonists.
Recently, there has been limited progress in the field of structural biology concerning PXR and CAR beyond the topics of agonist binding to the LBD and how the LBDs interact with the RXRα-LBD to form their respective complexes. In one recent trend, the structure of the PXR-LBD when bound to ligand has been used to aid the design of a compound that will have no off-target effect on PXR [36, 56, 58, 59]. For CAR, no new crystallographic model has been published since the initial four structures were published simultaneously in 2004 [24, 25, 26]. Other than possible difficulties in obtaining a crystal, the reasons for this lack of new models for almost 15 years remain unclear, as several questions remain to be unanswered. For instance, although the mechanism of action of the inverse agonist androstenol on mCAR is well understood, it is unclear whether a similar mechanism operates with hCAR, as androstenol is mCAR selective. Many CAR inverse agonists are also agonists of PXR [69], and this is generally believed to be related to ligand size, as the LBP of PXRis large enough to accommodate almost any ligand that can bind in the CAR pocket. However, selective compounds for hCAR vs hPXR have been identified (e.g., CINPA1 [70]), and crystal structures of relevant ligands bound to the LBDs of both CAR and PXR could explain why this phenomenon occurs and provide possible strategies to avoid it when optimizing ligand specificity. Additionally, only one small peptide with one copy of the NR binding motif (i.e., the NR box) from the coactivator SRC-1 has been seen in crystal structures (excluding Adnectin-1), but coactivators/corepressors have multiple copies of NR binding motifs. What is the function of these other copies? A crystal structure with a longer coregulator peptide containing multiple NR binding motifs (NR boxes for coactivators, receptor interacting domain [ID] boxes for corepressors) could answer this. Another interesting question left largely unexplored lies in the differences in ligand response of PXR and CAR in other species. What structural features of these ortholog receptors contribute to their vastly different ligand specificity? Crystal structures of PXR and CAR from these other species with relevant ligands could help provide answers on these differences as sequence alignments can only provide so much information in terms of structural similarity.
Finally, full-length PXR and CAR have yet to be examined by structural studies. A crystal structure of the full-length receptor alone or in complex with DNA, RXR, a coactivator, a mediator, etc., would be extremely informative. As the size of the complexes increases, it will become more advantageous to use the recent advances in cryo-EM spectroscopy to determine their organization. For example, studying the full-length structures of PXR/RXR or CAR/RXR bound to DNA could provide answers to many questions: Do both dimer pairs in the PXR/RXR tetramer bind DNA? Do any portions of the LBDs interact with the DNA when in complex? Are there currently unknown allosteric sites, e.g., near the hinge region, that could be targeted for antagonism, thereby avoiding the troublesome selectivity issue presented by the LBPs? Do the LBD and DBD of the two receptors communicate with each other, and if so, how? Interaction between the LBD and DBD has been observed before in other NRs [41, 71], and HDX-MS of full-length proteins could provide information on the dynamic interplay between the LBD and DBD for CAR and PXR and on the potential role of the hinge region in allosteric communication. These types of questions and more will be addressable by a combination of X-ray crystallography, HXD-MS, and, possibly, cryo-EM spectroscopy in the coming years.
Article Highlights.
PXR and CAR are both characterized by a large, flexible, hydrophobic ligand-binding pocket that leads to ligand promiscuity and enables multiple binding positions for potential ligands, although there are residues that consistently interact with agonists.
Examination of the relative B factors of crystal structures reveals consistent regions of flexibility and allows for better visualization of the effect of ligand on H12 conformation in each receptor.
Both PXR and CAR can form homodimers, and unlike the CAR homodimer, the PXR homodimer is maintained when in complex with RXRα.
Crystallization techniques for CAR and PXR often involve the co-expression of a coactivator peptide or the tethering of a shorter peptide to the C-terminus of the receptor. The crystallization conditions for PXR are remarkably consistent across various laboratories.
The use of HDX-MS to study PXR reveals that the binding of ligand or coregulator can alter the conformation of PXR and aid in developing a hypothesis to explain SPA70 antagonism.
Acknowledgments
The authors thank Keith A. Laycock, PhD, ELS, for editing the manuscript.
Funding
This work was supported by ALSAC, St. Jude Children’s Research Hospital, the National Institutes of Health National Institute of General Medical Sciences (grants nos. RO1-GM110034 and R35-GM118041), and the National Cancer Institute (grant no. P30-CA21765).
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiological reviews. 2001;81(3):1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 2.Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2000;23(5):687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- 3*.Moore LB, Parks DJ, Jones SA, et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275(20):15122–15127. doi: 10.1074/jbc.M001215200. This study characterizes ligands that activate both PXR and CAR. [DOI] [PubMed] [Google Scholar]
- 4**.di Masi A, De Marinis E, Ascenzi P, et al. Nuclear receptors CAR and PXR: molecular, functional, and biomedical aspects. Mol Aspects Med. 2009;30(5):297–343. doi: 10.1016/j.mam.2009.04.002. A comprehensive review of PXR and CAR. [DOI] [PubMed] [Google Scholar]
- 5.Tolson AH, Wang H. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv Drug Deliv Rev. 2010;62(13):1238–1249. doi: 10.1016/j.addr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1(4):259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 7*.Wallace BD, Redinbo MR. Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective. Drug Metabol Rev. 2013;45(1):79–100. doi: 10.3109/03602532.2012.740049. This review addresses the structural biology of other nuclear receptors involved in drug metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faucette SR, Sueyoshi T, Smith CM, et al. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317(3):1200–1209. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352(21):2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin Pharmacokinet. 2003;42(15):1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- 11*.Kliewer SA, Moore JT, Wade L, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92(1):73–82. doi: 10.1016/s0092-8674(00)80900-9. This study first indentified PXR. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg B, Sabbagh W, Jr, Juguilon H, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12(20):3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumberg B, Kang H, Bolado J, Jr, et al. BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites. Genes Dev. 1998;12(9):1269–1277. doi: 10.1101/gad.12.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann JM, McKee DD, Watson MA, et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102(5):1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Baes M, Gulick T, Choi HS, et al. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14(3):1544–1552. doi: 10.1128/mcb.14.3.1544. This study first identified CAR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi HS, Chung M, Tzameli I, et al. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272(38):23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- 17.Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 18.Scheer N, Ross J, Rode A, et al. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. The Journal of clinical investigation. 2008;118(9):3228–3239. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72(3):231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie W, Barwick JL, Downes M, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406(6794):435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 21.Tzameli I, Pissios P, Schuetz EG, et al. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20(9):2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dussault I, Lin M, Hollister K, et al. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol. 2002;22(15):5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Watkins RE, Wisely GB, Moore LB, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292(5525):2329–2333. doi: 10.1126/science.1060762. This study was the first to publish a PXR-LBD structure. [DOI] [PubMed] [Google Scholar]
- 24*.Xu RX, Lambert MH, Wisely BB, et al. A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol Cell. 2004;16(6):919–928. doi: 10.1016/j.molcel.2004.11.042. This study was the first to publish a hCAR-LBD structure. [DOI] [PubMed] [Google Scholar]
- 25.Shan L, Vincent J, Brunzelle JS, et al. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16(6):907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suino K, Peng L, Reynolds R, et al. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16(6):893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 27.Watkins RE, Maglich JM, Moore LB, et al. 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry. 2003;42(6):1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 28.Watkins RE, Davis-Searles PR, Lambert MH, et al. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003;331(4):815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 29.Fleet JC. The role of vitamin D in the endocrinology controlling calcium homeostasis. Mol Cell Endocrinol. 2017;453:36–45. doi: 10.1016/j.mce.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rochel N, Wurtz JM, Mitschler A, et al. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5(1):173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 31.Shaffer PL, Gewirth DT. Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J. 2002;21(9):2242–2252. doi: 10.1093/emboj/21.9.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rochel N, Ciesielski F, Godet J, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18(5):564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 33.Orlov I, Rochel N, Moras D, et al. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 2012;31(2):291–300. doi: 10.1038/emboj.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochel N, Molnar F. Structural aspects of Vitamin D endocrinology. Mol Cell Endocrinol. 2017;453:22–35. doi: 10.1016/j.mce.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 35.Bailey S. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Khan JA, Camac DM, Low S, et al. Developing Adnectins that target SRC co-activator binding to PXR: a structural approach toward understanding promiscuity of PXR. J Mol Biol. 2015;427(4):924–942. doi: 10.1016/j.jmb.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 37.Shizu R, Osabe M, Perera L, et al. Phosphorylated nuclear receptor CAR forms a homodimer to repress its constitutive activity for ligand activation. Mol Cell Biol. 2017;37(10):e00969–16. doi: 10.1128/MCB.00649-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noble SM, Carnahan VE, Moore LB, et al. Human PXR forms a tryptophan zipper-mediated homodimer. Biochemistry. 2006;45(28):8579–8589. doi: 10.1021/bi0602821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace BD, Betts L, Talmage G, et al. Structural and functional analysis of the human nuclear xenobiotic receptor PXR in complex with RXRα. J Mol Biol. 2013;425(14):2561–2577. doi: 10.1016/j.jmb.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gampe RT, Jr, Montana VG, Lambert MH, et al. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000 Mar;5(3):545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 41**.Rastinejad F, Huang P, Chandra V, et al. Understanding nuclear receptor form and function using structural biology. J Mol Endocrinol. 2013;51(3):T1–T21. doi: 10.1530/JME-13-0173. This comprehensive review covers the structural biology of all nuclear receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15(6):708–715. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 44.McInerney EM, Rose DW, Flynn SE, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12(21):3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teotico DG, Frazier ML, Ding F, et al. Active nuclear receptors exhibit highly correlated AF-2 domain motions. PLoS Comput Biol. 2008;4(7):e1000111. doi: 10.1371/journal.pcbi.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navaratnarajah P, Steele BL, Redinbo MR, et al. Rifampicin-independent interactions between the pregnane X receptor ligand binding domain and peptide fragments of coactivator and corepressor proteins. Biochemistry. 2012;51(1):19–31. doi: 10.1021/bi2011674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Lin W, Wang YM, Chai SC, et al. SPA70 is a potent antagonist of human pregnane X receptor. Nat Commun. 2017;8(1):741. doi: 10.1038/s41467-017-00780-5. This article presents the first HDX-MS studies of PXR, including the characterization of the PXR antagonist SPA70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savkur RS, Bramlett KS, Clawson D, et al. Pharmacology of nuclear receptor-coregulator recognition. Vitam Horm. 2004;68:145–83. doi: 10.1016/S0083-6729(04)68005-8. [DOI] [PubMed] [Google Scholar]
- 49.Chai SC, Cherian MT, Wang YM, et al. Small-molecule modulators of PXR and CAR. Biochim Biophys Acta. 2016;1859(9):1141–1154. doi: 10.1016/j.bbagrm.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chrencik JE, Orans J, Moore LB, et al. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19(5):1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 51.Orans J, Teotico DG, Redinbo MR. The nuclear xenobiotic receptor pregnane X receptor: recent insights and new challenges. Mol Endocrinol. 2005;19(12):2891–2900. doi: 10.1210/me.2005-0156. [DOI] [PubMed] [Google Scholar]
- 52.Teotico DG, Bischof JJ, Peng L, et al. Structural basis of human pregnane X receptor activation by the hops constituent colupulone. Mol Pharmacol. 2008;74(6):1512–1520. doi: 10.1124/mol.108.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng Y, Redinbo MR. Activation of the human nuclear xenobiotic receptor PXR by the reverse transcriptase-targeted anti-HIV drug PNU-142721. Protein Sci. 2011;20(10):1713–1719. doi: 10.1002/pro.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delfosse V, Dendele B, Huet T, et al. Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat Commun. 2015;6:8089. doi: 10.1038/ncomms9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Prosise WW, Chen J, et al. Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability. Protein Eng Des Sel. 2008;21(7):425–433. doi: 10.1093/protein/gzn017. [DOI] [PubMed] [Google Scholar]
- 56.Hennessy EJ, Oza V, Adam A, et al. Identification and optimization of benzimidazole sulfonamides as orally bioavailable sphingosine 1-phosphate receptor 1 antagonists with in vivo activity. J Med Chem. 2015;58(17):7057–7075. doi: 10.1021/acs.jmedchem.5b01078. [DOI] [PubMed] [Google Scholar]
- 57.Xue Y, Chao E, Zuercher WJ, et al. Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg Med Chem. 2007;15(5):2156–2166. doi: 10.1016/j.bmc.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santella JB, 3rd, Gardner DS, Duncia JV, et al. Discovery of the CCR1 antagonist, BMS-817399, for the treatment of rheumatoid arthritis. J Med Chem. 2014;57(18):7550–7564. doi: 10.1021/jm5003167. [DOI] [PubMed] [Google Scholar]
- 59.Gong H, Weinstein DS, Lu Z, et al. Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (RORγ/RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity. Bioorg Med Chem Lett. 2018;28(2):85–93. doi: 10.1016/j.bmcl.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell T, Chao G, Sitkoff D, et al. Pharmacologic profile of the Adnectin BMS-962476, a small protein biologic alternative to PCSK9 antibodies for low-density lipoprotein lowering. J Pharmacol Exp Ther. 2014;350(2):412–424. doi: 10.1124/jpet.114.214221. [DOI] [PubMed] [Google Scholar]
- 61.Ramamurthy V, Krystek SR, Jr, Bush A, et al. Structures of adnectin/protein complexes reveal an expanded binding footprint. Structure. 2012;20(2):259–269. doi: 10.1016/j.str.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 62*.Yang L, Broderick D, Campbell Y, et al. Conformational modulation of the farnesoid X receptor by prenylflavonoids: Insights from hydrogen deuterium exchange mass spectrometry (HDX-MS), fluorescence titration and molecular docking studies. Biochim Biophys Acta. 2016;1864(12):1667–1677. doi: 10.1016/j.bbapap.2016.08.019. This study uses HDX-MS to examine graded agonism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frego L, Davidson W. Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2006;15(4):722–730. doi: 10.1110/ps.051781406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Hamuro Y, Coales SJ, Morrow JA, et al. Hydrogen/deuterium-exchange (H/D-Ex) of PPARγ LBD in the presence of various modulators. Protein Sci. 2006;15(8):1883–1892. doi: 10.1110/ps.062103006. This study uses HDX-MS to examine graded agonsim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan X, Perez E, Leid M, et al. Deuterium exchange and mass spectrometry reveal the interaction differences of two synthetic modulators of RXRα LBD. Protein Sci. 2007;16(11):2491–2501. doi: 10.1110/ps.073019707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Chalmers MJ, Stayrook KR, et al. Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure. 2010;18(10):1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cherian MT, Chai SC, Wright WC, et al. CINPA1 binds directly to constitutive androstane receptor and inhibits its activity. Biochem Pharmacol. 2018;152:211–223. doi: 10.1016/j.bcp.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Redinbo MR, Venkatesh M, et al. Novel yeast-based strategy unveils antagonist binding regions on the nuclear xenobiotic receptor PXR. J Biol Chem. 2013;288(19):13655–13668. doi: 10.1074/jbc.M113.455485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cherian MT, Chai SC, Chen T. Small-molecule modulators of the constitutive androstane receptor. Expert Opin Drug Metab Toxicol. 2015 Jul;11(7):1099–1114. doi: 10.1517/17425255.2015.1043887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cherian MT, Lin W, Wu J, et al. CINPA1 is an inhibitor of constitutive androstane receptor that does not activate pregnane X receptor. Mol Pharmacol. 2015;87(5):878–889. doi: 10.1124/mol.115.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Vera IMS, Zheng J, Novick S, et al. Synergistic regulation of coregulator/nuclear receptor interaction by ligand and DNA. Structure. 2017;25(10):1506–1518e4. doi: 10.1016/j.str.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]