Abstract
Newcastle disease virus (NDV) belongs to genus Avulavirus and family Paramyxoviridae. There are thirteen serotypes named APMV-I (Avian Paramyxovirus-I) to APMV-13 of which NDV has been designated as APMV-1. The disease has been reported worldwide affecting both domestic and wild avian species. Morbidity and mortality rates up to 100% have been reported in cases of unvaccinated flocks. Stringent vaccination schedule is practiced in endemic/disease prone areas in order to prevent the disease. Despite this, NDV outbreaks have been reported even in cases of vaccinated populations. In this study we describe detailed pathological and molecular investigation that were undertaken in an organized poultry farm from Bareilly region, Uttar Pradesh, India, involving layer flocks which succumbed to ND outbreak in spite of following strict vaccination protocol. The mortality rate ranged from 76.80 to 84.41% in different flocks with an average mortality of 79.50%. Necropsied birds had gross lesions suggestive of viscerotropic ND including petechial hemorrhages on the proventricular tips, intestinal lumen with necrotic areas covered with hemorrhages, hemorrhagic cecal tonsils, para-tracheal edema and mottling of spleen. The characteristic histopathological lesions were mainly seen in the blood vessels and lymphoid tissues. Vascular changes characterized by congestion, edema, and hemorrhage were found in majority of the organs. Lymphocytolysis in spleen and cecal tonsils was evident. Immunohistochemical studies revealed positive signals mostly in macrophage and lymphocytes. PCR assay was done to confirm the NDV genome, which revealed an amplicon size of 356 bp. The phylogenetic analysis revealed the resemblance of the present isolate (ADS01) with class II genotype NDV XIIIA. The isolate belonged to velogenic NDV as the Minimum Lethal Dose (MLD) and Mean Death Time (MDT) for the present isolate were 10−8 and 41 h, respectively. Thus this study clearly demonstrates that in spite of strict vaccination regime and biosecurity procedures, ND continues to be rampant. Hence it is important to effectively administer the present vaccine in addition to strains matching to the field isolates to provide longer and optimal protection against spreading of virus by means of reducing the extent of viral shedding.
Keywords: Genotype, Molecular characterization, Pathology, Poultry, Vaccine mismatch, Velogenic viscerotropic Newcastle disease virus
Introduction
There are many infectious diseases causing severe mortality among the birds and in turn causing severe economic losses to the farmers and poultry industry. Owing to its highly infectious nature, Newcastle Disease (ND) is one of the OIE (Office International des epizootics) notifiable diseases amongst avian infectious diseases. Newcastle disease virus (NDV) belongs to the genus Avulavirus in the family Paramyxoviridae. There are thirteen serotypes under Paramyxoviridae named APMV-I (Avian Paramyxovirus-I) to APMV-13 and Newcastle disease virus (NDV) has been designated as APMV-1. The disease has been reported worldwide and it is currently controlled in some western European countries, the United States and Canada but continues to be a threat in South America, Asia and parts of Africa [1]. Both domestic and wild species are affected but with varying degrees of mortality and morbidity depending upon the host species and also the strain of virus [2]. The duration of incubation period is very short, ranging from 2 to 15 days with an average of 5–6 days. Morbidity and mortality rates up to 100% have been reported in cases of unvaccinated flocks [3]. Direct contact with diseased or carrier birds is the main mode of transmission followed by virus shedding by the infected birds in their feces, thus contaminating the environment. The disease is very contagious in nature as it takes only 2–6 days to affect the entire birds in the susceptible flock.
Virus infectivity is mainly dependent on the cleavage of Fusion (F) protein of the virus, which in virulent strains has been reported to be polybasic in nature, and thus this can be easily cleaved by furin-like proteases appearing in all organs in the host enabling easier and faster viral spread [2].
Phylogenetic analysis of the NDV isolates circulating in India revealed the presence of various genotypes including II, III, IV, VI, XIII, XIIIa and XIIIb. To delineate the genotype involved in the current ND outbreak, the partial genome of ADS01 isolate was sequenced and phylogenetic analysis was done on the basis of partial Fusion gene sequencing. The isolate (ADS01) clustered with other genotype XIIIA isolates, within Class II. ADS01 isolate was categorized as a velogenic NDV strain assessed biologically by the MLD, MDT and polybasic amino acid residues at the F protein cleavage site.
Stringent vaccination schedule are being practiced in endemic/disease prone areas in order to prevent the disease. NDV vaccine strains belonging to genotype I and II are generally being used in vaccines to control the clinical disease [2]. Despite of the stringent vaccination protocol, NDV outbreaks have been reported even in cases of vaccinated populations [4]. In this study, we describe detailed pathological and molecular investigation that were undertaken in an organized poultry farm involving layers of three different age groups belonging to white leghorn breed, which succumbed to ND outbreak in spite of following strict vaccination protocol.
Materials and methods
Farm history, necropsy examination and collection of samples
A disease outbreak among vaccinated layer flocks belonging to White leghorn strain was investigated in an organized poultry farm in Bareilly region, Uttar Pradesh India. The birds belonged to three different age groups of 10–18 weeks (n = 1200), 19–35 weeks (n = 600) and 36–72 weeks (n = 3000). Clinical signs, vaccination history and mortality were recorded. The birds were vaccinated against major viral diseases (Table 1). Necropsy examination of the dead birds (n ≃ 500) was carried out randomly and the gross findings were recorded. Representative tissue samples (n = 25) from all the flocks were collected and stored in 10% formalin for histopathological studies and pooled tissue samples (spleen, trachea and caecal tonsils: n = 10) in − 80 °C for molecular studies.
Table 1.
Vaccination regime followed in the farm
| S. No | Day | Vaccine | Route |
|---|---|---|---|
| 1. | 2 | MD | S/C |
| 2. | 6 | IB + LaSota | Occular |
| 4. | 15 | IBD intermediate | Occular |
| 5. | 24 | IBD intermediate | Occular |
| 6. | 30 | Fowl pox | Wingweb |
| 8. | 8th week | IB + LaSota | Occular |
| 10. | 11th week | R2B | I/M |
| 12. | 16th week | IB + LoSota | Occular |
| ND killed | S/C | ||
| 14. | Once in 4–6 weeks | ND LaSota | Occular |
Histopathology
The tissue samples were fixed in 10% neutral buffered formalin and processed by routine paraffin-embedding technique. Briefly, 4–5 µm thick sections were taken followed by Haematoxylin and Eosin (H&E) staining for detailed microscopic studies [5].
Immunohistochemistry
Immunohistochemical staining was performed on the tissues selected from tissue blocks based on the screening of H&E stained sections. Immunohistochemical staining was conducted using an avidin–biotin complex immunoperoxidase method following antigen retrieval by microwave based method [6]. Anti-NDV polyclonal antibody (ABCAM, USA) raised in poultry was used as the primary antibody. Briefly, duplicate sections of each tissue sections were stained with NDV primary antibody at a dilution of 1:100. After washing the sections thrice with PBS-T (Phosphate Buffered Saline-Tween 20), these were treated with biotylated anti-chicken antibodies raised in rabbit (1:100, Sigma Aldrich, USA) for 1 h, followed by three washings with PBS-T. Finally, the sections were subjected to 1:100 dilution of HRP-conjugated avidin (Sigma Aldrich, USA) for 1 h. Post-washing with PBS-T, the enzyme reactions were developed in AEC staining kit (Sigma Aldrich, USA) to produce a dark red precipitate and the sections were counterstained with Mayer’s hematoxylin. Tissue sections were processed for negative control as above by omitting primary antibody in the procedure.
Genome detection studies (RNA isolation, cDNA synthesis, RT-PCR and nested PCR)
The tissue samples stored in − 80 °C were subjected to total RNA extraction with TRIzol reagent (Invitrogen, USA) as per the manufacturer’s instructions. The extracted RNA was used to synthesize cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) following manufacturer’s instructions. RT-PCR was performed with the primers targeting the fusion gene of NDV (Forward primer: GCAGCTGCAGGGATTGTGGT; reverse primer TCTTTGAGCAGGAGGATGTTG) [7]. The RT-PCR was performed in a 25 μl reaction mixture containing 3.0 μl cDNA (5 ng/μl), 12.5 μl DreamTaq PCR Master mix 2× (Thermo Scientific, USA), 1.0 μl forward primer (10 p mol/μl), 1.0 μl reverse primer (10 p mol/μl) and 7.5 μl nuclease free water. The cyclic conditions for primary amplification were initial denaturation at 95 °C for 15 min, followed by 35 cycles of 94 °C for 45 s, 52 °C for 45 s, 72 °C for 45 s and final extension at 72 °C for 10 min.
Nucleotide sequencing and phylogenetic analysis
Partial fusion protein gene of 535 bp size was amplified [8]. The PCR products were purified using GeneJET PCR purification kit (Thermo Fisher Scientific, USA) and then sequenced in both directions using commercial sequencing services (Eurofins, Bangalore). Forward and reverse nucleotide sequences were curated and aligned using MEGA 7.0 software followed by BLAST analysis (http://blast.ncbi.nlm.nih.gov/) to find the most related sequences. The evolutionary history of the isolate ADS01 isolate of NDV was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [9]. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed [10]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+ G, parameter = 1.7867)). The rate variation model allowed for some sites to be evolutionarily invariable ([+ I], 22.4665% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 59 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 523 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [11]. The sequence analysis of the predicted amino acids of the amplicon of the present ADS01 isolate was carried out to determine the presence of virulent amino acid motif in the F protein cleavage site [12]. Lentogenic strains of NDV have been shown to depict the FPCS cleavage site motif 112G/E-K/R-Q-G/E-R-L117 with few basic amino acid residues at carboxyl terminus of the F2 protein while the virulent strains (mesogenic and velogenic) possess 112R/K-R-Q-R/K-R-F117 motif with two pairs of basic amino acids and a Phenylalanine in the N terminus of the F1 (residue 117). In ADS01 strain the sequence of the FPCS motif is 112R-R-Q-K-R-F117.
GenBank accession numbers
The sequenced data was submitted to GenBank and the accession number obtained was MG013984.
Virus isolation and identification
Virus propagation was carried out in 9 days old embryonated eggs procured from Hatchery Unit of ICAR-CARI, Izatnagar. The pooled samples from the trachea, brain and spleen stored in − 80 °C were triturated, filtered using 45 μm syringe filter and the filtrates were used to inoculate the embryonated eggs for virus isolation via allantoic route following standard protocol [1]. The presence of NDV in the allantoic fluid (AF) was identified by means of Haemagglutination (HA) and Haemagglutination inhibition (HI) test. The tests were performed in V-bottom/shaped microtiter plates as described by earlier workers [1, 13]. RT-PCR was carried following cDNA synthesis from the AF to confirm the presence of NDV [14].
Calculation of minimum lethal dose (MLD) and mean death time (MDT)
MLD was performed in 9 days old embryonated eggs. Serial tenfold dilution of the harvested allantoic fluid was prepared and 0.1 ml of the dilutions (10−6, 10−7, 10−8 and 10−9) was inoculated into the allantoic cavity (5 eggs/dilution). The MLD corresponds to the highest virus dilution that causes all the embryos inoculated to die. Thereafter, mean death time (MDT) was calculated based on the mean time in hours for the minimum lethal dose to kill all the inoculated embryos [13].
Results
Clinical signs and mortality
Clinical signs appeared suddenly with marked depression, anorexia, sharp drop in egg production, respiratory rales, greenish diarrhea, edematous swellings of the head, cyanosis of the combs, and conjunctivitis with ocular discharge. Prostration was observed in many birds followed by death within a span of 10 days. The mortality rate recorded in different age groups were 84.41, 83.00 and 76.80%, respectively, with an average mortality of 79.50% (Table 2).
Table 2.
Flock details of the investigated farm
| Flocks | Age group (weeks) | Number of birds | Number of birds dead | Mortality rate (%) |
|---|---|---|---|---|
| Layers shed I | 10–18 | 1200 | 1013 | 84.41 |
| Layers shed II | 19–35 | 600 | 498 | 83.00 |
| Layers shed III | 36–72 | 3000 | 2305 | 76.80 |
Gross findings
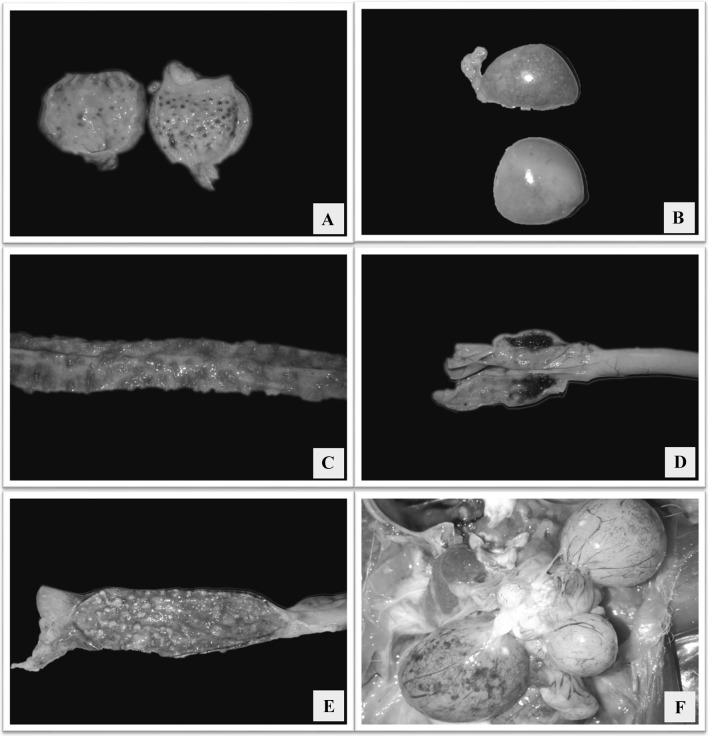
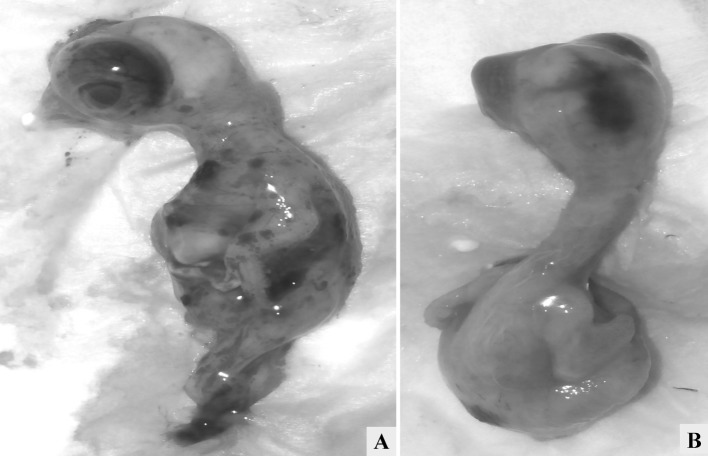
Necropsied birds had facial swelling with mucoid to catarrhal exudate from the nares. Hemorrhages in the lower eyelid conjunctiva were evident in majority of the cases. Gross lesions were prominent in the mucosa of the proventriculus, small and large intestine. Proventriculus showed range of lesions from hemorrhagic line at the junction between esophagus and proventriculus, petechial hemorrhages on the proventricular tips (Fig. 1a), ecchymotic to suffusions covering the mucosa. Mottling of spleen was evident characterized by numerous multifocal pale areas on the surface (Fig. 1b). Grossly, the intestines revealed discolored areas of necrosis from the serosal surface. On opening the intestines, the lumen contained numerous necrotic areas covered with hemorrhages and mucoid content (Fig. 1c, e), and on washing the necrotic areas they revealed raised button type of ulcers which were yellowish in color. These lesions were seen either diffusely throughout the intestine or seen spread in multifocal areas. Cecal tonsils were swollen, prominent and hemorrhagic in all the cases examined (Fig. 1d). In few cases there were pale necrotic foci in the pancreas. Respiratory tract lesions were predominantly of mucosal hemorrhage and marked congestion of the trachea and para-tracheal edema. Airsacculitis was present in majority of the cases, and thickening of the air sacs with catarrhal or caseous exudates was observed. Layer chickens had free egg yolk in the abdominal cavity. The ovarian follicles were often flaccid, highly congested, with petechiae on their surfaces and appeared degenerative (Fig. 1f). The bursa of Fabricius was edematous.
Fig. 1.
a Proventriculus showing hemorrhages in the glandular tips; b Spleen with numerous multifocal pale areas giving a mottling appearance; c Mucosa of duodenum showing multifocal to coalescing raised hemorrhagic to necrotizing ulcerated areas; d Caecal tonsils with hemorrhages; e Mucosa of rectum showing multifocal to coalescing raised necrotic ulcerated areas; f Congested ovarian follicles with petechial to ecchymotic hemorrhages
Histopathological findings
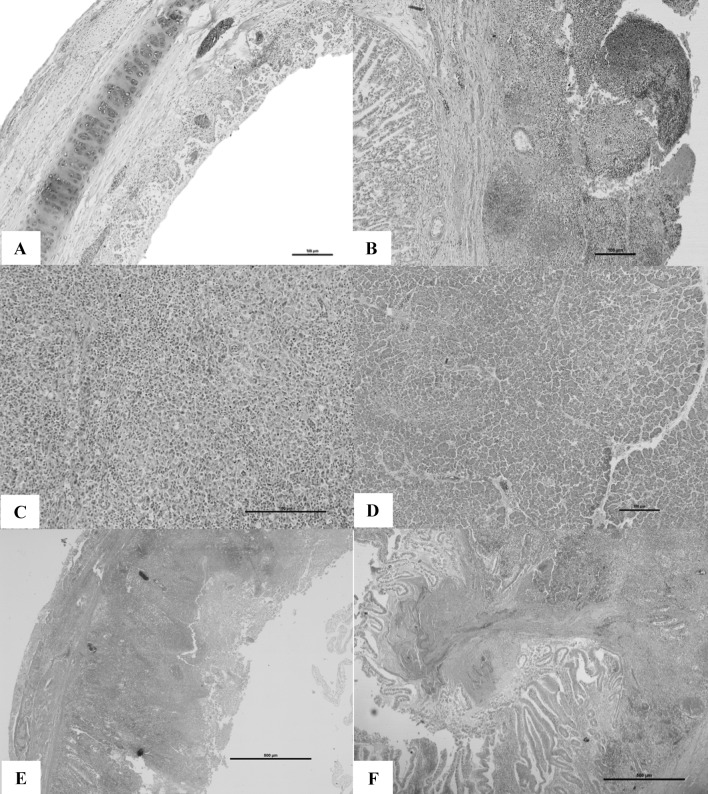
The characteristic histopathological lesions were mainly seen in the blood vessels and lymphoid tissues. Vascular changes characterized by congestion, edema, and hemorrhages were found in majority of the organs. The tunica media of the arterioles were hyalinized, endothelial cells of the blood vessels were swollen and necrosed. Few blood vessels contained thrombosis in the lumen. Depletion of lymphoid tissue was the prominent finding in majority of the lymphoid organs including spleen, bursa, intestinal lymphoid aggregates like Peyer’s patches and cecal tonsils. In upper respiratory tract, the lesions were seen throughout the length of the trachea. Cilia were lost. In the mucosa of the upper respiratory tract, congestion, edema, and dense cellular infiltration of lymphocytes and macrophages were seen (Fig. 2a). Marked lesions of the lung were seen with severe congestion and edema of the parabronchi. Air sacs revealed mild to moderate edema with mixed cell type infiltration. Sections from proventriculus revealed necrotic and denuded surface epithelium with severe hemorrhages (Fig. 2b). Pancreas revealed multifocal areas of necrosis. Multi focal necrotic areas were found throughout the spleen (Fig. 2c). Focal vacuolation and destruction of lymphocytes was seen in the cortical areas and germinal centers of the spleen. Inclusion bodies were observed in the spleen. Marked degeneration of lymphocytes in the medullary region was seen in the bursa of Fabricius. Spleen also contained severe reticular cell hyperplasia. Pancreas exhibited multi-focal areas of necrosis and interstitial edema (Fig. 2d). Intestinal mucosal epithelium was denuded completely and the structure of the villi was lost. The mucosa was necrotic with severe inflammatory reaction with mixed cell populations in the lamina propria (Fig. 2e). Bacterial colonies were found adhered to the necrotic debri attached to the ulcerated area. In case of cecal tonsils, there was severe lymphocytolysis, loss of lymphocytes severe necrosis and hemorrhages. Necrotic debri was observed in the lumen of the caecum (Fig. 2f).
Fig. 2.
a Trachea with necrotic and denuded surface epithelium with edematous submucosa and mononuclear cell infiltration. H&E 10×; b Proventriculus showing necrotic and denuded surface epithelium with severe hemorrhages H&E 10×; c Spleen showing focal to coalescing lymphoid depletion with associated reticular cell hyperplasia H&E 20×; d Pancreas with multi-focal areas of necrotic foci and interstitial edema. H&E 10×; e Duodenum with fusion of villi and the surface epithelium showing necrotic changes and sub-mucosa reveal severe infiltration with mononuclear cells and hemorrhage. H&E 4×; f Caecal tonsil showing severe lymphocytolysis, necrotic epithelium and eosinophilic exudates in the lumen admixed with fibrin and inflammatory cells. H&E 4×
Immunohistochemistry
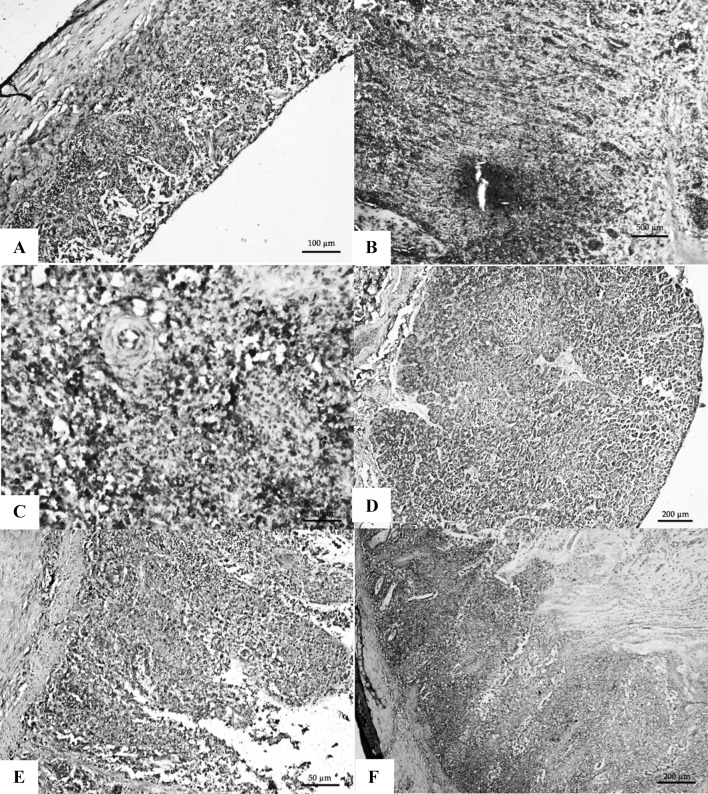
Tracheal epithelium showed strong positive signals, submucosal macrophages showed positive signals and the lymphocytes showed the same results (Fig. 3a). Epithelial cells and cells in lamina propria of proventriculus papillae also showed positive signals (Fig. 3b). In spleen, positive signals were present diffusely and mostly in macrophages and lymphocytes surrounding the central arteriole (Fig. 3c). Pancreas showed strong positive signals from the necrotic areas (Fig. 3d). Intestinal lymphoid aggregates and macrophages in sub mucosal layer and in lamina propria showed intense positive signals (Fig. 3e, f). The denuded and necrotic contents in the lumen revealed strong positive signals.
Fig. 3.
a Trachea with strong positive signals from the necrotic mucosa, sub-mucosa and inflammatory cells. IHC 20×; b Proventriculus showing strong positive signals from the necrotic mucosa and inflammatory cells IHC 40×; c Spleen showing strong positive signals in the RE cells and macrophages from the areas surrounding the central arteriole in the PALS area IHC 40×; d Pancreas with strong positive signals from the necrotic areas. IHC 10×; e Duodenum showing strong positive signals from the inflammatory cells in the lamina propria and sub-mucosa IHC 40×; f Caecal tonsil with strong positive signals from lymphocytes and other inflammatory cells in the sub-mucosa. H&E 20×
Genome detection studies
An amplicon of size 356 bp on 1.5% agarose gel was visualized, indicating the presence of NDV genome in the samples (Fig. 4).
Fig. 4.
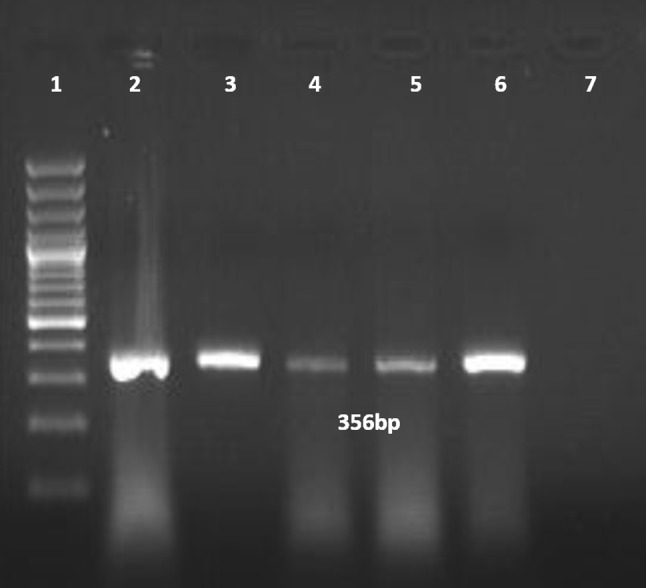
Agarose gel electrophoretic picture showing 356 bp PCR amplified product of fusion protein gene (Lane 1–DNA ladder; lane 2–positive control; lanes 3 to 6–field test samples; 7–non template control
Sequencing and phylogenetic analysis
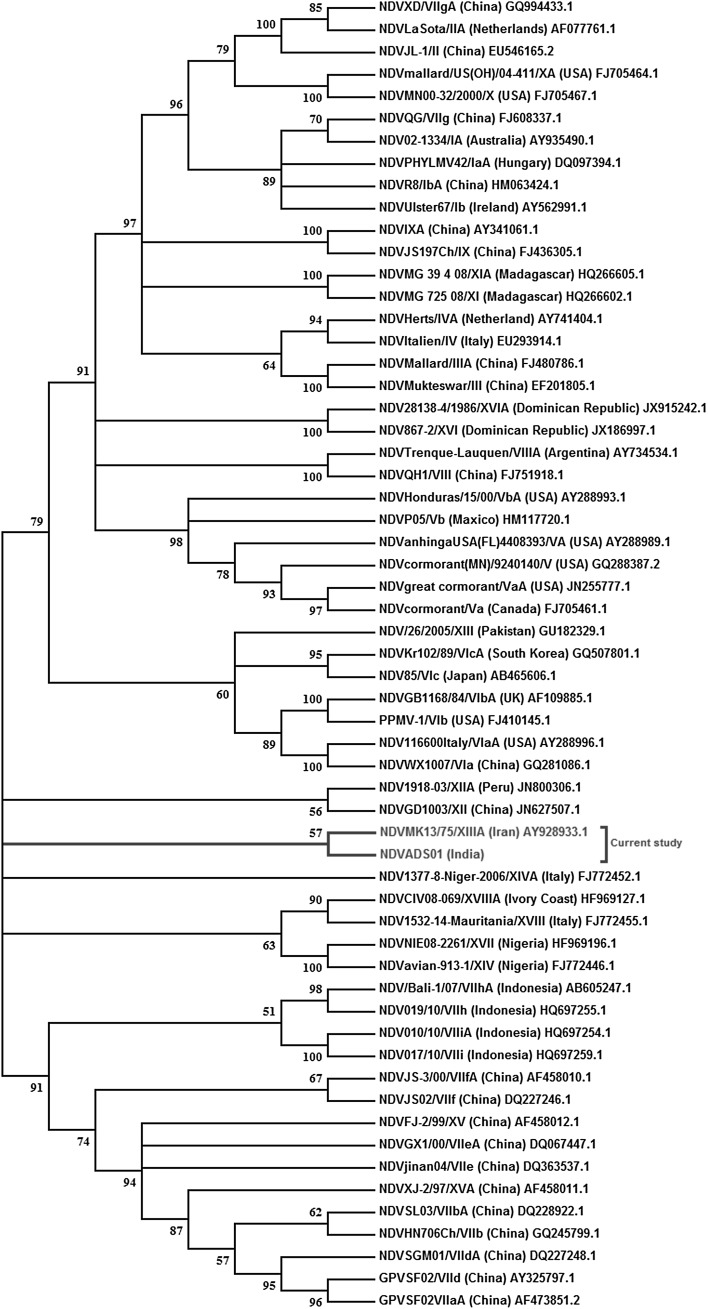
Partial fusion protein gene of size 535 bp was amplified and sequencing of the PCR amplicon was done followed by phylogenetic tree construction. The phylogenetic analysis revealed the resemblance of ADS01 isolate with genotype NDV XIIIA (Fig. 5). The ‘F’ protein cleavage site amino acid motif for ADS01 isolate was 111G-R-R-Q-K-R-F117 which is a virulent amino acid motif and thus the isolate was classified as velogenic.
Fig. 5.
The phylogenetic analysis of the sequenced partial Fusion protein gene showing the resemblance of ADS01 isolate with genotype NDV XIIIA
Isolation and identification
The chicken embryos that died before 36 h post-inoculation with tissue triturate were discarded and the allantioc fluid (AF) was found negative for HA, HI and RT-PCR. Most of the embryos died between 36 to 48 h post inoculation and their AF were found positive for HA, HI and RT-PCR testing. Grossly, the virus infected embryos were edematous, had typical hemorrhage in the occipital area of the embryos along with hemorrhages throughout the body surface (Fig. 6a, b). The pooled AF showed HA titre of 1:512 and HI titre of 210 RT-PCR revealed an amplicon size of 350 bp indicating the presence of NDV in the harvested AF.
Fig. 6.
a Embryos from embryonated eggs infected with tissue triturate showing petechial to ecchymotic hemorrhages throughout the body surface; b Typical hemorrhage in the occipital area of the infected embryo
Calculation of minimum lethal dose (MLD) and mean death time (MDT)
The MLD and MDT for the present isolate (ADS01) were found to be 10−8 and 41 h respectively.
Discussion
Newcastle disease exhibits a range of pathogenicity in chickens depending on its virulence that varies from peracute disease with nearly 100% mortality to subclinical disease with no outward lesions. The MDT is a measure of viral pathogenicity where it determines the virulence of an isolate by the duration between inoculation of virus and time of death in embryonated chicken eggs. Velogenic strains cause the chicken embryos to die in less than 60 h, lentogenic strain fails to kill the embryos even after 90 h, and the strains that cause embryo mortality between 60 and 90 h are mesogenic [3]. In the present study, the MDT for the virus isolate (ADS01) was found to be 41 h, thus the isolate is pathogenic and falls in velogenic category.
Among velogenic category there are two forms based on visceral or nervous signs viz., viscerotropic (VVNDV) and neurotropic (VNNDV). Grossly, viscerotropic form is characterized by the presence of necrosis, ulceration of the lymphoid tissues in the intestines, splenomegaly, necrosis in the spleen, hemorrhages and necrosis of caecal tonsils [15–18]. Microscopically, in the viscerotropic form, the lymphoid tissues seen scattered throughout the body from intestines to lymphoid organs reveal severe necrosis [17, 18]. Immunohistochemistry has been used to study the distribution of virus in the tissues of NDV infected birds. By immunohistochemistry, VVNDV could be detected throughout the body in all the primary and secondary lymphoid tissues including thymus, bursa, spleen, and cecal tonsils. In the spleen, macrophages surrounding the penicilliary arteries showed intense positive signals, while in other lymphoid organs the positive signals emanated in the middle of the lymphoid follicles containing macrophages and lymphocytes. In the cecal tonsil, NDV signals were noticed from macrophages within the lamina propria. Thus, the gross, microscopical and immunohistochemical findings recorded in the present study were in corroboration with findings noticed in viscerotropic NDV category.
Phylogenetic analysis of NDV isolates reported during 1989–2013 has revealed the presence of various genotypes circulating in India including Genotype XIII genotype XIIIa, genotype VI, genotype III and genotype II in cranes and chicken [12]. Apart from various genotypes reported, pathotypic and sequence characterization of NDV from vaccinated chickens revealed circulation of genotype II, IV and XIII in the country [19]. Genotype XIII (XIIIb) was isolated in commercial vaccinated layer farms [20] and from Emu [21]. In the present study, partial sequencing of fusion protein gene followed by phylogenetic analysis revealed the present isolate (ADS01) that was of class II genotype NDV XIII and the finding was subsequently supported by deduced amino acid sequence analysis of F protein cleavage site which revealed a virulent amino acid motif.
The mortality in the investigated layer flock was 79.50%. Mortality rate varies among the affected flocks depending upon the strain of the virus, vaccination status, genetic resistance and species of the birds [2]. ND Outbreaks in fully susceptible chickens are generally devastating with mortality up to 100% [22]. But in the present study, there was a substantial mortality among the vaccinated birds. Severe mortality due to ND among vaccinated birds have also been reported earlier [23, 24]. The one reason may be attributed to the virus shedding of the healthy birds post-vaccination. It has been reported that the vaccination against ND helps in protecting the poultry against mortality and clinical outcome, and it’s quite imperative that these vaccines could not stop shedding of viral particles which may act as a prime source of infection in well vaccinated flocks [4, 25, 26]. It is well established that the amount of viral shedding depends on host’s immunity, affected host species, the dose and infectivity of the challenge virus, the type and dose of ND vaccine used and the duration between vaccination and challenge [27, 28]. Usage of vaccines that match the field genotype significantly have been reported to reduce viral shedding compared to mismatched vaccines [29, 30]. The other reason for disease outbreak in vaccinated flock may be due to inadequate protection conferred by the present vaccine strain to the vaccinated flocks, and few researchers also described that it is need of the hour for newer vaccination strategies against ND [4, 31, 32]. Hu et al. [29] and Miller et al. [30] claimed that matched vaccines generated from field isolates provide enhanced protection to the chickens and reduced the viral shedding in turn reducing the transmission to the uninfected flock. LaSota vaccinated commercial broilers showed varying protection levels against two strains viz., AF2240-I (genotype VIII) and IBS002 (genotype VII) during challenge studies. But, the inactivated genotype VII NDV recombinant vaccine conferred full protection against both NDV isolates along with reduction in virus shedding [33], proving the effectiveness of matched genotype vaccines in conferring full protection.
On contrary to the above findings, Bwala et al. [34] and Liu et al. [35] showed that genotype II vaccine (LaSota) conferred 100% cross-protection against heterologous viruses of genotypes V, VIb, VIg, VIId and IX, thus excluding the role of genotype-matched vaccines in ND outbreak. Dortmans et al. [36] proved that administration of LaSota strain protected chickens when challenged with a velogenic virus (NL/93, genotype VII) completely without any clinical disease and absence of viral shedding. The adapted vaccine from NL/93 strain yielded similar results compared to the LaSota based vaccine. The authors concluded that the currently practiced routes for vaccine application like aerosol (spraying) or oral administration via drinking water do not guarantee that every bird in the flock receive an optimum vaccine dose, which is the reason behind vaccination failure. Susta et al. [37] reported that LaSota vaccine strain successfully protected chickens against the genotype VII and XVII NDV strain with non-significant virus shedding. Efficacy of vaccines was not affected even after repeated challenge with virulent NDV [38].
Thus it is very clear that apart from vaccination, other electrifying factors that predispose for vaccine failure include live vaccine virus being neutralized by the maternal antibodies, vaccines being inappropriately handled and administered thus causing inactivation of the live vaccine virus, and infection with other agents like infectious bursal disease virus, inclusion body hepatitis, Marek’s disease and mycotoxins [14, 39].
As reported, vaccine failure may be because of inadequate application procedures of vaccines and vaccine mismatch. Thus, as India poultry population is endemic to ND, it will be very appropriate to incorporate local virus strain along with including genotype II vaccine in the vaccination regime and by following appropriate application procedures, which would enhance post-vaccinal immunity, antibody specificity, prevention of clinical disease and mortality, and reduce viral shedding from vaccinated birds. By following such matched vaccine practices and adopting strict bio-security measures, several deadly infectious poultry diseases can be contained in a better way thus preventing morbidity and mortality in the poultry flocks and in turn enhancing the productivity of the birds.
Acknowledgements
We thank the Director, ICAR-Indian Veterinary Research Institute for providing necessary facilities and support.
References
- 1.OIE. Manual of diagnostic tests and vaccines for terrestial animal. 2012. http://www.oie.int/eng/normes/manual
- 2.Bulbule NR, Madale DS, Meshram CD, Pardeshi RB, Chawak MM. Virulence of newcastle disease virus and diagnostic challenges. Adv Anim Vet Sci. 2015;3:14–21. doi: 10.14737/journal.aavs/2015/3.5s.14.21. [DOI] [Google Scholar]
- 3.Cattoli G, Susta L, Terregino C, Brown C. Newcastle disease: a review of field recognition and current methods of laboratory detection. J Vet Diagn Investig. 2011;23:637–656. doi: 10.1177/1040638711407887. [DOI] [PubMed] [Google Scholar]
- 4.van Boven M, Bouma A, Fabri TH, Katsma E, Hartog L, Koch G. Herd immunity to Newcastle disease virus in poultry by vaccination. Avian Pathol. 2008;37:1–5. doi: 10.1080/03079450701772391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luna LG. Manual of histologic staining methods of the armed forces institute of pathology. New York: Mc Graw Hill; 1968. [Google Scholar]
- 6.Swanson P. Microwave antigen retrieval in citrate buffer. Lab. Med. 1994;25:520–522. doi: 10.1093/labmed/25.8.520. [DOI] [Google Scholar]
- 7.Jindal N, Chander Y, Primus A, Redig P, Goyal S. Isolation and molecular characterization of Newcastle disease viruses from raptors. Avian Pathol. 2010;39:441–445. doi: 10.1080/03079457.2010.517249. [DOI] [PubMed] [Google Scholar]
- 8.Jaganathan S, Ooi PT, Phang LY, Allaudin ZNB, Yip LS, Choo PY, Lim BK, Lemiere S, Audonnet JC. Observation of risk factors, clinical manifestations and genetic characterization of recent Newcastle Disease Virus outbreak in West Malaysia. BMC Vet Res. 2015;11:219. doi: 10.1186/s12917-015-0537-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desingu P, Singh S, Dhama K, Karthik K, Vinodh Kumar O, Malik Y. Phylogenetic analysis of Newcastle disease virus isolates occurring in India during 1989–2013. Virus Dis. 2016;27:203–206. doi: 10.1007/s13337-016-0320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilal ES, Elnasri IM, Alhassan AM, Khalifa KA, Elhag JI, Ahmed SO. Biological pathotyping of Newcastle disease viruses in Sudan 2008–2013. J Vet Med. 2014;2014:209357. doi: 10.1155/2014/209357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdel-Glil MY, Mor SK, Sharafeldin TA, Porter RE, Goyal SM. Detection and characterization of Newcastle disease virus in formalin-fixed, paraffin-embedded tissues from commercial broilers in Egypt. Avian Dis. 2014;58(1):118–123. doi: 10.1637/10616-071813-Reg.1. [DOI] [PubMed] [Google Scholar]
- 15.Alexander DJ. Newcastle disease. Gordon memorial lecture. Br Poult Sci. 2001;42:5–22. doi: 10.1080/713655022. [DOI] [PubMed] [Google Scholar]
- 16.Alexander DJ, Senne DA, et al. Newcastle disease and other avian paramyxoviruses. In: Dufour-Zavala L, Senne DA, Glisson JR, et al., editors. A laboratory manual for the isolation, identification and characterization of avian pathogens. 5. Athens: American Association of Avian Pathologists; 2008. pp. 135–141. [Google Scholar]
- 17.Kommers GD, King DJ, Seal BS, Brown CC. Pathogenesis of chicken-passaged Newcastle disease viruses isolated from chickens and wild and exotic birds. Avian Dis. 2003;47:319–329. doi: 10.1637/0005-2086(2003)047[0319:POCNDV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 18.Susta L, Miller PJ, Afonso CL, Brown CC. Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet Pathol. 2011;48(2):349–360. doi: 10.1177/0300985810375806. [DOI] [PubMed] [Google Scholar]
- 19.Jakhesara S, Prasad V, Pal J, Jhala M, Prajapati K, Joshi C. Pathotypic and sequence characterization of newcastle disease viruses from vaccinated chickens reveals circulation of genotype II, IV and XIII and in India. Transbound Emerg Dis. 2014;63:523–539. doi: 10.1111/tbed.12294. [DOI] [PubMed] [Google Scholar]
- 20.Khorajiya J, Pandey S, Ghodasara P, Joshi B, Prajapati K, Ghodasara D, Mathakiya R. Patho-epidemiological study on Genotype-XIII Newcastle disease virus infection in commercial vaccinated layer farms. Vet World. 2015;8:372–381. doi: 10.14202/vetworld.2015.372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowthaman V, Singh S, Dhama K, Desingu P, Kumar A, Malik Y, Munir M. Isolation and characterization of genotype XIII Newcastle disease virus from Emu in India. Virus Dis. 2016;27:315–318. doi: 10.1007/s13337-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DJ, Elizabeth WA, Chad MF. The long view: a selective review of 40 years of Newcastle disease research. Avian Pathol. 2012;41:329–335. doi: 10.1080/03079457.2012.697991. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, Ohtsu N, Nakamura T, Yamamoto Y, Yamada M, Mase M, Imai K. Pathologic and immunohistochemical studies of Newcastle disease (ND) in broiler chickens vaccinated with ND: severe non-purulent encephalitis and necrotizing pancreatitis. Vet Pathol. 2008;45:928–933. doi: 10.1354/vp.45-6-928. [DOI] [PubMed] [Google Scholar]
- 24.Perozo F, Marcano R, Afonso CL. Biological and phylogenetic characterization of a genotype VII Newcastle disease virus from Venezuela: efficacy of field vaccination. J Clin Microbiol. 2012;50:1204–1208. doi: 10.1128/JCM.06506-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapczynski DR, King DJ. Protection of chickens against overt clinical disease and determination of viral shedding following vaccination with commercially available Newcastle disease virus vaccines upon challenge with highly virulent virus from the California 2002 exotic Newcastle disease outbreak. Vaccine. 2005;23:3424–3433. doi: 10.1016/j.vaccine.2005.01.140. [DOI] [PubMed] [Google Scholar]
- 26.Miller PJ, King DJ, Afonso CL, Suarez DL. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine. 2007;25:7238–7246. doi: 10.1016/j.vaccine.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Alexander DJ, Manvell RJ, Banks J, Collins MS, Parsons G, Cox B, Frost KM, Speidel EC, Ashman S, Aldous EW. Experimental assessment of the pathogenicity of the Newcastle disease viruses from outbreaks in Great Britian in 1997 for chickens and turkeys and the protection afforded by vaccination. Avian Pathol. 1999;28:501–512. doi: 10.1080/03079459994542. [DOI] [PubMed] [Google Scholar]
- 28.King DJ. Avian paramyxovirus type 1 from pigeons: isolate characterization and pathogenicity after chicken or embryo passage of selected isolates. Avian Dis. 1996;40:707–714. doi: 10.2307/1592284. [DOI] [PubMed] [Google Scholar]
- 29.Hu S, Ma H, Wu Y, Liu W, Wang X, Liu Y, Liu X. A vaccine candidate of attenuated genotype VII Newcastle disease virus generated by reverse genetics. Vaccine. 2009;27(6):904–910. doi: 10.1016/j.vaccine.2008.11.091. [DOI] [PubMed] [Google Scholar]
- 30.Miller PJ, Estevez C, Yu Q, Suarez DL, King DJ. Comparison of viral shedding following vaccination with inactivated and live Newcastle disease vaccines formulated with wild-type and recombinant viruses. Avian Dis. 2009;53:39–49. doi: 10.1637/8407-071208-Reg.1. [DOI] [PubMed] [Google Scholar]
- 31.Rauw F, Gardin Y, Palya V, Anbari S, Lemaire S, Boschmans M, van den Berg T, Lambrecht B. Improved vaccination against Newcastle disease by an in ovo recombinant HVT-ND combined with an adjuvanted live vaccine at day-old. Vaccine. 2010;28:823–833. doi: 10.1016/j.vaccine.2009.10.049. [DOI] [PubMed] [Google Scholar]
- 32.Palya V, Kiss I, Tatar-Kis T, Mato T, Felfoldi B, Gardin Y. Advancement in vaccination against Newcastle disease: recombinant HVT NDV provides high clinical protection and reduces challenge virus shedding with the absence of vaccine reactions. Avian Dis. 2012;56:282–287. doi: 10.1637/9935-091511-Reg.1. [DOI] [PubMed] [Google Scholar]
- 33.Roohani K, Tan S, Yeap S, Ideris A, Bejo M, Omar A. Characterisation of genotype VII Newcastle disease virus (NDV) isolated from NDV vaccinated chickens, and the efficacy of LaSota and recombinant genotype VII vaccines against challenge with velogenic NDV. J Vet Sci. 2015;16:447. doi: 10.4142/jvs.2015.16.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bwala DG, Abolnik C, van Wyk A, Cornelius E, Bisschop SP. Efficacy of a genotype 2 Newcastle disease vaccine (Avinew) against challenge with highly virulent genotypes 5 and 3d. J S Afr Vet Assoc. 2009;80:174–178. doi: 10.4102/jsava.v80i3.197. [DOI] [PubMed] [Google Scholar]
- 35.Liu XF, Wan HQ, Ni XX, Wu YT, Liu WB. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. 2003. Arch Virol. 2003;148:1387–1403. doi: 10.1007/s00705-003-0014-z. [DOI] [PubMed] [Google Scholar]
- 36.Dortmans J, Peeters B, Koch G. Newcastle disease virus outbreaks: vaccine mismatch or inadequate application? Vet Microbiol. 2012;160:17–22. doi: 10.1016/j.vetmic.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Susta L, Jones M, Cattoli G, Cardenas-Garcia S, Miller P, Brown C, Afonso C. Pathologic characterization of genotypes XIV and XVII Newcastle disease viruses and efficacy of classical vaccination on specific pathogen-free birds. Vet Pathol. 2014;52:120–131. doi: 10.1177/0300985814521247. [DOI] [PubMed] [Google Scholar]
- 38.Taylor TL, Miller PJ, Olivier TL, Montiel E, Cardenas Garcia S, Dimitrov KM, Williams-Coplin D, Afonso CL. Repeated challenge with virulent Newcastle disease virus does not decrease the efficacy of vaccines. Avian Dis. 2017;61(2):245–249. doi: 10.1637/11555-120816-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 39.Jeon WJ, Lee EK, Lee YJ, Jeong OM, Kim YJ, Kwon JH, Choi KS. Protective efficacy of commercial inactivated Newcastle disease virus vaccines in chickens against a recent Korean epizootic strain. J Vet Sci. 2008;9:295–300. doi: 10.4142/jvs.2008.9.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]