Abstract
Motility of cilia (also known as flagella in some eukaryotes) is based on axonemal doublet microtubule sliding that is driven by the dynein molecular motors. Dyneins are organized into intricately patterned inner and outer rows of arms, whose collective activity is to produce inter-microtubule movement. However, to generate a ciliary bend, not all dyneins can be active simultaneously. The switch point model accounts, in part, for how dynein motors are regulated during ciliary movement. On the basis of this model, supported by key direct experimental observations as well as more recent theoretical and structural studies, we are now poised to understand the mechanics of how ciliary dynein coordination controls axonemal bend formation and propagation.
MICROTUBULE SLIDING AND CILIARY/FLAGELLAR BENDING
Fifty years ago, Peter Satir provided the first experimental evidence that microtubules slide during ciliary bending (Satir, 1968) (Figure 1A). Satir tested the sliding microtubule model for ciliary bending (Gibbons, 1981) by rapid fixation of beating cilia in clam gills and capturing the cilia at known positions in the bending cycle. He then performed electron microscopy on serial sections from the tips of cilia, captured in either the effective or reverse bends, and determined that the microtubules are not contractile, but rather slide relative to one another (Figure 1A). Based on observations that ciliary bends are in the form of circular arcs (Brokaw, 1965), the data quantitatively fit a sliding model as illustrated in Figure 1A. Satir’s work was founded in part on early electron microscopy describing the 9 + 2 axoneme (Satir et al., 2014), and its substructures, including the “outer doublet arms” that were shown to be the dynein ATPases (Gibbons and Rowe, 1965). As discussed below, the dynein arms were found to power microtubule sliding and the oscillatory bending that characterizes ciliary and eukaryotic flagellar motility, and this led to the first computer simulations for ciliary and flagellar bending movement (Brokaw, 1972a, b).
FIGURE 1:
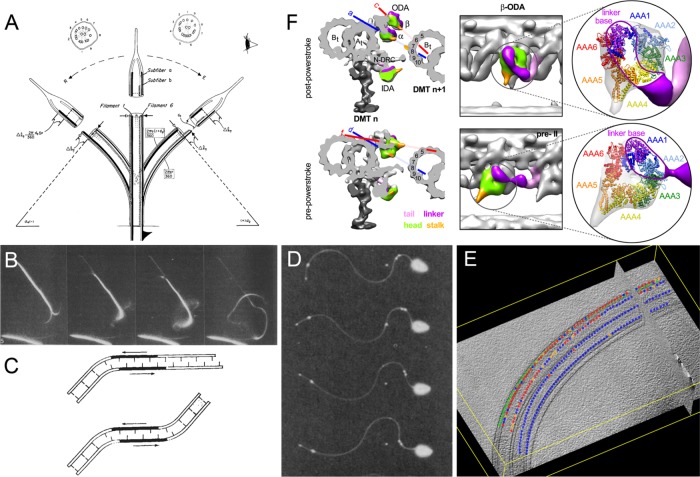
Microtubule sliding during axonemal bending. (A) The sliding microtubule (“sliding filament”) hypothesis (reproduced from Satir, 1968). (B) Dark-field microscopy of ATP-driven outer doublet microtubule sliding in trypsin-treated axonemes (reproduced from Summers and Gibbons, 1971). (C) Local application of ATP causing two equal but opposite bends in the axoneme (reproduced from Shingyoji et al., 1977). (D) Microtubule sliding during bending revealed by the oscillatory motion of colloidal gold particles attached to beating axonemes (reproduced from Brokaw, 1989). (E) Differential activation of dyneins on opposite sides of the axoneme (“switching”) during bending (reproduced from Lin et al., 2014). (F) Three-dimensional isosurface renderings of outer doublet cross and long views showing conformational changes in outer dynein arms between the postpower stoke and prepower stoke states (reproduced from Lin et al., 2014).
Active doublet microtubule sliding was unequivocally and directly visualized by Summers and Gibbons (1971) using dark-field microscopy (Figure 1B). Their results were based on the discovery that isolated axonemes could be reactivated to beat by addition of ATP and that the in vitro reactivated movement closely mimicked the motion of cilia and flagella in live cells (Gibbons and Gibbons, 1972). This important advance demonstrated that bend initiation and propagation resided in the 9+2 axoneme. Thus, although the ciliary membrane is highly specialized and allows the concentration of ATP and various ions to be maintained at levels required for motility, ciliary motility itself is a fundamental property of, and controlled by, the axoneme. In their groundbreaking experiment, Summers and Gibbons briefly digested isolated sea urchin sperm axonemes with trypsin before adding ATP. Rather than reactivating axonemal beating, the axonemes now slid apart by dynein-driven translocation of individual doublet microtubules moving past each other. In addition, electron microcopy of the results of this microtubule sliding revealed that dyneins generate force in a single direction and move toward the minus end of the axonemal microtubules (Sale and Satir, 1977). In vitro motility assays using isolated axonemal dyneins to move polarity marked microtubules confirmed that they are minus-end–directed motors (Vale and Toyoshima, 1988). As discussed further below, a major consequence of the uniform polarity of dynein force generation is that during a ciliary beat only some doublets or portions of the doublets can have active arms at any one time. Otherwise, with all dyneins active at once on each outer doublet, the axoneme would behave as a rigid rod. The hypothesis is called the switch point model for ciliary bending (Wais-Steider and Satir, 1979).
Further support for the sliding microtubule model of axonemal bending came from the ingenious experimentation of Chikako Shingyoji in the Takahashi laboratory at the University of Tokyo (Shingyoji et al., 1977). Shingyoji attached a demembranated straight sea urchin axoneme to a microneedle and added a small amount of ATP to the middle portion of the axoneme. Since the axoneme was straight and its ends in rigor (i.e., with dyneins tightly bound to the adjacent doublet), when dynein arms became activated they produced equal and opposite bends as the microtubules slid (the results are illustrated in Figure 1C). These observations supported the microtubule sliding model for axonemal bending and implied that mechanical strain produced by dynein arm activity at one position can transmit the strain along the axoneme to activate dynein arm activity at a more distant location, thereby generating a bend in the opposite direction. These experiments were later expanded on to reveal that microtubule curvature can regulate axonemal dynein-driven microtubule sliding (Morita and Shingyoji, 2004; Hayashi and Shingyoji, 2009) and supported a switching model for opposing ciliary bends. Brokaw also took advantage of ATP reactivation of sperm flagellar axonemes to analyze microtubule sliding during active bending (Brokaw, 1989, 1991). In this case, Brokaw attached colloidal gold particles to the surface of the doublet microtubules and, using dark-field microscopy, observed that the distance between gold particles attached to different doublet microtubules changed as the axoneme underwent rhythmic bends (Figure 1D). The results directly revealed microtubule sliding during bending.
CURRENT CHALLENGES AND OPPORTUNITIES
The major challenge is to understand regulation and coordination of dynein activity during ciliary bending. As illustrated above, many early studies on the biochemistry of dyneins and mechanism of axonemal bending made use of sea urchin sperm flagella. Through studies in model organisms such as Chlamydomonas, we now know that there are at least 15 different axonemal dyneins that are distinct in both composition and location within the axonemal 96-nm repeat structure; while some dyneins are found along the entire axoneme, others are present only in the proximal or distal regions or assemble only onto specific doublets (Kamiya and Yagi, 2014; King, 2018). From analyses of mutant cells missing subsets of dyneins, we also know that the outer dynein arms are responsible for controlling the axonemal beat frequency and that the inner dynein arms control the form of the beat (Brokaw and Kamiya, 1987; King and Kamiya, 2009). Further analyses of mutant phenotypes due to the absence of specific inner arm dyneins will allow for the role of these individual motors to be more precisely defined (for example, see Yagi et al., 2005). In addition, analysis of the composition of axonemal dyneins has begun to reveal potential regulators of dynein activity, including calcium-binding proteins, redox-active components, and proteins associated with the microtubule-binding domain (Kamiya and Yagi, 2014; King, 2018).
A major question is how the 96-nm repeat pattern along the doublet microtubules is assembled. The key to understanding this will be to define the mechanisms localizing each dynein and other axonemal structures, including the radial spokes, calmodulin-spoke complex, and nexin-dynein regulatory complex, at precise sites. Exciting progress in understanding this complex molecular patterning is being made, and, for example, a recent study revealed that two highly extended molecules act as molecular rulers and help precisely define the 96-nm repeat and location of radial spokes and possibly some inner dynein arms (Oda et al., 2014). However, with the exception of the outer dynein arm, which is localized by a trimeric docking complex (Owa et al., 2014), we do not yet know the precise basis for localization of other structures within the 96-nm repeat.
Dyneins are enormous protein complexes that provide immense experimental challenges. For example, the outer arm dyneins in Chlamydomonas axonemes are composed of three heavy chains, two intermediate chains, and 10 light chains with a combined mass of ∼2 MDa. However, recent advances have led to an atomic-scale understanding of purified dynein structure and motor activation and of the alterations that occur during the mechanochemical cycle (Kikkawa, 2013; Roberts et al., 2013; Lin et al., 2014; Bhabha et al., 2016; Pigino and King, 2017). Biophysical analysis of single dyneins will continue to provide ever more precise insight into dynein force generation and regulation (Kikkawa, 2013; Roberts et al., 2013; Cianfrocco et al., 2015; Bhabha et al., 2016). When combined with modeling (Brokaw, 1972a; Lindemann, 2011; Bayly and Dutcher, 2016; Sartori et al., 2016), biophysical and structural studies will lead to clearer understanding of axonemal bend formation and propagation.
In axonemal biology, some of the most notable recent progress has come from cryoelectron tomography (cryo-ET), which began with the studies of Nicastro et al. (2006). Cryo-ET is now the standard for structural analysis of the axoneme (for example, see Nicastro et al., 2006, 2009; Heuser et al., 2012; Oda et al., 2014; Ishikawa, 2015) and has enabled analysis of structural changes that occur during axonemal bending and switching at unprecedented resolution (Lin et al., 2014, and Figure 1, E and F). In a switching model for ciliary bend formation, the prediction is that dyneins on opposite sides of the axis of the axoneme would be in different functional states and that these states could be revealed as structural changes in the axonemal dynein motors (for example, see Goodenough and Heuser, 1982). To test this, Lin et al. (2014) rapidly captured actively swimming sea urchin sperm by freezing and analyzed the bent axonemes by cryo-ET. To assess the functional state of the motors, they then examined the structure of dyneins on individual doublet microtubules from opposite sides of the axoneme (Figure 1E, reproduced from Supplemental Figure 1 in Lin et al., 2014). As shown in Figure 1F, they were able to identify different outer dynein arm conformations including the “postpower stroke” (red dots, Figure 1E) and “prepower stroke” (blue dots, Figure 1E) states. Notably, the authors observed outer dynein arms in different states on opposite sides of the axonemal bend, exactly as predicted from the switching model (Figure 1E).
In the 50 years since Satir provided the first experimental evidence for the sliding microtubule model, enormous progress has been made in understanding the molecular mechanisms of dynein-driven ciliary beating. With continuing technical advances allowing for ever-higher-resolution imagery, we now appear poised to examine in exquisite detail the changes in axonemal structure associated with microtubule sliding and bending.
Acknowledgments
We are grateful to the authors and journals for permission to reprint the images in Figure 1 and to Jianfeng Lin and Daniela Nicastro, University of Texas Southwestern Medical Center, for sharing their unpublished data on changes in dynein structure during flagellar bending. This work was supported by grants from the National Institutes of Health (GM051293 to S.M.K. and GM051173 to W.S.S.).
Abbreviation used:
- cryo-ET
cryoelectron tomography.
Footnotes
REFERENCES
- Bayly PV, Dutcher SK. (2016). Steady dynein forces induce flutter instability and propagating waves in mathematical models of flagella. J R Soc Interface , 20160523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Johnson GT, Schroeder CM, Vale RD. (2016). How dynein moves along microtubules. Trends Biochem Sci , 94–105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ. (1965). Non-sinusoidal bending waves of sperm flagella. J Exp Biol , 155–169. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. (1972a). Computer simulation of flagellar movement. I. Demonstration of stable bend propagation and bend initiation by the sliding filament model. Biophys J , 564–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ. (1972b). Flagellar movement: a sliding filament model. Science , 455–462. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. (1989). Direct measurements of sliding between outer doublet microtubules in swimming sperm flagella. Science , 1593–1596. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. (1991). Microtubule sliding in swimming sperm flagella: direct and indirect measurements on sea urchin and tunicate spermatozoa. J Cell Biol , 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. (1987). Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskel , 68–75. [DOI] [PubMed] [Google Scholar]
- Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. (2015). Mechanism and regulation of cytoplasmic dynein. Annu Rev Cell Dev Biol , 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Gibbons IR. (1972). Flagellar movement and adenosine triphosphatase activity in sea urchin sperm extracted with Triton X-100. J Cell Biol , 75–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. (1981). Cilia and flagella of eukaryotes. J Cell Biol , 107s–124s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. (1965). Dynein: a protein with adenosine triphosphatase activity from cilia. Science , 424–426. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. (1982). Substructure of the outer dynein arm. J Cell Biol , 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Shingyoji C. (2009). Bending-induced switching of dynein activity in elastase-treated axonemes of sea urchin sperm—roles of Ca2+ and ADP. Cell Motil Cytoskel , 292–301. [DOI] [PubMed] [Google Scholar]
- Heuser T, Barber CF, Lin J, Krell J, Rebesco M, Porter ME, Nicastro D. (2012). Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc Natl Acad Sci USA , E2067–E2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T. (2015). Cryo-electron tomography of motile cilia and flagella. Cilia , 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, Yagi T. (2014). Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zool Sci , 633–644. [DOI] [PubMed] [Google Scholar]
- Kikkawa M. (2013). Big steps toward understanding dynein. J Cell Biol , 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM. (2018). Composition and assembly of axonemal dyneins. In: Dyneins: Structure, Biology and Disease, Vol. 1, The Biology of Dynein Motors, ed. King SM, Oxford, UK: Elsevier, Academic Press, 163–201. [Google Scholar]
- King SM, Kamiya R. (2009).Axonemal dyneins: assembly, structure and force generation. In: The Chlamydomonas Source Book, Vol. 3, 2nd ed., Cell Motility and Behavior, ed. Witman GB, San Diego: Elsevier, 131–208. [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. (2014). Structural mechanism of the dynein power stroke. Nat Cell Biol , 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB. (2011). Experimental evidence for the geometric clutch hypothesis. Curr Top Dev Biol , 1–31. [DOI] [PubMed] [Google Scholar]
- Morita Y, Shingyoji C. (2004). Effects of imposed bending on microtubule sliding in sperm flagella. Curr Biol , 2113–2118. [DOI] [PubMed] [Google Scholar]
- Nicastro D. (2009). Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol , 1–39. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science , 944–948. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. (2014). A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science , 857–860. [DOI] [PubMed] [Google Scholar]
- Owa M, Furuta A, Usukura J, Arisaka F, King SM, Witman GB, Kamiya R, Wakabayashi K. (2014). Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc Natl Acad Sci USA , 9461–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, King SM. (2017). Switching dynein motors on and off. Nat Struct Mol Biol , 557–559. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. (2013). Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol , 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. (1977). Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci USA , 2045–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori P, Geyer VF, Scholich A, Julicher F, Howard J. (2016). Dynamic curvature regulation accounts for the symmetric and asymmetric beats of Chlamydomonas flagella. Elife , e13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. (1968). Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol , 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Heuser T, Sale WS. (2014). A structural basis for how motile cilia beat. Bioscience , 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingyoji C, Murakami A, Takahashi K. (1977). Local reactivation of Triton-extracted flagella by iontophoretic application of ATP. Nature , 269–270. [DOI] [PubMed] [Google Scholar]
- Summers KE, Gibbons IR. (1971). Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc Natl Acad Sci USA , 3092–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Toyoshima YY. (1988). Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell , 459–469. [DOI] [PubMed] [Google Scholar]
- Wais-Steider J, Satir P. (1979). Effect of vanadate on gill cilia: switching mechanism in ciliary beat. J Supramol Struct , 339–347. [DOI] [PubMed] [Google Scholar]
- Yagi T, Minoura I, Fujiwara A, Saito R, Yasunaga T, Hirono M, Kamiya R. (2005). An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J Biol Chem , 41412–41420. [DOI] [PubMed] [Google Scholar]