Abstract
Centrioles are microtubule-based organelles that organize the microtubule network and seed the formation of cilia and flagella. New centrioles assemble through a stepwise process dependent notably on the centriolar protein SAS-5 in Caenorhabditis elegans. SAS-5 and its functional homologues in other species form oligomers that bind the centriolar proteins SAS-6 and SAS-4, thereby forming an evolutionarily conserved structural core at the onset of organelle assembly. Here, we report a novel interaction of SAS-5 with microtubules. Microtubule binding requires SAS-5 oligomerization and a disordered protein segment that overlaps with the SAS-4 binding site. Combined in vitro and in vivo analysis of select mutants reveals that the SAS-5–microtubule interaction facilitates centriole assembly in C. elegans embryos. Our findings lead us to propose that the interdependence of SAS-5 oligomerization and microtubule binding reflects an avidity mechanism, which also strengthens SAS-5 associations with other centriole components and, thus, promotes organelle assembly.
INTRODUCTION
Centrioles are microtubule-based organelles fundamental for a number of critical processes in eukaryotic cells. Centrioles direct the formation of flagella, motile and sensory cilia, as well as of centrosomes, which in animal cells organize the microtubule network, including the mitotic spindle (reviewed in Bornens, 2012; Arquint et al., 2014; Conduit et al., 2015). As a result, proper centriole function is essential for crucial cellular processes, including locomotion, signaling and equitable segregation of the genetic material during mitosis. The importance of centrioles in cell and organism physiology is also illustrated by the fact that mutations in genes coding for major centriole and ciliary components are associated with human diseases, including primary microcephaly, primordial dwarfism, and multisystemic ciliopathies, while aberrations in centriole structure or numbers may lead to cancer (reviewed in Nigg and Raff, 2009; Thornton and Woods, 2009; Bettencourt-Dias et al., 2011; Chavali et al., 2014; Gönczy, 2015; Venghateri et al., 2015).
Uncovering the molecular mechanisms that underpin the formation of functional centrioles is, thus, a fundamental question in cell biology. Centrioles are ninefold radially symmetric cylindrical assemblies typically of ∼500 nm in length and ∼200 nm in diameter (reviewed in Winey and O’Toole, 2014), with an exterior microtubule “wall” that assembles around a centrally located “cartwheel” scaffold (Nakazawa et al., 2007; Guichard et al., 2012, 2013). Centriole assembly takes place once per cycle in most proliferating cells, starting approximately at the G1 to S transition, and involves the sequential recruitment of dedicated protein components (reviewed in Azimzadeh and Marshall, 2010; Firat-Karalar and Stearns, 2014). In particular, three evolutionary conserved structural proteins, SAS-6, SAS-5, and SAS-4, are key for initiating centriole assembly as they control essential elements of the organelle architecture (reviewed in Gönczy, 2012; Jana et al., 2014; Dong, 2015; Banterle and Gonczy, 2017).
SAS-6 homodimers from most species self-assemble into cartwheel-like structures with an inherent ninefold radial symmetry (Kitagawa et al., 2011; van Breugel et al., 2011, 2014; Guichard et al., 2017), thereby facilitating the formation of likewise symmetric centrioles (Hilbert et al., 2016). Drosophila SAS-4 and its vertebrate homologue CPAP interact directly with tubulin and microtubules to control centriole length (Hsu et al., 2008; Cormier et al., 2009; Sharma et al., 2016; Zheng et al., 2016); furthermore, SAS-4/CPAP forms oligomers that may provide structural support for centriole elongation (Cottee et al., 2013; Hatzopoulos et al., 2013; Zheng et al., 2014; Cutts et al., 2015; Alvarez-Cabrera et al., 2017). Caenorhabditis elegans SAS-5, as well as its Drosophila (Ana2) and vertebrate (STIL) related proteins, provide structural bridges between SAS-6 and SAS-4/CPAP as they directly interact with both of these components (Tang et al., 2011; Qiao et al., 2012; Cottee et al., 2013; Hatzopoulos et al., 2013; Hilbert et al., 2013; Lettman et al., 2013; Ohta et al., 2014; Moyer et al., 2015). Notably, SAS-6 and SAS-5 localize in an interdependent manner at the site of centriole assembly in C. elegans (Leidel et al., 2005), where they are essential for the formation of the “central tube,” a structure equivalent to the cartwheel in centrioles of other species (Pelletier et al., 2006; Sugioka et al., 2017), and to which SAS-4 and microtubules are recruited (Delattre et al., 2006; Pelletier et al., 2006).
Recent structural and functional studies demonstrated that SAS-5, Ana2, and STIL comprise homo-oligomers (Shimanovskaya et al., 2013; Slevin et al., 2014; Cottee et al., 2015; Rogala et al., 2015; David et al., 2016; Cottee et al., 2017), and suggested that formation of these oligomers promotes centriole assembly likely via an avidity mechanism (Shimanovskaya et al., 2013; Slevin et al., 2014; Cottee et al., 2015; Rogala et al., 2015), whereby intermolecular interactions are enhanced through the cooperative engagement of multiple binding sites (Mammen et al., 1998). For instance, SAS-5 features a highly stable globular dimeric domain, termed Implico (“entangled”), and a coiled-coil domain that forms trimers in a concentration-dependent manner (Rogala et al., 2015). Together, these two domains drive the formation of higher-order SAS-5 oligomers that are essential for function, as disruption of either the Implico or coiled-coil domains by mutagenesis impairs SAS-5 localization at the centriole and abrogates centriole assembly in C. elegans embryos (Rogala et al., 2015). Here, we report a novel direct interaction between SAS-5 and microtubules that crucially relies on SAS-5 higher-order oligomerization and that assists SAS-5 function in centriole duplication. We propose that the SAS-5–microtubule association may provide a molecular mechanism for microtubule binding to centriolar components complementary to that offered by SAS-4. As both SAS-4/CPAP (Hsu et al., 2008; Cormier et al., 2009; Sharma et al., 2016; Zheng et al., 2016) and SAS-6 (Gupta et al., 2015) also bind microtubules, our discovery raises the prospect that microtubule binding is a conserved feature among structural proteins acting at the onset of centriole formation.
RESULTS
SAS-5 directly associates with microtubules
We previously generated transgenic C. elegans animals expressing GFP-SAS-5 protein during oogenesis and in the resulting embryos (Rogala et al., 2015). Upon live-cell imaging of early embryos from these animals, we noted that, in addition to the previously reported centriolar localization, GFP-SAS-5 is also present transiently on the mitotic spindle during metaphase and early anaphase (white arrows in Figure 1, A and B; Supplemental Movie 1). We addressed whether endogenous SAS-5 exhibits a similar spindle localization using immunofluorescence analysis of wild-type embryos and found this to be indeed the case (white arrows in Figure 1, C and D). This distribution is specific to SAS-5 since we found it to be absent in embryos from which the endogenous protein has been depleted by RNA interference (RNAi) (Figure 1, E and F).
FIGURE 1:
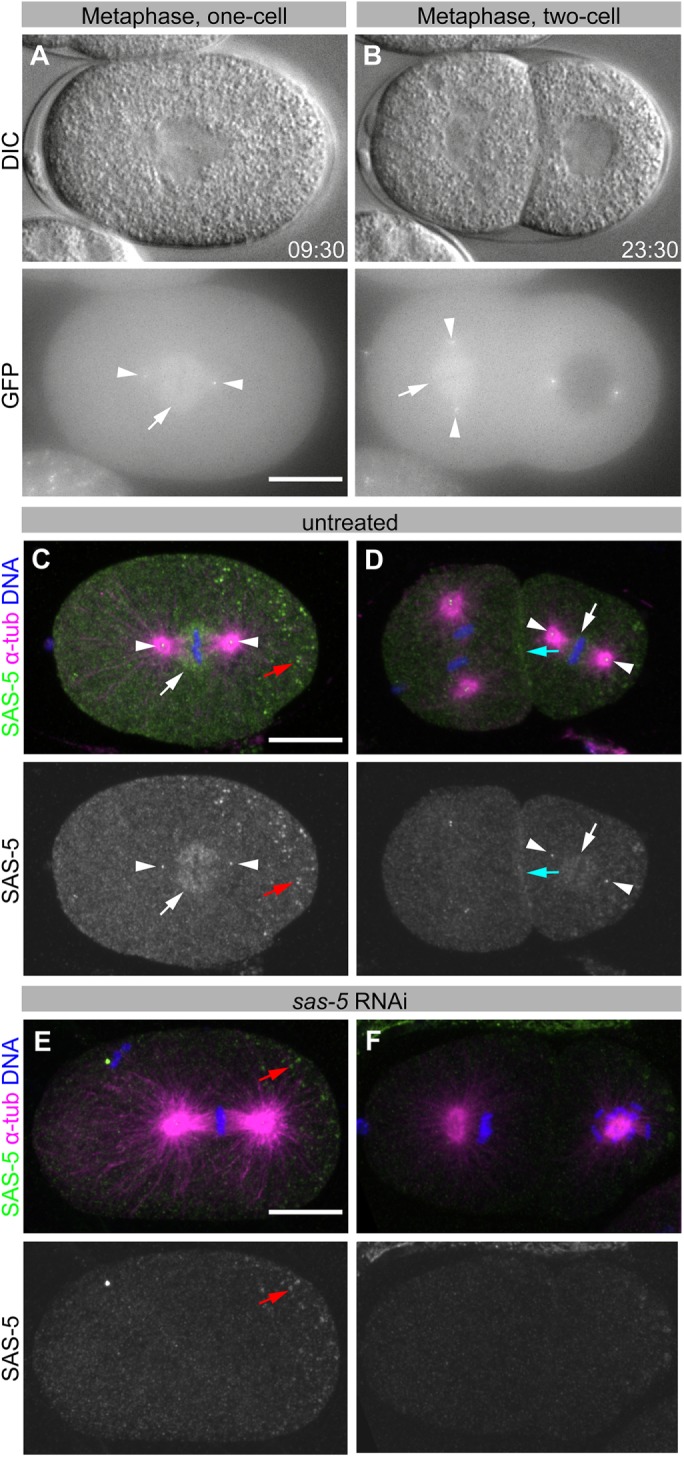
SAS-5 weakly localizes to the mitotic spindle. (A, B) DIC (top panels) and GFP fluorescence (bottom panels) images of an embryo expressing GFP-SAS-5, at metaphase of the one-cell stage (A) and at metaphase of the AB blastomere in the two-cell stage (B, left cell). See Supplemental Movie 1. (C–F) Immunofluorescence of wild-type (N2) embryos from worms grown without RNAi (C, D) or upon sas-5(RNAi) (E, F). Top panels show SAS-5 (green), α-tubulin (magenta), and DNA (blue); bottom panels show SAS-5 only. Images are maximum-intensity z-projections. White arrows indicate SAS-5 spindle staining; white arrowheads, centrioles; cyan arrows, membrane/cortical staining; and red arrow, nonspecific P-granule staining. SAS-5 localizes to centrioles, as well as to the mitotic spindle, most notably proximal to kinetochores, and at the cell cortex, together suggesting that SAS-5 exhibits affinity for microtubule plus ends. Scale bars, 10 μm. All embryos are oriented with the anterior on the left and the posterior on the right.
Movie S1.
Association of SAS-5 with the mitotic spindle. DIC (left) and GFP maximum intensity z-projection (right) of a GFP-SAS-5 WT embryo (approximately 50 μm-long). Images were taken every 30 seconds, gathering 21 z-sections at 0.5 μm intervals. Time indicated in mm:ss.
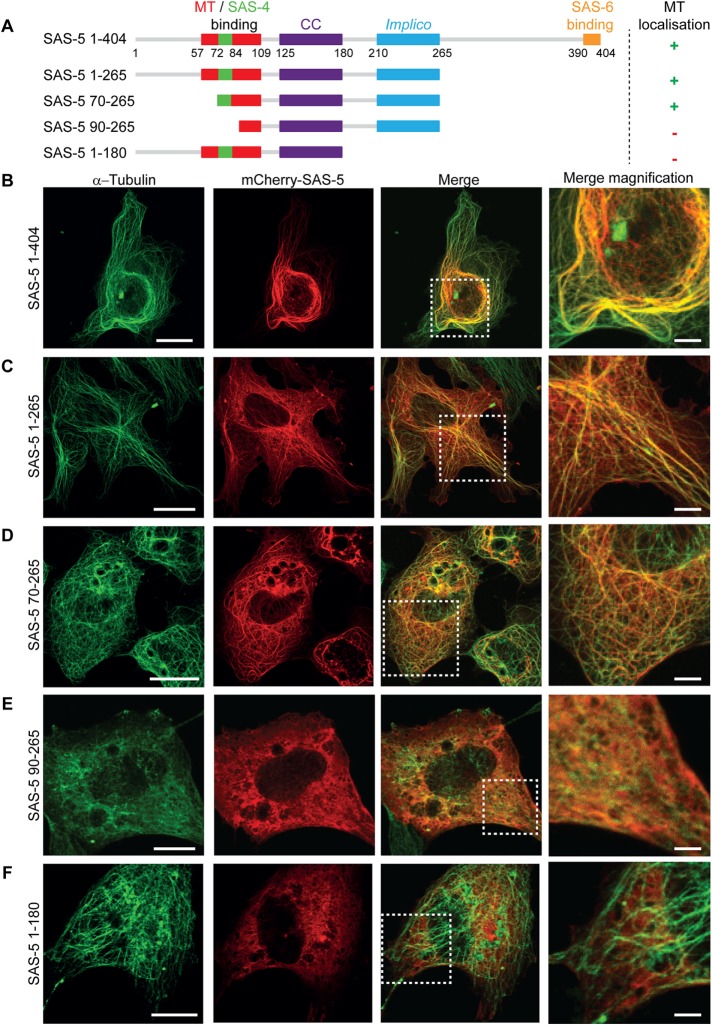
Although the SAS-5 related protein Ana2 is involved in regulating mitotic spindle orientation in Drosophila neuroblasts via direct interactions with a light chain of dynein (Wang et al., 2011; Slevin et al., 2014), SAS-5 does not appear to play a role in mitotic spindle assembly or function in C. elegans, as the spindle apparatus forms and separates sister chromatids normally in sas-5(RNAi) one-cell stage embryos (Dammermann et al., 2004; Delattre et al., 2004; Rogala et al., 2015). Nevertheless, the transient spindle localization raises the possibility that SAS-5 may interact with microtubules in a manner apparent at high microtubule density. Compatible with this possibility, overexpression of mCherry-SAS-5 in vertebrate COS-7 cells via transient transfection led to strong colocalization of mCherry-SAS-5 with microtubules (Figure 2B; a summary of interaction properties of SAS-5 constructs in this study is presented in Table 1). Similar results were obtained with HA-tagged SAS-5 in COS-7 cells and mCherry-SAS-5 in HEK293T cells (Supplemental Figure S1, A and B), suggesting that this interaction is neither tag- nor cell type–specific. To determine whether SAS-5 association leads to microtubule stabilization, we assessed the persistence of microtubules following cold treatment of COS-7 cells. As shown in Supplemental Figure S2, upon such treatment, tubulin became diffuse in untransfected control cells, whereas cells that expressed mCherry-SAS-5 retained microtubules, to which SAS-5 colocalized. We conclude that SAS-5 can associate with cytoplasmic microtubules and stabilize them in the cellular context.
FIGURE 2:
SAS-5 colocalizes with microtubules in mammalian cells. (A) Schematic representation of SAS-5 constructs used in transfection assays, showing the coiled-coil and Implico oligomerization domains of SAS-5, as well as microtubule (MT)-, SAS-4-, and SAS-6-interaction regions. Note that the SAS-4 and microtubule-binding sites partly overlap. The microtubule localization in COS-7 cells of each construct is indicated on the right. (B–F) Representative fluorescence images of COS-7 cells transiently expressing mCherry-SAS-5 full length (B) or truncations as indicated (C–F). Microtubules were visualized using immunofluorescence with α-tubulin antibodies; SAS-5 was visualized by mCherry fluorescence. Scale bar, 20 μm. The rightmost column corresponds to digital magnification of boxed areas in merged images. Scale bar, 5 μm in magnified images. Assays were performed once. The diffused microtubule fluorescence in cells expressing mCherry-SAS-5 90–265 (E) was seen throughout this particular transfection but not in parallel transfections of HEK293T cells (Supplemental Figure S1E), indicating that such diffused appearance is not a property of the SAS-5 construct but a transfection artifact.
TABLE 1:
Microtubule-binding properties of SAS-5 constructs and variants.
| Constructa | Microtubule colocalization in COS-7 cells | Microtubule-pelleting assays | DrCPAPG-box interaction by ITC |
|---|---|---|---|
| 1–404 | + | NDb | ND |
| 1/2-265 (SAS-52–265) | + | +++ | ND |
| 70–265 | + | ND | ND |
| 90–265 | – | ND | ND |
| 1–180 | – | ND | ND |
| SAS-52–265 L141E | – | + | ND |
| SAS-52–265 I247E | – | + | ND |
| SAS-52–265 L141/I247E | – | – | ND |
| SAS-52–265 ∆72–80 | ND | ++ | ND |
| SAS-52–265 ∆81–90 | ND | + | ND |
| SAS-52–265 ∆91–100 | ND | ++ | ND |
| SAS-52–265 ∆101–110 | ND | +++ | ND |
| SAS-52–265 K65E/K66E/K67E | ND | – | ND |
| SAS-52–265 R86E/K88E/K94E | ND | – | ND |
| 2–122 (SAS-5N) | ND | +/− | +++ |
| SAS-5N ∆72–80 | ND | ND | ++ |
| SAS-5N ∆81–90 | ND | ND | +++ |
| SAS-5N ∆91–100 | ND | ND | +++ |
| SAS-5N ∆101–110 | ND | ND | +++ |
| SAS-5N K65E/K66E/K67E | ND | ND | ++ |
| SAS-5N R86E/K88E/K94E | ND | ND | + |
aDenotes SAS-5 amino acid residue ranges, construct acronyms, amino acid substitutions and/or deletions.
bND, not determined.
We proceeded to identify the minimal SAS-5 fragment that colocalizes with microtubules. mCherry-SAS-5 constructs comprising amino acids 1–265 (Figure 2C) or amino acids 70–265 (Figure 2D) retained SAS-5 colocalization with microtubules in both COS-7 and HEK293T (Supplemental Figure S1, C and D) cells. In contrast, mCherry-SAS-5 constructs comprising amino acids 90–265 (Figure 2E and Supplemental Figure S1E) or amino acids 1–180 (Figure 2F and Supplemental Figure S2F) yielded diffuse cytoplasmic mCherry-SAS-5 fluorescence and no specific microtubule colocalization. We conclude that a SAS-5 fragment that spans residues 70–265, which includes both SAS-5 oligomerization domains, is required for strong microtubule colocalization in cells.
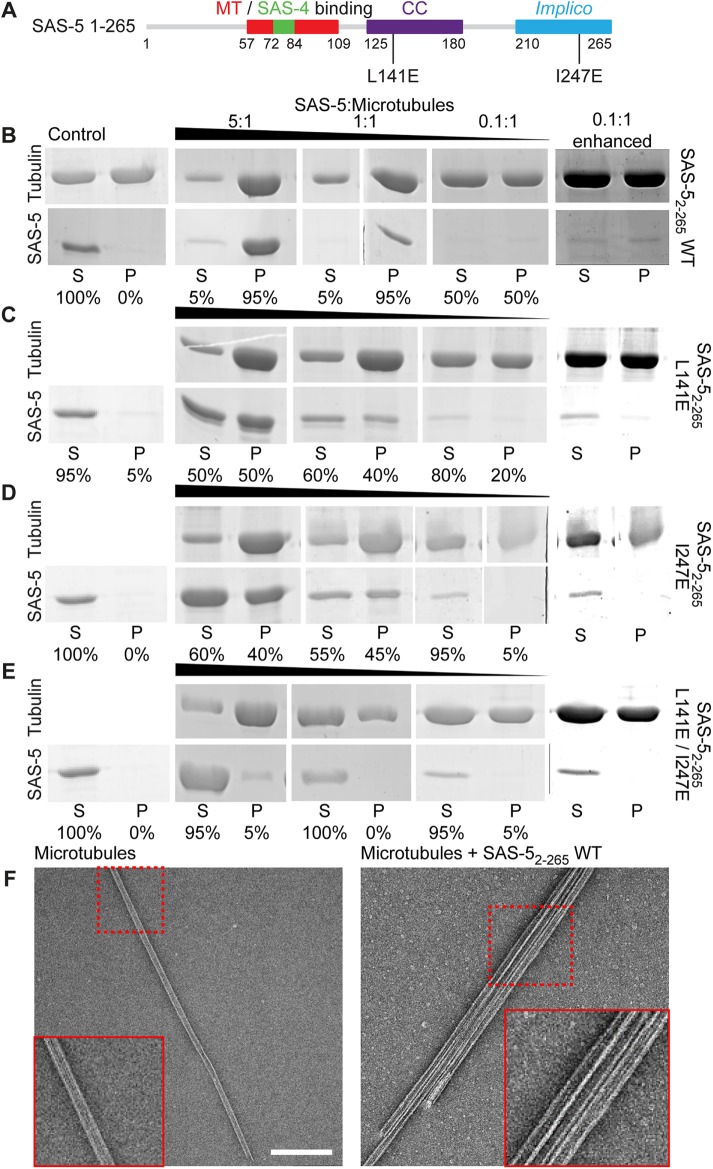
Given that SAS-5 interacts with SAS-4 (Cottee et al., 2013; Hatzopoulos et al., 2013), and that variants of the SAS-4 vertebrate homologue CPAP bind microtubules (Hsu et al., 2008; Cormier et al., 2009; Sharma et al., 2016; Zheng et al., 2016), we evaluated whether the microtubule colocalization of SAS-5 in cells represents a direct protein interaction, as opposed to one that occurs indirectly through CPAP. To that end, we assayed the direct binding of a recombinant SAS-5 fragment spanning residues 2–265 (SAS-52–265) to Taxol-stabilized microtubules using low-speed pelleting assays. We observed strong copelleting of microtubules with SAS-52–265 but no pelleting of SAS-52–265 alone (Figure 3B). Given the concentrations of SAS-52–265 and microtubules used in these assays, the apparent SAS-5–microtubule affinity was estimated in the low micromolar (1–5 μM Kd) range. Examination of pellets by negative stain electron microscopy revealed only single microtubules in the absence of SAS-5 (n = 10 observations). By contrast, in ∼40% of microtubule assemblies observed in the presence of SAS-52–265 comprised large bundles (Figure 3F; n = 31 observations). Taken together, we conclude that SAS-5 associates with microtubules directly and can cause their bundling.
FIGURE 3:
SAS-5 interacts directly with microtubules in an oligomerization-dependent manner. (A) Schematic representation of SAS-5 constructs used in pelleting assays, annotated as in Figure 2A, and further showing amino acid substitutions abrogating SAS-5 oligomerization via the coiled-coil (L141E) and Implico (I247E) domains. (B–E) Shown are relevant sections of Coomassie-stained SDS–PAGE from the supernatant (S) and pellet (P) fractions of low-speed microtubule-pelleting assays performed twice using three different stoichiometric ratios of purified SAS-52–265 vs. tubulin, as indicated. Row A corresponds to SAS-52–265 WT; rows B–D correspond to dimeric (B; L141E), trimeric (C; I247E), or monomeric (D; L141E/I247E) SAS-52–265. The rightmost panels of row A are contrast-enhanced variants of the 0.1:1 SAS-5 to microtubule ratio panels. Fractional values under each lane correspond to the percentage of SAS-5 constructs present in the supernatant vs. the pellet of assays. Assembled tubulin concentration was 3 μM throughout. Control assays were performed using either 3 μM tubulin in microtubules (top) or 3 μM SAS-52–265 (bottom) alone. (E) Negative-stain electron micrographs of pellet fractions from assays of microtubules alone (left) or in the presence of SAS-52–265 WT (right). Scale bar, 200 nm. Insets correspond to twofold magnified views of the regions indicated in dashed boxes.
SAS-5 oligomerization strengthens its microtubule interaction
The smallest SAS-5 fragment that strongly colocalizes with microtubules (Figure 2, C and D) includes both SAS-5 oligomerization domains and, thus, can form higher-order oligomers (Rogala et al., 2015). We examined the dependence of microtubule binding on SAS-5 higher-order oligomerizsation by using single amino acid substitutions, L141E or I247E, that selectively disrupt the coiled-coil or Implico domains, and limit SAS-5 oligomerization to dimers or trimers, respectively (Rogala et al., 2015). Pelleting assays showed reduced association with Taxol-stabilized microtubules of higher-order oligomerization-defective SAS-52–265 variants (Figure 3, C–E). Thus, whereas SAS-52–265 wild type (WT) pelleted fully with microtubules when present in a 1:1 stoichiometric ratio with tubulin (Figure 3B), SAS-52–265 L141E and SAS-52–265 I247E displayed only half as much pelleting (Figure 3, C and D), and the monomeric SAS-52–265 L141E/I247E showed no detectable pelleting under the same conditions (Figure 3E). However, we noted that SAS-52–265 L141E/I247E did show weak copelleting with microtubules when used in high (5:1) stoichiometric ratios with tubulin, suggesting that it retains very weak microtubule affinity. To address the impact of SAS-5 oligomerization on microtubule binding in the cellular context, we expressed in COS-7 cells mCherry-SAS-5 variants spanning residues 1–265 that harbor L141E, I247E, or L141E/I247E substitutions. We found that all these SAS-5 variants showed weaker microtubule colocalization (Supplemental Figure S3, C–E) compared with the WT protein (Supplemental Figure S3B), confirming that the SAS-5–microtubule interaction is attenuated.
The above observations taken together lead us to hypothesize that direct microtubule binding involves a protein epitope located at the SAS-5 N-terminus, potentially between amino acids 70–90. To test this hypothesis, we performed microtubule-pelleting assays using a SAS-5 fragment corresponding to amino acids 2–122 (SAS-5N), which is disordered and monomeric in solution (Rogala et al., 2015). We observed partial pelleting of this construct with Taxol-stabilized microtubules when used in high (20:1) stoichiometric ratios compared with tubulin (Supplemental Figure S4A). We interpret these results as evidence of a weak SAS-5–microtubule interaction mediated by the flexible protein’s N-terminus, which is strengthened upon inclusion of the structured SAS-5 oligomerization domains.
Localizing the interactions of SAS-5 with microtubules and SAS-4
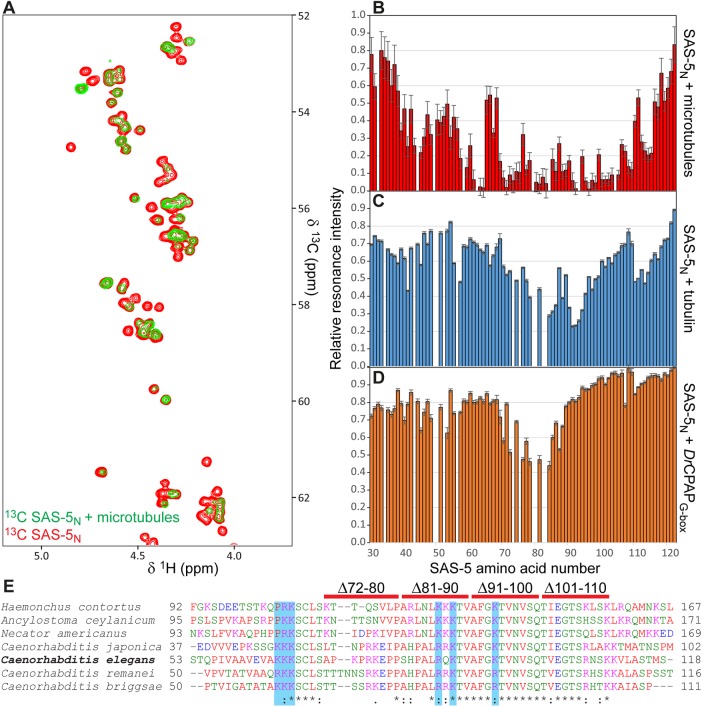
We proceeded to precisely locate the microtubule-interaction epitope of SAS-5 within its N-terminal fragment. To that end, we titrated uniformly 13C-labeled SAS-5N at 33 μM concentration with an equimolar amount of unlabeled Taxol-stabilized microtubules, and monitored the interaction by 13C-heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) experiments. As shown in Figure 4A, the NMR resonances of SAS-5N Hα atoms displayed poor chemical shift dispersion, as anticipated due to disorder in this protein fragment (Rogala et al., 2015). Upon microtubule addition, we observed significant attenuation of SAS-5N Hα-Cα resonance intensities, which is consistent with association of the flexible SAS-5N with a high molecular weight counterpart. Attenuation of resonance intensity was strongest for a nearly continuous stretch of SAS-5N comprising amino acids 57–64 and 69–109 (Figure 4B). This region of SAS-5 is highly conserved among nematodes, with more than 45% sequence identity and 64% sequence similarity (Figure 4E). We conclude that a stretch of SAS-5 residues (amino acids 57–109) forms the direct microtubule interaction epitope, which is consistent with the loss of microtubule colocalization in COS-7 cells when using a SAS-5 fragment lacking the first 89 amino acids (Figure 2E). Similar NMR 15N-HSQC assays using 33 μM SAS-5N and an equal concentration of unpolymerized tubulin showed significantly lower attenuation of SAS-5 resonances compared with that seen for microtubules, and over a narrower SAS-5 span (amino acids 84–94; Figure 4C), suggesting that SAS-5 interacts preferentially with microtubules compared with free tubulin.
FIGURE 4:
NMR assays to localize SAS-5 interactions. (A) Shown in the overlay are representative sections of 13C-HSQC NMR experiments corresponding to Hα-Cα resonances of labeled SAS-5 residues 2–122 (SAS-5N) alone (red) or in 1:1 complex with Taxol-stabilized microtubules (green). Attenuation of specific Hα-Cα SAS-5N resonances is indicative of direct microtubule binding. (B–D) Quantification of SAS-5N NMR resonance intensities upon binding to protein partners. Plotted are fractional resonance intensities of SAS-5N in complex vs. alone as function of SAS-5 amino acid number. Panel B represents SAS-5 binding to microtubules and derives from Hα-Cα resonances. Panels C and D represent SAS-5 binding to unpolymerized tubulin and DrCPAPG-box, respectively, and derive from HN-N resonances in 15N-HSQC experiments. Error bars are derived from the noise level of HSQC spectra. Assays were performed once. (E) Clustal omega (Sievers et al., 2011) multiple sequence alignment of nematode SAS-5 sequences spanning the microtubule binding identified in C. elegans SAS-5. Blue highlights and red bars demote C. elegans SAS-5 amino acid residues substituted or regions deleted, respectively, aiming to abrogate the microtubule interaction.
SAS-5 associates with the centriolar protein SAS-4 (Cottee et al., 2013), and previous work on the SAS-5 relatives STIL and Ana2 established that they bind the SAS-4/CPAP G-box domain through a linear epitope that includes a proline-rich segment (Cottee et al., 2013; Hatzopoulos et al., 2013). Sequence alignment suggested that this proline-rich site corresponds to SAS-5 residues 75–84 (Hatzopoulos et al., 2013); thus, the microtubule and SAS-4 interaction epitopes of SAS-5 may partly overlap. Despite repeated efforts, we were unable to produce recombinant C. elegans SAS-4 G-box domain in a state suitable for biophysical studies. However, isothermal titration calorimetry (ITC) experiments showed that SAS-5N can associate with the homologous Danio rerio CPAP G-box domain (DrCPAPG-box) with a dissociation constant, Kd, of 34 ± 4 μM (Supplemental Figure S5). Thus, we utilized the SAS-5N–DrCPAPG-box interaction as a proxy for the SAS-5–SAS-4 association. NMR 15N-HSQC assays using labeled SAS-5N and unlabeled DrCPAPG-box confirmed the presence of an interaction between these proteins, judging by the extent of attenuation of resonance intensities, over amino acids 72–84 of SAS-5 (Figure 4D). This SAS-5 segment matches well to the proline-rich site predicted to be important for SAS-4 binding by sequence alignment (Hatzopoulos et al., 2013). Hence, we conclude that the microtubule and SAS-4 interaction epitopes of SAS-5 indeed partly overlap. Microtubule-pelleting assays combining SAS-52–265 with DrCPAPG-box showed that CPAP cannot displace SAS-5 from microtubules, even when used in high (10:1) stoichiometric ratios (Supplemental Figure S4B); however, it should be noted that SAS-5 may have substantially higher affinity for the C. elegans SAS-4 G-box domain compared with DrCPAPG-box.
Selective disruption of the SAS-5–microtubule interaction
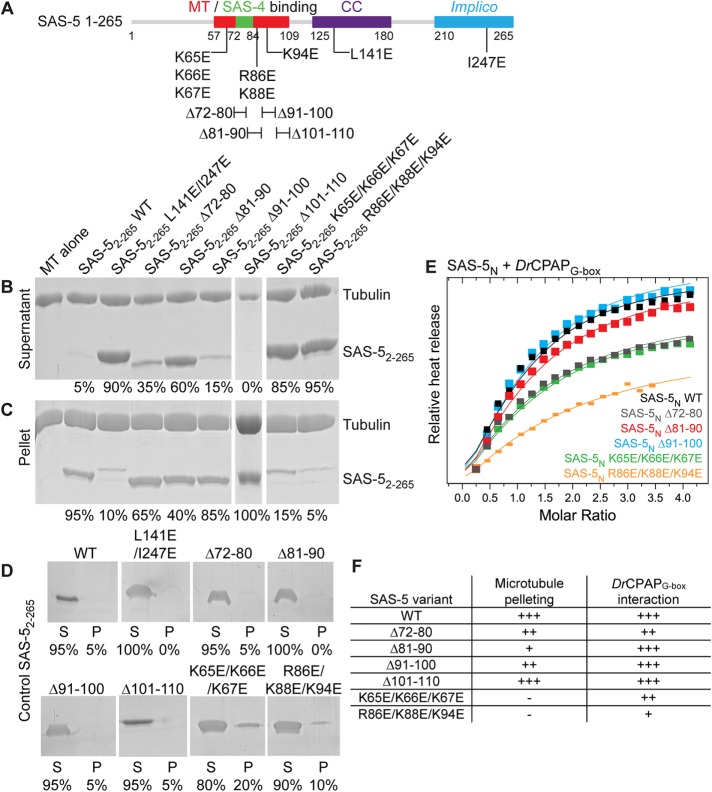
We sought to disrupt the SAS-5–microtubule interaction while minimally affecting the SAS-5–SAS-4 association and retaining the SAS-5 oligomeric structure. The segment of SAS-5 directly interacting with microtubules as shown by NMR, amino acids 57–109, is highly positively charged (calculated pI of ∼10.4). Largely positively charged sequence stretches are common among microtubule associated proteins as they form favorable electrostatic interactions with the negatively charged surface of microtubules (Baker et al., 2001). Thus, we derived a series of SAS-5 variants that reversed the charge of conserved positively charged residues in the microtubule-interaction epitope (Figure 4E; K65E/K66E/K67E, R86E/K88E/K94E) or removed parts of this site (Δ72–80, Δ81–90, Δ91–100, Δ101–110), and proceeded to simultaneously assess the microtubule and DrCPAPG-box interactions of these variants.
Pelleting assays indicated that SAS-52–265 variants K65E/K66E/K67E, R86E/K88E/K94E, and Δ81–90 substantially attenuated the microtubule-binding affinity compared with the WT protein (Figure 5, A–C and E), as evidenced by the relative amount of SAS-5 remaining in the supernatant fraction. In contrast, SAS-52–265 Δ72–80, Δ91–100, and Δ101–110 derivatives showed minimal or no perturbation of microtubule binding. To assess the impact of SAS-5 microtubule-binding variants on the SAS-4 interaction, we quantified the binding of SAS-5N variants harboring the same amino acid substitutions and deletions to DrCPAPG-box using ITC (Figure 5, D and E, and Supplemental Figure S5). Our results suggest that three SAS-5N variants, Δ81–90, Δ91–100, and Δ101–110, retained DrCPAPG-box affinities comparable to the WT protein; variants Δ72–80 and K65E/K66E/K67E produced approximately twofold reduced affinity; and variant R86E/K88E/K94E displayed approximately fourfold reduced affinity.
FIGURE 5:
Decoupling microtubule and SAS-4 binding activity in SAS-5. (A–C) Relevant sections of Coomassie-stained SDS–PAGE from the supernatant (A) and pellet (B) fractions of pelleting assays performed using Taxol-stabilized microtubules alone and with SAS-52–265 variants designed to disrupt the microtubule interaction. 3 μM of SAS-52–265 and tubulin were used throughout. SAS-52–265 WT and monomeric L141E/I247E variant comprise microtubule-binding positive and negative controls, respectively. Panel C shows control pelleting assays of 3 μM SAS-52–265 WT and variants without microtubules; S represents the supernatant fraction and P denotes the pellet fraction. Fractional values under each lane correspond to the percentage of SAS-5 constructs present in the supernatant vs. the pellet of assays. (D) Superimposed ITC heat released upon injection of SAS-5N WT and select variants to DrCPAPG-box. Heat released upon injection (closed squares) was scaled relative to the per mole enthalpy change derived from fitting ITC data to a single association model; fits are shown as solid lines. Complete, unscaled ITC data of all SAS-5N variants are shown in Supplemental Figure S6. Assays were performed once. (E) Tabular representation of microtubule and DrCPAPG-box binding affinities of SAS-5 variants.
The SAS-5–microtubule interaction contributes to centriole duplication in C. elegans embryos
We next sought to assess the role of microtubule binding by SAS-5 in centriole duplication. To this end, we generated C. elegans transgenic animals expressing under the control of a germline promoter GFP-SAS-5 variants Δ81–90, which attenuates microtubule binding in vitro but not the interaction with SAS-4, as well as K65E/K66E/K67E, which disrupts the association with microtubules but also slightly perturbs the SAS-4 interaction. All exons of the sas-5 gene were recoded to confer resistance to RNAi directed against endogenous sas-5. Sperm is not affected by RNAi in these experiments, such that WT self-fertilizing hermaphrodites treated with sas-5(RNAi) contribute a normal pair of centrioles to the newly fertilized embryo, which direct formation of a bipolar mitotic spindle during the first division (Delattre et al., 2004). However, due to depletion of the maternal pool of endogenous SAS-5, no new centrioles can assemble in the embryo, leading to monopolar spindles at the second cell cycle and failure to reach the four-cell stage (Supplemental Figure S6 and Supplemental Movie 2). In contrast, embryos from animals expressing RNAi-resistant GFP-SAS-5 WT form centrioles normally in the embryo, leading to bipolar spindle assembly in each blastomere at the second cell cycle and resulting in a four-cell stage configuration similar to the non-RNAi treated WT (Supplemental Figure S6 and Supplemental Movie 3).
Movie S2.
2- to 4-cell stage progression of C. elegans embryos with microtubule-binding deficient SAS-5 variants. Shown are DIC movies of embryos (approximately 50 μm-long) from non-transgenic wild-type worms (M2), or worms expressing GFP-SAS-5 WT (M3), Δ81-90 (M4) or K65E/K66E/K67E (M5) variants. All strains were subjected to sas-5(RNAi) for 24 hours prior to extracting embryos for analysis. Images were taken every 5 seconds, frame rate 12 fps. Time indicated in mm:ss.
Movie S3.
2- to 4-cell stage progression of C. elegans embryos with microtubule-binding deficient SAS-5 variants. Shown are DIC movies of embryos (approximately 50 μm-long) from non-transgenic wild-type worms (M2), or worms expressing GFP-SAS-5 WT (M3), Δ81-90 (M4) or K65E/K66E/K67E (M5) variants. All strains were subjected to sas-5(RNAi) for 24 hours prior to extracting embryos for analysis. Images were taken every 5 seconds, frame rate 12 fps. Time indicated in mm:ss.
We found by Western blot analysis that both GFP-SAS-5 protein variants are overexpressed compared with endogenous SAS-5 (Supplemental Figure S7A). Analysis by dual fluorescence and time-lapse differential interference contrast (DIC) microscopy showed that these constructs efficiently rescued the sas-5(RNAi) phenotype, allowing embryos to progress to the four-cell stage (Supplemental Figure S7, B and C, and Supplemental Movies 4 and 5). We conclude that GFP-SAS-5 variants that weaken the microtubule interaction in vitro can nevertheless sustain centriole assembly when present in excess. Further, overexpression of the same GFP-SAS-5 variants yielded transient SAS-5 localization on the mitotic spindle during metaphase and early anaphase (Supplemental Figure S7D), similar to that observed for WT GFP-SAS-5 (Figure 1, A and B).
Movie S4.
2- to 4-cell stage progression of C. elegans embryos with microtubule-binding deficient SAS-5 variants. Shown are DIC movies of embryos (approximately 50 μm-long) from non-transgenic wild-type worms (M2), or worms expressing GFP-SAS-5 WT (M3), Δ81-90 (M4) or K65E/K66E/K67E (M5) variants. All strains were subjected to sas-5(RNAi) for 24 hours prior to extracting embryos for analysis. Images were taken every 5 seconds, frame rate 12 fps. Time indicated in mm:ss.
Movie S5.
2- to 4-cell stage progression of C. elegans embryos with microtubule-binding deficient SAS-5 variants. Shown are DIC movies of embryos (approximately 50 μm-long) from non-transgenic wild-type worms (M2), or worms expressing GFP-SAS-5 WT (M3), Δ81-90 (M4) or K65E/K66E/K67E (M5) variants. All strains were subjected to sas-5(RNAi) for 24 hours prior to extracting embryos for analysis. Images were taken every 5 seconds, frame rate 12 fps. Time indicated in mm:ss.
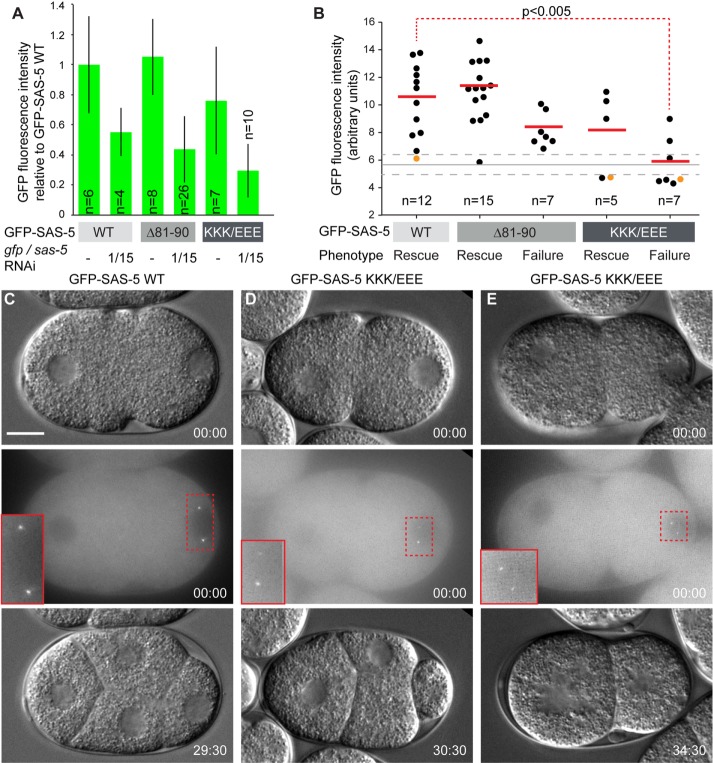
Next, we addressed whether differences between GFP-SAS-5 WT and microtubule-binding deficient variants might be revealed when expression levels were diminished. To this end, we subjected transgenic animals not only to sas-5 RNAi to deplete the endogenous protein, but also to gfp RNAi to reduce the levels of the GFP-SAS-5 fusion proteins. Analysis of embryos from animals expressing GFP-SAS-5 WT, Δ81–90, or K65E/K66E/K67E variants under such dual RNAi conditions showed GFP fluorescence intensity reduced by ∼50% compared with non-RNAi controls (Figure 6A). Nevertheless, both GFP-SAS-5 variants correctly localized to centrioles in all embryos examined (Figure 6, C–E). Analysis by time-lapse microscopy showed that, despite reduced levels of the fusion protein, GFP-SAS-5 WT consistently rescued the sas-5(RNAi) phenotype (Figure 6, B and C). In contrast, expression of GFP-SAS-5 Δ81–90 showed rescue in only ∼68% of embryos (Figure 6, B and D). It is worth noting that the partial RNAi rescue by this variant is in line with its partial microtubule-binding affinity demonstrated in vitro (Figure 5B). Pairwise Wilcoxon rank sum analysis showed no statistically significant difference (p > 0.05) between GFP fluorescence intensities of embryos with GFP-SAS-5 WT and embryos with GFP-SAS-5 Δ81–90 that failed to rescue the sas-5 RNAi phenotype, suggesting that failure to rescue is not related to sas-5 expression levels but rather due to the altered microtubule-interaction properties of GFP-SAS-5 Δ81–90. Expression of the GFP-SAS-5 K65E/K66E/K67E variant, which strongly weakens microtubule binding and slightly impairs the interaction with SAS-4, showed rescue in only ∼42% of embryos under these conditions (Figure 6, B and E). However, in this case, there was a statistical difference in GFP fluorescence intensities when comparing embryos with GFP-SAS-5 WT to those with GFP-SAS-5 K65E/K66E/K67E that failed to rescue (Figure 6B). As a result, we cannot ascertain whether the further impairment in function of GFP-SAS-5 K65E/K66E/K67E when compared with GFP-SAS-5 Δ81–90 reflects the additional reduction in microtubule binding, the weakened interaction with SAS-4, or merely the fact that overall protein levels are lower. Nevertheless, analysis of the GFP-SAS-5 Δ81–90 variant allows us to conclude that microtubule binding contributes to SAS-5 function in centriole duplication.
FIGURE 6:
SAS-5 microtubule-binding variants affect centriole duplication. (A) Average normalized GFP fluorescence intensities of transgenic embryos resulting from worms expressing GFP-SAS-5 WT or microtubule-binding deficient variants in the absence of RNAi or after sas-5 and gfp RNAi treatment, as indicated. Error bars correspond to one SD of measured cytoplasmic fluorescence intensities from multiple embryos as shown. GFP-SAS-5 K65E/K66E/K67E is abbreviated as KKK/EEE. (B) Dot plot of transgenic embryos expressing GFP-SAS-5 WT or microtubule-binding deficient variants under dual RNAi conditions; shown here are GFP fluorescence intensities of individual embryos and phenotypic outcomes (rescue or failure to rescue the sas-5 RNAi phenotype). The number of embryos monitored is indicated. There was a strong (R2 = 82%) correlation between GFP intensity levels in the cytoplasm and centrioles of embryos in which this could be reliably measured (n = 38). Red solid lines correspond to average GFP fluorescence intensity in each outcome class, the gray solid line to average background fluorescence measured in nontransgenic N2 embryos (n = 18), and gray dashed lines to one SD confidence intervals for background fluorescence. The red dashed line denotes pairwise Wilcoxon rank sum statistical analysis. The probability that GFP intensities are similar between the two indicated embryo classes is shown. No other pairwise analysis of WT vs. mutant sas-5 transgene GFP intensities expression approached statistical significance (p < 0.05); however, we acknowledge that given the limited embryo numbers, our ability to discern potential differences in protein levels, for instance in the subset of GFP-SAS-5 Δ81–90 embryos that failed to rescue, is restricted. (C–E) DIC (top panel) and GFP fluorescence images (middle panel, maximum-intensity z-projections) are shown for three embryos indicated in panel B by orange dots, and DIC images of the same embryos are shown ∼ 30 min later when the four-cell stage should have been attained (bottom panels). Panel C derives from embryos expressing GFP-SAS-5 WT; panels D and E derive from embryos expressing GFP-SAS-5 variant K65E/K66E/K67E and showing rescue of the sas-5 RNAi phenotype (D) or failure to do so (E). Insets in the middle panels are twofold magnifications of the indicated image areas. Time stamps are indicated in mm:ss. Scale bar, 10 μm. Embryos are oriented with the anterior on the left and the posterior on the right.
DISCUSSION
Microtubules are the most prominent structural feature of centrioles. Thus, it is not surprising that microtubule association is a relatively common feature of centriole components as illustrated by the identification of microtubule-binding sites in the centriole proteins Cep120 (Lin et al., 2013b), Bld10p/Cep135 (Carvalho-Santos et al., 2012; Lin et al., 2013a; Kraatz et al., 2016), Cep162 (Wang et al., 2013), and SAXO1 (Dacheux et al., 2015), among others. In this context, it is remarkable that all three evolutionarily conserved structural proteins essential for initiating centriole assembly, SAS-6 (Gupta et al., 2015), SAS-4/CPAP (Hsu et al., 2008; Sharma et al., 2016; Zheng et al., 2016), and now SAS-5, harbor microtubule-binding epitopes. This underscores both the importance of microtubule binding in general and the fact that microtubules are recruited very early during centriole formation.
Previous work and this study suggest some commonalities between the microtubule-binding properties of SAS-6, SAS-5, and SAS-4. In all three cases, the microtubule-binding epitopes comprise intrinsically disordered and positively charged protein segments, thereby enabling favorable electrostatic interactions with the negatively charged surface of the microtubule (Baker et al., 2001). Apparent affinities for microtubules are, in all cases, in the micromolar range (Hsu et al., 2008; Gupta et al., 2015), with SAS-4/CPAP also binding free tubulin with a tens of nanomolar dissociation constant (Sharma et al., 2016; Zheng et al., 2016). However, the mechanism by which SAS-5 gains microtubule affinity differs from that of SAS-4 and SAS-6. In contrast to the two latter components, the binding epitope of SAS-5 provides weak affinity in isolation, with strong microtubule association being possible only through SAS-5 oligomerization. Complexes between protein oligomers benefit from the cooperative engagement of multiple interaction epitopes, leading to strengthened associations in a process known as avidity (Mammen et al., 1998). Thus, we posit that formation of SAS-5 oligomers allows for the simultaneous interaction of multiple, independently weak binding epitopes with the microtubule lattice, thereby enhancing the overall strength of the SAS-5–microtubule association.
Avidity mechanisms may be of particular relevance to centriole formation, as all structural proteins at the beginning of the organelle assembly process form large oligomers. Indeed, previous biophysical analysis of SAS-5 and Ana2 had highlighted avidity arising from protein oligomerization as potentially beneficial to intermolecular interactions (Shimanovskaya et al., 2013; Slevin et al., 2014; Cottee et al., 2015; Rogala et al., 2015). Our present analysis clearly demonstrates the role of avidity on intermolecular interactions between centriole components. Further, our work raises the possibility that SAS-5 binding to SAS-6 (Qiao et al., 2012; Hilbert et al., 2013) and SAS-4 oligomers (Cottee et al., 2013; Hatzopoulos et al., 2013) may be influenced by avidity in a manner similar to the SAS-5–microtubule association. The central role of SAS-5 oligomerization for function has been demonstrated in vivo, where SAS-5 single amino acid substitutions that disrupt higher-order oligomerization completely abrogate centriole duplication (Rogala et al., 2015). It is likely that this effect reflects the simultaneous weakening of SAS-5 affinity for SAS-6, SAS-4, and microtubules and, hence, the perturbation of multiple processes involving SAS-5 interactions in centriole assembly.
We found that the SAS-4- and microtubule-binding epitopes of SAS-5 partly overlap, and that the vertebrate SAS-4 homologue CPAP cannot displace SAS-5 from microtubules in vitro. However, it should be noted that the estimated SAS-5–microtubule affinity (1–5 μM Kd) is comparable to the affinity of CPAP for the vertebrate SAS-5 relative STIL (∼0.5 μM Kd; Cottee et al., 2013; Hatzopoulos et al., 2013). Thus, we anticipate that in cells SAS-5 may associate with SAS-4 or microtubules depending on the local concentration of interaction sites and the structural context. In this manner SAS-5 could, for example, bind SAS-4 in the cytoplasm and assist its centriole localization, while preferentially binding microtubules in the context of centriole architecture.
We were able to derive a SAS-5 variant, Δ81–90, that attenuates microtubule affinity in vitro while preserving binding to the SAS-4 vertebrate homologue CPAP. Functional assays of SAS-5 Δ81–90 in C. elegans embryos upon depletion of endogenous SAS-5 revealed that at low protein levels this variant localized correctly at the site of centriole formation, but failed to assemble centrioles in ∼30% of cases. These data suggest that the partial disruption of microtubule binding in SAS-5 Δ81–90 leads to stochastic failure in correct centriole assembly. Consistent with this interpretation, overexpression of SAS-5 Δ81–90, which would counteract any reduction in the microtubule-interaction affinity, led to complete rescue of endogenous SAS-5 depletion. Thus, we conclude that the SAS-5–microtubule interaction contributes to robust centriole assembly.
A second SAS-5 variant, K65E/K66E/K67E, demonstrated stronger attenuation of the microtubule interaction compared with SAS-5 Δ81–90, with this association nearly abrogated in vitro. However, SAS-5 K65E/K66E/K67E also reduced SAS-4/CPAP binding by approximately twofold. Earlier analysis of the SAS-4/CPAP interaction with the SAS-5 relatives STIL and Ana2 showed little perturbation of centriole formation in Drosophila embryos even when this interaction was attenuated by ∼10-fold (Cottee et al., 2013); hence, we considered the approximately twofold reduction in CPAP affinity by SAS-5 K65E/K66E/K67E to be an acceptable compromise. Functional evaluation of this SAS-5 variant demonstrated failure of centriole formation in ∼60% of C. elegans embryos when exogenous protein expression levels were low. However, our assays could not distinguish whether the increased rate of failure in centriole assembly with SAS-5 K65E/K66E/K67E compared with SAS-5 Δ81–90 was due to the stronger effect on microtubule association or, alternatively, because of overattenuated protein levels in embryos. Hence, conclusive functional evaluation of SAS-5 K65E/K66E/K67E will necessitate finer control of protein expression levels in the transgenic organism; indeed, use of a transgenic expression system providing low but stable protein amounts would benefit the phenotypic analysis of all SAS-5 mutants.
How may microtubule binding assist the role of SAS-5 in centriole assembly? SAS-4/CPAP is a key protein for the formation of centriolar microtubules (Pelletier et al., 2006; Dammermann et al., 2008; Hsu et al., 2008; Sharma et al., 2016; Zheng et al., 2016). However, in most species, microtubule binding by SAS-4/CPAP is assisted by the centriolar protein Cep135/Bld10p (Ohta et al., 2002; Matsuura et al., 2004; Mottier-Pavie and Megraw, 2009). Cep135/Bld10p interacts with SAS-6 (Lin et al., 2013a) and microtubules (Carvalho-Santos et al., 2012; Lin et al., 2013a; Kraatz et al., 2016), and is thought to bridge the inner cartwheel with the exterior microtubule wall by being part of a structured assembly known as the “pinhead” (Hiraki et al., 2007; Guichard et al., 2013). Interestingly, no Cep135/Bld10p homologue has been identified in C. elegans (Carvalho-Santos et al., 2010; Hodges et al., 2010). Thus, our findings lead us to propose that SAS-5 assumes part of the Cep135/Bld10p role by contributing to the connection of a central SAS-6 scaffold with peripheral microtubules in nematodes.
A model of centriole formation in C. elegans thus emerges, whereby organelle assembly is initiated by the interdependent colocalization of SAS-6 and SAS-5 (Leidel et al., 2005; Pelletier et al., 2006; Lettman et al., 2013). SAS-6 may adopt an elongated ninefold symmetric spiral conformation (Hilbert et al., 2013) stabilized by direct interactions with a subpopulation of SAS-5 molecules (Qiao et al., 2012; Hilbert et al., 2013; Lettman et al., 2013; Rogala et al., 2015), while other SAS-5 copies continuously shuttle in and out of centrosomes (Delattre et al., 2004). Together, SAS-6 and SAS-5 form the central tube scaffold—the first visible centriole substructure in C. elegans (Pelletier et al., 2006; Sugioka et al., 2017). SAS-4, in turn, binds SAS-5 (Cottee et al., 2013; Hatzopoulos et al., 2013) and is recruited to the central tube (Pelletier et al., 2006), thereby enabling microtubule polymerization and centriole elongation. Finally, SAS-5 may contribute to the exceptional stability of C. elegans centrioles (Balestra et al., 2015) by forming further supporting links between SAS-6, SAS-4, and microtubules.
In summary, we report here a novel mechanism by which SAS-5 assists centriole formation. The SAS-5–microtubule interaction is enhanced by protein oligomerization via an avidity mechanism compromised in SAS-5 variants that disrupt protein oligomerization, thus impairing centriole assembly. Although sequence similarity between SAS-5 and its insect and vertebrate relatives is limited, SAS-5, Ana2, and STIL have thus far been shown to act in a comparable manner in centriole assembly and to feature similar interaction properties. Thus, it would be intriguing to examine whether Ana2 and STIL interact with microtubules as SAS-5 does.
MATERIALS AND METHODS
Protein recombinant expression and purification
C. elegans SAS-5 fragments were produced and purified as described previously (Rogala et al., 2015). Briefly, gene fragments encoding SAS-5 WT or variant constructs were cloned in a pFloat vector with a human rhinovirus (HRV) 3C protease-cleavable N-terminal hexahistidine-SUMO tag, and transformed into Escherichia coli strain BL21(DE3) Rosetta2 (Novagen). Cells were grown in lysogeny broth (LB) or M9 media supplemented with 13C-enriched glucose and 15N-enriched ammonium chloride at 37°C. At OD600 of 0.6, growth temperature was shifted to 18°C and protein expression was induced by isopropyl-β-d-1-thiogalactopyranoside (IPTG; 0.5 mM final concentration). Cells were harvested after 16 h, and resuspended in phosphate-buffered saline (PBS) solution (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) supplemented with 350 mM NaCl, disrupted by sonication, and the resulting cell lysates were clarified by centrifugation and filtration. Clarified lysates were applied to Ni-NTA (GE Healthcare) metal affinity resin, washed with PBS supplemented with 20 mM imidazole and eluted with PBS supplemented with 350 mM NaCl, 1 mM β-mercaptoethanol, and 500 mM imidazole. His6-tags were cleaved with homemade HRV 3C protease. Further purification was performed by anion-exchange and size exclusion chromatography. Pure protein fractions were buffer exchanged by dialysis, and concentrated by centrifugal ultrafiltration. Protein concentration was estimated by UV absorption at 280 nm, and protein identity confirmed by electrospray ionization mass-spectroscopy.
Danio rerio CPAPG-box (amino acids 943–1121) was produced and purified as described (Hatzopoulos et al., 2013). Briefly, gene fragments encoding the DrCPAPG-box domain were cloned in a pGEX-6P-2 vector (GE Healthcare) providing an N-terminal glutathione S-transferase (GST)-tag, and transformed into E. coli strain Rosetta(DE3)-pLysS. Cells were grown in LB media and protein expression induced by 0.3 mM IPTG at 18°C. Cells were resuspended in 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5% (vol/vol) glycerol buffer and lysed by lysozyme treatment and sonication. Lysates were clarified by centrifugation, incubated with glutathione-sepharose beads (GE Healthcare), washed and eluted in 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 12 mM reduced glutathione buffer. The GST-tag was cleaved with HRV 3C protease and DrCPAPG-box was further purified by size exclusion chromatography.
Microtubule-pelleting assays
Microtubule-pelleting assays were adapted from Campbell and Slep (2011). Bovine brain tubulin was purchased from the Centro de Investigaciones Biológicas (Microtubule Stabilizing Agents Group), CSIC, Madrid, Spain. Tubulin stock (10 mg/ml) was diluted to 1.1 mg/ml in BRB80 buffer (80 mM K-PIPES, pH 6.8, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid, 1 mM MgCl2) supplemented with 0.5 mM GTP and 1.25 mM dithiothreitol and kept on ice for 5 min. Microtubule polymerization was initiated by incubating the tubulin mix for 10 min at 37°C, followed by a stepwise addition of 0.1, 1, and 10 μM paclitaxel dissolved in dimethyl sulfoxide (Sigma-Aldrich) with 5 min incubation intervals between each step and a final incubation step of 10 min at 37°C. Polymerized microtubules (3 μM) were mixed with purified SAS-5 constructs predialyzed against BRB80 buffer, and incubated for 10 min at room temperature. Microtubules and SAS-5 constructs at corresponding concentrations were included in isolation as controls. Following centrifugation at 100,000 × g for 12 min, 25 μl of the supernatant was removed and supplemented with 5 μl 5× SDS loading dye. The remaining supernatant was discarded and pellets were resuspended in 120 μl BRB80 buffer and 25 μl 5× SDS loading dye. Supernatant and pellet fractions (30 μl) were analyzed by 12.5–15% (wt/vol) SDS–PAGE. Quantification of relative SAS-5 amounts in supernatant and pellet fractions was done using ImageJ (Schneider et al., 2012).
Electron microscopy
Microtubule-pelleting supernatant and pellet fractions were used for negative staining electron microscopy. Aliquots (5 μl) from dilution series were transferred to freshly UV activated homemade carbon-coated copper grids. After 20 s of incubation, excess liquid was removed by side blotting, and the grids were washed twice with BRB80 buffer and once with double-distilled H2O. Subsequently, the grid was stained three times with freshly prepared uranyl acetate (1–2% [wt/vol]) solution. Micrographs were taken using a JEM2200FS (JEOL) electron microscope operated at 200 kV and equipped with a TVIPS F416 CCD camera.
NMR assignments and binding assays
Sequence-specific NMR resonance assignments were performed as described previously (Mayer et al., 2012). Briefly, NMR experiments were performed using a home-built spectrometer with room-temperature inverse probe head and 17.6 T magnetic field strength. Samples of 13C/15N-enriched SAS-5N at 0.5 mM concentration in BRB80 buffer were supplemented with 5% (vol/vol) D2O, 0.02% (wt/vol) NaN3, and 50 μM 4,4-dimethyl-4-silapentane-1-sulfonic acid. Assignment experiments were performed at 10°C using 3D CBCA(CO)NH, CBCANH, HNCO, HBHA(CO)NH, and HBHANH pulse sequences. NMR data were processed using NMRpipe (Delaglio et al., 1995) and analyzed using CCPN analysis (Vranken et al., 2005). Assignments were deposited in BioMagResBank under accession number 27056. Spectra overlays were prepared with Sparky 3 (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Perturbations of resonance intensities were mapped at 10°C using 15N-HSQC experiments and samples in BRB80 buffer of 33 μM SAS-5N alone, in the presence of 33 μM tubulin or in the presence of 66 μM DrCPAPG-box. Perturbations of resonance intensities upon microtubule binding were mapped at 37°C using 13C-HSQC experiments and samples in BRB80 buffer of 33 μM SAS-5N alone or in the presence of 33 μM Taxol-stabilized microtubules prepared as described above. SAS-5N Hα-Cα resonances were tracked between 10 and 37°C using a series of 13C-HSQC variable temperature experiments collected in intervals of 5°C.
ITC
All recombinant proteins for ITC were prepared in 20 mM Na2HPO4, pH 7.5, 0.5 mM 2-mercaptoethanol buffer by extensive dialysis. ITC experiments using an iTC200 instrument (MicroCal) were performed at 25°C as follows: the cell (volume ∼200 μl) contained DrCPAPG-box at 35 μM concentration and the syringe (volume ∼40 μl) contained SAS-5N variants at 700 μM concentration. A first injection of 0.5 μl was followed by 19 injections of 2.0 μl with a delay between injections of 250 s. Blank titration was performed by injecting SAS-5N variants into buffer and resulting heats of dilution were subtracted from those of binding. Raw data were processed and fitted with MicroCal Origin software using a one-site model.
Cloning for mammalian cell culture
SAS-5 mammalian expression constructs were generated as previously described (Kriz et al., 2010; Mansouri et al., 2016). Briefly, all constructs except SAS-5 1–265 Δ91–100, K65E/K66E/K67E, and R86E/K88E/K94E were cloned with a hexahistidine tag followed by a thrombin cleavage site and thioredoxin into pSI-AKR1 vector (N-terminal mCherry-tag, Kanamycin resistance) using the SapI insertion site. SAS-5 1–265 Δ91–100, K65E/K66E/K67E, and R86E/K88E/K94E constructs were similarly cloned but derived from a synthetically produced codon-optimized DNA fragment that did not include hexahistidine and thioredoxin tags. Full-length SAS-5 was also cloned in a pHA-L11 vector providing an N-terminal hemagglutinin (HA) tag.
Mammalian cell culture, DNA transfection, fixation, and staining
COS-7 cells were incubated at 37°C in a humidified atmosphere containing 5% (vol/vol) CO2 and cultured in DMEM (Amimed) containing 10% (vol/vol) fetal calf serum (FCS; Life Technologies) without antibiotics. HEK293T cells were cultured at 37°C in 5% (vol/vol) CO2 in DMEM/Hams-F10 (50/50%) containing 10% (vol/vol) FCS and 1% (wt/vol) penicillin/streptomycin. One day before transfection, cells were transferred into six well plates at a density of ∼40% per well on glass coverslips. Cells were transfected with 0.02 μg/μl plasmid DNA using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s recommendation.
Cells were fixed 24–30 h after transfection by transferring the coverslips into 4% (wt/vol) formaldehyde in PBS for 20 min, and then washing three times with PBS. Cells were permeabilized with 1% (wt/vol) NP40 in PBS for 10 min and then incubated with mouse anti–α-tubulin (Sigma-Aldrich; 1:300 dilution) primary antibody for 2 h. All antibodies were diluted in PBS. After three washing steps with PBS goat anti-mouse DyLight488 or Cy5 (Jackson ImmunoResearch, Suffolk, UK; 1:150 dilution), secondary antibodies were incubated with the cells for 1.5 h. Alternatively, cells were incubated with rabbit anti-HA (Santa Cruz; 1:200 dilution) in addition to mouse anti–α-tubulin as primary antibodies, and goat anti-mouse Alexa488 (Molecular Probes; 1:1000 dilution) and donkey anti-rabbit Cy3 (Dianova; 1:1000 dilution) as secondary antibodies. Slides were counterstained with 1 μg/μl Hoechst 33258 (Sigma-Aldrich) to reveal DNA.
Samples were mounted in Gelvatol and 10–30 cells analyzed on a confocal laser-scanning microscope (SP5; Leica Microsystems, Germany) using a 63× HCX PL APO CS oil objective with a 1.4 NA. The sequential scanning mode was chosen and the number of overexposed pixels was kept at a minimum. Representative maximum projections of each channel and a merge image were done using ImageJ (Schneider et al., 2012).
Cold treatment of cells for microtubule depolymerization was performed 24 h posttransfection by incubating cells on ice for 30 min, followed by fixation and staining as described above.
Nematode strains and RNA interference
Nematode culture was according to standard procedures (Brenner, 1974). RNAi-resistant sas-5 variants were generated and cloned in pIC26 vector as described previously (Rogala et al., 2015). pIC26 contains a pie-1 promoter and 3′ untranslated region, as well as a GFP coding sequence fused upstream of and in frame with sas-5 (Cheeseman and Desai, 2005). Transgenic animals were generated by bombardment (Praitis et al., 2001). Integrated lines were recovered after bombardment with gfp::sas-5 Δ81–90 (strain GZ1332; isIs54{pie-1::gfp::sas-5[recΔ81–90]}) and K65E/K66E/K67E (strain GZ1335; isIs53{pie-1::gfp::sas-5[rec5K65E/K66E/K67E]}). The nonintegrated line expressing gfp::sas-5 WT (strain GZ1300; isEx5{pie-1::gfp::sas-5[rec]}) has been described previously (Rogala et al., 2015). RNAi was carried out by selecting L3-L4 WT hermaphrodites and feeding them for 26 h at 24°C using the sas-5(RNAi) feeding strain targeting nucleotides 301–1170 of the genomic sequence (Delattre et al., 2004), or a gfp(RNAi) feeding strain.
C. elegans immunofluorescence
Adult worms were dissected, and embryo fixation and immunofluorescence was carried out as previously described (Rogala et al., 2015). Briefly, rabbit anti–SAS-5 (Delattre et al., 2004) and mouse anti–α-tubulin (DM1A; Sigma-Aldrich) antibodies were used at 1/200 dilutions, followed by appropriate corresponding secondary antibodies and counterstaining with 1 μg/μl Hoechst 33258 to reveal DNA. Confocal imaging was carried out using a Zeiss LSM 700 microscope with a Plan-Apochromat 63× oil-immersion objective, NA 1.40. Z-sections were imaged at an interval of ∼0.3 μm. All images shown are maximum-intensity projections and were processed using Fiji (Schindelin et al., 2012) maintaining relative intensities within a series.
C. elegans live imaging
Time-lapse DIC microscopy of early C. elegans embryos was carried out as described (Gönczy et al., 1999), recording one image every 5 s at 24°C. For Supplemental Figure S7C, GFP z-stacks were taken at 0.5 μm intervals over 21 sections, using a Zeiss Axioplan 2 microscope (Bellanger and Gönczy, 2003). The motorized filter wheel, two external shutters, and the 1392 × 1040 pixel, 12-bit Photometrics CoolSNAP ES2 CCD camera (Photometrics, Tucson, AZ) were controlled by μManager (Edelstein et al., 2010). Images were acquired with an exposure time of 100 ms using the Zeiss Filter Set 10 (GFP). For quantification of fluorescence intensities, the mean intensity was measured from a single plane in the middle of each embryo. All images were processed and fluorescence intensities measured using ImageJ (Schneider et al., 2012). For Figure 6, embryos were imaged using a Zeiss AX10 microscope equipped with a Zeiss Plan-Apochromat 63× oil-immersion objective, NA 1.40, controlled by μManager. GFP z-stacks were taken at 0.5 μm intervals over 21 sections, and stacks were captured at 30 s intervals for time-lapse recordings. Quantification of fluorescence intensities in Figure 6 was carried out by taking the average projection of the GFP z-stack, and measuring the mean gray value over the whole embryo.
Western blotting
L4 worms were picked and matured for 16 h at 24°C. One hundred adult worms were then collected manually, boiled in Laemmli buffer for 5 min, sonicated, and loaded on a 4–12% (wt/vol) SDS–PAGE (Biorad, Hercules, CA). Proteins were transferred to a 0.45 μm nitrocellulose membrane, blocked with 5% (wt/vol) milk powder in PBS, 0.1% (vol/vol) Tween-20, and blotted with rabbit anti–SAS-5 (full length, 1/1000 dilution [ Delattre et al., 2004]). Immunocomplexes were visualized using anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibodies at 1/5000 dilution (Promega, Madison, WI), chemiluminescence kit (Roche), and x-ray films (Fujifilm, Tokyo, Japan).
Supplementary Material
Acknowledgments
We are grateful to David Staunton and Nick Soffe for maintenance of supporting facilities at Oxford Biochemistry, Coralie Busso for the generation of transgenic lines at École Polytechnique Fédérale de Lausanne, and Georgios Hatzopoulos, Natacha Olieric, and Virginie Hamel for critical reading of the manuscript. The pHA-L11 vector was a kind gift from S. Schmidt. We thank the Biotechnology and Biological Sciences Research Council UK (Grant no. BB/J008265/1 to I.V.), the Medical Research Council UK (Grant no. MR/N009274/1 to I.V.), the Swiss National Science Foundation (Grant no. 31003A_166608 to M.O.S.), and the European Research Council (Grant no. AdG 340227 to P.G.) for their support. K.B.R. was funded by a PhD studentship from the Engineering and Physical Sciences Research Council UK (Centre for Doctoral Training in Systems Biology, Oxford). The Oxford Biochemistry NMR facility was supported by the Wellcome Trust (Grant No. 094872/Z/10/Z), the Wellcome Institutional Strategic Support Fund, the EPA Cephalosporin Trust, and the John Fell OUP Research Fund.
Abbreviations used:
- DIC
differential interference contrast
- DrCPAPG-box
G-box domain of Danio rerio CPAP
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- KKK/EEE
GFP-SAS-5 K65E/K66E/K67E
- NMR
nuclear magnetic resonance
- RNAi
RNA interference
- SAS-52-265
SAS-5 amino acids 2–265
- SAS-5N
SAS-5 amino acids 2–122.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0412) on January 24, 2018.
REFERENCES
- Alvarez-Cabrera AL, Delgado S, Gil-Carton D, Mortuza GB, Montoya G, Sorzano CO, Tang TK, Carazo JM. (2017). Electron microscopy structural insights into CPAP oligomeric behavior: a plausible assembly process of a supramolecular scaffold of the centrosome. Front Mol Biosci , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Nigg EA. (2014). Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci , doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. (2010). Building the centriole. Curr Biol , R816–R825.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA , 10037–10041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra FR, von Tobel L, Gönczy P. (2015). Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res , 642–644.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banterle N, Gonczy P. (2017). Centriole biogenesis: from identifying the characters to understanding the plot. Annu Rev Cell Dev Biol , 23–49.. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Gönczy P. (2003). TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol , 1488–1498.. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. (2011). Centrosomes and cilia in human disease. Trends Genet , 307–315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. (2012). The centrosome in cells and organisms. Science , 422–426. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics , 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Slep KC. (2011). αβ-Tubulin and microtubule-binding assays. Methods Mol Biol , 87–97.. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Alvarez-Martins I, Gouveia SM, Jana SC, Duarte P, Amado T, Branco P, Freitas MC, Silva ST, et al. (2012). BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell , 412–424.. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. (2010). Stepwise evolution of the centriole-assembly pathway. J Cell Sci , 1414–1426.. [DOI] [PubMed] [Google Scholar]
- Chavali PL, Putz M, Gergely F. (2014). Small organelle, big responsibility: the role of centrosomes in development and disease. Philos Trans R Soc Lond B Biol Sci , doi: 10.1098/rstb.2013.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. (2005). A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE , pl1. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW. (2015). Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol , 611–624.. [DOI] [PubMed] [Google Scholar]
- Cormier A, Clement MJ, Knossow M, Lachkar S, Savarin P, Toma F, Sobel A, Gigant B, Curmi PA. (2009). The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J Biol Chem , 6909–6917.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Johnson S, Raff JW, Lea SM. (2017). A key centriole assembly interaction interface between human Plk4 and STIL appears to not be conserved in flies. Biol Open , 381–389.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Johnson S, Leveson J, Raff JW, Lea SM. (2015). The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. Elife , e07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, Andreeva A, Oegema K, Lea SM, Raff JW, van Breugel M. (2013). Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife , e01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts EE, Inglis A, Stansfeld PJ, Vakonakis I, Hatzopoulos GN. (2015). The centriolar protein CPAP G-box: an amyloid fibril in a single domain. Biochem Soc Trans , 838–843.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux D, Roger B, Bosc C, Landrein N, Roche E, Chansel L, Trian T, Andrieux A, Papaxanthos-Roche A, Marthan R, et al. (2015). Human FAM154A (SAXO1) is a microtubule-stabilizing protein specific to cilia and related structures. J Cell Sci , 1294–1307.. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. (2008). SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J Cell Biol , 771–785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell , 815–829.. [DOI] [PubMed] [Google Scholar]
- David A, Amartely H, Rabinowicz N, Shamir M, Friedler A, Izraeli S. (2016). Molecular basis of the STIL coiled coil oligomerization explains its requirement for de-novo formation of centrosomes in mammalian cells. Sci Rep , 24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR , 277–293.. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gönczy P. (2006). Sequential protein recruitment in C. elegans centriole formation. Curr Biol , 1844–1849.. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gönczy P. (2004). Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol , 656–664.. [DOI] [PubMed] [Google Scholar]
- Dong G. (2015). Building a ninefold symmetrical barrel: structural dissections of centriole assembly. Open Biol , doi: 10.1098/rsob.150082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. (2010). Computer control of microscopes using microManager. Curr Protoc Mol Biol, doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Stearns T. (2014). The centriole duplication cycle. Philos Trans R Soc Lond B Biol Sci , doi: 10.1098/rstb.2013.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2012). Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol , 425–435. [DOI] [PubMed] [Google Scholar]
- Gönczy P. (2015). Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer , 639–652. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. (1999). Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol , 927–946.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gönczy P. (2012). Cartwheel architecture of Trichonympha basal body. Science , 553. [DOI] [PubMed] [Google Scholar]
- Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Flückiger I, Yamada A, Kihara K, Nishida Y, et al. (2013). Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr Biol , 1620–1628.. [DOI] [PubMed] [Google Scholar]
- Guichard P, Hamel V, Le Guennec M, Banterle N, Iacovache I, Nemcikova V, Fluckiger I, Goldie KN, Stahlberg H, Levy D, et al. (2017). Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Nat Commun , 14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta H, Badarudeen B, George A, Thomas GE, Gireesh KK, Manna TK. (2015). Human SAS-6 C-terminus nucleates and promotes microtubule assembly in vitro by binding to microtubules. Biochemistry , 6413–6422.. [DOI] [PubMed] [Google Scholar]
- Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, Stansfeld PJ, Vakonakis I. (2013). Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure , 2069–2077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert M, Erat MC, Hachet V, Guichard P, Blank ID, Flückiger I, Slater L, Lowe ED, Hatzopoulos GN, Steinmetz MO, et al. (2013). Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc Natl Acad Sci USA , 11373–11378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Flückiger I, Jaussi R, et al. (2016). SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat Cell Biol , 393–403.. [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. (2007). Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol , 1778–1783.. [DOI] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. (2010). Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci , 1407–1413.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WB, Hung LY, Tang CJ, Su CL, Chang Y, Tang TK. (2008). Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp Cell Res , 2591–2602.. [DOI] [PubMed] [Google Scholar]
- Jana SC, Marteil G, Bettencourt-Dias M. (2014). Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr Opin Cell Biol , 96–106.. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, Steinmetz MO. (2011). Structural basis of the 9-fold symmetry of centrioles. Cell , 364–375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraatz S, Guichard P, Obbineni JM, Olieric N, Hatzopoulos GN, Hilbert M, Sen I, Missimer J, Gönczy P, Steinmetz MO. (2016). The human centriolar protein CEP135 contains a two-stranded coiled-coil domain critical for microtubule binding. Structure , 1358–1371.. [DOI] [PubMed] [Google Scholar]
- Kriz A, Schmid K, Baumgartner N, Ziegler U, Berger I, Ballmer-Hofer K, Berger P. (2010). A plasmid-based multigene expression system for mammalian cells. Nat Commun , 120. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. (2005). SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol , 115–125.. [DOI] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. (2013). Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell , 284–298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK. (2013a). Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J , 1141–1154.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJ, Chang CW, Tang TK. (2013b). CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol , 211–219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen M, Choi SK, Whitesides GM. (1998). Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed , 2755–2794.. [DOI] [PubMed] [Google Scholar]
- Mansouri M, Bellon-Echeverria I, Rizk A, Ehsaei Z, Cianciolo Cosentino C, Silva CS, Xie Y, Boyce FM, Davis MW, Neuhauss SC, et al. (2016). Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat Commun , 11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M. (2004). Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol , 663–671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Slater L, Erat MC, Konrat R, Vakonakis I. (2012). Structural analysis of the plasmodium falciparum Erythrocyte membrane protein 1 (PfEMP1) intracellular domain reveals a conserved interaction epitope. J Biol Chem , 7182–7189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL. (2009). Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell , 2605–2614.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. (2015). Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol , 863–878.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. (2007). SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol , 2169–2174.. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell , 663–678.. [DOI] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. (2014). Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun , 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. (2002). Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol , 87–99.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. (2006). Centriole assembly in Caenorhabditis elegans. Nature , 619–623.. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics , 1217–1226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. (2012). SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. EMBO J , 4334–4347.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala KB, Dynes NJ, Hatzopoulos GN, Yan J, Pong SK, Robinson CV, Deane CM, Gönczy P, Vakonakis I. (2015). The Caenorhabditis elegans protein SAS-5 forms large oligomeric assemblies critical for centriole formation. Elife , e07410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, Jaussi R, Grigoriev I, Croisier M, Kammerer RA, Akhmanova A, et al. (2016). Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev Cell , 362–376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanovskaya E, Qiao R, Lesigang J, Dong G. (2013). The SAS-5 N-terminal domain is a tetramer, with implications for centriole assembly in C. elegans. Worm , e25214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol , 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin LK, Romes EM, Dandulakis MG, Slep KC. (2014). The mechanism of dynein light chain LC8-mediated oligomerization of the Ana2 centriole duplication factor. J Biol Chem , 20727–20739.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka K, Hamill DR, Lowry JB, McNeely ME, Enrick M, Richter AC, Kiebler LE, Priess JR, Bowerman B. (2017). Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. Elife , e20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. (2011). The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J , 4790–4804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. (2009). Primary microcephaly: do all roads lead to Rome. Trends Genet , 501–510.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. (2011). Structures of SAS-6 suggest its organization in centrioles. Science , 1196–1199.. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Wilcken R, McLaughlin SH, Rutherford TJ, Johnson CM. (2014). Structure of the SAS-6 cartwheel hub from Leishmania major. Elife , e01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venghateri JB, Jindal B, Panda D. (2015). The centrosome: a prospective entrant in cancer therapy. Expert Opin Ther Targets , 957–972.. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins , 687–696.. [DOI] [PubMed] [Google Scholar]
- Wang C, Li S, Januschke J, Rossi F, Izumi Y, Garcia-Alvarez G, Gwee SS, Soon SB, Sidhu HK, Yu F, et al. (2011). An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev Cell , 520–533.. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Tsou MF. (2013). CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat Cell Biol , 591–601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O’Toole E. (2014). Centriole structure. Philos Trans R Soc Lond B Biol Sci , doi: 10.1098/rstb.2013.0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gooi LM, Wason A, Gabriel E, Mehrjardi NZ, Yang Q, Zhang X, Debec A, Basiri ML, Avidor-Reiss T, et al. (2014). Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc Natl Acad Sci USA , E354–E363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Ramani A, Soni K, Gottardo M, Zheng S, Ming Gooi L, Li W, Feng S, Mariappan A, Wason A, et al. (2016). Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat Commun , 11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.