Abstract
In Schizosaccharomyces pombe, cytokinesis requires the assembly and constriction of an actomyosin-based contractile ring (CR). A single essential formin, Cdc12, localizes to the cell middle upon mitotic onset and nucleates the F-actin of the CR. Cdc12 medial recruitment is mediated in part by its direct binding to the F-BAR scaffold Cdc15. Given that Cdc12 is hyperphosphorylated in M phase, we explored whether Cdc12 phosphoregulation impacts its association with Cdc15 during mitosis. We found that Cdk1, a major mitotic kinase, phosphorylates Cdc12 on six N-terminal residues near the Cdc15-binding site, and phosphorylation on these sites inhibits its interaction with the Cdc15 F-BAR domain. Consistent with this finding, a cdc12 mutant with all six Cdk1 sites changed to phosphomimetic residues (cdc12-6D) displays phenotypes similar to cdc12-P31A, in which the Cdc15-binding motif is disrupted; both show reduced Cdc12 at the CR and delayed CR formation. Together, these results indicate that Cdk1 phosphorylation of formin Cdc12 antagonizes its interaction with Cdc15 and thereby opposes Cdc12’s CR localization. These results are consistent with a general role for Cdk1 in inhibiting cytokinesis until chromosome segregation is complete.
INTRODUCTION
Cytokinesis, the final stage in cell division, results in the physical separation of two daughter cells. This event is accomplished in many eukaryotic cells by an actin- and myosin-based contractile ring (CR) that forms and constricts between the two segregated genomes. In Schizosaccharomyces pombe, CR assembly depends on a single formin, Cdc12, which nucleates, elongates, and bundles F-actin of the CR (Nurse et al., 1976; Chang et al., 1997; Kovar et al., 2003; Kovar and Pollard, 2004; Bohnert et al., 2013).
Cdc12 targeting to the CR at mitotic onset depends on two redundant genetic modules (Wachtler et al., 2006; Laporte et al., 2011). The first module consists of IQGAP Rng2 and non-muscle myosin II (Myo2, Cdc4, and Rlc1), while the second module consists of the F-BAR scaffold Cdc15 (Laporte et al., 2011). Although the molecular mechanism by which the first module recruits Cdc12 is still unknown, Cdc15’s F-BAR domain interacts directly with Cdc12 and recruits it to the CR. We identified the Cdc15-binding motif within the Cdc12 N-terminus (residues 24–36) and also found that a cdc12-P31A mutant, in which the Cdc15-binding motif on Cdc12 is disrupted, had reduced Cdc12 CR localization, delayed medial F-actin accumulation and CR formation, and decreased viability upon CR perturbation (Willet et al., 2015). Thus, the Cdc12–Cdc15 interaction is an important contributor to Cdc12 localization and CR formation.
Cdc15 is a member of the F-BAR family of proteins, which oligomerize and bind membranes through their F-BAR domains (Tsujita et al., 2006; McDonald et al., 2015; McDonald and Gould, 2016) and link the membrane to other proteins (Roberts-Galbraith et al., 2009; Bohnert and Gould, 2012; Ren et al., 2015). Cdc15 activity is under strong cell cycle–dependent phosphoregulation: Cdc15 is hyperphosphorylated during interphase, but hypophosphorylated in mitosis (Fankhauser et al., 1995). Cdc15 hyperphosphorylation inhibits its membrane binding, oligomerization, and binding to protein partners. Cdc15 dephosphorylation, in contrast, induces an open conformation that supports these activities (Roberts-Galbraith et al., 2010; McDonald et al., 2015). Cdc12 and Cdc15 associate when Cdc15 is hypophosphorylated; interestingly, cdc15 alleles with phospho-abolishing mutations precociously recruit Cdc12 and other interacting proteins to the cell middle (Roberts-Galbraith et al., 2010). Thus, while Cdc15 phosphoregulation controls its interaction with Cdc12 and other binding partners, whether or not Cdc12 is also regulated to control its interaction with Cdc15 was unknown.
Like Cdc15, Cdc12 is phosphorylated in a cell cycle–dependent manner; but, unlike Cdc15, its hyperphosphorylation occurs during mitosis, not interphase (Bohnert et al., 2013). Sid2, the terminal kinase in the septation initiation network (SIN), phosphorylates Cdc12 on four residues to regulate a C-terminal oligomerization domain (Bohnert et al., 2013). When all four Sid2 phospho-sites are mutated to phospho-abolishing residues, Cdc12 is still phosphorylated in vivo (Bohnert et al., 2013), suggesting that other kinases must also phosphorylate Cdc12.
Cdk1 (cyclin-dependent kinase) is a master controller of the cell cycle. Though Cdk1 is required for mitotic commitment, cytokinesis is not initiated until later in mitosis when Cdk1 activity is low (He et al., 1997; Wheatley et al., 1997; Guertin et al., 2000; Niiya et al., 2005; Potapova et al., 2006; Wolf et al., 2007; Dischinger et al., 2008; Bloom et al., 2011). As low Cdk1 activity is a hallmark of mitotic exit, Cdk1 is commonly regarded as a cytokinetic inhibitor (reviewed in Wolf et al., 2007; Bohnert and Gould, 2011). A recent study identified threonine 95 on Cdc12 as a Cdk1 target site (Swaffer et al., 2016), yet the role of this phosphorylation was unknown.
Here we show that Cdk1 phosphorylates Cdc12 on six N-terminal residues, inhibiting its interaction with the Cdc15 F-BAR domain. Cells producing only Cdc12-6D, which has phospho-mimetic residues at the six Cdk1 sites, display phenotypes similar to those of cdc12-P31A cells, which lack the Cdc15-binding motif on Cdc12; both have reduced Cdc12 accumulation at the cell division site and delayed CR formation. These results underscore the multitiered regulation of formin activity during cytokinesis and are consistent with a role for Cdk1 in inhibiting cytokinesis until chromosome segregation is complete.
RESULTS and discussion
Cdc12 is a Cdk1 substrate
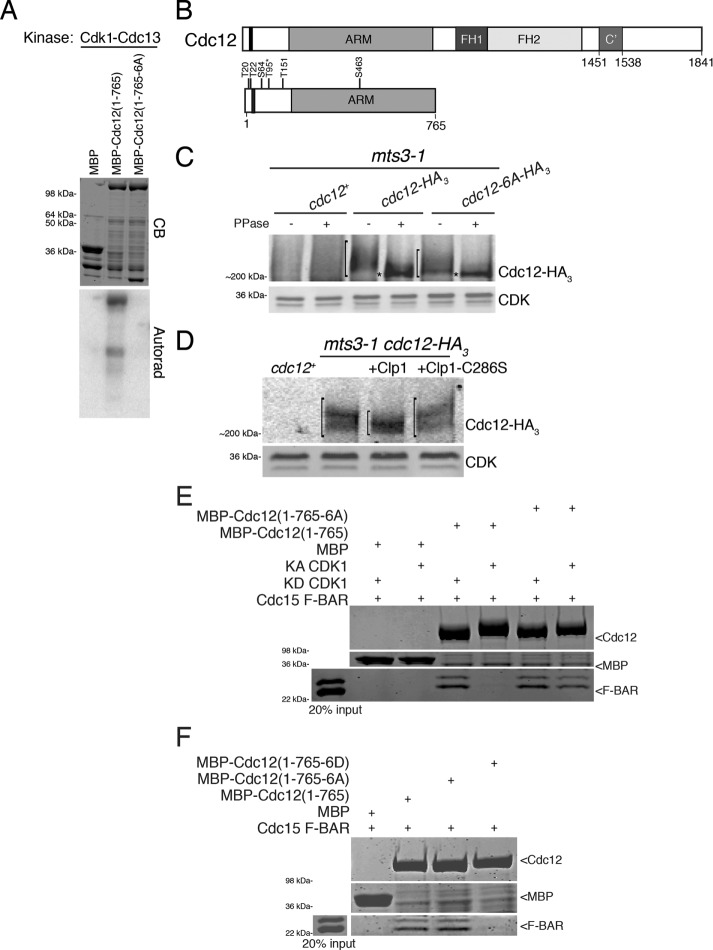
Loss of Sid2 activity only partially eliminated Cdc12 phosphorylation (Bohnert et al., 2013), indicating that other kinases also phosphorylate Cdc12. Peak Cdc12 phosphorylation correlates temporally with high Cdk1 activity, and threonine 95 on Cdc12 was identified in a large-scale phosphoproteomics screen designed to identify Cdk1 substrates (Swaffer et al., 2016). Another phosphoproteomics screen identified a host of mitotic Cdc12 phosphopeptides (Koch et al., 2011). Thus, we investigated whether Cdk1 is involved in Cdc12 phosphoregulation. The Cdc12 N-terminus was robustly phosphorylated by Cdk1 in vitro (Figure 1, A and B). In accord with the known Cdk1 consensus site, all targeted residues were within S/T-P motifs (Supplemental Figure S1). Individual mutation of each Cdk1 consensus site identified residues T20, T22, S64, T151, and S463 and confirmed T95 as major Cdk1-targeted residues (Supplemental Figure S1, B and C). When all six phosphosites were mutated to alanine, Cdk1-mediated Cdc12 phosphorylation was abolished (Figure 1, A and B). We constructed an endogenously expressed cdc12 allele in which all six phosphorylated residues were mutated to alanine (cdc12-6A). When immunoprecipitated from mitotically arrested cells, we detected a reduced mobility shift for Cdc12-6A-HA3 in comparison to Cdc12-HA3 (Figure 1C), indicating that these residues are in vivo phosphosites. Consistent with Cdk1 phosphorylating Cdc12 in vivo, Cdc12-HA3 showed increased gel mobility when incubated with recombinant MBP-Clp1 in comparison with Cdc12-HA3 not incubated with protein or incubated with an inactive form of Clp1 (Figure 1D). Clp1 is a phosphatase that dephosphorylates Cdk1 phosphosites (Gray et al., 2003; Stegmeier and Amon, 2004; Clifford et al., 2008; Mocciaro et al., 2010; Chen et al., 2013). Taking these results together, we conclude that Cdk1 phosphorylates Cdc12 during mitosis.
FIGURE 1:
Cdc12 is a Cdk1 substrate. (A) In vitro kinase assay using Cdk1–Cdc13 purified from insect cells and bacterially produced MBP, MBP-Cdc12(1–765), and MBP-Cdc12(1–765-6A) fragments. The gel was stained with Coomassie blue (CB) and proteins labeled by γ–P32 were detected by autoradiography. (B) Schematic of Cdc12 with formin homology (FH) domains 1 and 2, armadillo repeats (ARM), oligomerization domain (C’), and Cdk1-targeted residues labeled. The Cdc15-binding motif (residues 24–36) is indicated by the black bar. T95, identified by Swaffer et al. (2016), is labeled with an asterisk. (C, D) Denatured cell lysates were prepared from the indicated mitotically arrested mts3-1 strains. Anti-HA immunoprecipitates of the samples were (C) subjected to lambda phosphatase treatment or buffer control or (D) incubated with MBP-Clp1, MBP-Clp1-C286S, or buffer control. Samples were resolved by SDS–PAGE and immunoblotted. CDK served as loading control. Brackets span phosphorylated species and asterisks mark hypophosphorylated species of Cdc12. (E) In vitro binding assay of bead-bound recombinant MBP, MBP-Cdc12(1–765), or MBP-Cdc12(1–765-6A) with recombinant Cdc15 F-BAR(19–312) incubated with either kinase active (KA) or kinase dead (KD) Cdk1–Cdc13. Uncropped images are in Supplemental Figure S2B. (F) In vitro binding assay of bead-bound recombinant MBP, MBP-Cdc12(1–765), MBP-Cdc12(1–765-6A), or MBP-Cdc12(1–765-6D) with recombinant Cdc15 F-BAR(19–312). Uncropped images are in Supplemental Figure S2C. (E, F) Samples were washed, resolved by SDS–PAGE, and stained with CB.
Cdk1 phosphorylation of Cdc12 inhibits the Cdc12–Cdc15 interaction
Because Cdk1 phosphorylation sites on Cdc12 are near the Cdc15-binding motif (Figure 1B; Willet et al., 2015), we investigated whether Cdc12 phosphorylation affects its binding to Cdc15 F-BAR. In vitro, MBP-Cdc12(1–765) bound Cdc15 F-BAR (Figure 1E; Carnahan and Gould, 2003; Willet et al., 2015), which runs as a double band due to incomplete His6-tag cleavage (Supplemental Figure S2A). However, incubation of MBP-Cdc12(1–765) with Cdk1 prevented Cdc15 F-BAR binding (Figure 1E; Supplemental Figure S2B), suggesting that phosphorylation at the Cdk1-targeted residues blocks this interaction. Consistent with this interpretation, Cdk1 no longer affected the interaction when all six Cdk1 phosphosites on Cdc12 were mutated to alanine (Figure 1E; Supplemental Figure S2B). Further, a Cdc12 fragment with all six Cdk1 sites mutated to phosphomimetic aspartate residues did not bind Cdc15 F-BAR in vitro (Figure 1F; Supplemental Figure S2C). These results indicate that Cdk1 phosphorylates Cdc12 on six residues to preclude Cdc15 binding.
Cells with constitutive inhibition of the Cdc12–Cdc15 interaction are prone to cytokinesis failure
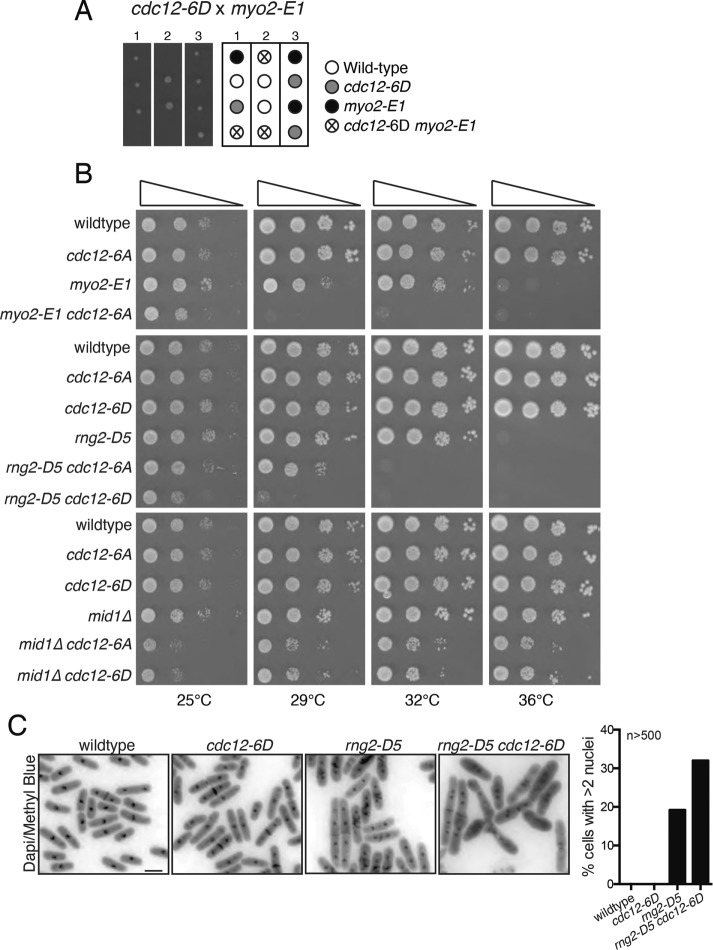
To determine the functional consequence of abolishing or constitutively mimicking Cdc12 N-terminal phosphorylation by Cdk1 in cells, we examined cdc12 alleles in which Cdc12’s six N-terminal Cdk1 phosphorylation sites were mutated to either alanines (cdc12-6A) or aspartates (cdc12-6D). On the basis of the in vitro results, we expected that the cdc12-6D allele might cause phenotypes similar to the cdc12-P31A allele, which disrupted Cdc12’s association with Cdc15 and was synthetically lethal with myo2-E1, rng2-D5, and mid1∆ (Willet et al., 2015). As expected, cdc12-6D was synthetically lethal with myo2-E1 (Figure 2A) and synthetically sick with rng2-D5 and mid1∆ (Figure 2B). DAPI staining of wildtype, cdc12-6D, rng2-D5, and cdc12-6D rng2-D5 revealed that the double mutant had a higher percentage of multiple nuclei indicative of cytokinesis failure than the wild type and single mutants (Figure 2C). Contrary to expectation, cdc12-6A also displayed negative genetic interactions with myo2-E1, rng2-D5, and mid1∆ (Figure 2B), although these were much milder than those of cdc12-6D. It could be that 1) improper temporal regulation of the Cdc12–Cdc15 interaction is detrimental to cytokinesis, 2) Cdk1-mediated phosphorylation at these sites must be dynamic to support correct Cdc12 function during mitosis, 3) phosphorylation affects another unknown Cdc12 interaction, or 4) the 6A mutants negatively affect the overall structure and function of full-length Cdc12.
FIGURE 2:
Genetic interactions of cdc12-6A and cdc12-6D. (A) Tetrads from cdc12-6D crossed to myo2-E1 shown with a schematic of relevant genotypes. (B) Cells of the indicated genotypes were spotted on YE media in 10-fold serial dilution, and plates were imaged after incubation for 3 d at the indicated temperatures. (C) The indicated strains were grown at 25°C and shifted to 36°C for 4 h before fixing and staining. Representative images are shown on the left and the percentage of cells with more than two nuclei is quantified on the right. n ≥ 500 for each strain. Bar, 5 μm.
Cdk1-dependent regulation of the Cdc12–Cdc15 interaction is important for Cdc12 recruitment
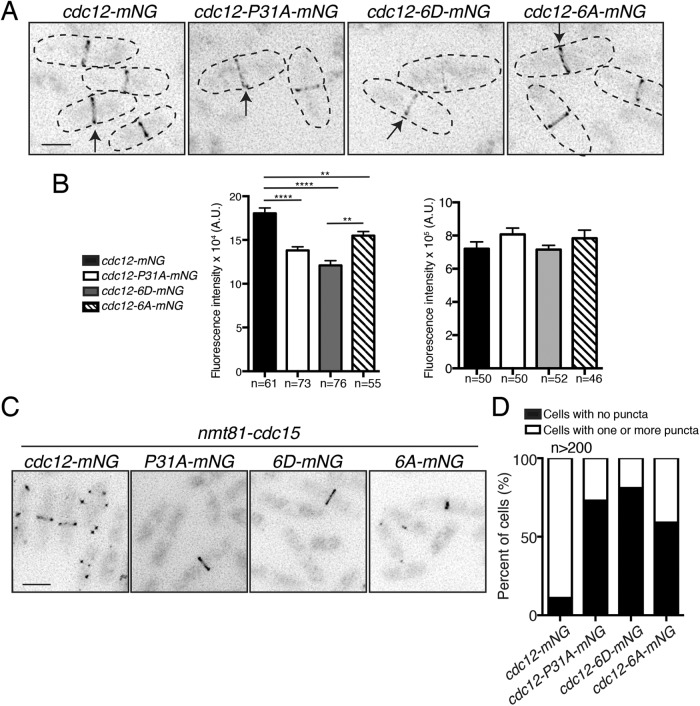
Cdc12–Cdc15 binding is important in recruiting Cdc12 to the CR (Laporte et al., 2011; Willet et al., 2015). To test whether Cdk1 phosphorylation of Cdc12 influences Cdc12’s CR localization, we tagged wild-type and mutant alleles with a single copy of mNeonGreen (mNG) (Shaner et al., 2013). As expected, Cdc12-6D-mNG had substantially decreased CR localization, even compared with Cdc12-P31A-mNG (Figure 3, A and B), and Cdc12-6A-mNG CR localization was intermediate between wild type and Cdc12-P31A-mNG (Figure 3, A and B). However, there was no difference among strains in total Cdc12 protein levels in mitotic cells (Figure 3, A and B). These data are consistent with Cdk1 phosphorylation of Cdc12 modulating Cdc12 targeting to the CR in vivo.
FIGURE 3:
Cdk1 phosphorylation of Cdc12 inhibits its CR localization. (A) Live-cell imaging of endogenously tagged cdc12-mNG, cdc12-P31A-mNG, cdc12-6D-mNG, and cdc12-6A-mNG. Arrows indicate CR localization of Cdc12. (B) Quantification of Cdc12 fluorescence intensity in the CR (left) and whole cell (right) from images of the indicated genotypes. A.U. = arbitrary units. (In right graph, cdc12 vs. cdc12-P31A p = 0.13; wt vs. cdc12-6D p = 0.92; and wt vs. cdc12-6A p = 0.33.) Measurements from three biological replicates. In left graph: **p ≤ 0.01 and ****p ≤ 0.0001, one-way ANOVA. Error bars represent SEM. (C) Cdc12-mNG localization in cells overexpressing cdc15 from the nmt81 promoter for 20 h at 32°C. (D) Quantification of the images from C. Bars in A and C, 5 μm.
Cdc15 also influences Cdc12 localization in abnormal cell cycle situations (Carnahan and Gould, 2003; Roberts-Galbraith et al., 2010). For example, cdc15 overexpression results in the formation of large puncta of Cdc12 (Carnahan and Gould, 2003). As previously reported, the P31A mutation in cdc12 prevents puncta formation because it disrupts the Cdc15–Cdc12 interaction (Figure 3, C and D; Willet et al., 2015). Consistent with Cdk1 phosphorylation inhibiting Cdc12 medial recruitment, Cdc12-6D-mNG formed puncta in only 4% of cdc15-overexpressing cells. cdc12-6A-mNG cells displayed puncta more commonly, but less than cdc12-mNG cells (Figure 3, C and D). All strains overexpressed Cdc15 to approximately the same level (Supplemental Figure S2D). Thus, under both normal and abnormal conditions, Cdk1-dependent phosphorylation modulates the medial recruitment of Cdc12 by affecting its interaction with Cdc15.
Cdk1-dependent regulation of the Cdc12–Cdc15 interaction is important in the initial formation of F-actin
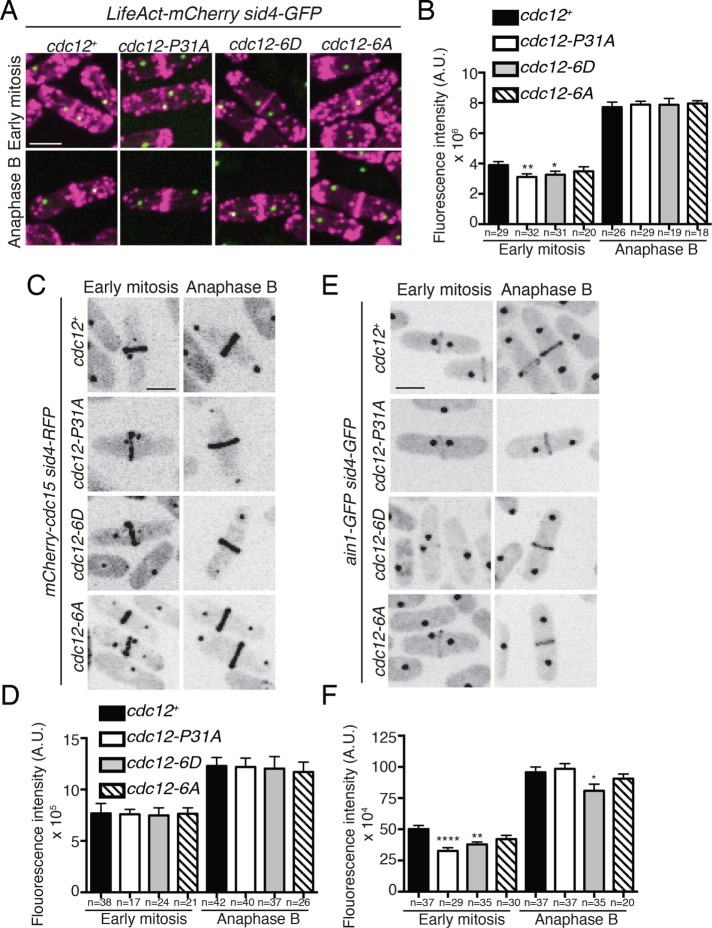
Previous findings showed that reduced Cdc12 recruitment to the division site results in less F-actin during early mitosis, but not anaphase B (Willet et al., 2015). We thus visualized F-actin with LifeAct-mCherry and compared the amount of F-actin in the CR of cdc12 phosphomutant cells during early mitosis and anaphase B. In early mitosis, there was 20% less F-actin in the CR of cdc12-P31A cells compared with wild type, and 16% less F-actin in the CR of cdc12-6D cells (Figure 4, A and B). However, there was no statistically significant difference of cdc12-6A cells from wild type (p = 0.29) during early mitosis (Figure 4, A and B). In addition, there was no statistically significant difference in the amount of F-actin in the CR between any of the strains during anaphase B (Figure 4, A and B). Thus, Cdc12-6D, similarly to Cdc12-P31A, initially produces less F-actin in the forming CR, but with time is able to reestablish typical F-actin levels.
FIGURE 4:
Mimicking constitutive Cdk1 phosphorylation on Cdc12 reduces F-actin in the CR. (A) Live-cell imaging of cdc12+, cdc12-P31A, cdc12-6D, and cdc12-6A cells expressing LifeAct-mCherry Sid4-GFP. (B) Quantification of fluorescence intensity of LifeAct-mCherry CRs in the indicated genotypes and cell cycle stage (cdc12+ vs. cdc12-6A early mitosis p = 0.29; cdc12+ vs. cdc12-P31A anaphase B p = 0.67; cdc12+ vs. cdc12-6A anaphase B p = 0.56; cdc12+ vs. cdc12-6D anaphase B p = 0.76). (C, E) Live-cell imaging of cdc12+, cdc12-P31A, cdc12-6D, and cdc12-6A cells expressing endogenously tagged mCherry-Cdc15 Sid4-RFP (C) or Ain1-GFP Sid4-GFP (E). (D, F) Quantification of fluorescence intensity of mCherry-Cdc15 (D) and Ain1-GFP (F) CRs in the indicated strain and cell cycle stage. (D) cdc12+ vs. cdc12-P31A anaphase B p = 0.63; cdc12+ vs. cdc12-6A anaphase B p = 0.37. (F) cdc12+ vs. cdc12-P31A early mitosis p = 0.95; cdc12+ vs. cdc12-6A early mitosis p = 0.97; cdc12+ vs. cdc12-6D early mitosis p = 0.88; cdc12+ vs. cdc12-P31A anaphase B p = 0.92; cdc12+ vs. cdc12-6A anaphase B p = 0.63; cdc12+ vs. cdc12-6D anaphase B p = 0.84. Measurements in the graphs from B, D, and F represent three biological replicates. Bars, 5 μm. *p ≤ 0.05, **p ≤ 0.01, and ****p ≤ 0.0001, one-way ANOVA. Error bars represent SEM. A.U. in panels B, D, and F = arbitrary units.
We expected that proteins targeted to the CR independently of Cdc12 and F-actin, such as Cdc15 (Wu et al., 2006), would not be affected in cdc12 phosphomutants. Indeed, mCherry-Cdc15 did not have altered protein levels in CRs of mutant cells compared to those of wild type at any mitotic stage (Figure 4, C and D). In contrast, proteins that localize to the CR in an F-actin–dependent manner might exhibit decreased CR abundance in early mitotic cdc12 phosphomutants. Indeed, during early mitosis, there was a 30% decrease in Ain1-GFP (alpha-actinin, Wu et al., 2001) abundance in the CRs of cdc12-P31A cells and a 24% decrease in cdc12-6D cells (Figure 4, E and F). A significant decrease was not observed for Ain1-GFP in early mitotic cdc12-6A cells (Figure 4, E and F). Like F-actin, Ain1-GFP recovered to wild-type levels during anaphase B in cdc12-P31A cells, but remained slightly reduced in cdc12-6D cells (Figure 4, E and F).
Cdk1-dependent phosphorylation of Cdc12 is important during CR formation
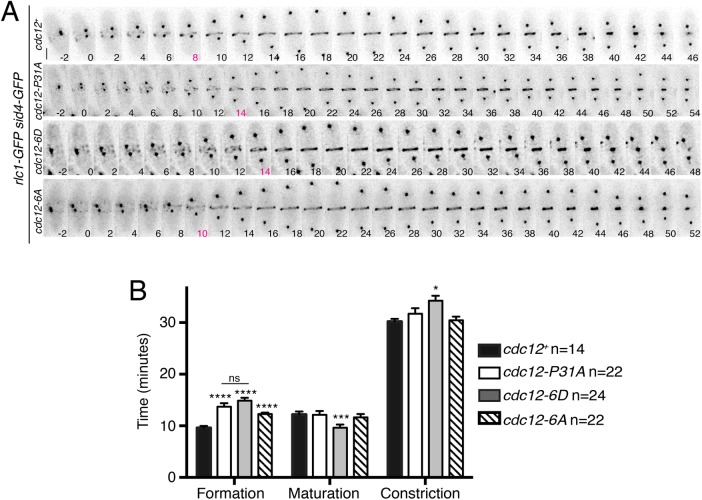
Because our results indicated that phosphomodulation of the Cdc15–Cdc12 interaction adversely affects CR assembly, we used time-lapse microscopy to examine cytokinesis in its entirety. Rlc1-GFP and Sid4-GFP served as markers for the CR and mitotic progression, respectively. As reported previously, CR formation (the time from SPB separation to the appearance of a CR) in cdc12-P31A cells was delayed by ∼4 min (29% slower than for wild type). A similar delay in CR formation was seen for cdc12-6D cells (35% slower; Figure 5, A and B), consistent with the reduction in F-actin. cdc12-6D also displayed a shorter maturation period (the time from CR formation to constriction onset) and slightly longer CR constriction (Figure 5, A and B). Consistent with other phenotypes of the cdc12-6A allele manifesting as an intermediate between wild type and cdc12-6D, it had a relatively slight delay in CR formation (21% slower; Figure 5, A and B).
FIGURE 5:
Mimicking constitutive Cdc12 phosphorylation delays CR formation. (A) Live-cell imaging of cdc12+, cdc12-P31A, cdc12-6D, or cdc12-6A cells expressing Rlc1-GFP Sid4-GFP. Images were acquired every 2 min. Bar, 5 μm. The time point indicated in magenta is the first in which a complete ring is detected. (B) Quantification of cytokinesis event timing for each strain. Measurements are from at least three biological replicates. *p ≤ 0.05, ***p ≤ 0.001, and ****p ≤ 0.0001, one-way ANOVA. Error bars represent SEM.
In summary, this study confirms that the cytokinetic formin Cdc12 is a Cdk1 substrate. In vitro, Cdk1 phosphorylation of six N-terminal residues on Cdc12 inhibits its interaction with the F-BAR domain of Cdc15. Cells expressing a cdc12 phospho-mimetic 6D mutant showed phenotypes similar to those of cells in which the Cdc15-binding motif on Cdc12 had been mutated; both exhibit reduced Cdc12 CR localization and delayed F-actin accumulation in the CR, leading to defects in CR formation. Thus, the in vitro inhibition of Cdc12–Cdc15 binding by Cdk1 is consistent with what happens in vivo. The phenotypes of the Cdc12-6A mutant, however, do not lend themselves to such a straightforward interpretation. Though alanine substitutions of the Cdk1 sites in the Cdc12 N-terminus do not impact Cdc15 F-BAR interaction in vitro, they do modify protein function in vivo. One possible explanation is that the 6 N-terminal alanine substitutions impact an unknown Cdc12 binding partner or regulator. Another binding partner clearly exists, based on genetic evidence (Laporte et al., 2011) and the fact that Cdc12 localizes to the CR in the absence of Cdc15 F-BAR binding and F-actin (Willet et al., 2015). Another potential explanation for the slight loss of function of Cdc12-6A is a need for dynamic Cdc12–Cdc15 interactions during CR assembly. FRAP experiments indicate that Cdc12 turns over rapidly in the CR (Yonetani et al., 2008); altering Cdc12 interactions may affect its dynamics and be detrimental to CR function. Last, the 6A mutations might simply disturb the structure of Cdc12. In addition to phosphorylating Cdc12’s N-terminus, Cdk1 phosphorylates at least one additional site in Cdc12’s C-terminus in vivo (Ser 1798) (Swaffer et al., 2016). It will be interesting to define a function for this phosphorylation site and to determine whether it cooperates with SIN-dependent phosphorylation to control the nearby Cdc12 multimerization domain (Bohnert et al., 2013).
At least some Cdk1 phosphosites on Cdc12 are dephosphorylated by Clp1, a member of the Cdc14 phosphatase family, thereby allowing maximal Cdc12 accumulation at the CR. Indeed, Cdc12 localizes to the CR, concomitant with Clp1 (Clifford et al., 2008). Bni1 and Bnr1, two redundant cytokinetic formins in budding yeast, are direct Cdc14 targets whose localization to the cell division site is dependent on Cdc14-mediated dephosphorylation (Bloom et al., 2011). Therefore, formin-dependent modification by Cdk1/Cdc14 may be a conserved mechanism by which formin subcellular localization is controlled during cytokinesis.
Multiple CR components may be phosphorylated and regulated by Cdk1. Saccharomyces cerevisiae Iqg1 (Rng2 in S. pombe and IQGAPs in humans) was identified as a direct Cdk1 target (Holt et al., 2009; Naylor and Morgan, 2014; Miller et al., 2015). Abolishing Cdk1-mediated phosphorylation of Iqg1 results in precocious CR formation prior to anaphase. Thus, similarly to our study, these findings suggest that Cdk1 phosphorylation of CR components counteracts early CR formation. Interestingly, Rng2 binds Clp1, indicating that it is also a Cdk1 target (Chen et al., 2013). Thus, it will be interesting to determine whether Rng2 phosphorylation similarly inhibits its CR localization, and how such regulation, or that of other CR components, is coordinated with Cdk1-mediated phosphoinhibition of formin localization to fine-tune assembly of the cell division apparatus.
MATERIAL AND METHODS
Yeast methods
S. pombe strains (Supplemental Table S1) were grown in yeast extract (YE). To make endogenous cdc12-6A and cdc12-6D alleles, a pSK vector (pBluescript backbone) was constructed that contained, in the following order, 5′ cdc12 flank including its promoter, full-length cdc12+, kanR cassette, and 3′ cdc12 flank (pKG 5431) (Bohnert et al., 2013). The cdc12-6A and cdc12-6D mutations were created by site-directed mutagenesis of pKG 5431 by PCR and confirmed with sequencing. The mutant constructs were then released from the vector by digestion with XbaI and SacI and transformed into wild-type S. pombe cells using a lithium acetate method (Keeney and Boeke, 1994). G418-resistant cells were selected and the cdc12 locus was sequenced to identify transformants containing the desired and correct mutations. cdc12+, cdc12-6A, and cdc12-6D were tagged endogenously at the 3′ end with HA3:kanR or HA3:hygR using pFA6 cassettes as previously described (Wach et al., 1994; Bähler et al., 1998).
cdc12+, cdc12-6A, and cdc12-6D endogenously tagged mNG alleles were made using a pSK vector that contained a 5′ cdc12 flank including its promoter, full-length cdc12+, and sequences encoding mNG, kanR cassette, and 3′ cdc12 flank (pKG 7922). mNG is a green fluorescent protein derived from the lancelet Branchiostoma lanceolatum that was chosen for imaging experiments because of its superior brightness (Shaner et al., 2013) (Allele Biotechnology). The constructs were then released from the vector by digestion with XbaI and SacI and transformed into wild-type S. pombe cells using a lithium acetate method (Keeney and Boeke, 1994). G418-resistant cells were selected and the cdc12 locus was sequenced to identify transformants containing the desired and correct mutations and tag.
For growth assays, cells were grown to log phase at 25°C in YE, 10 million cells were resuspended in 1 ml of YE, and 10-fold serial dilutions were made. Then 2.5 μl of each dilution was spotted on YE plates and the plates were incubated at the indicated temperatures.
Protein expression and purification
Cdc12(1–765), Clp1, and Clp1-C286S were cloned into pMAL-c2 for expression as an MBP fusion (Carnahan and Gould, 2003; Clifford et al., 2008; Chen et al., 2013). Cdc15 F-BAR (19–312) was cloned into pET15b for expression as a His6 fusion (Willet et al., 2015). Proteins were induced in Escherichia coli Rosetta2(DE3)pLysS cells with 0.4 mM IPTG overnight at 18°C. Proteins were purified on amylose beads (New England Biolabs) or cOmplete His-Tag resin (Roche) according to the manufacturer’s protocols. Cdc15(19–312)’s His6 tag was removed with thrombin protease and the protein was further purified on a HiTrap Q SP anion exchange column (GE Healthcare) and concentrated.
Protein methods
Cell pellets were snap-frozen in dry ice–ethanol baths. Lysates were prepared using a Fastprep cell homogenizer (MP Biomedicals). Immunoprecipitations were performed as previously described (Gould et al., 1991) in NP-40 buffer containing SDS for denatured lysates. Protein samples were resolved by SDS–PAGE and transferred to polyvinylidene fluoride (PVDF) membrane (Immobilon P; EMD Millipore). Anti-HA (12CA5), anti-Cdc2 (PSTAIRE; Sigma), or anti-Cdc15 (Roberts-Galbraith et al., 2009) was used in immunoprecipitations and/or as a primary antibody in immunoblotting. Secondary antibodies were conjugated to IRDye800 or IRDye680 (LI-COR Biosciences). Blotted proteins were detected via an Odyssey Classic (LI-COR Biosciences). For gel shifts, denatured lysates were treated with λ-phosphatase (New England Biolabs) in 25 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, and 1 mM MnCl2 and incubated for 30 min at 30°C with shaking. The Clp1 phosphatase assay was performed as previously described (Chen et al., 2013) with minor modifications. Briefly, Cdc12-HA3 was immunoprecipitated from 180 ODs of cells (6 30 OD pellets per sample) from a 3-h mts3-1 arrest using 4 µg of 12CA5 antibody and 20 µl of PureProteome protein G magnetic beads (Millipore). Beads were washed twice with 1 ml NP-40 buffer and once with 1 ml of Clp1 phosphatase buffer (50 mM imidazole, pH 6.9, 1 mM EDTA, and 1 mM dithiothreitol [DTT]), divided into three equal parts. The first was treated with buffer alone, the second with 200 ng of MBP-Clp1, and the third with 200 ng of MBP-Clp1-C286S in 20-μl reactions composed of 1 × Clp1 phosphatase buffer. The samples were then incubated for 30 min at 30°C with shaking before the reaction was quenched with 5 μl of 5x SDS sample buffer. Where indicated, samples were resolved by SDS–PAGE in the presence of 5 μM PhosTag acrylamide per the manufacturer’s protocol (Wako Chemical USA).
Kinase reactions were performed in protein kinase buffer (10 mM Tris, pH 7.4, 10 mM MgCl2, and 1 mM DTT) with 10 μM cold ATP, 3 μCi of [32P]ATP, and 100 ng of kinase-active or kinase-dead insect cell–produced Cdk1-Cdc13 in 20 μl reactions that were incubated at 30°C for 30 min with shaking. MBP was used as a control substrate for Cdk1-Cdc13. Reactions were quenched by the addition of 5 μl of 5x SDS sample buffer. Proteins were separated by SDS–PAGE and transferred to PVDF membrane, and phosphorylated proteins were visualized by autoradiography. Kinase assays, phosphoamino acid analysis, and tryptic peptide mapping were performed as described in (Sparks et al., 1999; Feoktistova et al., 2012) and references therein. In vitro phosphorylation of recombinant proteins used in in vitro binding assays was performed via identical kinase assays, except that radioactive [γ32P]ATP was eliminated, and the final concentration of unlabeled ATP in reactions was increased to 2 mM.
In vitro binding
Recombinant proteins conjugated to amylose beads were incubated with recombinant Cdc15 F-BAR for 1 h at 4°C in 1 ml binding buffer (50 mM Tris-HCl, pH 7.0, 250 mM NaCl, 2 mM EDTA, 0.1% NP-40). Following extensive washing in binding buffer, samples were resolved by SDS–PAGE for CB staining.
Microscopy
Live-cell images of S. pombe cells were acquired using either a personal DeltaVision microscope system (Applied Precision) that includes an Olympus IX71 microscope, 60 × NA 1.42 PlanApo, fixed- and live-cell filter wheels, a Photometrics CoolSnap HQ2 camera, and softWoRx imaging software or a spinning disk confocal microscope (Ultraview LCI; PerkinElmer, Waltham, MA) equipped with a 63 × 1.46 NA PlanApochromat oil immersion objective (Zeiss), an EM-CCD ImagEM X2 camera (Hamamatsu) and µManager software (Edelstein et al., 2014).
Images were acquired at 25–29°C and cells were imaged in YE media. Images were acquired using 0.5 µm z spacing. Representative images in Figure 3 were deconvolved with 10 iterations and are maximum-intensity projections. Representative images in Figure 4 are maximum-intensity projections. Images used for quantification in Figures 3 and 4 were not deconvolved and were sum projected.
Time-lapse imaging was performed on cells in log phase using an ONIX microfluidics perfusion system (CellASIC). Cells were loaded into Y04C plates for 5 s at 8 psi, and YE liquid media was flowed into the chamber at 5 psi throughout imaging. For Figure 5, CR formation is the time from SPB separation to CR formation, maturation is the time from CR formation to the start of CR constriction, and CR constriction is the time from the first frame of CR constriction until the frame where the CR is completely constricted and has disassembled.
Intensity measurements were made with ImageJ software (http://rsweb.nih.gov/ij/; Schindelin et al., 2012). For all intensity measurements, the background was subtracted by creating a region of interest (ROI) in the same image where there were no cells. The raw intensity of the background was divided by the area of the background, which was multiplied by the area of the ROI. This number was subtracted from the raw integrated intensity of that ROI. For CR intensity quantification, an ROI was drawn around the CR and measured for raw integrated density, and for whole cell intensity quantification an ROI was drawn around the entire cell. To compare populations of cells for all genotypes, cells were imaged on the same day with the same microscope parameters.
For DAPI and methyl blue staining, cells were grown to log phase at 25°C, shifted to 36°C, and then fixed in 70% ethanol for at least 30 min.
All statistical analyses of variance (ANOVAs) used Tukey’s post hoc analysis.
Supplementary Material
Acknowledgments
We thank C. Snider, M. Mangione, and M. Wos for critical reading and editing of the manuscript. A.W. was supported by American Heart Association Grant 14PRE19740000, K.A.B. was supported by National Institutes of Health Grant T32-CA119925, and this work was supported by National Institutes of Health Grant GM101035 to K.L.G.
Abbreviations used:
- Cdk1
cyclin-dependent kinase 1
- CR
contractile ring
- F-actin
filamentous actin
- MBP
maltose-binding protein
- mNG
mNeonGreen.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-11-0646) on January 17, 2018.
REFERENCES
- Bähler J, Wu J-Q, Longtine MS, Shah NG, Mckenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast , 943–951 [DOI] [PubMed] [Google Scholar]
- Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. (2011). Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem , 5434–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. (2011). On the cutting edge: post-translational modifications in cytokinesis. Trends Cell Biol , 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. (2012). Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet , e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Grzegorzewska AP, Willet AH, Vander Kooi CW, Kovar DR, Gould KL. (2013). SIN-dependent phosphoinhibition of formin multimerization controls fission yeast cytokinesis. Genes Dev , 2164–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. (2003). The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol , 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. (1997). Cdc12P, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol , 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-S, Broadus MR, McLean JR, Feoktistova A, Ren L, Gould KL. (2013). Comprehensive proteomics analysis reveals new substrates and regulators of the fission yeast Clp1/Cdc14 phosphatase. Mol Cell Proteomics , 1074–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford DM, Chen C-T, Roberts RH, Feoktistova A, Wolfe BA, Chen J-S, McCollum D, Gould KL. (2008). The role of Cdc14 phosphatases in the control of cell division. Biochem Soc Trans , 436–438 [DOI] [PubMed] [Google Scholar]
- Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V. (2008). Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J Cell Sci , 843–853 [DOI] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. (2014). Advanced methods of microscope control using μManager software. J Biol Methods , 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. (1995). The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell , 435–444 [DOI] [PubMed] [Google Scholar]
- Feoktistova A, Morrell-Falvey J, Chen J-S, Singh NS, Balasubramanian MK, Gould KL. (2012). The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol Biol Cell , 1636–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J , 3297–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray CH, Good VM, Tonks NK, Barford D. (2003). The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J , 3524–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. (2000). The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J , 1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. (1997). The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Cell Biol , 7965–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science , 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics , 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Krug K, Pengelley S, Macek B, Hauf S. (2011). Mitotic substrates of the kinase Aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal , rs6. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol , 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. (2004). Progressing actin: formin as a processive elongation machine. Nat Cell Biol , 1158–1159 [DOI] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. (2011). Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol , 1005–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Gould KL. (2016). Linking up at the BAR: oligomerization and F-BAR protein function. Cell Cycle , 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. (2015). Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev Cell , 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DP, Hall H, Chaparian R, Mara M, Mueller A, Hall MC, Shannon KB. (2015). Dephosphorylation of Iqg1 by Cdc14 regulates cytokinesis in budding yeast. Mol Biol Cell , 2913–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, Gillespie D, Jallepalli P, Schiebel E. (2010). Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol , 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SG, Morgan DO. (2014). Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci , 1128–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niiya F, Xie X, Lee KS, Inoue H, Miki T. (2005). Inhibition of cyclin-dependent kinase 1 induces cytokinesis without chromosome segregation in an ECT2 and MgcRacGAP-dependent manner. J Biol Chem , 36502–36509 [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet , 167–178 [DOI] [PubMed] [Google Scholar]
- Potapova TA, Daum JR, Pittman BD, Hudson JR, Jones TN, Satinover DL, Stukenberg PT, Gorbsky GJ. (2006). The reversibility of mitotic exit in vertebrate cells. Nature , 954–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al. (2015). The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell , 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen J-S, Wang J, Gould KL. (2009). The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol , 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen J-S, McLeod I, McDonald WH, Gygi SP, Yates JR, Gould KL. (2010). Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell , 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods , 407–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. (1999). Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol , 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. (2004). Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet , 203–232 [DOI] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. (2016). CDK substrate phosphorylation and ordering the cell cycle. Cell , 1750e16–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. (2006). Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol , 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast , 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. (2006). Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell , 3254–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Hinchcliffe EH, Glotzer M, Hyman AA, Sluder G, Wang YL. (1997). CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J Cell Biol , 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. (2015). The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol , 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F, Sigl R, Geley S. (2007). “… The end of the beginning”: cdk1 thresholds and exit from mitosis. Cell Cycle , 1408–1411 [PubMed] [Google Scholar]
- Wu JQ, Bähler J, Pringle JR. (2001). Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell , 1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol , 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. (2008). Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. , 2208–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.