Abstract
Background
Liver cancer is considered one of the main causes of cancer related deaths across the globe. Moreover, the incidence of liver cancer in developed countries is likely to increase in future. The increase in the incidence of liver cancer, the limited availability of standard treatments, and the side effects of the existing drugs demands exploration and identification of new targets and treatment strategies for liver cancer. In this context, the present study investigated the potential of miR-21 as the therapeutic target for the management of liver cancer.
Material/Methods
Total RNA was extracted by RNA isolation kit (RNeasy) as per the manufacturer’s instructions. The cDNA synthesis was carried out with the help of RevertAid cDNA synthesis kit (Fermentas). Expression analysis was performed by quantitative RT-PCR. Cell proliferation was examined by CellTiter 96 aqueous one cell proliferation assay kit (Promega) as per manufacturer’s guidelines. Apoptosis was detected by DAPI and Annexin V/PI staining. Cell migration was assessed by wound healing assay. MicroRNA-383 target was delimited by TargetScan software. Protein expression analysis was evaluated by western blotting.
Results
Our results revealed that miR-21 was significantly upregulated in liver cancer cells. However, downregulation of miR-21 inhibited cancer cell proliferation, promoted apoptosis, inhibited cell migration, and triggered cell cycle arrest in KYN-2 liver cancer cells. Additionally, in silico analysis revealed PTEN to be the downstream target of miR-21, which was further confirmed by expression analysis through western blotting.
Conclusions
Our results reveal that miR-21 might prove to be an important target for the management of liver cancer.
MeSH Keywords: Apoptosis; Carcinoma, Hepatocellular; MicroRNAs; Transcellular Cell Migration
Background
Liver cancer is considered one of the major causes of cancer related deaths worldwide and is the fifth most prevalent type of malignancy in men and the eight most prevalent type in woman. Annually, more than 0.55 million liver cancer cases are diagnosed, which includes around 0.4 million men and more than 0.15 million women [1]. It has been reported that liver cancer occurs before the age of 20 years in high-risk countries and occurs around or after the age of 50 years in low-risk countries [2]. As evident from these estimates, the incidence of liver cancer in men is more than two times that in women [2]. Moreover, studies have reported that the incidence of liver cancer in developed countries is likely to increase in the future [3]. The increase in the incidence of liver cancer, the limited availability of standard treatments, and the stern side effects of the existing drugs demands exploration and identification of new targets and treatment strategies for liver cancer. MicroRNAs (miRNAs) include a large family of small non-coding RNAs that are involved in the inhibition of protein synthesis by binding to different sites on 3′-untranslated region (3′-UTR) of target genes. Therefore, miRNAs exhibit remarkable biological functions in a diversity of cellular process [4,5]. Among several human miRNAs, miR-21 has been reported to be principally upregulated in several types of cancers. MiR-21 has been reported to promote progression of several types of cancers, which includes, but is not limited to, gastric cancer and glioma [6,7]. In the present study, the expression of miRNA-21 was determined in different liver cancer cell lines and it was found to be significantly upregulated in cancer cells compared to controls. Inhibition of miR-21 in liver cancer cells suppressed their proliferation and migration. In silico analysis revealed that miR-21 targeted tensin homolog (PTEN) tumor suppressor, which was further confirmed by determining the expression of PTEN in miRNA-21 inhibited liver cancer cells. Taken together we proposed that miRNA-21 can prove to be an important therapeutic target for the management of liver cancer and deserves further research.
Material and Methods
Chemicals, reagents, and cell cultures
DAPI (4′,6-diamidino-2-phenylindole), RNase A, Triton X-100, and dimethyl and sulfoxide (DMSO) were obtained from Sigma-Aldrich Co. Primary and secondary antibodies were procured from Santa Cruz Biotechnology Inc. Fetal bovine serum (FBS), RPMI-1640 medium, L-glutamine, and antibiotics were obtained from Invitrogen Life Technologies. Human liver cancer cell lines KIM-1, KYN-1, KYN-2, KYN-3, HAK-1A, and HAK-1B, and one normal liver cell line LO2 were purchased from Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. The cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum, 100 U/mL each of both penicillin and streptomycin; cells were maintained in a humidified atmosphere containing 5% CO2.
Isolation of RNA, cDNA synthesis, and expression analysis
For isolation of RNA, RNeasy RNA isolation kit was used and the whole procedure was carried out as per the manufacturer’s instructions. Thereafter, cDNA was synthesized with the help of RevertAid cDNA synthesis kit (Fermentas) as per the manufacturer’s protocol. To carry out the qRT-PCR, the cDNA was diluted 20 times and qRT-PCR was carried out thrice in triplicate in ABI StepOne Real time using SYBR Green Master Mix (Fermentas). The relative quantification method (ΔΔ−CT) was employed to determine quantitative variation between the replicates examined. β-actin was used as a positive control.
Inhibition of miRNA-21 in liver cancer cells
The inhibitor of human miR-21 (miR21-In, 107 units/mL), and its non-specific miRNA lentivirus (miR-C, 107 units/mL) were obtained from RiboBio (China). KYN-2 cells were incubated with lentiviral particles (10 mL/1,000 cells, multiplicity of infection=10–15) and polybrene (8 mg/mL) for 48 hours. Thereafter, the media was changed, and liver cancer KYN-2 cells were kept for another 3–7 days for stabile transduction. The cells were then passaged and preserved in vitro for further analysis.
MTT proliferation assay
Liver cancer KYN-2 and miRNA-inhibited KYN-2 cancer cells were separately seeded in 96-well plates (5,000 cells/well) and permitted to grow for five days. Afterwards, cell proliferation was determined with the help of a CellTiter 96 aqueous one cell proliferation assay kit (Promega USA) as per manufacturer’s guidelines. In brief, at every 24-hour interval, 15 mL MTT solution was added to each well for two hours at 37°C. The absorbance was read at 570 nm.
Detection of apoptosis
Liver cancer KYN-2 and miRNA-inhibited KYN-2 cancer cells were separately seeded at the density of 2×105 cells/well in six-well plates and treated with different concentrations of cisplatin followed by an incubation period of 24 hours at 37°C. DAPI staining was performed by incubating the cells in six-well plates for 24 hours. The cells were then washed with PBS, fixed in formaldehyde (10%), and then again washed with PBS. The DAPI stained cells were then examined by fluorescence microscope. To estimate the apoptotic cell populations, KYN-2 and miRNA-inhibited KYN-2 cancer cells were seeded at a density of 1×106 cells/well in six-well plates and cultured for 24 hours. Thereafter, the cells were collected and washed with PBS. The cells were then incubated with Annexin V/FITC and PI for 15 minutes and the apoptotic cell populations were estimated by a flow cytometry (BD Biosciences, San Jose, CA, USA).
Cell migration assay
The cell migration potential of liver cancer KYN-2 and miR-21-inhibited KYN-2 cancer cells was investigated by wound healing assay. Briefly 5×104 cells/well were seeded in 96-well plates. Afterwards, the plates were incubated overnight at 37°C to allow the cells to adhere. Then a wound was scratched using a sterile pipette tip after the cells reached confluence. The cells were then washed with PBS to clear the detached cells. The cells were monitored after 20-hour intervals and photographed.
Identification of miRNA-21 target
TargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA. To find out the potential target of miR-21, it was subject to scanning in TargetScan search program.
Western blotting
Total protein from the liver cancer cells was isolated in RIPA lysis buffer. Equal protein extracts from each group were run on SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. This was followed by blocking with 5% non-fat milk and incubation at room temperature for one hour. Thereafter, the membranes were incubated with a specific primary antibody at 4°C overnight. This was followed by washing in washing buffer and incubation for one hour with the suitable secondary antibody. The protein bands of interest were visualized by an ECL Advanced Western Blot Detection Kit.
Statistical analysis
All experiments were carried out three times in triplicate and expressed as mean ±SD. One-way ANOVA and Tukey’s test were used for statistical analysis through GraphPad 7 software. The values were considered significant at p<0.01
Results
MiR-21 was overexpressed in liver cancer cell lines
The expression of miR-21 was determined in six liver cancer cell lines (KIM-1, KYN-1, KYN-2, KYN-3, HAK-1A, and HAK-1B) and one normal liver cell line (LO2) by qRT-PCR analysis (Figure 1). Our results revealed that the expression of miR-21 was highly upregulated in all the liver cancer cell lines compared to the normal non-cancerous LO2 liver cells. Moreover, the expression of miR-21 was found to be highest in liver cancer cell line KYN-2. Therefore, KYN-2 cell line was selected for further experimentation. Furthermore, these expression studies provided a clue about the potential of miR-21 as an important therapeutic target.
Figure 1.

Expression of miR-21 in different liver cancer and one normal liver cell line as determined by qRT-PCR. The experiments were carried out three times in triplicates and expressed as mean ±SD. The values were considered significant at p<0.01.
Suppression of miR-21 leads to inhibition of cancer cell proliferation
To gain insights about the role of miR-21 in the progression of liver cancer, we inhibited the expression of miR-21 via lentiviral transduction. The results revealed that the inhibition of miR-21 caused significant inhibition of growth of KYN-2 liver cancer cells as evident from the MTT proliferation assay (Figure 2).
Figure 2.

Effect of miR-21 inhibition of the cell viability of liver cancer cells as determined by absorbance at 570 (A50). The experiments were carried out three times in triplicates and expressed as mean ±SD. The values were considered significant at p<0.01.
Suppression of miR-21 expression promoted apoptosis
To investigate if the antiproliferative effects induced by suppression of miR-21 were due to induction of apoptosis, DAPI and annexin V/PI staining were performed. Interestingly, the results indicated that inhibition of miR-21 leads to triggering of apoptosis in liver KYN-2 cancer cells (Figure 3). Furthermore, V/PI staining results showed that the apoptotic cell populations were 37.4% in miR-21 inhibited cells compared to 3.5% in the controls (Figure 4).
Figure 3.
Effect of miR-21 on induction of apoptosis in KYN-2 liver cancer cells as depicted by DAPI staining. The experiments were carried out three times in triplicates and expressed as mean ±SD. The values were considered significant at p<0.01.
Figure 4.
Estimation of apoptotic cell populations in KYN-2 control and miR-21 inhibited KYN2 cells by Annexin V/PI followed by flow cytometry. The experiments were carried out three times in triplicates.
Inhibition of miR-21 triggered cell cycle arrest
The results of the cell migration assay revealed that inhibition of miR-21 promoted G2/M cell cycle arrest as evident from the accumulation of higher numbers of KYN-2 liver cancer cells in G2 phase of the cell cycle (Figure 5). In the G2 phase, control miR-21 suppressed KYN-2 cells were found to be 42.68% compared to 2.45% in control KYN-2 cells.
Figure 5.
Effect of miR-21 on cell cycle phase distribution of KYN-2 liver cancer cells as depicted by flow cytometry. The experiments were carried out three times in triplicates.
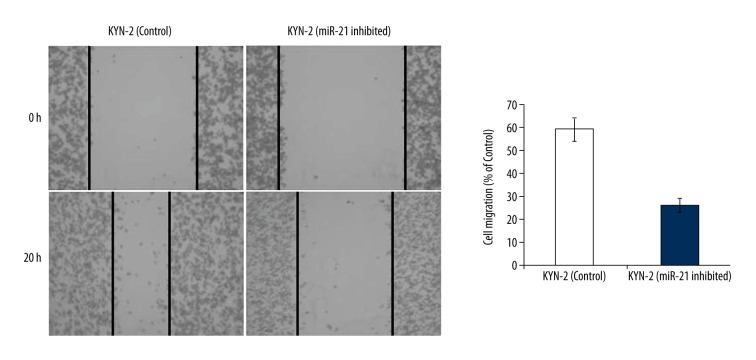
Suppression of miR-21 leads to inhibition of cell migration
To examine if miR-21 caused inhibition of cell migration, we used the wound healing assay. The results of our study indicated that inhibition of miR-21 expression in KYN-2 cells caused significant suppression of cell migration (Figure 6). In miR-21 silenced KYN-2 cells, cell migration was significantly lower compared to KYN-2 cells. The cell migration in miR-21 suppressed KYN-2 cells was found to be 26% compared to 58% in control KYN-2 cells.
Figure 6.
Wound healing assay showed the effect of miR-21 inhibition on cell migration of KYN2 liver cells. The experiments were carried out three times in triplicates and expressed as mean ±SD.
MiR-21 targeted PTEN gene
To find out the downstream target gene of microRNA-383, we used a target scan (www.targetscan.org) server. We observed that PTEN was the potential target gene of miRNA-21. The 3′-UTR DNA segment of PTEN was found to be complimentary to the binding of miR-21 (Figure 7A). To further confirm the target, the expression of PTEN in miR-21-inhibited KYN-2 and KYN-2 cancer cells was determined by western blotting. Our results revealed that miR-21suppression significantly upregulated PTEN expression in KYN-2 cells (Figure 7B), further indicating PTEN as the potential target of miR-21 in liver cancer cells.
Figure 7.
MiR-21 targets PTEN as confirmed by (A) TargetScan software and (B) western blotting. The experiments were carried out three times in triplicates.
Discussion
Liver cancer is considered one of the major causes of cancer related deaths worldwide. The incidence of liver cancer in men is more than two times higher than in women [1,2]. Moreover, studies have reported that the incidence of liver cancer in developed countries is likely to increase in the future [3]. Therefore, there is pressing need to develop novel strategies for the treatment of liver cancer. It is now well-established in the literature that miRNAs play a vital role in human cancer development and progression. The expression of miRNAs might be either enhanced or suppressed depending on whether they promote or suppress tumorigenesis and progression of cancers. Recently, miRNAs have become important therapeutic targets for the management of several types of cancers [8]. Consistently, miRNA-21 has been reported to play a vital role in human cancers such as glioma, gastric cancer, and ovarian cancer [9–12]. However, the exact role and mechanism of miRNA-21 in the development of progression of liver cancer has not been thoroughly studied. In our study, we determined the expression of miR-21 in six liver cancer cell lines and one normal liver cell line. The results indicated that the expression of miR-21 was highly upregulated in liver cancer cell lines. Our study confirmed previous study results, wherein the expression of miR-21 was reported to be highly upregulated in several types of cancers [11]. Our results, as well as the results from previous studies, clearly indicated that miR-21 might prove to be an important therapeutic target for the management of cancers. To confirm the role of miR-21 in liver cancer, we silenced the expression of miR-21 in KYN-2 liver cancer cells by lentiviral transduction. We observed that inhibition of miR-21 expression suppressed the cell proliferation of KYN cells compared to controls. To gain insights about the mechanism by which miR-21 promotes inhibition of cell proliferation, we carried out DAPI staining. The results indicated that the inhibition of miR-21 triggered apoptosis in KYN cells. This was further confirmed by Annexin V/PI staining, which revealed that miR-21 caused 37.4% of apoptosis compared to 3.5% in controls. Apoptosis is an important mechanism by which cancerous cells are removed from the body [13]. Several anticancer agents have been reported to exhibit anticancer activities via induction of apoptosis. In addition, apoptosis is an important mechanism to avoid the development of drug resistance [14]. Apoptosis, cell cycle arrest is also considered an important mechanism by which anticancer agents exert their effects [15].
Our results indicated that inhibition of miR-21 leads to G2/M cell cycle arrest in liver cancer KYN-2 cells as was evident from the increased number of cells in the G2 phase of the cell cycle. Cell migration is also considered an important factor for cancer progression and metastasis [16]. It has been previously reported that suppression of cell migration is essential to prevent cancer metastasis in vivo [17]. We investigated if suppression of miR-21 had any effect on the migration of liver cancer cells. We observed that suppression of miR-21 leads to significant inhibition of KYN-2 cell migration. Next, we used online software TargetScan to delimit the potential target of miR-21. The results showed that PTEN, a tumor suppressor gene [18], was the downstream target gene of miR-21. Recent studies have demonstrated that PTEN protein suppresses tumorigenesis and progression of several human cancers [19]. To further confirm the involvement of PTEN in miR-21 prompted antiproliferative effects, we investigated the expression of PTEN in KYN-2 with silenced miR-21. The results indicated the expression of PTEN was highly enhanced in these cells, confirming the involvement of PTEN in miR-21 triggered cell death in liver cancer cells.
MiR-21 is among the first miRNAs to be overexpressed in cancer and has been considered a therapeutic target [20]. Downstream targets of miR-21 have been identified, including PTEN [20]. The present study established that miR-21 could prove to be essential in the treatment of liver cancer. Drugs can be designed to suppress the expression of miR-21, which in turn will cause apoptosis of cancer cells. Hence, miR-21 inhibition appears a promising therapeutic strategy.
Conclusions
We concluded that miR-21 was highly upregulated in liver cancer cells, and inhibition of miR-21 promoted inhibition of cell proliferation and migration and triggered G2/M cell cycle arrest by targeting PTEN. These results clearly indicated that miR-21 can be a critical target in the treatment and management of liver cancer.
Footnotes
Source of support: This study was supported by Guiding Project of Science and Technology Research Project of Hubei Education Department (Project No: B20121305)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ryerson AB, Eheman CR, Altekruse SF, et al. Annual report to the nation on the status of cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–37. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: Profile, profile, profile. Int J Cancer. 2008;122(5):969–77. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 5.Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–88. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105(36):13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 10.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 11.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12(12):2171–76. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriyama T, Ohuchida K, Mizumoto K, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8(5):1067–74. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 13.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21(3):485–95. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 14.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–48. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256(1):42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 16.Tamura M, Gu J, Matsumoto K, Aota SI, et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280(5369):1614–17. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 17.Jones DH, Nakashima T, Sanchez OH, et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature. 2006;440(7084):692–96. doi: 10.1038/nature04524. [DOI] [PubMed] [Google Scholar]
- 18.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22(14):2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 19.Tamura M, Gu J, Takino T, Yamada KM. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: differential involvement of focal adhesion kinase and p130Cas. Cancer Res. 1999;59(2):442–49. [PubMed] [Google Scholar]
- 20.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]