Significance
Viruses that infect bacteria (phages) represent the most abundant living entities on the planet, and many aspects of our fundamental knowledge of phage–bacteria relationships have been derived in the context of exponentially growing bacteria. In the case of the prototypical Escherichia coli phage T7, specific inhibition of the housekeeping form of the RNA polymerase (Eσ70) by a T7 protein, called Gp2, is essential for the development of viral progeny. We now reveal that T7 uses a second specific inhibitor that selectively inhibits the stationary phase RNA polymerase (EσS), which enables T7 to develop well in exponentially growing and stationary phase bacteria. The results have broad implications for our understanding of phage–bacteria relationships and the therapeutic application of phages.
Keywords: Escherichia coli, RNA polymerase, T7 phage, transcription regulation, stationary phase
Abstract
T7 development in Escherichia coli requires the inhibition of the housekeeping form of the bacterial RNA polymerase (RNAP), Eσ70, by two T7 proteins: Gp2 and Gp5.7. Although the biological role of Gp2 is well understood, that of Gp5.7 remains to be fully deciphered. Here, we present results from functional and structural analyses to reveal that Gp5.7 primarily serves to inhibit EσS, the predominant form of the RNAP in the stationary phase of growth, which accumulates in exponentially growing E. coli as a consequence of the buildup of guanosine pentaphosphate [(p)ppGpp] during T7 development. We further demonstrate a requirement of Gp5.7 for T7 development in E. coli cells in the stationary phase of growth. Our finding represents a paradigm for how some lytic phages have evolved distinct mechanisms to inhibit the bacterial transcription machinery to facilitate phage development in bacteria in the exponential and stationary phases of growth.
Viruses of bacteria, phages, have evolved diverse and sophisticated mechanisms to take over essential host processes to facilitate the successful development of phage progeny. Many such host takeover mechanisms involve small proteins that interact with and repurpose, inhibit, or modulate the activity of essential bacterial enzymes, which as a consequence, often result in the demise of the bacterial cell (1). Thus, a detailed understanding of phage-encoded antibacterial small proteins and their bacterial targets at a molecular level not only will unravel new phage biology but also may inform and inspire the discovery of novel antibacterial targets and antibacterial compounds. Unsurprisingly, the acquisition of the bacterial transcription machinery, the RNA polymerase (RNAP), is a major mechanism by which phages reprogram bacterial cellular processes to mount a successful infection (2, 3). The prototypical lytic phage of Escherichia coli, T7, synthesizes three proteins, Gp0.7, Gp2, and Gp5.7, which interact with host RNAP to facilitate the temporal coordinated expression of its genome. The genes of T7 are categorized as early, middle, and late to reflect the timing of their expression during the infection process. Early and middle genes generally encode proteins required for phage RNA synthesis, DNA replication, and host takeover, whereas the late genes specify T7 virion assembly and structural proteins. The translocation of the T7 genome into E. coli is a transcription-coupled process and requires the housekeeping form of the host RNAP (Eσ70) to transcribe the early genes from three strong early gene promoters, T7 A1, A2, and A3, and catalyze the entry of T7 DNA into the cell (4). The coordinated action of the early gene product Gp0.7 and the essential middle gene product Gp2 subsequently shuts off Eσ70 activity on the T7 genome. The viral single-subunit RNAP (T7 RNAP, Gp1, a product of an early gene) transcribes the middle and late viral genes. The shutting down of host RNAP is crucial for the coordination of the activities of bacterial and phage RNAPs on the phage genome, and thus, as a consequence, for successful completion of the infection cycle: Gp0.7 is a protein kinase that phosphorylates Eσ70, leading to increased transcription termination at sites located between the early and middle genes on the T7 genome (5, 6), and Gp2 binds in the main DNA binding channel of Eσ70 and thereby prevents the formation of the transcriptionally proficient open promoter complex (RPO) at the T7 A1-3 promoters (7). Gp2 is indispensable for T7 growth. In a T7 Δgp2 phage, aberrant transcription of middle and late T7 genes (which are normally transcribed by the T7 RNAP) by Eσ70 results in interference between the two RNAPs and, consequently, in aborted infection (5). Recently, a T7 middle gene product, Gp5.7, was identified as a repressor of RPO formation specifically on the T7 A1-3 promoters by Eσ70 molecules, which might have escaped inhibition by Gp2 (8). However, as phage genomes tend to be compact and efficient, it is puzzling that T7 has evolved two markedly different proteins to inhibit Eσ70, especially as Gp5.7, unlike Gp2, is a relatively poor inhibitor of Eσ70 (8). In this study, we unveil additional biological roles for Gp5.7 during T7 development in E. coli.
Results
Gp5.7 Is an Inhibitor of the E. coli Stationary Phase RNAP, EσS.
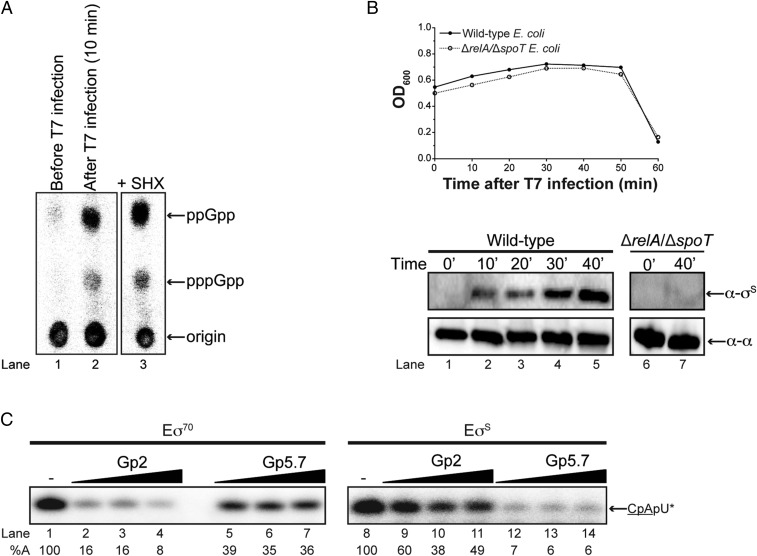
Previously, we posited that Gp5.7 prevents transcription initiation from T7 A1–A3 promoters by Eσ70 that might have escaped inhibition by Gp2 (8). Although this still remains a role for Gp5.7 in T7 development in E. coli, we noted a report by Friesen and Fill describing the accumulation of the stress-signaling nucleotide guanosine pentaphosphate, (p)ppGpp, in a valine auxotroph strain of E. coli during T7 development (9). Because (p)ppGpp simultaneously induces σS transcription and accumulation of σS (the predominant σ factor active in stationary phase E. coli), and considering that Gp5.7 is an inefficient inhibitor of Eσ70 compared with Gp2 (8), we contemplated whether Gp5.7 might preferentially inhibit EσS over Eσ70. Initially, we established that (p)ppGpp does indeed accumulate during T7 development in exponentially growing wild-type E. coli cells (Fig. 1A). We further demonstrated that the accumulation of (p)ppGpp is accompanied by an increase in the intracellular levels of σS during T7 development in exponentially growing E. coli (Fig. 1B). Control experiments with a relA/spoT mutant E. coli strain confirmed that the accumulation of σS during T7 infection was indeed (p)ppGpp-dependent (Fig. 1B).
Fig. 1.
Gp5.7 is an inhibitor of the E. coli stationary phase RNAP, EσS. (A) Polyethylenimine cellulose showing (p)ppGpp production during T7 infection. Lane 1: before T7 infection; lane 2: 10 min after infection with T7; and lane 3: positive control showing (p)ppGpp production in response to addition of sodium hydroxamate (SHX). The migration positions of ppGpp and pppGpp and the origin where the samples were spotted are indicated. (B) Expression of σS during T7 infection. (Top) Graph showing the optical density (OD600nm) of wild-type and ΔrelA/ΔspoT E. coli cultures as a function of time after infection with T7 phage. (Bottom) Image of a Western blot probed with anti-σS and anti-RNAP α-subunit (loading control) antibodies. Lanes 1–5 contain whole-cell extracts of wild-type E. coli cells at 0, 10, 20, 30, and 40 min after infection with T7; lanes 6 and 7 contain whole-cell extracts of ΔrelA/ΔspoT E. coli cells at 0 and 40 min after infection with T7. (C) Autoradiograph of denaturing gels comparing the ability of EσS and Eσ70 to synthesize a dinucleotide-primed RNA product from the T7 A1 promoter in the absence and presence of Gp2 and Gp5.7. The dinucleotide used in the assay is underlined and the asterisks indicate the radiolabeled nucleotide. The concentration of EσS and Eσ70 was 75 nM, and Gp2 and Gp5.7 were present at 75, 150, and 300 and 1,200, 1,500, and 1,875 nM, respectively. The percentage of RNA transcript synthesized (%A) in the reactions containing Gp5.7 or Gp2 with respect to reactions with no Gp5.7 or Gp2 added is given at the bottom of the gel, and the value obtained in at least three independent experiments fell within 3–5% of the %A value shown.
Next, we tested whether EσS could initiate transcription from the T7 A1 promoter as efficiently as Eσ70. To do this, we conducted an in vitro transcription assay using a 65-bp DNA fragment containing the T7 A1 promoter sequence as the template. Under the conditions used here, this assay reports the ability of Eσ70 or EσS to bind to the promoter, initiate DNA strand separation, and synthesize a trinucleotide RNA transcript, CpApU, which is complementary to the first three nucleotides (+1 to +3) of the sequence of the template strand of the T7 A1 promoter. The results shown in Fig. 1C revealed that EσS could initiate transcription from the T7 A1 promoter as efficiently as Eσ70 (Fig. 1C, cf. lanes 1 and 8). Consistent with previous results (7, 10), Gp2 inhibited Eσ70 activity by >80% when present at a molar ratio of 1:1 with respect to Eσ70; in contrast, Gp2 inhibited EσS by only 40% when present at a molar ratio of 1:1 with respect to EσS (Fig. 1C, cf. lanes 2 and 9). However, as previously shown (8), ∼16-fold more Gp5.7 than Gp2 was required to obtain a ∼60% inhibition of Eσ70 (Fig. 1C, cf. lanes 2 and 5). Strikingly, with the same concentration of Gp5.7, we observed >90% inhibition of EσS activity (Fig. 1C, lane 12). Control experiments with a functionally defective mutant of Gp5.7 [Gp5.7-L42A (8)] confirmed that the inhibition of EσS activity on the T7 A1 promoter by Gp5.7 was specific (SI Appendix, Fig. S1). It thus seems that Gp5.7 is a more efficient inhibitor of EσS than of Eσ70 (Fig. 1C). In contrast and consistent with previous observations (7), Gp2 is a more effective inhibitor of Eσ70 than EσS (Fig. 1C). Overall, we conclude that Gp5.7 and Gp2 are both required to fully shutdown the Eσ70 and EσS to allow optimal T7 development in E. coli cells during the exponential phase of growth.
Gp5.7 Interacts with Region 4 of σS.
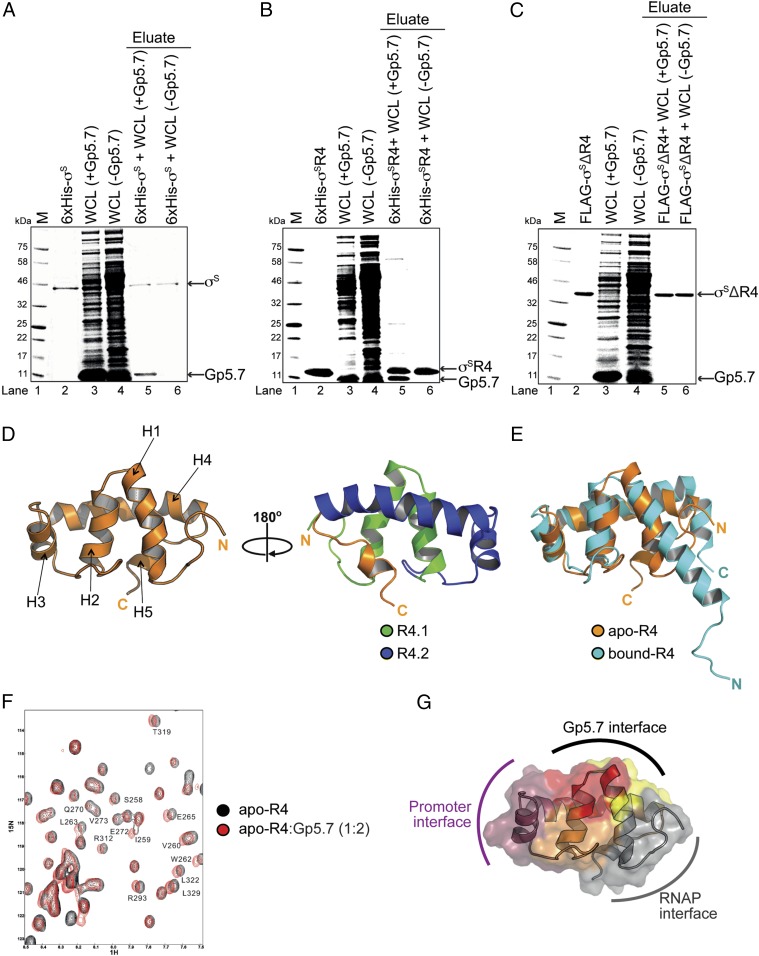
Although Gp5.7 interacts with the core subunits of the RNAP (8), our results indicate that σS would likely constitute a major interacting site of Gp5.7. Therefore, we next focused on identifying the Gp5.7 interacting site on σS. Nickel affinity pull-down experiments with hexa-histidine-tagged σS (6xHis-σS) and untagged Gp5.7 were conducted. As shown in Fig. 2A, incubation of 6xHis-σS (lane 2) with whole E. coli cell extracts in which untagged Gp5.7 is overexpressed from a plasmid (lane 3) led to the copurification of untagged Gp5.7 with 6xHis-σS (lane 5). Control experiments with E. coli whole-cell extracts with an empty plasmid (Fig. 2A, lanes 4 and 6) confirmed that Gp5.7 interacts specifically with 6xHis-σS and copurifies with it. Because we showed previously that Gp5.7 interacts with promoter DNA proximal to or overlapping the consensus −35 motif of the T7 A1 promoter (8), which is also bound by the conserved region 4 (R4) domain of σ factors, we considered whether R4 domain of σS could be a binding site for Gp5.7. To test this, we conducted affinity pull-down experiments, as in Fig. 2A, using a hexa-histidine-tagged version of the R4 domain (amino acid residues 245–330) of σS (6xHis-σSR4). As shown in Fig. 2B (lane 5), we detected untagged Gp5.7 copurifying with the 6xHis-σSR4 domain. In the converse experiment, we repeated affinity pull-down experiments, as in Fig. 2A, using FLAG epitope-tagged σS lacking the R4 domain (FLAG-σSΔR4). As indicated in Fig. 2C, and as expected, we failed to detect untagged Gp5.7 copurifying with the FLAG-σSΔR4 protein (lane 5). Based on the affinity pull-down experiments shown in Fig. 2 A–C, we conclude that the R4 domain of σS constitutes the binding site for Gp5.7.
Fig. 2.
Structural insights into the interaction between R4 domain of σS and Gp5.7. (A) Image of a denaturing gel showing that 6xHis-σS pulls down overexpressed untagged Gp5.7 from E. coli whole-cell lysate. The migration positions of the Gp5.7 and 6xHis-σS are indicated. (B) As in A, but using 6xHis-σSR4 domain. (C) As in A, but using FLAG-σSΔR4. (D, Left) Cartoon representation of apo-σSR4 solution structure. (Right) The image on the left rotated 180° clockwise with the α-helices corresponding to R4 subregions 4.1 (in green) and 4.2 (in blue) indicated. (E) Overlay of cartoon representations of apo-σSR4 domain (orange) and bound-σSR4 domain (cyan). (F) Overlay of 2D 1H-15N HSQC spectra of the apo-σSR4 without (black) and with Gp5.7 (red) recorded at pH 6.0, 300 K. Peaks that experienced broadening or chemical shift perturbation are labeled according to the amino acid residues in σS. (G) A surface representation of apo-σSR4 showing the regions interfacing with Gp5.7 (amino acid residues with significant peak broadening and chemical shift perturbation are shown in red, whereas those that experience moderate peak broadening and chemical shift perturbation are shown in orange and those that experience weak peak broadening and chemical shift perturbation are shown in yellow); amino acid residues associated with interacting with or proximal to the promoter DNA (raspberry) and RNAP subunits (gray) are also shown.
Structural Insights into the Interaction Between the R4 Domain of σS and Gp5.7.
To independently verify that the R4 domain of σS constitutes the binding site for Gp5.7 within EσS, and to map the Gp5.7 interface within R4 of σS, we solved the solution structure of the isolated 6xHis-σSR4 domain by NMR spectroscopy (Fig. 2D and SI Appendix, Table S1). The structure demonstrates that the R4 domain of σS (hereafter referred to as the apo-R4 domain) is able to fold as an isolated subdomain, consisting of five α helices (H1–H5). The α helices H1–H5 superpose well with the equivalent region from the crystal structure of EσS transcription initiation complex, in which the R4 domain (hereafter referred to as the bound-R4 domain) is connected to RNAP subunits via a long and flexible linker (11). Interestingly, the apo-R4 domain exhibits some conformational differences in the carboxyl (C) and amino (N) termini compared with the bound-R4 domain (Fig. 2E). The N terminus of the apo-R4 domain, which in the bound-R4 domain is connected to EσS via a flexible linker, appears more disordered. The C terminus of the apo-R4 domain contacts the core of the structure, making the apo-R4 domain more compact and stable in the absence of the remaining σS domains, RNAP subunits, or promoter DNA. We next recorded the 2D 1H-15N HSQC NMR spectra to monitor the backbone amide chemical shift and line-width perturbations for a 15N-labeled apo-R4 domain in the presence of up to a twofold molar excess of unlabeled Gp5.7. As shown in Fig. 2F, several peaks exhibited measurable broadening effects, and the extent of broadening correlated with the amount of Gp5.7 added (Fig. 2F). The Gp5.7 interaction surface was mapped on the structure of the apo-R4 domain (Fig. 2G), revealing that the main interacting residues localize to the C-terminal part of H1 and N-terminal part of H2. This analysis suggests that Gp5.7 binds between the RNAP-facing surface and the promoter-facing surface of R4 of σS (notably H3 of the 4.2 subregion of R4; Fig. 2D). Overall, the results from the affinity pull-down and structural analyses unambiguously indicate that the R4 of σS constitutes the binding site for Gp5.7 on σS.
Gp5.7 Inhibits RPO Formation by EσS on the T7 A1 Promoter.
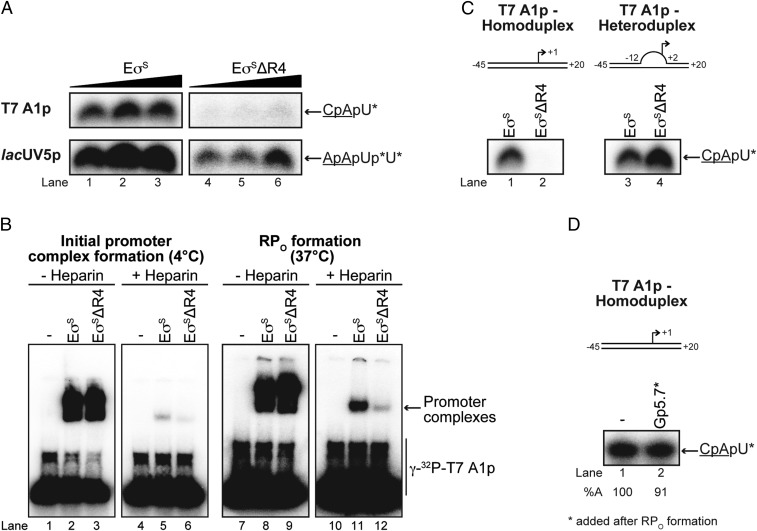
The location of the Gp5.7 binding surface on σS implies that Gp5.7 has evolved to target a σS domain potentially important for transcription initiation from the T7 A1 promoter. Previous reports from several groups have implied that the interaction between R4 of σ70 and the consensus −35 promoter region is required for the stabilization of early intermediate promoter complexes en route to the RPO at the T7 A1 promoter (12–15). Therefore, we considered whether Gp5.7 inhibits EσS-dependent transcription from the T7 A1 promoter by antagonizing RPO formation. In agreement with this view, whereas EσS reconstituted with σSΔR4 was able to initiate transcription from a prototypical Eσ70-dependent promoter [i.e., lacUV5 (albeit at a lower efficiency compared with reactions with wild-type EσS)], we failed to detect any transcription by EσSΔR4 from the T7 A1 promoter (Fig. 3A). We then conducted electrophoretic gel mobility shift assays at 4 °C (to detect initial RNAP-promoter complex formation) and at 37 °C (to detect RPo formation) with EσSΔR4 and a γ-32P-labeled T7 A1 probe to determine whether EσSΔR4 was able to interact with the T7 A1 promoter to form the initial promoter complex, or whether the initial promoter complex formed by EσSΔR4 on the T7 A1 promoter was unable to isomerize into the RPo, respectively. Results, shown in Fig. 3B, demonstrated that although EσS and EσSΔR4 formed the initial promoter complex on the T7 A1 promoter equally well (lanes 2–3 and lanes 5–6), those formed by EσSΔR4 seem unable to isomerize to form the transcriptionally proficient RPo (lanes 8–9 and lanes 11–12). Consistent with this conclusion, in vitro transcription assays with a T7 A1 promoter probe containing a heteroduplex segment between positions −12 and +2 (to mimic the RPO) revealed that EσSΔR4 is able to synthesize the CpApU transcript (Fig. 3C, cf. lanes 1 and 2 with lanes 3 and 4). In further support of the view that Gp5.7 inhibits RPO formation at the T7 A1 promoter, EσS was able to synthesize the CpApU transcript in the presence of Gp5.7 when the latter was added to a preformed RPO (i.e., when Gp5.7 was added to the reaction after preincubation of EσS and the homoduplex T7 A1 promoter at 37 °C; Fig. 3D). To better understand how Gp5.7 inhibits RPO formation at the T7 A1 promoter, we used the 2D 1H-15N HSQC NMR data of the interaction between Gp5.7 and R4 of σS (Fig. 2 F and G), the solution structure of Gp5.7 (8) and the X-ray crystal structures of EσS-transcription initiation complex (TIC; in which the interaction between R4 of σS and the consensus −35 promoter region is absent; ref. 11), and the E. coli Eσ70 TIC (in which the interaction between the consensus −35 promoter region and the R4 of σ70 is present; ref. 16) to construct a model of Gp5.7-bound EσS TIC, using HADDOCK (17). This model, shown in SI Appendix, Fig. S2, suggests that Gp5.7 binds to EσS in such an orientation that the positively changed side chains of amino acid residues R24 and R47 face the DNA region immediately adjacent to the consensus −35 motif of the T7 A1 promoter. Because efficient RPO formation at the T7 A1 promoter depends on the interaction between R4 of σ70 (12–15) and σS (Fig. 3 A and B) and the consensus −35 promoter region, we envisage a scenario in which the interaction of Gp5.7 with this region of the T7 A1 promoter antagonizes the interactions between R4 and the consensus −35 motif that are required for efficient RPO formation at this promoter. This view is also supported by our previous observation that apo Gp5.7 interacts, albeit weakly, with the region immediate upstream of the −35 motif of the T7 A1 promoter (8), and an alanine substitution at R24 (but not R47) renders Gp5.7 inactive in vivo (8). Further, the model suggests that Gp5.7 is also proximal to core RNAP subunits (notably the β subunit), consistent with the finding that Gp5.7 can interact with the RNAP in the absence of any σ factors (8). Overall, we conclude that R4 of σS is important for RPO formation at the T7 A1 promoter and Gp5.7 inhibits RPO formation by EσS during T7 development by interfering with the R4 of σS.
Fig. 3.
Gp5.7 inhibits RPO formation by EσS on the T7 A1 promoter. (A) Autoradiograph of denaturing gels showing the ability of the EσSΔR4 to synthesize a dinucleotide-primed RNA product from the T7 A1 and lacUV5 promoters. The dinucleotide used in the assay is underlined and the asterisks indicate the radiolabeled nucleotide. (B) Autoradiographs of nondenaturing gels showing the ability of EσS and EσSΔR4 to form the initial promoter complex (<4 °C) and the RPO (at 37 °C) on the T7 A1 promoter. The migration positions of promoter complexes and free DNA are indicated. Reactions to which heparin were added are indicated. See Materials and Methods for details. (C) As in A, but using the T7 A1 homoduplex and heteroduplex (−12 to +2) promoters. (D) As in A, but Gp5.7 (1,875 nM) was added to the preformed RPO [formed using 75 nM EσS (see text for details)].
Role for Gp5.7 in Managing σS During T7 Development in Stationary Phase E. coli.
The results so far indicate that EσS accumulates as a consequence of (p)ppGpp buildup during T7 development in exponentially growing E. coli cells and that Gp5.7 is required to preferentially inhibit EσS activity on the T7 A1 promoter. However, when E. coli cells are in the stationary phase of growth, the major species of RNAP molecules will contain σS [also because of the buildup of (p)ppGpp in response to the stresses encountered by E. coli cells in the stationary phase of growth; reviewed in ref. 18]. In addition, (p)ppGpp, together with EσS, also contributes to the shutting-down of cellular activities in the stationary phase of growth. Therefore, the development of a phage can be affected by changes in the growth state, and thus cellular activities, of the bacterial cell. Consistent with this view, Nowicki et al. (19) recently reported that progeny production by Shiga toxin converting lamboid phages was significantly more efficient in a ΔrelA/ΔspoT E. coli [which is unable to synthesize (p)ppGpp] than in its isogenic wild-type strain.
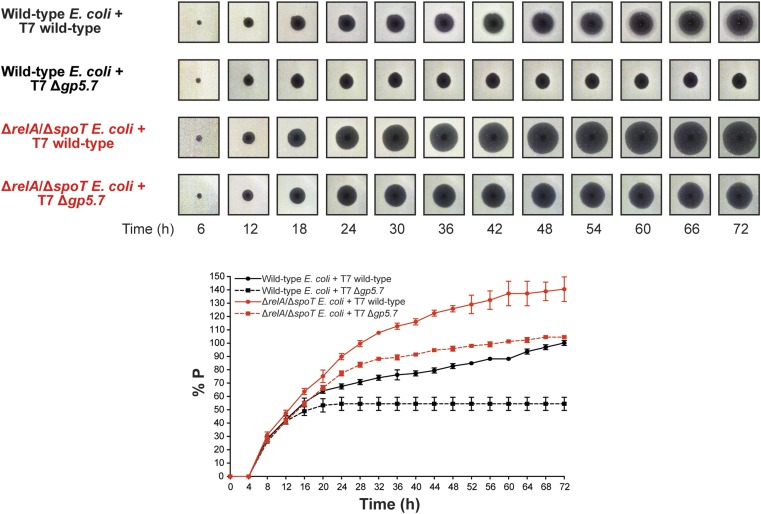
A phage plaque is a clearing in a bacterial lawn, and plaques form via an outward diffusion of phage progeny virions that prey on surrounding bacteria. Therefore, the rate of plaque enlargement can serve as a proxy for how efficiently a phage develops and produces progeny within an infected bacterial cell. Further, although the aging of the bacterial lawn often represents a major barrier for plaque enlargement, T7 plaques have been reported to enlarge continually on matured E. coli lawns (20), suggesting that T7 has evolved specific mechanisms to infect and develop in the stationary phase of E. coli growth. Therefore, we investigated whether Gp5.7 is required for T7 development in E. coli in the stationary phase of growth by measuring plaque size formed on a lawn of E. coli as a function of incubation time in the context of wild-type and ΔrelA/ΔspoT E. coli strains [recall accumulation of σS will be compromised in the mutant strain because of the absence of (p)ppGpp; see Fig. 1B (18)]. As shown in Fig. 4, the rate of plaque enlargement and plaque size on a lawn of wild-type E. coli infected with T7 wild-type and Δgp5.7 phage was indistinguishable for the first 12 h (Fig. 4 and Movie S1). However, whereas T7 wild-type plaques continued to enlarge, the rate at which the plaques formed by T7 Δgp5.7 enlarged significantly slowed after ∼12 h of incubation and completely ceased after ∼20 h of incubation (Fig. 4 and Movie S1). Hence, after 72 h of incubation, the size of the plaque formed by the T7 Δgp5.7 phage was ∼50% smaller than the plaque formed by T7 wild-type phage on a lawn of wild-type E. coli cells. We were able to partially, yet specifically, revert the rate of plaque enlargement and plaque size of the T7 Δgp5.7 phage to that of T7 wild-type phage by exogenously providing Gp5.7 from an inducible plasmid in E. coli (SI Appendix, Fig. S3). The results overall imply that Gp5.7 is required for T7 development in E. coli in the stationary phase of growth. To independently verify this view, the relative efficiency of plaque formation (E.O.P) by the T7 Δgp5.7 phage on exponentially growing E. coli was compared with that on E. coli in the stationary phase of growth. Results shown in SI Appendix, Fig. S4 indicated that the relative E.O.P by the T7 Δgp5.7 phage was almost three times lower than that of the wild-type phage (SI Appendix, Fig. S4). This observation further underscores the view that T7 development in E. coli in the stationary phase of growth is compromised in the absence of Gp5.7.
Fig. 4.
A role for Gp5.7 in managing σS during T7 development in stationary phase E. coli. (Top) Representative scanned images of T7 wild-type and T7 Δgp5.7 plaques formed on a lawn of wild-type E. coli and ΔrelA/ΔspoT E. coli over a 72 h incubation period. (Bottom) Graph showing plaque size (%P) as percentage of final plaque size formed by T7 wild-type phage on a lawn of wild-type E. coli after 72 h of incubation (set at 100%) as a function of incubation time.
In marked contrast, although the rate of enlargement of the plaques formed by T7 wild-type phage was similar on a lawn of ΔrelA/ΔspoT E. coli to that observed on a lawn of wild-type E. coli for the first 8 h of incubation, the plaques formed by T7 wild-type phage continued to enlarge at a faster rate on a lawn of ΔrelA/ΔspoT E. coli than on a lawn of wild-type E. coli (Fig. 4). For example, after 48 h of incubation, the size of the plaques formed by the T7 wild-type phage on a lawn of ΔrelA/ΔspoT E. coli was ∼twofold larger than those formed on a lawn of wild-type E. coli (Fig. 4). Strikingly, whereas the plaques formed by the T7 Δgp5.7 phage ceased enlarging after ∼20 h of incubation on a lawn of wild-type E. coli, they continued to enlarge (albeit at a slower rate than that of T7 wild-type phage) on a lawn of ΔrelA/ΔspoT E. coli (Fig. 4). Overall, although we cannot fully exclude the possibility that the absence of (p)ppGpp in ΔrelA/ΔspoT E. coli will generally provide more favorable intracellular conditions for T7 development than in wild-type E. coli cells, the results clearly demonstrate that the accumulation of (p)ppGpp during T7 infection antagonizes T7 development in E. coli, and Gp5.7 represents a mechanism by which T7 overcomes the antagonistic effect of (p)ppGpp-mediated accumulation of σS on its development, and therefore Gp5.7 is also a T7 factor required for T7 development in E. coli cells in the stationary phase of growth.
Discussion
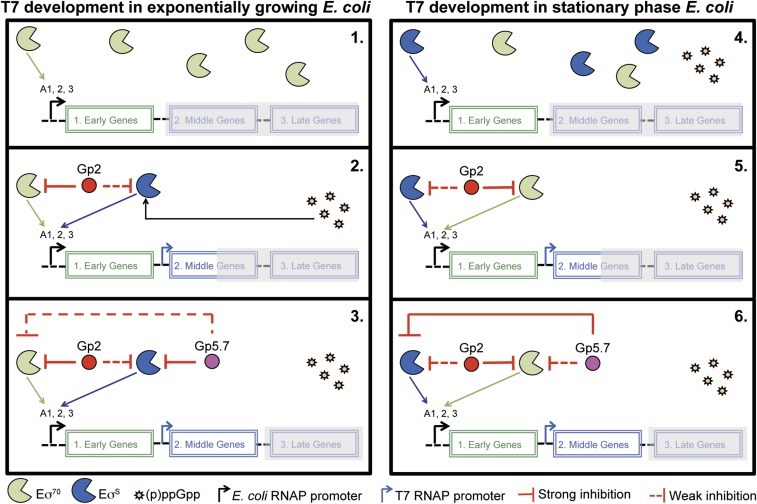
The inhibition of the host transcription machinery, the RNAP, is a central theme in the strategies used by phages to acquire their bacterial prey. In the prototypical E. coli phage T7, the switching from using the host RNAP for transcription of early T7 genes to the T7 RNAP for transcription of middle and late T7 genes is tightly regulated by two bacterial RNAP inhibitors, Gp2 and Gp5.7. Dysregulation of this process, for example, because of the absence of any of these factors, is believed to lead to steric interference between the host and T7 RNAP molecules on the T7 genome and results in compromised or aborted development of phage progeny (5, 8). In an earlier study, we proposed that Gp5.7 acts as a last line-of-defense molecule to prevent aberrant transcription of middle and late T7 genes by host RNAP molecules that may have escaped inhibition by Gp2 (8). The present study has uncovered additional biological roles for Gp5.7 in the T7 development cycle. The biological roles for Gp5.7 in the T7 developmental cycle uncovered in the present study are summarized in the model in Fig. 5, which is partly supported by experimental evidence but is also based on several assumptions (e.g., the differences in the intracellular levels of phage proteins and σ factors) that may not hold up as more evidence emerges. The results from the in vitro experiments presented here imply that during infection of exponentially growing E. coli cells by T7 phage, Gp5.7 serves to inhibit transcription initiation from T7 A1-3 promoters by EσS (Fig. 1C), which accumulates (Fig. 1B), possibly as a consequence of the (p)ppGpp-mediated stress response mechanism (Fig. 1A) to T7 infection (Fig. 5, box 1–3). Gp5.7 is also required for T7 development in E. coli in the stationary phase of growth (Fig. 5, box 4–6). In this case, we envisage that Gp5.7 will be absent when the transcription (of early T7 genes)-dependent translocation of the T7 genome by EσS occurs during infection of E. coli in the stationary phase of growth (Fig. 5, box 4), but becomes available when the EσS is no longer required (i.e., when the T7 RNAP takes over the transcription of the middle and late genes; Fig. 5, box 5). The fact that Gp2 only poorly inhibits EσS (7) further supports the need for Gp5.7 for T7 development in both exponentially growing and stationary phase E. coli cells. Thus, to the best of our knowledge, Gp5.7 is the only phage-encoded host RNAP inhibitor (or phage factor) described to date that is required for successful phage development in stationary phase bacteria. Intriguingly, we are unable to rescue the T7 Δgp5.7 phage in a ΔrpoS E. coli strain in the context of the plaque-enlargement assay shown in Fig. 4. As shown in SI Appendix, Fig. S5, wild-type and Δgp5.7 T7 phages are equally compromised to efficiently develop in the ΔrpoS E. coli strain. However, based on the assumption that the intracellular levels of Eσ70 will be higher in ΔrpoS E. coli than in wild-type E. coli because of the absence of the competing σS (21), we propose that T7 is unable to efficiently develop in ΔrpoS E. coli because of the inadequate ability of Gp2 and Gp5.7 (and Gp0.7) to inhibit the excess Eσ70 molecules, which will presumably dilute the intracellular pool of Gp2 and Gp5.7 (and Gp0.7) available to fully inhibit Eσ70 to allow optimal T7 development. Overall, our results indicate that although T7 development in E. coli depends on the host RNAP (for transcription-dependent translocation of T7 genome into the bacterial cell and transcription of early T7 genes), efficient management of host RNAP activity is clearly obligatory for T7 to optimally develop in both exponentially growing and stationary phase E. coli cells. As a consequence, any perturbations in RNAP levels or activity can have adverse effects on T7 development.
Fig. 5.
Model proposing how the host RNAP is managed during T7 development in exponentially growing and stationary phase E. coli cells.
We further note that although Gp2 and Gp5.7 bind to sites located at different faces of the RNAP [with respect to the active center of the RNAP, the Gp2 binding site is located on the β′ jaw domain at the downstream face of the RNAP (10), whereas the Gp5.7 binding site is located at the upstream face of the RNAP (this study)], both T7 proteins seem to inhibit RPO formation by misappropriation of essential domains of the σ factor [region 1 of σ70 in case of Gp2 (7, 22), and R4 of σS in the case of Gp5.7 (this study)]. Because the R4 domain of σ70 is also targeted by a T4 phage protein, called AsiA, to recruit the host RNAP to transcribe phage genes (reviewed in ref. 23), it is interesting to speculate whether phages, regardless of their dependence on the host RNAP (unlike the T7 phage, the T4 phage fully relies on the host RNAP for the transcription of its genes), have evolved to misappropriate essential bacterial σ factor domains to inhibit (e.g., T7) or redirect (e.g., T4) host RNAP activity to serve phage developmental requirements.
This study unambiguously shows that (p)ppGpp accumulates in T7 infected E. coli cells. The involvement of (p)ppGpp in phage development has been previously documented. For example, (p)ppGpp is required for the replication of phage Mu in E. coli (24), and in phage lambda it contributes to the switching between the lytic and lysogenic cycles (25). However, the role of (p)ppGpp in T7 development and the signaling pathway or pathways that results in its synthesis are unknown. In E. coli, two different pathways are involved in the production of (p)ppGpp: the RelA- and SpoT-dependent pathways (reviewed in ref. 26). RelA is associated with ribosomes and produces (p)ppGpp in response to uncharged tRNA in the ribosomal A-site during amino acid starvation. In contrast, SpoT is primarily responsible for the accumulation of (p)ppGpp in response to most stresses (e.g., fatty acid or iron starvation) and nutrient limitations (e.g., carbon starvation) apart from amino acid starvation. However, it seems paradoxical that T7-infected E. coli cells experience amino acid starvation, as cellular translation becomes increased during T7 development (to serve phage gene expression needs) through the phosphorylation of translation elongation factors G and F and the ribosomal protein S6 (27). Although this study describes a strategy T7 uses to mitigate the effect of accumulation of (p)ppGpp during T7 development in E. coli, clearly, the role of (p)ppGpp in T7 development and the signaling pathway or pathways that induce its synthesis warrant further investigation.
In summary, our study has uncovered the distinct strategies used by this phage to shut down bacterial RNAP for an optimal infection outcome in E. coli in an exponentially growing and stationary phase of growth. The latter is clearly relevant to bacteria encountered by T7 in the natural environment, which are often in a starved, and thus in a growth-attenuated or slow-growing, state. Therefore, the insights from this study also have implications for the emerging interest in the use of phages and phage-derived antibacterial compounds and their bacterial targets to treat bacterial infections where bacteria largely exist in a stressed state and mostly depend on EσS-dependent gene expression for survival (28).
Materials and Methods
(p)ppGpp Measurements.
A culture of E. coli MG1655 rpoC-FLAG was setup from an overnight culture in 5 mL potassium morpholinopropane sulfonate minimal media with a starting OD600 of 0.05 at 37 °C. At OD600 0.1, 20 μCi/mL [32P] H3PO4 was added as phosphate source, and the culture was left to grow to an OD600 of 0.45. The culture was then infected with T7 wild-type (ratio of 10:1, T7:E. coli) in the presence of 1 mM CaCl2. To detect (p)ppGpp production, 500 μL of the cultures before infection (time 0) and at 10 min after infection were added to 100 μL ice cold 2 M formic acid and incubated on ice for 30 min. The samples were then centrifuged for 5 min at 17,000 × g, and 10 μL supernatant was spotted on TLC polyethylenimine cellulose F (dimensions 20 cm × 20 cm; Merck Millipore). The spot was left to migrate to the top of the sheet in a TLC tank in the presence of 1.5 M KH2PO4 at pH 3.6, dried before exposing overnight onto phosphor screen, and viewed using PhosphorImager. For a positive control, 100 μg/mL serine hydroxamate was added to MG1655 rpoC-FLAG at OD600 of 0.45 and a sample taken after 10 min and processed as described earlier.
Western Blotting.
E. coli MG1655 rpoC-FLAG strain was grown in LB at 30 °C to an OD600 of ∼0.45. The culture was then infected with T7 wild-type (ratio of 0.1:1, T7:E. coli) in the presence of 1 mM CaCl2. To detect σS production, 20 mL of the culture before infection (time 0) and at 10-min intervals after infection were taken until complete lysis was obtained. Experiments with the MG1655 ΔrelA/ΔspoT strain (ref. 29; kindly provided by Kenn Gerdes, Centre for Bacterial Stress Response and Persistence, University of Copenhagen, Copenhagen) were conducted exactly as described here, but samples were taken at 0 and 40 min after T7 infection. Cultures were centrifuged and cell pellets resuspended in 500 μL of 20 mM Na2PO4, 50 mM NaCl, and 5% glycerol and sonicated. The cleared cell lysate was then loaded on 4–20% SDS/PAGE and ran at 200 V for 30 min. The SDS/PAGE gel was transferred onto polyvinylidene difluoride membrane (0.2 μm), using a Trans-Blot Turbo Transfer System (Bio-Rad) device, and processed according to standard molecular biology protocols. The primary antibodies were used at the following titers: anti-E. coli RNAP σS antibody at 1:500 (1RS1; Biolegend) and anti-E. coli RNAP α-subunit antibody at 1:1,000 (4AR2; Biolegend). The secondary antibody Rabbit Anti-Mouse IgG H&L [horseradish peroxidase (HRP)] was used at 1:2,500 (ab97046; Abcam). Bands were detected using an Amersham ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences) and analyzed on a ChemiDoc, using the Image Lab Software.
Protein Expression and Purification.
FLAG-tagged E. coli σ70 and σS were PCR amplified from E. coli genome and cloned into the pT7-FLAG-1 vector (Sigma-Aldrich). Recombinant vectors pT7-FLAG::rpoD and pT7-FLAG::rpoS were confirmed by DNA sequencing. For the biochemical experiments, recombinant FLAG-tagged E. coli σ70 and σS were made by FLAG affinity purification from E. coli strain BL21 (DE3). Briefly, the culture of BL21 (DE3) cells containing pT7-FLAG::rpoD was grown at 37 °C to an OD600 of ∼0.4 and cold shocked on ice for 15 min before protein expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were left to grow at 16 °C overnight before harvesting. For FLAG-tagged E. coli σS expression, BL21 (DE3) containing pT7-FLAG::rpoS were grown at 37 °C to an OD600 of ∼0.4, and protein expression was induced with 0.1 mM IPTG. The cells were left to continue growing at 37 °C for 3 h before harvesting. The cell pellets for both FLAG-tagged E. coli σ70 and σS were resuspended in binding buffer (50 mM Tris⋅HCl, 150 mM NaCl at pH 7.4) containing a mixture of protease inhibitors and lysed by sonication. The cleared cell lysate was loaded to a column containing anti-FLAG M2 affinity gel (Sigma-Aldrich), and the purified proteins were obtained by adding elution buffer [100 μg/mL 3XFLAG peptide (Sigma-Aldrich) in binding buffer] for 30 min at 4 °C. The purified proteins were dialyzed into storage buffer (10 mM Tris⋅HCl at pH 8.0, 50 mM NaCl, 50% glycerol, 0.1 mM EDTA, and 1 mM DTT) and stored in aliquots at −80 °C. The FLAG-tagged σSΔR4 (amino acid residues 1–262) was made by introducing a stop codon into pT7-FLAG::rpoS by site-directed mutagenesis, and its expression and purification were done as described for the full-length protein. The 6xHis-σSR4 (amino acid residues 245–330) was amplified from E. coli genomic DNA by Gibson assembly and ligated into the pET-46 Ek/LIC vector (Merck Millipore) and expressed in E. coli strain BL21 (DE3) for the pull-down experiments and structural studies. The cells were grown in either LB (for pull-down experiments) or M9 Minimal medium labeled with 15N and 13C (for the structural studies) and induced with 0.5 mM IPTG when the OD600 reached 0.6, and incubated overnight at 18 °C before harvesting by centrifugation. The cells were lysed by sonication in 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole at pH 8, and purified using Ni-NTA beads (Qiagen). The eluate was then dialyzed against 50 mM NaH2PO4 and 350 mM NaCl at pH 6, and subsequently concentrated down for NMR experiments, whereas for pull-down experiments, the 6xHis-σSR4 protein was kept in 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole pH 8. The 6xHis-σS was amplified from E. coli genomic DNA, cloned into pET-46 Ek/LIC vector (Merck Millipore) by Gibson assembly, expressed and purified as described for 6xHis-σSR4. The cells were lysed by sonication under denaturing condition containing 8 M urea, purified with Ni-NTA beads under denaturing conditions, and the denatured 6xHis-σs was refolded in 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole at pH 8. The 6xHis-Gp5.7 was amplified from pBAD18::gp5.7 (8) and cloned into pET-46 plasmid. The Histidine tag on Gp5.7 was deleted to express tag-free Gp5.7, using a Q5 site-directed mutagenesis kit (NEB). The protein was expressed under the same condition as 6xHis-σSR4. Recombinant 6xHis-Gp5.7 and 6xHis-Gp2 expression and purification were performed exactly as previously described (8, 10). Sequences of all oligonucleotides used in the construction of the expression vectors are available in SI Appendix, Table S2.
In Vitro Transcription Assays.
In vitro transcription assays were conducted exactly as previously described (8) in 10 mM Tris at pH 7.9, 40 mM KCl, 10 mM MgCl2, using E. coli core RNAP from NEB, and FLAG-tagged versions of σ70 and σS were purified exactly as described here. Reactions in Fig. 3 C and D were conducted in 100 mM K-glutamate, 40 mM Hepes at pH 8, 10 mM MgCl2 and 100 μg/mL BSA. In all reactions, Gp5.7 or Gp2 was preincubated with EσS or Eσ70 at the indicated concentrations before adding promoter DNA to the reaction. However, in the reactions shown in Fig. 3D, Gp5.7 was added to the preformed RPO (i.e., after preincubation of EσS and the promoter DNA). Sequences of all oligonucleotides used to generate promoter probes are available in ref. 8 or on request.
Pull-Down Assays.
For the pull-down assays shown in Fig. 2 A and B, Ni-NTA beads (Qiagen) were used. Approximately 0.02 mg of recombinant 6xHis-σS or 6xHis-σSR4 in binding buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole at pH 8) was added to beads and incubated at 4 °C for 30 min. E. coli whole-cell lysate containing overexpressed untagged Gp5.7 was added to resin containing sigma and incubated for 1 h at 4 °C. The beads were washed three times in 1 mL wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole at pH 8) for 10 min to remove any nonspecific protein–protein interaction. To elute samples from beads, elution buffer containing 250 mM imidazole was added. For FLAG-tag protein pull-down assay (Fig. 2C), 0.02 mg FLAG-σSΔR4 was incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) in 50 mM Tris⋅HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA at 4 °C for 2 h. E. coli whole-cell lysate containing overexpressed untagged Gp5.7 was added to resin containing sigma and incubated for 2 h at 4 °C. The beads were washed three times in 50 mM Tris⋅HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 for 10 min to remove any nonspecific protein–protein interaction. To elute samples from beads, elution buffer containing 100 µg/mL 3XFLAG peptide (Sigma-Aldrich) was added. Ten microliters of samples together with Laemmli 2× concentrate SDS Sample Buffer was loaded on a 10–15% SDS/PAGE alongside Protein standard Marker and stained with Coomassie Brilliant Blue.
NMR Structure Determination.
NMR spectra were collected at 310 K on Bruker DRX600 and DRX800 spectrometers equipped with cryo-probes. Spectral assignments were completed using our in-house, semiautomated assignment algorithms and standard triple-resonance assignment methodology (30). Hα and Hβ assignments were obtained using HBHA (CBCACO)NH, and the full side-chain assignments were extended using HCCH-total correlation (TOCSY) spectroscopy and (H)CC(CO)NH TOCSY. 3D 1H-15N/13C NOESY-HSQC (mixing time, 100 ms at 800 MHz) experiments provided the distance restraints used in the final structure calculation (31). The ARIA protocol was used for completion of the NOE assignment and structure calculation. The frequency window tolerance for assigning NOEs was ±0.025 ppm and ±0.03 ppm for direct and indirect proton dimensions and ±0.6 ppm for both nitrogen and carbon dimensions. The ARIA parameters p, Tv, and Nv were set to default values. One hundred eight dihedral angle restraints derived from TALOS+ were also implemented. The 10 lowest energy structures had no NOE violations greater than 0.5 Å and dihedral angle violations greater than 5°. The structural statistics are shown in SI Appendix, Table S1.
NMR Titration.
Unlabeled Gp5.7 was added to 15N-labeled 6xHis-σSR4, according to stoichiometric ratio to perform NMR titration. Maximum fivefold Gp5.7 was added to 6xHisσSR4 to broad out the entire spectra.
Electrophoretic Mobility Shift Assays.
These were conducted exactly as previously described to distinguish between initial promoter complex and RPO formation (10). Briefly, 75 nM E. coli core RNAP (NEB) was incubated with 300 nM σS or σSΔR4 either on ice (to monitor initial promoter complex formation) or at 37 °C (to monitor RPO formation) for 5 min in 100 mM K-glutamate, 40 mM Hepes at pH 8, 10 mM MgCl2, and 100 μg/mL BSA. Twenty nanomolars of 32P-labeled T7 A1p was added and incubated for 5 min. Because initial T7 A1 promoter complexes (formed at temperatures <4 °C) are sensitive to heparin and, conversely, RPO (formed at 37 °C) are resistant to heparin (32), the reactions were challenged with 100 μg/mL heparin before separating the RNAP bound and free promoter DNA by native gel electrophoresis on a 4.5% (wt/vol) native polyacrylamide gel run at 100 V for 100 min at 4 °C (to monitor initial promoter complex formation) or for 60 min at room temperature (to monitor RPO formation). The dried gel was then analyzed by autoradiography.
Plaque-Enlargement Assay.
T7 phage plaques were formed as described in ref. 8. To obtain images of plaques, E. coli MG1655 cultures and MG1655 ΔrelA/ΔspoT were grown to an OD600 of 0.45 in LB at 30 °C and 300 μL aliquots of the culture were taken out and either T7 wild-type or T7 Δgp5.7 lysate (sufficient to produce ∼10 plaques) were added together with 1 mM CaCl2 and incubated at 37 °C for 10 min to allow the phage to adsorb to the bacteria. Three milliliters of 0.7% (wt/vol) top agar was added to each sample and plated onto plates containing exactly 20 mL of 1.5% (wt/vol) LB agar. The plates were then put in a Epson perfection V370 photo scanner (Model J232D) inside a 30 °C incubator, and images of the plates were taken every 2 h during a 72-h period for analysis. For complementation experiments, E. coli MG1655 cells containing pBAD18::empty or pBAD18::gp5.7 or pBAD18::gp5.7-L42A (8) were used, and the plaque-enlargement assay was carried out as above on plates containing 100 μg/mL ampicillin and 0.04% (wt/vol) l-arabinose to induce gp5.7 expression. The MG1655 ΔrpoS strain was obtained by phage transduction from ΔrpoS mutant in Keio library (33).
Supplementary Material
Acknowledgments
We thank Daniel Brown for help with the (p)ppGpp measurements. This work was supported by Wellcome Trust awards (Investigator Award 100280 and Multiuser Equipment Grant 104833 to S.J.M. and Investigator Award 100958 to S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6FI7), and NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 34234).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800429115/-/DCSupplemental.
References
- 1.De Smet J, Hendrix H, Blasdel BG, Danis-Wlodarczyk K, Lavigne R. Pseudomonas predators: Understanding and exploiting phage-host interactions. Nat Rev Microbiol. 2017;15:517–530. doi: 10.1038/nrmicro.2017.61. [DOI] [PubMed] [Google Scholar]
- 2.Nechaev S, Severinov K. The elusive object of desire–Interactions of bacteriophages and their hosts. Curr Opin Microbiol. 2008;11:186–193. doi: 10.1016/j.mib.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 4.Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 5.Savalia D, Robins W, Nechaev S, Molineux I, Severinov K. The role of the T7 Gp2 inhibitor of host RNA polymerase in phage development. J Mol Biol. 2010;402:118–126. doi: 10.1016/j.jmb.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Severinova E, Severinov K. Localization of the Escherichia coli RNA polymerase beta’ subunit residue phosphorylated by bacteriophage T7 kinase Gp0.7. J Bacteriol. 2006;188:3470–3476. doi: 10.1128/JB.188.10.3470-3476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James E, et al. Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2. Mol Cell. 2012;47:755–766. doi: 10.1016/j.molcel.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabib-Salazar A, et al. Full shut-off of Escherichia coli RNA-polymerase by T7 phage requires a small phage-encoded DNA-binding protein. Nucleic Acids Res. 2017;45:7697–7707. doi: 10.1093/nar/gkx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen JD, Fiil N. Accumulation of guanosine tetraphosphate in T7 bacteriophage-infected Escherichia coli. J Bacteriol. 1973;113:697–703. doi: 10.1128/jb.113.2.697-703.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cámara B, et al. T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc Natl Acad Sci USA. 2010;107:2247–2252. doi: 10.1073/pnas.0907908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Zuo Y, Steitz TA. Structures of E. coli σS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci USA. 2016;113:4051–4056. doi: 10.1073/pnas.1520555113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Tang H, Ebright RH. Functional interaction between RNA polymerase alpha subunit C-terminal domain and sigma70 in UP-element- and activator-dependent transcription. Mol Cell. 2003;11:1621–1633. doi: 10.1016/s1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 13.Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. An intersubunit contact stimulating transcription initiation by E coli RNA polymerase: Interaction of the alpha C-terminal domain and sigma region 4. Genes Dev. 2003;17:1293–1307. doi: 10.1101/gad.1079403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase sigma 70 region 4.2 and alpha-subunit C-terminal domains in promoter complex formation on the extended -10 galP1 promoter. J Biol Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 15.Sclavi B, et al. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc Natl Acad Sci USA. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 18.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowicki D, Kobiela W, Węgrzyn A, Wegrzyn G, Szalewska-Pałasz A. ppGpp-dependent negative control of DNA replication of Shiga toxin-converting bacteriophages in Escherichia coli. J Bacteriol. 2013;195:5007–5015. doi: 10.1128/JB.00592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin J. Evolution of bacteriophage T7 in a growing plaque. J Bacteriol. 1993;175:1272–1277. doi: 10.1128/jb.175.5.1272-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: A case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 22.Bae B, et al. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of σ70 domain 1.1. Proc Natl Acad Sci USA. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinton DM. Transcriptional control in the prereplicative phase of T4 development. Virol J. 2010;7:289. doi: 10.1186/1743-422X-7-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.North SH, Kirtland SE, Nakai H. Translation factor IF2 at the interface of transposition and replication by the PriA-PriC pathway. Mol Microbiol. 2007;66:1566–1578. doi: 10.1111/j.1365-2958.2007.06022.x. [DOI] [PubMed] [Google Scholar]
- 25.Slomińska M, Neubauer P, Wegrzyn G. Regulation of bacteriophage lambda development by guanosine 5′-diphosphate-3′-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- 26.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson ES, Aggison LA, Nicholson AW. Phosphorylation of elongation factor G and ribosomal protein S6 in bacteriophage T7-infected Escherichia coli. Mol Microbiol. 1994;11:1045–1057. doi: 10.1111/j.1365-2958.1994.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 28.Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens. Infect Immun. 2010;78:887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchant J, Sawmynaden K, Saouros S, Simpson P, Matthews S. Complete resonance assignment of the first and second apple domains of MIC4 from Toxoplasma gondii, using a new NMRView-based assignment aid. Biomol NMR Assign. 2008;2:119–121. doi: 10.1007/s12104-008-9100-1. [DOI] [PubMed] [Google Scholar]
- 31.Pardi A. Multidimensional heteronuclear NMR experiments for structure determination of isotopically labeled RNA. Methods Enzymol. 1995;261:350–380. doi: 10.1016/s0076-6879(95)61017-0. [DOI] [PubMed] [Google Scholar]
- 32.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.