Significance
Cotton is an important crop, and terpenoids form the largest group of natural products. Gossypol and related sesquiterpene aldehydes in cotton function as phytoalexins against pathogens and pests but pose human health concerns, as cotton oil is still widely used as vegetable oil. We report the isolation and identification of four enzymes and the recharacterization of one previously reported P450. We are now close to the completion of the gossypol pathway, an important progress in agricultural and plant sciences, and the data are beneficial to improving food safety. Among the six compounds (intermediates) isolated following gene silencing, one affected plant disease resistance significantly. Thus, these “hidden natural products” harbor interesting biological activities worthy of exploration.
Keywords: cotton, sesquiterpene, gossypol biosynthesis, P450, secondary metabolism
Abstract
Gossypol and related sesquiterpene aldehydes in cotton function as defense compounds but are antinutritional in cottonseed products. By transcriptome comparison and coexpression analyses, we identified 146 candidates linked to gossypol biosynthesis. Analysis of metabolites accumulated in plants subjected to virus-induced gene silencing (VIGS) led to the identification of four enzymes and their supposed substrates. In vitro enzymatic assay and reconstitution in tobacco leaves elucidated a series of oxidative reactions of the gossypol biosynthesis pathway. The four functionally characterized enzymes, together with (+)-δ-cadinene synthase and the P450 involved in 7-hydroxy-(+)-δ-cadinene formation, convert farnesyl diphosphate (FPP) to hemigossypol, with two gaps left that each involves aromatization. Of six intermediates identified from the VIGS-treated leaves, 8-hydroxy-7-keto-δ-cadinene exerted a deleterious effect in dampening plant disease resistance if accumulated. Notably, CYP71BE79, the enzyme responsible for converting this phytotoxic intermediate, exhibited the highest catalytic activity among the five enzymes of the pathway assayed. In addition, despite their dispersed distribution in the cotton genome, all of the enzyme genes identified show a tight correlation of expression. Our data suggest that the enzymatic steps in the gossypol pathway are highly coordinated to ensure efficient substrate conversion.
Humans have domesticated wild plants to develop them as a safe food source. Most plants produce specialized (secondary) metabolites that confer resistance to pathogens (1) and herbivores (2) (including insects and mammals). In addition to their toxicity, specialized metabolites possess undesirable antinutritional properties that have been reduced or removed from human and domestic-animal foods during domestication. For example, potato (Solanum tuberosum) (3) and tomato (S. lycopersicum) (4, 5) have been bred for low levels of toxic steroidal glycoalkaloids, and cucumber (Cucumis sativus) cultivars contain low levels of bitter cucurbitacins (6, 7).
In the case of cotton species that have been cultivated mainly for spinnable fiber to produce clothing, their specialized metabolites may not have been under the negative selection pressure in the course of domestication, compared with food crops. Plants of cotton synthesize a group of cadinene-type sesquiterpene aldehydes as defense compounds (phytoalexins), represented by gossypol (8–10). Cottonseeds are valuable since they are good sources of protein (∼23%) and oil (∼21%). Cottonseed meal is widely used as animal feed, and cotton oil is still the major cooking oil in some developing countries, such as Pakistan (11, 12). As a result, high gossypol content in cottonseeds poses a health concern (13) for both domestic-animal and human uses.
Elucidation of the gossypol biosynthetic pathway started decades ago. Early 14C tracing experiments proved that (+)-δ-cadinene is a precursor to all cadinene-type sesquiterpenoids in cotton, including both 7- and 8-hydroxylated derivatives (14, 15). Sesquiterpene synthases convert farnesyl diphosphate (FPP) into differently structured products. The (+)-δ-cadinene synthase (CDN) activity in cotton (15, 16) and the cDNAs encoding two subfamilies of CDNs (CDNA and CDNC) were then reported (17, 18). Later, a cytochrome P450 monooxygenase (CYP706B1) was demonstrated to catalyze the hydroxylation of (+)-δ-cadinene, presumably at the 8- position (19). In addition, a desoxyhemigossypol methyltransferase was characterized (20). Gossypol is formed through dimerization of hemigossypol (21–23). Comparison of (+)-δ-cadinene and hemigossypol structures suggests several hydroxylation, desaturation, and cyclic ether formation steps in the pathway. However, until now, neither the enzymes nor the reactions downstream of (+)-δ-cadinene have been characterized, except a tentative identification of CYP706B1, and even the biosynthetic intermediates remain largely unknown.
All cotton species bear the lysigenous glands located in the subepidermal layer of aerial organs, in which sesquiterpene aldehydes (such as gossypol and hemigossypolone) are stored. There are also glandless cultivars which do not produce these phytoalexins in aerial parts (17, 24, 25) (Fig. 1 A and B). Recently, the gene responsible for gland formation, GoPGF, was cloned, which encodes a basic helix–loop–helix transcription factor (25). By transcriptome-based comparison of the glandular and the glandless cultivars and coexpression analyses, in combination with virus-induced gene silencing (VIGS) and partial reconstitutions of the pathway in heterologous system, we isolated four enzymes and identified five steps of the pathway, covering the first four consecutive steps and most of the hydroxylation reactions of gossypol biosynthesis.
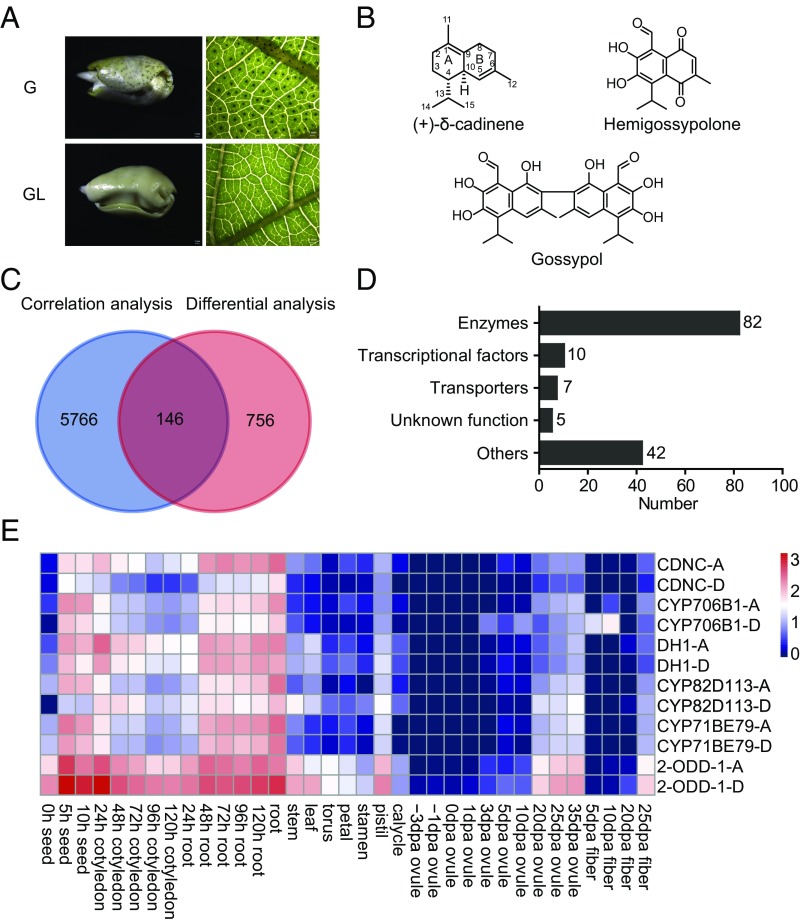
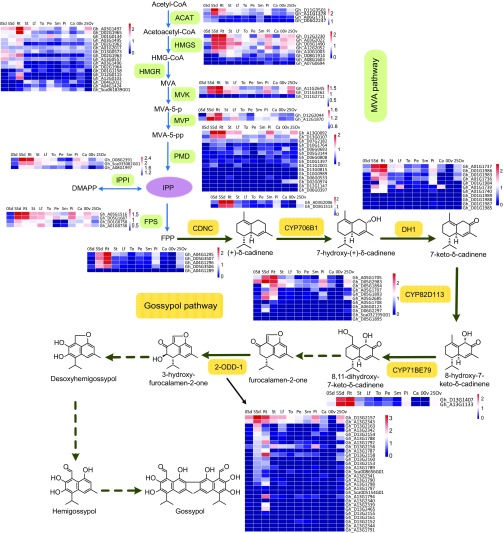
Fig. 1.
Transcriptomics-based mining of gossypol pathway genes. (A) View of the seed and leaf of glandular (G) and glandless (GL) cultivars of G. hirsutum. (B) Structure of (+)-δ-cadinene, gossypol, and hemigossypolone. (C and D) Venn diagram (C) showing the numbers of genes identified by correlation analysis using CDNC as a bait (correlation ≥0.5) and by differential analysis (down-regulated in glandless cotton leaf). In total, 146 genes were retrieved by both methods, and their numbers in each category are shown in D. (E) Global heatmap of transcript abundances of indicated genes in different organs or in ovule (seed) at different stages. In the expression cluster, DH1, CYP82D113, CYP71BE79, and 2-ODD-1 are most correlated to the reported gossypol pathway genes CDNC and CYP706B1. The heatmap was drawn by the R pheatmap package. Hours (h) postgermination and days postanthesis (dpa) are indicated.
Results
Isolation of Gossypol Pathway Genes.
Upland cotton, Gossypium hirsutum, is an allotetraploid species widely cultivated around the world (26). Analyses by HPLC detected a high level of sesquiterpene aldehydes in the leaf, seed (cotyledon), and floral organs of G. hirsutum cv. CCRI12, but not the glandless mutant CCRI12gl (SI Appendix, Fig. S1A). Although the sesquiterpenes are widely distributed throughout the glandular cotton plant, their level and composition in different organs vary: while gossypol is predominant in seed and root, hemigossypolone is abundant in leaf (SI Appendix, Fig. S1A).
In cotton CDN, a sesquiterpene cyclase and the cytochrome P450 monooxygenase CYP706B1 catalyze the first two steps of gossypol biosynthesis (17, 19). To further characterize the pathway, we adopted an integrative approach combining two-stage transcriptome analyses and VIGS to isolate genes encoding the downstream enzymes. Comparison of the transcript abundances in the leaves of glandular and glandless cotton uncovered 902 genes significantly down-regulated in the latter (Fig. 1C). Next, correlation analysis using the correlation value of ≥0.5 grouped 5,912 transcripts with the bait CDNC of the CDN family (Fig. 1C). Combination of these two datasets disclosed 146 genes in total that were potentially linked to gossypol biosynthesis, among which 82 encode enzymes, including the previously reported CDNC and CYP706B1, and the mevalonate (MVA) pathway genes (Fig. 1D). Subsequent analysis of spatial expression patterns using the R pheatmap package identified seven enzymes that form the most likely gene expression cluster related to gossypol biosynthesis (Fig. 1E and SI Appendix, Table S1), of which four have not been investigated before.
Real-time quantitative PCR confirmed the RNA-sequencing data: the four enzyme genes were tightly coexpressed with CDNC and CYP706B1, with their transcript levels high in glandular leaves but low or undetectable in glandless leaves (Fig. 2A). During development, young ovules (seeds) do not produce gossypol until 20 d postanthesis (SI Appendix, Fig. S1B), when CDNC and CYP706B1 as well as the four candidate genes were coordinately activated, concomitant with gossypol accumulation (Fig. 2B).
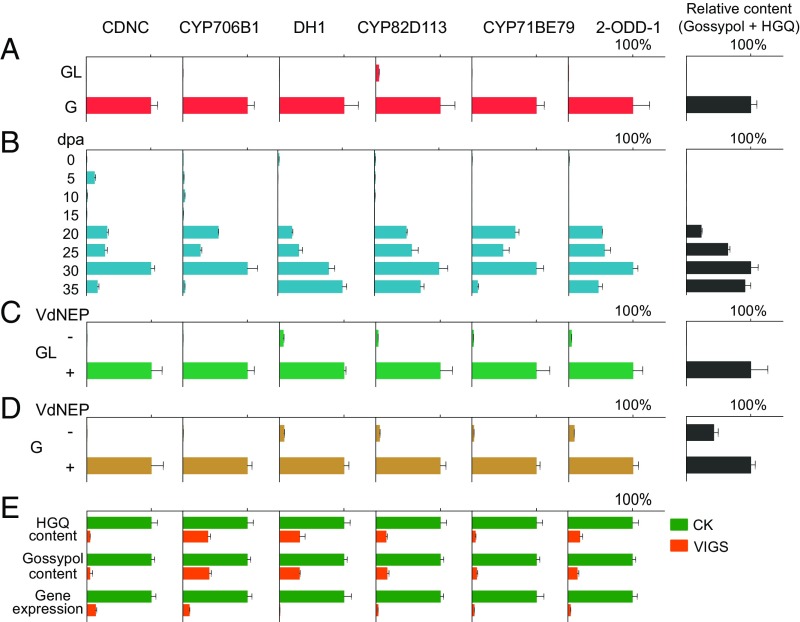
Fig. 2.
Relative gene expressions of the enzymes in relation to accumulation of gossypol and hemigossypolone (HGQ). Six enzymes were analyzed, including the previously reported CDNC and CYP706B1, and the four isolated in this study: DH1, CYP82D113, CYP71BE79, and 2-ODD-1. (A) Down-regulation of the genes in leaves of the glandless cotton cultivar CCRI12gl (GL) compared with the glanded cultivar CCRI12 (G) (means ± SD, n = 3). (B) Relative gene expressions in developing ovule (seed) collected at different days postanthesis (dpa) (means ± SD, n = 3). (C and D) Induced gene expression in GL (C) and G (D) cotyledons after treatment with fungal elicitor VdNEP (means ± SD, n = 3). (E) Decreased gene expression level and gossypol/HGQ content in leaves after VIGS of the gene as indicated. Value of the empty tobacco rattle virus (TRV) vector control (CK) was set to 1 (means ± SD, n = 6 independent experiments). See also SI Appendix, Figs. S1 and S2.
Previous investigations demonstrated that biosynthesis of sesquiterpene phytoalexins in cotton cells can be induced by the pathogenic fungus Verticillium dahliae (17, 20). HPLC analysis showed that treatment of cotton cotyledons by the V. dahliae elicitor VdNEP (27) led to increased production of gossypol and hemigossypolone, whereas in glandless cotyledons, in which the sesquiterpene aldehydes were undetectable before elicitation, hemigossypolone was induced to accumulate (SI Appendix, Fig. S2). Consistently, the six enzyme genes were all up-regulated by elicitation (Fig. 2 C and D).
Selected candidate genes were submitted to VIGS, and silenced genes were then monitored by metabolite analysis of cotton leaves (28). Silencing of CDNC decreased hemigossypolone and gossypol levels by 95.1% and 96.7%, respectively, and silencing of CYP706B1 decreased the sesquiterpene levels by 59.4% and 61.2%, respectively, compared with empty vector controls (Fig. 2E). An extended assay showed that silencing of four enzymes, including two cytochromes P450 (CYP82D113 and CYP71BE79), one alcohol dehydrogenase (DH1), and one 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD-1), each reduced the level of gossypol and hemigossypolone by more than 50% (Fig. 2E). These data strongly suggested the involvement of the candidate genes in gossypol biosynthesis.
Identification of Biosynthetic Intermediates.
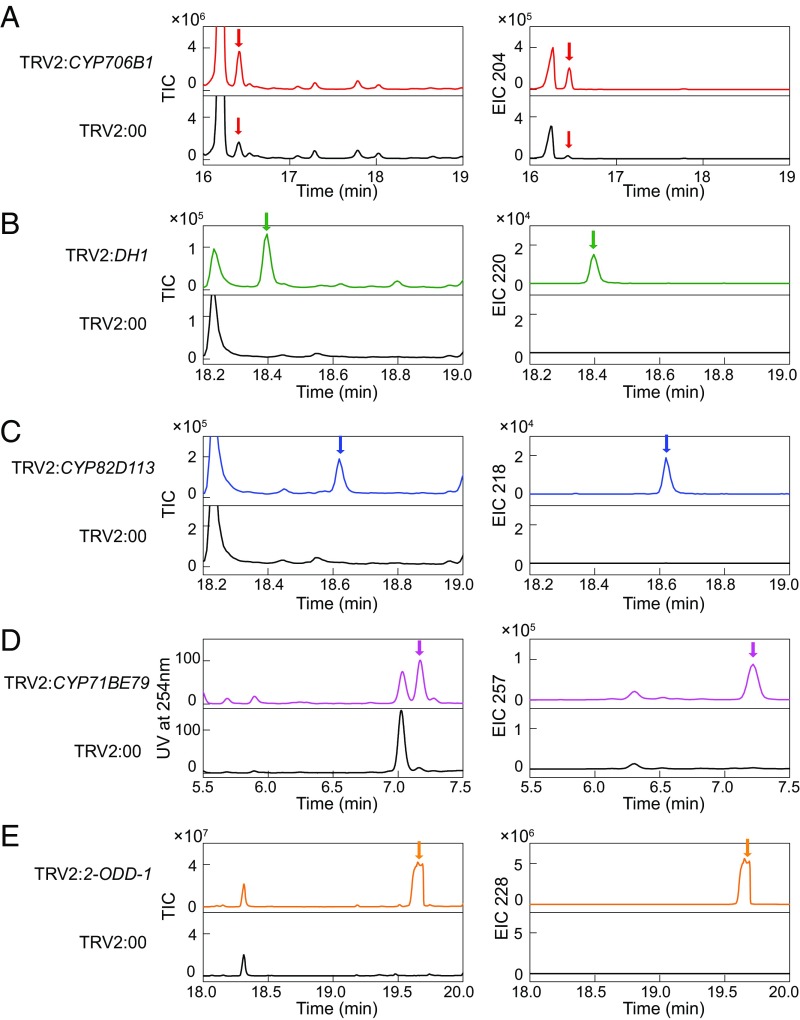
As silencing of CYP706B1 resulted in an accumulation of its substrate (+)-δ-cadinene in cotton leaves (Fig. 3A), we further analyzed the leaf extracts of the VIGS-treated plant by GC-MS and LC-MS to explore clues to the enzyme activity. We found that the CYP706B1 product, which has an m/z of 220, accumulated in the VIGS-DH1, but not the control leaves, suggesting that DH1 may be functional in reducing the CYP706B1 product (Fig. 3B). Silencing of CYP82D113 led to the accumulation of a compound that has an m/z of 218 (Fig. 3C); thus, this P450 may act immediately after DH1.
Fig. 3.
Identification of enzyme genes of gossypol biosynthesis by VIGS. Silencing of the candidate enzyme genes by VIGS led to accumulation of the putative substrates in leaf. (A–C and E) GC-MS profiles of the extracts prepared from the cotton leaves harboring TRV2:CYP706B1 (A), TRV2:DH1 (B), TRV2:CYP82D113 (C), TRV2:2-ODD-1 (E), or empty vector (TRV2:00). The peaks of the substrates, indicated by arrows, are shown (electron ionization in positive-ion mode). Total-ion chromatograms (TIC) and extracted-ion chromatogram (EIC) of the substrate of the enzyme, as indicated, at m/z 204 (A), m/z 220 (B), m/z 218 (C), and m/z 228 (E). (D) LC-MS analysis of the extracts from the cotton leaves harboring TRV2:CYP71BE79 or empty vector (TRV2:00). The peak of the CYP71BE79 substrate [with UV and EIC of the parent ions at m/z 257 [M + Na]+ on positive mode] is shown.
By LC-MS, we found that a peak with m/z (+) 257 [M + Na]+ appeared in the extract of the CYP71BE79-silenced leaves, which could be the substrate of CYP71BE79 (Fig. 3D). In addition, GC-MS identified that silencing of 2-ODD-1 resulted in accumulation of an upstream intermediate with an m/z of 228 (Fig. 3E).
We also noted that the VIGS-CYP71BE79 plants grown in the greenhouse frequently developed disease phenotypes (brown sunken lesions covering the hypocotyl–root junction) (SI Appendix, Fig. S3 A and B), similar to the symptoms caused by the soilborne necrotrophic fungus Rhizoctonia solani (29), whereas the control and other VIGS-treated plants did not. As PGF silencing blocked the whole gossypol biosynthesis pathway (25), the decreased amount of sesquiterpene phytoalexins in VIGS-CYP71BE79 plants was unlikely responsible for the enhanced susceptibility. Determination by LC-MS revealed that the substrate of CYP71BE79 accumulated in the hypocotyl–root junction after the gene silencing (SI Appendix, Fig. S3C).
Functional Characterization of Enzymes.
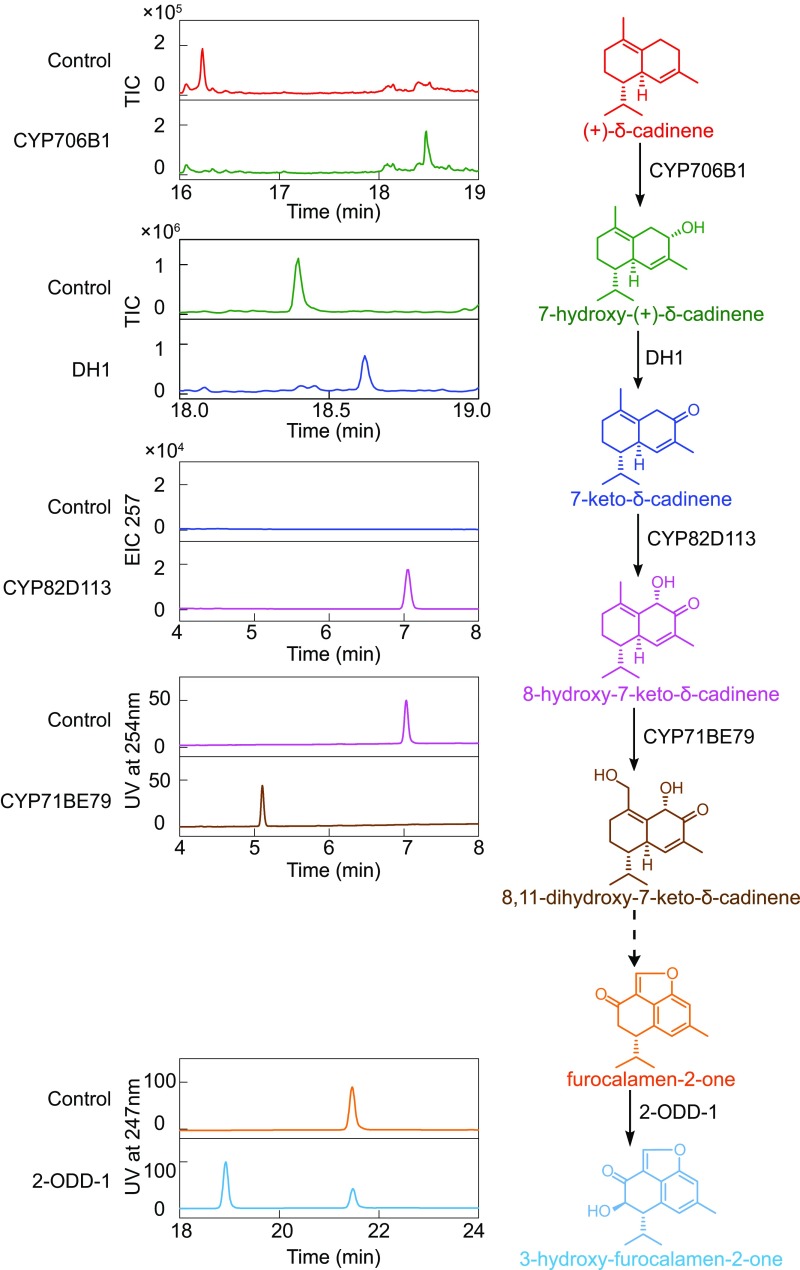
To obtain intermediate standards for structure elucidation and to perform enzyme assays in vitro, we expressed the three cytochromes P450 in Saccharomyces cerevisiae and other enzymes in Escherichia coli. As determined by GC-MS, incubation of the starting substrate FPP with CDNC produced (+)-δ-cadinene, and further reaction with CYP706B1 gave rise to a hydroxylated product (Fig. 4) that was previously proposed to be 8-hydroxy-(+)-δ-cadinene (19). Subsequent incubation revealed that DH1 converted the CYP706B1 product into a compound of Mr 218 (Fig. 4), suggesting a dehydrogenation reaction. NMR spectroscopy detected a ketonic group at the C-7 position; thus, the product is 7-keto-δ-cadinene (Fig. 4).
Fig. 4.
Functional characterization of enzymes by in vitro assays and determination of the products. (+)-δ-Cadinene, 7-hydroxy-(+)-δ-cadinene, and 7-keto-δ-cadinene were detected by GC-MS, and metabolite profiles were monitored as total-ion chromatograms (TIC), whereas 8-hydroxy-7-keto-δ-cadinene, 8,11-dihydroxy-7-keto-δ-cadinene, furocalamen-2-one, and 3-hydroxy-furocalamen-2-one were detected by LC-MS with UV, as indicated. The sample without the relevant protein served as negative control. Structures of all compounds, except (+)-δ-cadinene, were further determined by MS/MS and NMR spectroscopy (SI Appendix, Figs. S4–S16 and Tables S2 and S3). The purified recombinant proteins of DH1 and 2-ODD-1 expressed in E. coli and the microsomes of yeast cells expressing the respective cytochromes P450 were assayed.
Formation of 7-keto-δ-cadinene cast doubt on the previous identification of the CYP706B1 product as 8-hydroxy-(+)-δ-cadinene based on 1H-NMR spectroscopy (19). Indeed, both 13C NMR and heteronuclear multiple-bond correlation spectra revealed the compound as 7-hydroxy-(+)-δ-cadinene (SI Appendix, Figs. S4–S6). Thus, CYP706B1 is reassigned as (+)-δ-cadinene-7-hydroxylase, and DH1 is 7-keto-δ-cadinene synthase (Fig. 4).
The compound 7-keto-δ-cadinene was first identified from G. hirsutum plants engineered to express an RNAi construct targeting CYP82D109, which was named (4aR, 5S)-δ-cadinen-2-one (24), but the activity of CYP82D109 has remained unknown. CYP82D113 is 92% identical to CYP82D109. To determine the enzyme activity of CYP82D113, yeast microsomes enriched with CYP82D113 were incubated with 7-keto-δ-cadinene. LC-MS identified an expected peak of the product having an m/z of (+) 257. MS and NMR analyses indicated that, in the presence of NADPH, CYP82D113 transferred a hydroxyl group to C-8 of 7-keto-δ-cadinene, generating 8-hydroxy-7-keto-δ-cadinene (Fig. 4 and SI Appendix, Figs. S7–S9).
The CYP82D113 product has an MS spectrum identical to that of the proposed substrate of CYP71BE79 (Fig. 3D). To test whether CYP71BE79 is involved in further decoration of the (+)-δ-cadinene backbone, we incubated it with 8-hydroxy-7-keto-δ-cadinene, which was then efficiently converted into a product with an m/z of (+) 273 [M + Na]+ (Fig. 4). NMR analysis identified that CYP71BE79 transferred a new hydroxyl group to C-11 to form 8,11-dihydroxy-7-keto-δ-cadinene (SI Appendix, Figs. S10–S12).
Lastly, the metabolite accumulated in the 2-ODD-1–silenced leaves (Fig. 3E) was identified to be furocalamen-2-one (SI Appendix, Figs. S13–S14). As expected, incubation with 2-ODD-1 converted it to a new compound, 3-hydroxy-furocalamen-2-one (Fig. 4 and SI Appendix, Figs. S15–S16).
We next measured the kinetic parameters of the five enzymes (Table 1). Notably, CYP71BE79 exhibited a much higher maximum activity (Vmax) than other enzymes tested, including two upstream cytochromes P450 (CYP706B1 and CYP82D113), and its catalytic efficiency (Vmax/Km) was also clearly higher. To test substrate specificity, the five enzymes were assayed with available intermediates possessing similar structures. Most enzymes showed little activity toward alternative substrates under identical assay conditions (SI Appendix, Fig. S17). However, in addition to 7-hydroxy-(+)-δ-cadinene, DH1 also accepted 8-hydroxy-7-keto-δ-cadinene and 8,11-dihydroxy-7-keto-δ-cadinene as substrates, although with lower efficiency (SI Appendix, Fig. S17). Thus, DH1 is, to some extent, promiscuous in dehydrogenation of the hydroxyl group-containing metabolites.
Table 1.
Kinetic analyses of the enzymes determined in vitro
| Enzyme | Substrate | Km, μM | Vmax, nmol·min−1·mg−1 |
| CYP706B1 | (+)-δ-Cadinene | 7.57 ± 1.14 | 31.26 ± 1.56 |
| DH1 | 7-Hydroxy-(+)-δ-cadinene | 0.48 ± 0.04 | 10.42 ± 0.21 |
| CYP82D113 | 7-Keto-δ-cadinene | 1.02 ± 0.13 | 22.00 ± 0.73 |
| CYP71BE79 | 8-Hydroxy-7-keto-δ-cadinene | 9.67 ± 1.34 | 304.90 ± 10.88 |
| 2-ODD-1 | Furocalamen-2-one | 1.81 ± 0.21 | 49.54 ± 1.11 |
Each dataset represents means ± SD (n = 3 independent experiments).
Partial Reconstitution of Gossypol Pathway in Tobacco Leaf.
Along with in vitro assays of enzyme activities, we utilized the Agrobacterium-mediated transient expression system to reconstitute the gossypol pathway reactions in Nicotiana benthamiana leaves. The 35S promoter was used to express each of the six enzymes, including an FPP synthase (AtFPS2) from Arabidopsis thaliana (AT4G17190), as well as CDNC, CYP706B1, DH1, CYP82D113, and CYP71BE79 from cotton, which catalyze the six consecutive steps of gossypol biosynthesis starting from isopentenyl diphosphate/dimethylallyl diphosphate. Four metabolic intermediates, (+)-δ-cadinene, 7-hydroxy-(+)-δ-cadinene, 7-keto-δ-cadinene, and 8-hydroxy-7-keto-δ-cadinene, were detected in the leaves expressing the respective enzymes (SI Appendix, Fig. S18 A–D). Following CYP71BE79 expression with the upstream enzymes, a glycosylated product, rather than 8,11-dihydroxy-7-keto-δ-cadinene itself, was formed (SI Appendix, Fig. S18 E–G).
Together, data from VIGS and in vitro and tobacco leaf transient expression assays suggest that CYP706B1, DH1, CYP82D113, and CYP71BE79 catalyze four consecutive oxidative reactions on (+)-δ-cadinene, and 2-ODD-1 is responsible for a later hydroxylation step in the biosynthetic pathway leading to sesquiterpene aldehydes (Fig. 5).
Fig. 5.
Genes of gossypol pathway enzymes and their expressions. Genes of the enzymes catalyzing the defined steps in MVA and gossypol pathways and their homologs are shown. The expressions are indicated by heatmap, estimated using Cuffdiff by computing the FPKM value (fragments per kilobase of transcript per million reads sequenced) for each transcript. Genes encoding the identified enzymes or showing an expression pattern correlated to gossypol biosynthesis are on the TOP. Dashed arrows indicate unidentified reaction(s). 0Ov, 0-dpa ovule; 25Ov, 25-dpa ovule; 0Sd, 0-h postgermination seed; 5Sd, 5-h postgermination seed; ACAT, acyl CoA-cholesterol acyltransferase; Ca, calyx; DMAPP, dimethylallyl diphosphate; FPS, FPP synthase; HMGR, HMG-CoA reductase; HMGS, 3-hydroxy-3-methylglutaryl-coenzyme-A (HMG-CoA) synthase; IPP, isopentenyl diphosphate; IPPI, IPP isomerase; Lf, leaf; MVK, mevalonate kinase; MVP, phosphomevalonate kinase; Pe, petal; Pi, pistil; PMD, diphosphomevalonate decarboxylase; Rt, root; Sm, stamen; St, stem; To, torus. Distributions of the genes in G. hirsutum genome are indicated by their accession numbers and also shown in the genome atlas (SI Appendix, Fig. S19).
Gossypol Pathway Genes Are Dispersed in the Cotton Genome.
Several examples exist where genes encoding biosynthetic pathway enzymes of specialized metabolites, including terpenoids and alkaloids, tend to be clustered together in the plant genome (3, 6, 30, 31). In cotton, however, the gossypol pathway genes are dispersed among different chromosomes (Fig. 5 and SI Appendix, Fig. S19). On the other hand, the gene families of the gossypol as well as the core MVA pathways are often extensively expanded with tandem duplications (Fig. 5 and SI Appendix, Fig. S19). Most of the gossypol pathway enzymes identified, including CDN, DH1, CYP82D113, and 2-ODD-1, appear to have arisen from local duplications in the cotton genome. For example, in the allotetraploid genome of G. hirsutum, there are 11 genes encoding the alcohol dehydrogenase DH1 and homologs, all of which are tandemly arranged, with four genes (Gh_A01G1736, Gh_A01G1737, Gh_A01G1739, and Gh_A01G1740) on chromosome A1 (chromosome 1 of A subgenome) and seven (Gh_D01G1983 to Gh_D01G1989) on chromosome D1 (Fig. 5 and SI Appendix, Fig. S19).
Among the five enzymes catalyzing oxidative steps in the gossypol biosynthetic pathway, three are cytochromes P450 of different families. Members of CYP71 and CYP82 families are commonly involved in biosynthesis of specialized metabolites such as noscapine (32), podophyllotoxin (33), and artemisinin (34). As cotton CYP71BE79 is distinct in its high activity (Table 1), we analyzed it further.
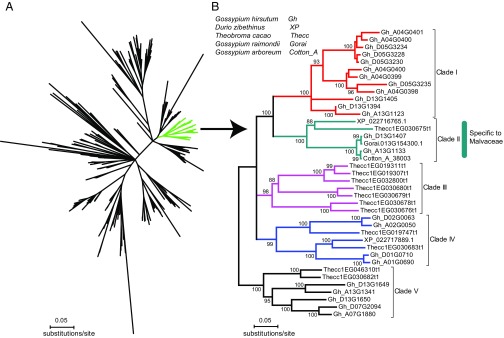
Using CYP71BE79 as query, we performed a bioinformatic blast search of CYP71 family proteins from publicly available genomes of nine plant species, including three species from the family Malvaceae: G. hirsutum, Durio zibethinus, and Theobroma cacao. In total, 312 CYP71 proteins were retrieved (SI Appendix, Fig. S20). We found that the CYP71BE proteins form a Malvaceae-specific subfamily (green in Fig. 6A), which contained 37 members clustered into five clades. Clade II was composed of six CYP71BEs, including the two CYP71BE79 homologs of G. hirsutum (Gh_A13G1133 and Gh_D13G1407). Notably, CYP71BE genes have been maintained as a truly single copy in diploid genomes or subgenomes (Fig. 6B).
Fig. 6.
Maximum-likelihood phylogenetic trees of the CYP71 family. (A) Members of the CYP71 family from nine land plants with sequence identity >40% are included (CYP71BE79 as a seed query). CYP71BE79 is located in the green branch. Plants analyzed are G. hirsutum, D. zibethinus, T. cacao, Aquilegia coerulea, A. thaliana, Oryza sativa subsp. Japonica, Amborella trichopoda, Selaginella moellendorffii, and Physcomitrella patens. The National Center for Biotechnology Information and Phytozome databases (51) were searched. (B) Members of the CYP71BE subfamily from five species of Malvaceae: G. hirsutum, G. raimondii, G. arboreum, T. cacao, and D. zibethinus. CYP71BEs are divided into five clades, and each diploid genome or subgenome harbors a single copy.
The nonsynonymous (Ka) and synonymous substitution rates (Ks) of three gossypol pathway cytochromes P450 (CYP706B1, CYP82D113, and CYP71BE79) in G. hirsutum were compared with their homologs in D. zibethinus (Table 2). The higher Ks values and the lower Ka/Ks ratios of CYP71BE79 indicate that this P450 has undergone less relaxed selection. Moreover, CYP71BE79 has a high Vmax value compared with other, identified cytochromes P450 of the gossypol pathway (Table 1), which supports an efficient transformation of its substrate (8-hydroxy-7-keto-δ-cadinene) that affects plant resistance to pathogens if accumulated (SI Appendix, Fig. S3). We propose that CYP71BE79 is functionally more conserved in Gossypium and in closely related genera in order to catalyze a highly controlled step to prevent the accumulation of the phytotoxic metabolite, along with gossypol pathway evolution.
Table 2.
The evolution rates and Ka/Ks values of three homologous P450 gene pairs between G. hirsutum and D. zibethinus
| Gene name | Genes in G. hirsutum | Homologs in D. zibethinus | Ka | Ks | Ka/Ks |
| CYP706B1_D | Gh_D03G1513 | XM_022882367.1 | 0.1271 | 0.4514 | 0.2816 |
| CYP706B1_A | Gh_A03G2006 | XM_022882367.1 | 0.1253 | 0.4342 | 0.2886 |
| CYP82D113_D | Gh_D05G1894 | XM_022910758.1 | 0.1093 | 0.5405 | 0.2022 |
| CYP82D113_A | Gh_A05G1705 | XM_022910758.1 | 0.105 | 0.5382 | 0.1951 |
| CYP71BE79_D | Gh_D13G1407 | XM_022861030.1 | 0.1201 | 0.9599 | 0.1251 |
| CYP71BE79_A | Gh_A13G1133 | XM_022861030.1 | 0.1165 | 0.9398 | 0.124 |
Discussion
Recent achievements in sequencing cotton genomes (26, 35–37) have facilitated the isolation and characterization of gossypol pathway enzymes through transcriptome mining. It is striking that the first oxidation reaction of (+)-δ-cadinene catalyzed by CYP706B1 toward gossypol biosynthesis occurs at the C-7 position, instead of C-8 as proposed previously. Besides gossypol and related sesquiterpene aldehydes that have a characteristic 8-hydroxyl group, there are other cadinene derivatives featuring oxidation at C-7 in cotton, such as 2-hydroxy-7-methoxycadalene (24). An earlier study showing that the tritiated CYP706B1 product was incorporated into gossypol (38) supported the involvement of this cytochrome P450 in gossypol biosynthesis. Here, we provide evidence that CYP706B1 produces 7-hydroxy-(+)-δ-cadinene, which is an upstream intermediate in the gossypol pathway.
Interestingly, 7-hydroxy-(+)-δ-cadinene is subjected to C-8 oxidation following C-7 carbonylation, and the C-7 carbonyl group seems indispensable for C-8 hydroxylation. The cadinene-type sesquiterpenes oxidized at both C-7 and C-8 have not been found before; subsequent oxidation at C-11 by CYP71BE79 presumes to react with a C-8 hydroxyl group to form a C-8–C-11 ether bridge in the structure of gossypol (Fig. 4). The fate of the C-7 carbonyl group awaits determination but could be deduced from structural comparison of 8,11-dihydroxy-7-keto-δ-cadinene and furocalamen-2-one, because the two intermediates leave a biosynthesis gap that may involve isomerization of carbonyl functionality to an enol group and the successive dehydration to form a benzene ring (ring B). Isomerization and dehydration are not uncommon in aromatization, such as the shikimate pathway rearrangement of chorismate to prephenate by chorismate mutase and the dehydration of arogenate to phenylalanine by arogenate dehydratase (39). Furthermore, ring B is also aromatized during desoxyhemigossypol formation from 3-hydroxy-furocalamen-2-one (Fig. 4). The present investigation resolves most of the oxidation reactions involved, leaving two remaining gaps that each involves similar aromatization reactions.
Notably, the reaction steps of gossypol formation are not randomly cascaded but rather accurately cascaded, from an energy point of view. The oxidation always occurs in the position much easier to take place, and the introduced oxidized group reduces the energy barrier of the next oxidation. For example, the first hydroxylation proceeds in the active C-7 allylic position, and then the newly formed carbonylation leaves its α position more active for subsequent hydroxylation; such is also the case of hydroxylations at positions 3 and 8, where there are preexisting carbonyl groups. Lastly, aromatizing provides the most stable napthalene ring. Thus, the gossypol pathway has evolved and been optimized through several low-energy intermediates.
The clear order and the strict substrate specificity of these biosynthetic reactions imply that the gossypol biosynthetic pathway may have evolved step by step, which might be a reason for discrete distributions of enzyme genes in the genome. We anticipate that in some plants of Malvaceae, such as cacao, okra, and roselle, the biosynthetic pathways of cadinene-type sesquiterpenes are not necessarily destined to be gossypol; the short-cut or diversified routes may result in a rich array of specialized metabolites. Comparative analyses of these pathways will enrich our knowledge on evolution of sesquiterpene biosynthetic pathways and provide valuable data for safe use and further exploration of food, oil, and vegetable crops in the Malvaceae and related families.
There are two lines of evidence that support a tight regulation of the gossypol biosynthetic pathway. First, although not clustered in the genome as frequently observed with other specialized pathways (3, 6, 18, 30), genes of all six enzymes characterized show highly similar expression patterns. This raises the possibility that all these genes are regulated by a common transcription factor complex, as seen from the MYB-bHLH-WD40 complex in the anthocyanin biosynthetic pathway (40, 41). Second, products of these gossypol pathway enzymes are mostly undetectable in plant tissues unless the downstream enzyme genes are silenced, suggesting a highly efficient conversion, which could be a result of substrate channeling (42). For example, the monoterpene indole alkaloid pathway in Catharanthus roseus involves a complex and highly regulated biosynthesis in which the upstream pathway enzymes are separated in different cellular compartments to prevent inappropriate accumulation of highly reactive strictosidine aglycone (43).
In addition to their function as phytoalexins in plants, gossypol and related sesquiterpene aldehydes also show anticancer (44, 45), antimicrobial (46, 47), and spermicidal (48) activities. We wonder whether the six intermediates identified here have similar or novel biological activities. In particular, the structure of 8-hydroxy-7-keto-δ-cadinene features an α, β-unsaturated ketone and an α-hydroxyl group next to the carbonyl, which may act as a Michael acceptor for biological nucleophiles; the similar enone group has been suggested as a general structural requirement for optimal cytotoxicity of quassinoids, a group of degraded triterpenes with promising antitumor and cytotoxic activity (49, 50), suggesting that this intermediate may harbor interesting biological activities. Cloning of the enzymes makes it possible to obtain these hidden natural products in large quantity for drug or agrochemical screening.
Methods
Details about plant materials and growth conditions are described in SI Appendix, SI Materials and Methods. Gene expression, elicitation, plant transformation, heterologous expression and purification of proteins, pathway reconstitution in N. benthamiana leaves, pathogen infection, enzymes assays, metabolites detection, and analysis were carried out according to protocols described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. Hu and Y. Shan for GC-MS and LC-MS analysis; S. Bu for NMR analysis; D. Chen, J. Chen, and X. Li for transcriptome analysis; and T. Liu, S. Wang, Z. He for discussions. The cytochromes P450 were named according to the alignment made by D. Nelson (drnelson.uthsc.edu/cytochromeP450.html). The research was supported by grants from the National Natural Science Foundation of China (31788103 and 31690092), the Chinese Academy of Sciences (XDB11030000 and QYZDY-SSW-SMC026), and the Ministry of Science and Technology of China and the Ministry of Agriculture of China (2013CB127000, 2016YFA0500800, 2016ZX08009001-009, and 2016ZX08005001-001).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805085115/-/DCSupplemental.
References
- 1.Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 2.Moghe GD, Leong BJ, Hurney SM, Daniel Jones A, Last RL. Evolutionary routes to biochemical innovation revealed by integrative analysis of a plant-defense related specialized metabolic pathway. eLife. 2017;6:e28468. doi: 10.7554/eLife.28468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonawane PD, et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat Plants. 2016;3:16205. doi: 10.1038/nplants.2016.205. [DOI] [PubMed] [Google Scholar]
- 4.Tieman D, et al. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- 5.Fan P, Miller AM, Liu X, Jones AD, Last RL. Evolution of a flipped pathway creates metabolic innovation in tomato trichomes through BAHD enzyme promiscuity. Nat Commun. 2017;8:2080. doi: 10.1038/s41467-017-02045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang Y, et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346:1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plants. 2016;2:16183. doi: 10.1038/nplants.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng YL, et al. Coordinated accumulation of (+)-δ-cadinene synthase mRNAs and gossypol in developing seeds of Gossypium hirsutum and a new member of the cad1 family from G. arboreum. J Nat Prod. 1999;62:248–252. doi: 10.1021/np980314o. [DOI] [PubMed] [Google Scholar]
- 9.Tan XP, et al. Expression pattern of (+)-δ-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta. 2000;210:644–651. doi: 10.1007/s004250050055. [DOI] [PubMed] [Google Scholar]
- 10.Bell AA, Stipanovic RD, O’Brien DH, Fryxell PA. Sesquiterpenoid aldehyde quinones and derivatives in pigment glands of Gossypium. Phytochemistry. 1978;17:1297–1305. [Google Scholar]
- 11.Shahid LA, Saeed MA, Amjad N. Present status and future prospects of mechanized production of oilseed crops in Pakistan–A review. Pak J Agric Res. 2010;23:83–93. [Google Scholar]
- 12.Ali M, Arifullah S, Manzoor H. Edible oil deficit and its impact on food expenditure in Pakistan. Pak Dev Rev. 2008;47:531–546. [Google Scholar]
- 13.Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci USA. 2006;103:18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinstein PF, Herman DL, Tove SB, Smith FH. Biosynthesis of gossypol. Incorporation of mevalonate-2-14C and isoprenyl pyrophosphates. J Biol Chem. 1970;245:4658–4665. [PubMed] [Google Scholar]
- 15.Davis GD, Essenberg M. (+)-δ-Cadinene is a product of sesquiterpene cyclase activity in cotton. Phytochemistry. 1995;39:553–567. doi: 10.1016/0031-9422(95)00771-7. [DOI] [PubMed] [Google Scholar]
- 16.Benedict CR, et al. The enzymatic formation of δ-cadinene from farnesyl diphosphate in extracts of cotton. Phytochemistry. 1995;39:327–331. [Google Scholar]
- 17.Chen XY, Chen Y, Heinstein P, Davisson VJ. Cloning, expression, and characterization of (+)-δ-cadinene synthase: A catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys. 1995;324:255–266. doi: 10.1006/abbi.1995.0038. [DOI] [PubMed] [Google Scholar]
- 18.Chen XY, Wang M, Chen Y, Davisson VJ, Heinstein P. Cloning and heterologous expression of a second (+)-δ-cadinene synthase from Gossypium arboreum. J Nat Prod. 1996;59:944–951. doi: 10.1021/np960344w. [DOI] [PubMed] [Google Scholar]
- 19.Luo P, Wang YH, Wang GD, Essenberg M, Chen XY. Molecular cloning and functional identification of (+)-δ-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 2001;28:95–104. doi: 10.1046/j.1365-313x.2001.01133.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Benedict CR, Stipanovic RD, Bell AA. Purification and characterization of S-adenosyl-l-methionine: Desoxyhemigossypol-6-O-methyltransferase from cotton plants. An enzyme capable of methylating the defense terpenoids of cotton. Plant Physiol. 1999;121:1017–1024. doi: 10.1104/pp.121.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veech JA, Stipanovic RD, Bell AA. Peroxidative conversion of hemigossypol to gossypol. A revised structure for isohemigossypol. J Chem Soc Chem Commun. 1976;4:144–145. [Google Scholar]
- 22.Benedict CR, Liu J, Stipanovic RD. The peroxidative coupling of hemigossypol to (+)- and (-)-gossypol in cottonseed extracts. Phytochemistry. 2006;67:356–361. doi: 10.1016/j.phytochem.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Effenberger I, et al. Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol. Angew Chem Int Ed Engl. 2015;54:14660–14663. doi: 10.1002/anie.201507543. [DOI] [PubMed] [Google Scholar]
- 24.Wagner TA, et al. RNAi construct of a cytochrome P450 gene CYP82D109 blocks an early step in the biosynthesis of hemigossypolone and gossypol in transgenic cotton plants. Phytochemistry. 2015;115:59–69. doi: 10.1016/j.phytochem.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Ma D, et al. Genetic basis for glandular trichome formation in cotton. Nat Commun. 2016;7:10456. doi: 10.1038/ncomms10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 27.Wang JY, et al. VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl Environ Microbiol. 2004;70:4989–4995. doi: 10.1128/AEM.70.8.4989-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, et al. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to verticillium wilt. Plant J. 2011;66:293–305. doi: 10.1111/j.1365-313X.2011.04491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, et al. iTRAQ-based proteomic analysis of defence responses triggered by the necrotrophic pathogen Rhizoctonia solani in cotton. J Proteomics. 2017;152:226–235. doi: 10.1016/j.jprot.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Osbourn A. Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 31.De Luca V, Salim V, Atsumi SM, Yu F. Mining the biodiversity of plants: A revolution in the making. Science. 2012;336:1658–1661. doi: 10.1126/science.1217410. [DOI] [PubMed] [Google Scholar]
- 32.Winzer T, et al. Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015;349:309–312. doi: 10.1126/science.aab1852. [DOI] [PubMed] [Google Scholar]
- 33.Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 36.Li F, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep. 2015;5:14139. doi: 10.1038/srep14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang YH, Davila-Huerta G, Essenberg M. 8-Hydroxy-(+)-δ-cadinene is a precursor to hemigossypol in Gossypium hirsutum. Phytochemistry. 2003;64:219–225. doi: 10.1016/s0031-9422(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 39.Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 40.Martin C, Glover BJ. Functional aspects of cell patterning in aerial epidermis. Curr Opin Plant Biol. 2007;10:70–82. doi: 10.1016/j.pbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Guo YH, et al. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009;183:62–75. doi: 10.1111/j.1469-8137.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- 43.Payne RM, et al. An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat Plants. 2017;3:16208. doi: 10.1038/nplants.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelley MD, Hartley L, Groundwater PW, Fish RG. Structure-activity studies on gossypol in tumor cell lines. Anticancer Drugs. 2000;11:209–216. doi: 10.1097/00001813-200003000-00009. [DOI] [PubMed] [Google Scholar]
- 45.Oliver CL, et al. (-)-Gossypol acts directly on the mitochondria to overcome Bcl-2- and Bcl-X(L)-mediated apoptosis resistance. Mol Cancer Ther. 2005;4:23–31. [PubMed] [Google Scholar]
- 46.Yildirim-Aksoy M, et al. In vitro inhibitory effect of gossypol from gossypol-acetic acid, and (+)- and (-)-isomers of gossypol on the growth of Edwardsiella ictaluri. J Appl Microbiol. 2004;97:87–92. doi: 10.1111/j.1365-2672.2004.02273.x. [DOI] [PubMed] [Google Scholar]
- 47.Mellon JE, Zelaya CA, Dowd MK. Inhibitory effects of gossypol-related compounds on growth of Aspergillus flavus. Lett Appl Microbiol. 2011;52:406–412. doi: 10.1111/j.1472-765X.2011.03020.x. [DOI] [PubMed] [Google Scholar]
- 48.Kim IC, et al. Comparative in vitro spermicidal effects of (+/-)-gossypol, (+)-gossypol, (-)-gossypol and gossypolone. Contraception. 1984;30:253–259. doi: 10.1016/0010-7824(84)90088-x. [DOI] [PubMed] [Google Scholar]
- 49.Guo Z, Vangapandu S, Sindelar RW, Walker LA, Sindelar RD. Biologically active quassinoids and their chemistry: Potential leads for drug design. Curr Med Chem. 2005;12:173–190. doi: 10.2174/0929867053363351. [DOI] [PubMed] [Google Scholar]
- 50.Fang X, et al. Unprecedented quassinoids with promising biological activity from Harrisonia perforata. Angew Chem Int Ed Engl. 2015;54:5592–5595. doi: 10.1002/anie.201412126. [DOI] [PubMed] [Google Scholar]
- 51.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.