Significance
It is commonly thought that each globular protein has a single 3D structure, or fold, that fosters its function. In contrast, recent studies have identified several fold-switching proteins whose secondary structures can be remodeled in response to cellular stimuli. Although thought to be rare, we found 96 literature-validated fold-switching proteins by exhaustively searching the database of protein structures [Protein Data Bank (PDB)]. Characterizing these proteins led us to hypothesize that their abundance may be underrepresented in the PDB. Thus, we developed a computational method that identifies fold-switching proteins and used it to estimate that 0.5–4% of PDB proteins switch folds. These results suggest that proteins switch folds with significant frequency, which has implications for cell biology, genomics, and human health.
Keywords: protein structure, protein fold switching, metamorphic proteins, conformational diversity, protein function
Abstract
A central tenet of biology is that globular proteins have a unique 3D structure under physiological conditions. Recent work has challenged this notion by demonstrating that some proteins switch folds, a process that involves remodeling of secondary structure in response to a few mutations (evolved fold switchers) or cellular stimuli (extant fold switchers). To date, extant fold switchers have been viewed as rare byproducts of evolution, but their frequency has been neither quantified nor estimated. By systematically and exhaustively searching the Protein Data Bank (PDB), we found ∼100 extant fold-switching proteins. Furthermore, we gathered multiple lines of evidence suggesting that these proteins are widespread in nature. Based on these lines of evidence, we hypothesized that the frequency of extant fold-switching proteins may be underrepresented by the structures in the PDB. Thus, we sought to identify other putative extant fold switchers with only one solved conformation. To do this, we identified two characteristic features of our ∼100 extant fold-switching proteins, incorrect secondary structure predictions and likely independent folding cooperativity, and searched the PDB for other proteins with similar features. Reassuringly, this method identified dozens of other proteins in the literature with indication of a structural change but only one solved conformation in the PDB. Thus, we used it to estimate that 0.5–4% of PDB proteins switch folds. These results demonstrate that extant fold-switching proteins are likely more common than the PDB reflects, which has implications for cell biology, genomics, and human health.
Living cells respond to stimuli by altering their internal chemical environments. These responses include ion influx and efflux, cofactor concentration changes, pH shifts, and redox potential adjustments. Such changes in the cellular environment can favor alternative protein conformations with modified functional capacities. For example, intrinsically disordered proteins (IDPs), which are flexible in their entirety, can undergo large disorder ↔ order transitions in response to cellular triggers (1). In contrast, structural changes within globular proteins are typically localized within termini, long loops, and short linkers (2, 3). Accordingly, changes in the cellular environment are thought to favor globular protein conformations whose functional capacities differ (4) because of localized structural variations on an essentially unchanged framework of anchoring secondary structure (3), although some solvent-exposed secondary structure elements can undergo localized unfolding and refolding (5).
Here, we hypothesize that globular proteins can also respond to cellular changes by remodeling their secondary structures, a phenomenon called “fold switching” (6). While it is widely accepted that this phenomenon is an evolutionary mechanism for generating new protein functions (7, 8), the increasing number of extant examples suggests that proteins might also switch folds to change functions in response to the cellular environment (9) and to enable tighter regulation. Supporting this hypothesis, an α-helical transcription factor can morph into a β-barrel translation factor, and both functional states can be observed within the cell (10). Additionally, some endolysins can switch from an inactive membrane-tethered conformation to an active cytosolic conformation with a different secondary structure (11).
Currently, extant fold-switching proteins are thought to be rare (12), with reviews of the subject focusing on a handful of important examples (6, 9, 13, 14). Consequently, no systematic analysis has been performed.
To gauge both the scope and the biological relevance of extant fold-switching proteins, we exhaustively searched for them in the Protein Data Bank (PDB), a repository of atomic-resolution protein structures. Specifically, we searched for instances of the same protein adopting two very different secondary structures. Contrary to the perception that these large conformational differences have been observed in “only very few proteins” (15), we find 96 unique literature-supported instances. These instances span every kingdom of life, perform dozens of disparate functions, and switch in response to many different triggers. Thus, fold switching appears to be a widespread mechanism to modulate protein function.
We find that solving the structures of extant fold-switching proteins often requires advanced techniques, such as cryo-EM, solid-state NMR, and handling of membrane proteins, which, combined with expectations of rareness, suggests that the natural abundance of these proteins might substantially exceed the proportion currently represented among solved structures in the PDB. Accordingly, we sought to computationally identify more extant proteins that were likely to switch folds. To do this, we identified two distinguishing features of extant fold-switching proteins: (i) cooperatively folding regions that are likely to unfold and refold independently in response to environmental triggers and (ii) discrepancies between predicted and experimentally determined secondary structure. We then searched the PDB for proteins with similar features and examined the literature for indications that these proteins actually switch folds. From this analysis, we estimated that 0.5–4% of proteins in the PDB could switch folds. Together, these results indicate that protein-fold switching is likely more common than currently believed, suggesting that many important functions of proteins remain unknown. This finding has implications for cell biology, genomics, and human health.
Results
Identifying Protein Fold Switches.
To test our hypothesis that a substantial number of proteins respond to cellular changes by switching folds, we searched the entire PDB for proteins with ≥90% sequence identity but different secondary structures (Methods and SI Appendix, Fig. S1A). Segments of protein structure with high levels of sequence identity but different folds were then excised from their parent PDBs and spatially aligned. When the root-mean-square deviations (rmsds) of these aligned segments exceeded 4.0 Å, parent PDBs were inspected manually. During inspection, we eliminated pairs differing solely by loop and linker motions or disorder ↔ order transitions (SI Appendix, Fig. S1B). To confirm biological relevance, we reviewed the literature reporting both structures in the pair and required that it (i) claims that the switch is biologically relevant and (ii) reports the trigger for the switch. This step also eliminates false positives resulting from weak electron density, crystal packing artifacts, insufficient data-derived constraints, and controversial structures. Because we were interested in extant fold switchers, those triggered by amino acid mutations were also excluded (SI Appendix, Fig. S1B). We allowed the 90% sequence identity threshold mentioned above to include natural proteins reported to switch folds whose alternative structures were stabilized through mutations. These proteins are reported to change conformation without mutation, although mutations facilitated transition to their alternative conformations.
Our search yielded 96 extant fold-switching proteins, which we call structurally validated fold switchers. We then looked for distinguishing characteristics of these proteins that would allow us to predict more.
Characterizing Protein Fold Switches.
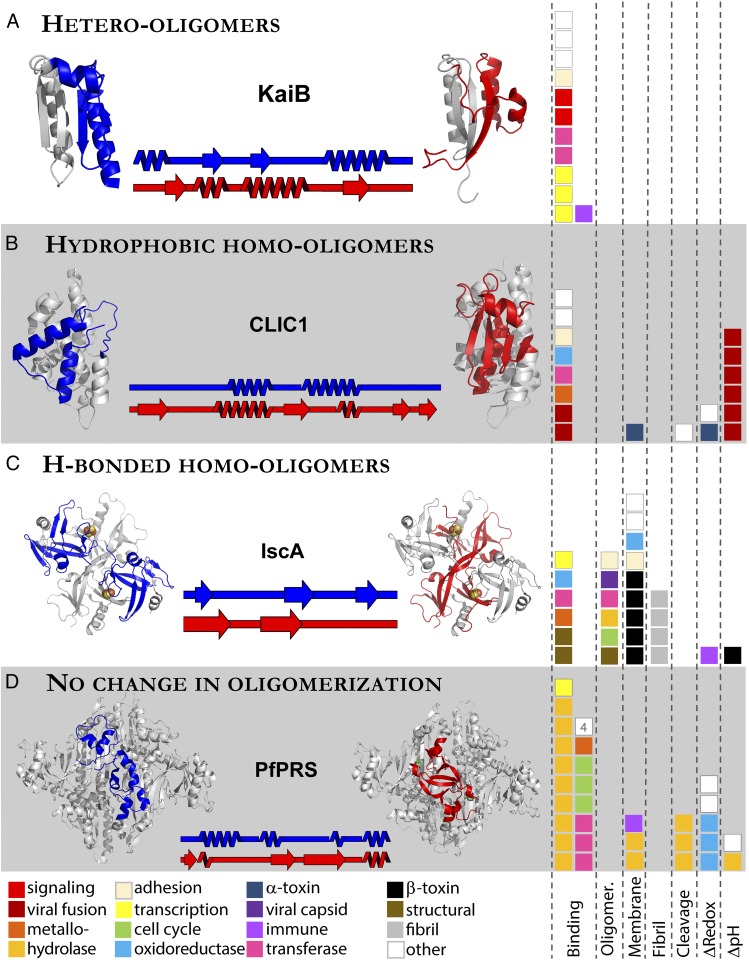
To better understand the types of conformational changes involved in protein fold switching, we classified our structurally validated fold switchers into four categories based on changes in their oligomeric states and hydrogen-bonding/hydrophobic interactions (Fig. 1). Class A includes proteins with one conformation whose fold-switching regions heterooligomerize (Fig. 1A). In every case but one, this oligomerization is through a hydrophobic interface. This category involves some of the most dramatic switching events currently known. For example, KaiB is a cyanobacterial circadian clock protein that populates two folds: its inactive homotetrameric βαββααβ fold and its rare active thioredoxin-like (βαβαββα) fold that binds to KaiC, another circadian clock protein (Fig. 1A). The slow interconversion between these two conformations helps to coordinate the cyanobacterial clock with the timing of the earth’s rotation (16). Class B groups proteins with one conformation that homooligomerizes through a hydrophobic interface (Fig. 1B). One example is human chloride intracellular channel 1 (CLIC1), a chloride channel that has both cytosolic and membrane-bound conformations (17). Under reducing conditions, this protein is a monomer that binds glutathione. Oxidation causes the cysteines in its N-terminal lobe to form a disulfide bond, stabilizing an alternative fold that forms a homodimeric interface. This protein can both insert into artificial membranes and function as a transporter under oxidizing, but not reducing, conditions. The dimeric form in the PDB is a likely intermediate to the membrane-inserted form (18). Class C comprises proteins with one conformation whose fold-switching regions homooligomerize through a hydrogen-bonded interface (Fig. 1C). For example, in its apo form, IscA forms a homodimer with two structurally identical subunits (blue conformation in Fig. 1C, as described in figure legend), but upon iron-sulfur binding, it forms a homotetramer with two distinct conformers. Cysteines from both conformers form covalent bonds with IscA’s ligand, [2Fe-2S]. These conformers differ by their β-strand registers and tertiary contacts. The conformational heterogeneity of this complex is thought to allow IscA to act as a scaffold that fosters Fe-S cluster biosynthesis (19). Class D contains proteins that switch folds while maintaining the same oligomeric state (Fig. 1D). Prolyl tRNA synthetase from Plasmodium falciparum is an antimalarial drug target. In its apo form, the N-terminal segment forms helices at the surface of its dimeric interface. Upon binding to the veterinary medicine halofuginone, this interface rearranges to a form with domain-swapped β-sheets. Activity in this form is substantially diminished, arresting parasite growth (20). The classifications of all 96 structurally validated fold switchers are reported in SI Appendix, Table S1, and more representative examples are shown in SI Appendix, Fig. S2.
Fig. 1.
(A–D, Left) Four categories of fold-switching proteins. Names of each category are in the upper left-hand corners of their respective white and gray boxes. Specific examples of each fold switch category are included. Blue and red regions of protein structure indicate regions that change folds. Secondary structure diagrams corresponding to the blue and red regions are placed between the two structures for comparison, with the names of the proteins above. Gray protein regions maintain essentially the same secondary structure in both conformations. PDB ID codes and chains (left to right) are as follows: 5jytA, 2qkeE (class A); 1rk4A, 1k0nA (class B); and 1x0gA&C, 1x0gB&D; 4twaA, 4ydqB (class C). Structures are meant to illustrate conformational differences, not the most biologically relevant conformations. Thus, both conformations of KaiB, CLIC1, and IscA are shown as monomers, monomers, and tetramers, respectively. Colored boxes at the bottom indicate the biological function of each protein; one box corresponds to one protein. All images of 3D protein structures in this figure and all others were made in PyMOL (38). (A–D, Right) Biological functions are grouped by triggers and separated by dashed lines. The numeral 4 in the white box in class D represents four proteins of the class “other” that did not all fit in the allotted space. Four proteins with other triggers are omitted from this figure.
Fig. 1 also demonstrates that fold-switching proteins perform a wide array of biological functions triggered by many different cellular signals. To date, proteins from 30 functional classes have been shown to switch folds. Furthermore, Fig. 1 shows seven different triggers, and there are several not depicted: illumination by light, viral capsid maturation, and a folding intermediate (SI Appendix, Table S1). Together, these observations suggest that fold-switching proteins might be more prevalent in nature than the PDB currently indicates.
Protein Fold Switching May Be Underrepresented in the PDB.
To probe how widespread fold switching might be, we first looked at its organismal distribution. If it were difficult for fold-switching proteins to evolve, then evolutionary theory would predict that fold switching would be observed more frequently in viruses and bacteria than in eukaryotes, because of the selective pressure to evolve quickly and maintain a compact genome (21), both of which favor multifunctional (and possibly multiconformational) proteins. In contrast, eukaryotes are not under the same stringent genomic size constraints, making it easier for a gene to be copied and evolved to perform a function different from the original [subfunctionalization (22)]. The calculated distribution is not consistent with this expectation. Instead, fold switching occurs as frequently in humans as in viruses, and less frequently in bacteria (SI Appendix, Fig. S3), at least as pertains to the proteins currently in the PDB. This suggests that fold switchers are not selected exclusively to save genome space. Thus, we hypothesized that a substantial number of amino acid sequences can adopt two or more stably folded conformations (i.e., different configurations of regular secondary structure, not disorder ↔ order transitions). Consistent with this hypothesis, several proteins with ≥80% sequence identity but different folds have been engineered successfully (23–25).
If a substantial number of proteins can populate two or more stably folded conformations, why are there not more instances in the PDB? One explanation is that these proteins could be difficult to characterize. To explore this possibility, we examined the methods used to solve the structure of each fold-switching pair and found that at least one conformation was frequently solved by noncrystallographic methods. Alternative methods were used significantly more often than in the PDB as a whole (27% vs. 10%; P < 0.05, Kolmogorov–Smirnov test). Most notably, 11% of these structures were solved by cryo-EM, while <1% of all structures in the PDB use this method. Similarly, 3% of these structures were solved by solid-state NMR, versus <0.1% of all structures in the PDB. The enrichment of these advanced methods is consistent with the observation that the number of fold switchers solved per year has increased recently, with >50% solved since 2013 (SI Appendix, Fig. S4). Furthermore, solution NMR, another method slightly overrepresented in this dataset (11% vs. 9% in the PDB), has fostered the discovery of fold switchers by revealing significant chemical shift changes under different conditions (26). Finally, in comparison to PDB proteins, fold-switching proteins have more membrane-bound conformations (14% vs. 3%), which are difficult to handle experimentally.
Characteristic Features of Protein Fold Switchers.
Subdomains with independent folding cooperativity.
Upon examining the structures of these fold-switching proteins, we noticed that most have both conformationally variable regions and structurally unchanged regions. These regions tended to be continuous and separate, suggesting that they may fold independently from one another. If true, these proteins’ functions could be modulated by localized refolding (14). One computational method for detecting independent folding cooperativity is the structure energy equivalence of domains (SEED) algorithm. Previously, the SEED algorithm correctly identified the boundaries of independent folding units in proteins with experimentally characterized folding intermediates (27, 28). This success suggested that the SEED algorithm might be able to identify whether fold-switching protein regions could unfold and refold independently from the remainder of the protein.
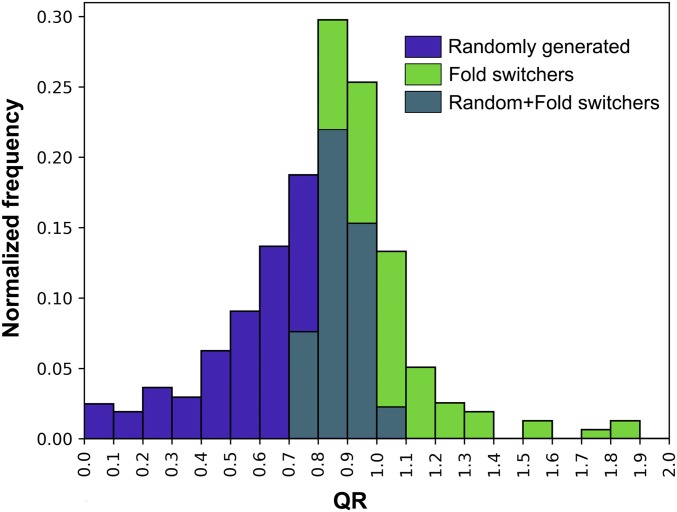
Using the SEED algorithm (27), we found that 90% (86 of 96) of fold-switching protein regions had at least one conformation predicted to fold independently from the rest of the protein (SI Appendix, Fig. S5 and Table S2). This result had very high statistical significance compared with cooperative units encompassing randomly selected protein fragments (P < 10−34, Kolmogorov–Smirnov test; Fig. 2 and Methods), suggesting that independent folding cooperativity is a characteristic feature of fold-switching protein regions.
Fig. 2.
Distributions of randomly generated and fold switch-derived QRs (measures of independent folding cooperativity from the SEED algorithm) differ significantly (P < 10−34, Kolmogorov–Smirnov test). The x axis is limited between 0 and 2, and the majority of both of these populations (98%, randomly generated; 89%, fold switch-derived) lie within these limits.
Discrepancies with secondary structure predictions.
By definition, protein fold switchers have regions whose secondary structures differ. Therefore, we reasoned that at least one member of each fold-switch pair would differ significantly from secondary structure predictions. We found that 85 of our fold-switching protein pairs had at least one member with substantial secondary structure discrepancies between experimentally determined and predicted secondary structure.
Identifying Additional Fold Switchers.
To identify additional proteins in the PDB that are likely to switch folds, we ran the SEED algorithm on protein regions with discrepancies between predicted and experimentally determined secondary structures. As a test case, we ran this calculation on all 85 fold-switching protein pairs with predicted secondary structure discrepancies. In 77 cases (91%), the fold-switching region of at least one conformer was predicted correctly; this included all fibril-forming proteins, suggesting that this method might identify other protein regions involved in misfolding diseases. Overall, these results are consistent with the performance of the SEED algorithm on specified regions experimentally known to switch folds (SI Appendix, Table S2).
We then ran the same calculations on a nonredundant subset of the PDB (Methods). The remaining putative hits comprised 32% (11,281 of 35,060) of the nonredundant PDB. We believe that this number significantly exceeds the true frequency of fold-switching proteins in the current PDB; however, we sought to quantify the power of our calculations in winnowing the search space of possible fold switchers.
To gauge the selective power of these calculations, we first performed a focused search of the PDB to identify other proteins with literature reports of fold switching but only one solved structure (Methods). We found a total of 92 additional fold-switching proteins (SI Appendix, Table S3A), which we call expected fold switchers. Of these 92 expected fold switchers, 15 were experimentally supported and 77 were inferred to switch folds either in the literature or because they were homologous to other fold-switching proteins. Combining these 92 expected fold switchers with the 96 structurally validated fold switchers, we estimate that at least 0.5% (188 of 35,060) of proteins in the PDB switch folds.
We then assessed whether these 92 expected fold switchers were enriched in our calculation-generated subset of the PDB and found that they were indeed significantly overrepresented. Specifically, our calculations correctly identified 63 of 92 expected fold switchers (P < 1.0 × 10−12, hypergeometric test; SI Appendix, Table S3A). Furthermore, 13 of 15 of the fold-switching regions of the experimentally supported expected fold switchers were identified correctly (P < 2.0 × 10−5, hypergeometric test; Table 1 and SI Appendix, Table S3B).
Table 1.
Predictions of experimentally supported fold switchers
| PDB ID codes + chains | Predicted fold-switching regions | Methods of experimental validation* |
| 2kxoA | 1–89 | NMR |
| 2lshA | 29–115 | NMR |
| 2mz7A | 267–312 | NMR |
| 4pmkA | 27–62 | NMR |
| 2n4oA | 16–69 | NMR |
| 2ktmA | 167–201 | NMR |
| 2le3A | Not predicted | NMR |
| 2x9cA | Not predicted | NMR |
| 4ov8A | 247–318 | Cryo-EM |
| 3j9eD | 2–71 | Cryo-EM |
| 5suzA | 474–509 or 415–509 | Cell-based assays |
| 4hlsA | 146–222 | Circular dichroism + size exclusion chromatography |
| 1s5pA | 48–107, 98–189, 208–274 | Isothermal titration calorimetry |
| 3tkaA | 236–313 | Small-angle X-ray scattering |
| 3gaxA | 48–120 | Fibrillar deposits identified in vivo |
SI Appendix, Table S3B contains literature justifications.
Having demonstrated that our calculations identify significantly more fold switchers than expected by chance, we used them to estimate an upper bound for fold switchers in the PDB. To determine the false-positive rate of our calculations, we selected 228 protein chains from 20 protein families not expected to switch folds (SI Appendix, Table S3C). Of those 228 protein chains, 66 (29%) were predicted to switch folds (i.e., false positives). Extrapolating this fraction to be the false-positive rate of our calculations as a whole and using a false-negative rate of 32% (29 of 92 expected fold switchers, as discussed above), we estimate that up to 4% [1.32 × (32–29%)] of proteins in the nonredundant PDB switch folds.
Discussion
Proteins are generally assumed to adopt one 3D structure that performs one well-defined function. Although protein structural dynamics are critical to their functions (4), observed changes in secondary structure are thought to be rare (discussion of ref. 15). Here, we report nearly 100 structurally validated extant fold-switching proteins, whose changes in activity are accompanied by secondary structure remodeling. We believe that all of them are biologically relevant, as we required a literature report of both the trigger of the structural change and how that change affects the protein’s function. These results suggest that protein fold switching could be an important mechanism by which proteins respond to the ever-changing cellular environment.
Protein fold switching differs from the disorder ↔ order transitions observed in IDPs. IDPs are commonly recognized as missing regions of electron density in X-ray crystal structures of proteins (29). In contrast, we require both conformations of protein fold switchers to be determined. Second, IDPs have characteristic amino acid compositions (1). We impose no sequence constraints in our search for structurally validated protein fold switchers (also SI Appendix, Fig. S6). Instead, we identify them based on discrepancies with secondary structure predictions and on their likelihood to fold cooperatively and independently. IDPs do not fold cooperatively in isolation (29). Therefore, fold-switching proteins are not IDPs, but rather a subset of globular proteins whose stably formed structures shift dramatically in response to their environments. Accordingly, growing evidence demonstrates that the cellular environment influences globular proteins both structurally and functionally (30, 31).
What advantages might fold switching confer to biological systems? Some fold-switching proteins have two disparate functions. This bifunctionality allows synergistic biological activities to be coupled quickly while obviating the need for additional cellular resources to transcribe and translate two proteins with different functions. One obvious example is RfaH, which functions as both a transcription factor and a translation factor (10). A less obvious example is human CLIC1, which can function as both a soluble glutathione reductase and a membrane-inserted chloride ion channel (17). CLIC1’s glutathione reductase activity suggests that its function might be modulated by changes in redox potential. Indeed, it inserts into lipid bilayers and functions as a chloride channel under oxidizing, but not reducing, conditions.
A second possible advantage of fold switching is stricter regulation. While many globular proteins can populate both active and inactive conformations under many sets of biologically relevant conditions, fold-switching proteins can be locked in an active or inactive state until a specific cellular trigger is present. One example is human mitochondrial HSP90N, which is proposed to exist in an autoinhibited state until it binds ATP and refolds (32). This would permit HSP90N to be present in the cell without being active. Thus, bifunctionality and tighter regulation might explain why eukaryotic proteins switch folds more frequently than genomic size constraints would predict.
To our knowledge, most fold-switching proteins in our dataset were discovered out of biological interest rather than expectation of conformational change. Many other proteins not present in the PDB might also switch folds. Thus, we developed a heuristic for identifying more fold-switching proteins. It uses two of their characteristic features: regions with independent folding cooperativity and discrepancies between predicted and experimentally determined secondary structures. Employing these features, we correctly identified the fold-switching regions of 13 of 15 experimentally supported fold switchers. This result has high statistical significance (P < 2.0 × 10−5, hypergeometric test), indicating that fold-switching protein regions indeed correspond to independent folding units (IFUs) with the ability to adopt multiple secondary structures, as evidenced by discrepancies between experimentally determined and predicted secondary structures. While independent folding cooperativity and secondary structure discrepancies are highly discriminatory in winnowing down the search space for potential fold switchers, our high false-positive rate indicates that there are probably other discriminatory features of fold switchers that we have not yet recognized. We are optimistic that as the structures of more fold switchers are solved and characterized, subsequent analysis will reveal more determining features.
A more thorough understanding of fold-switching proteins could potentially foster numerous scientific advances. For example, the antimalarial drug halofuginone arrests parasite growth by inactivating prolyl tRNA synthetase through a fold switch (20, 33). Therefore, fold-switching regions of proteins could be drug design targets. Furthermore, light causes a fold switch in the bacterial photosensory core (34). Better understanding of this transition could lead to the engineering of improved optogenetic reagents. Additionally, accurate predictions of fold-switching proteins could foster better protein structure predictions by homology modeling, which typically assumes that a protein sequence adopts a unique 3D topology that fosters one function. On a related note, our predictions suggest that the ∼80% plateau in secondary structure prediction accuracy (35) might be due, in part, to fold-switching regions in proteins. Finally, identifying putative fold switchers could help to reveal new functional roles for proteins with unexplained cellular localizations and binding partners. We hope that the observations presented here are a step toward realizing these potential advances.
Methods
All scripts, written in C++ and Python are available for download at https://github.com/llporter/Fold-Switch.git. Full details regarding our data are given in SI Appendix, Methods.
Identification of Fold Switchers.
All amino acid sequences in our database of protein primary and secondary structures (SI Appendix, Methods) were aligned with protein BLAST (36) using a library built from our sequence database. We allowed the resulting alignments to differ by up to 10 residues (regardless of alignment length) because some alternative conformations in crystal structures have been stabilized by designed mutations, even though the wild type can adopt both. The secondary structure annotations of alignments fitting these criteria were also aligned. Each letter in the alignment was assigned a score: 1.0 points for (α or turn) ↔ β changes, 0.6 points for coil ↔ (α, β or turn) changes, 0.25 points for helix ↔ turn, and 0 for everything else. The longest region of the alignment with a normalized score of >0.4 (sum of the alignment score normalized by length of the aligned region) was considered a putative fold switcher if its length was ≥10 residues. Fold-switching regions within the alignments were required to have either ≥90% sequence identity (93 of 96 cases) or ≥80% sequence identity and a literature report of a structural change for at least one conformation (three of 96 cases). Some fold-switching protein regions had one conformation covalently linked to additional protein chains and one conformation not linked. Cases in which both the linked and unlinked conformations had literature reports of biological relevance were accepted. All others were rejected.
Segments from protein pairs corresponding to differing secondary structure alignments were excised from their parent proteins, and their rmsds were calculated using Biopython (37). Pairs with rmsds of >4.0 Å were then inspected in PyMOL (38) to ensure that the conformational differences did not arise exclusively from changes in tertiary structure or inaccurate annotations of NMR structures. Upon satisfying this requirement, the published reports associated with both PDB structures were searched for both mention of a significant conformational change and the trigger of the change. Protein pairs satisfying all of these requirements were considered fold-switching proteins.
Secondary Structure Predictions.
A local install of SPIDER2 (39) was run on all 96 pairs of fold-switching proteins (SI Appendix, Table S2); the resulting secondary structure annotations were compared with annotations from PROSS (40). All continuous amino acid strings with ≥10 residues that satisfied the scoring function described in the previous section were considered potential fold-switch loci. These strings were aligned with the sequences of the fold switchers that we identified by means of our algorithm (SI Appendix, Table S2, bold sequences) using the pairwise2.align.localxs function from Biopython (37), with a −0.5/−0.1 point penalty for gap opening/extension. The minimum alignment score was 8.0, and all hits with an alignment score of <10.0 were examined manually. Those with fewer than seven consecutive aligned amino acids were discarded. A total of 108 of 192 proteins had fold switch loci that satisfied these alignment criteria, representing 85 of 96 fold-switch pairs (89%).
Identification of IFUs.
Calculations of IFUs were performed with the SEED algorithm (27). Because this method requires a lot of computational power, the protein regions corresponding to fold-switching segments were given as references for the IFU search. The SEED algorithm then searched for IFUs by calculating the qualifying ratio (QR; the measure by which the SEED algorithm determines independent folding cooperativity) of the reference sequence. The QR of the reference sequence was calculated and extended to maximize the QR in a constrained search space (SI Appendix, Methods).
To test the statistical significance of the QR values corresponding to the IFUs above, we calculated the maximal QRs of randomly selected protein segments whose lengths mapped one-to-one with those of the 96 IFUs corresponding to fold switches (SI Appendix, Methods). We chose this approach to preserve IFU length because QRs tend to increase with sequence length. The same 96-protein simulation was repeated 10 times, and the QRs from all 10 simulations were used to make the distribution in Fig. 2. Using the Kolmogorov–Smirnov test, we found that the randomly generated distribution and fold switch distribution differed significantly: P < 10−34.
Whole-PDB Fold Switcher Predictions.
SEED algorithm calculations were performed on all protein regions from a nonredundant subset of the PDB (SI Appendix, Methods) whose secondary structure annotations from SPIDER2 (39) differed substantially from their experimentally determined secondary structure annotations from PROSS, where substantial differences were the same as those defined in the previous section. This yielded 11,281 of 35,060 unique protein structures, or 32% of the PDB.
To get the lower bound estimate of possible fold-switching proteins in the PDB, we searched the nonredundant subset of the PDB for homologs of the 96 fold switchers and other proteins with related keywords in their PDB files (e.g., viral fusion proteins or pore-forming toxins) or literature support of a fold switch (SI Appendix, Methods). Combining these two approaches, we found a total of 92 expected fold switchers (SI Appendix, Table S3A). These 92 fold switchers combined with the 96 structurally validated switchers constituted 0.5% of the nonredundant PDB.retic test:
Enrichment statistics were performed using the hypergeometric test: , where n is the total number of fold switchers within a given category, k is the number of those fold switchers recognized by our code and i iterates from 0 to n–k in increments of 1, 35,060 is the total number of proteins in the nonredundant PDB, and 11,281 is the number of putative fold switchers recognized by our code.
Supplementary Material
Acknowledgments
We thank Eric Schreiter, George Rose, Brian English, Jacob Keller, Aaron Robinson, Carolyn Ott, Yan Zhang, Mike Harms, Joshua Porter, and our anonymous reviewers for helpful suggestions. This study was supported by NIH Grant F32GM10664901 (to L.L.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All software reported in this paper is available on GitHub at https://github.com/llporter/Fold-Switch.git.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800168115/-/DCSupplemental.
References
- 1.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kay LE. Protein dynamics from NMR. Biochem Cell Biol. 1998;76:145–152. doi: 10.1139/bcb-76-2-3-145. [DOI] [PubMed] [Google Scholar]
- 3.Li R, Woodward C. The hydrogen exchange core and protein folding. Protein Sci. 1999;8:1571–1590. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wand AJ, Roder H, Englander SW. Two-dimensional 1H NMR studies of cytochrome c: Hydrogen exchange in the N-terminal helix. Biochemistry. 1986;25:1107–1114. doi: 10.1021/bi00353a025. [DOI] [PubMed] [Google Scholar]
- 6.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordes MH, Stewart KL. The porous borders of the protein world. Structure. 2012;20:199–200. doi: 10.1016/j.str.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc Natl Acad Sci USA. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodchild SC, Curmi PMG, Brown LJ. Structural gymnastics of multifunctional metamorphic proteins. Biophys Rev. 2011;3:143. doi: 10.1007/s12551-011-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burmann BM, et al. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu M, et al. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 12.Tseng R, et al. Structural basis of the day-night transition in a bacterial circadian clock. Science. 2017;355:1174–1180. doi: 10.1126/science.aag2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lella M, Mahalakshmi R. Metamorphic proteins: Emergence of dual protein folds from one primary sequence. Biochemistry. 2017;56:2971–2984. doi: 10.1021/acs.biochem.7b00375. [DOI] [PubMed] [Google Scholar]
- 14.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 15.Giganti D, et al. Secondary structure reshuffling modulates glycosyltransferase function at the membrane. Nat Chem Biol. 2015;11:16–18. doi: 10.1038/nchembio.1694. [DOI] [PubMed] [Google Scholar]
- 16.Chang YG, et al. Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science. 2015;349:324–328. doi: 10.1126/science.1260031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littler DR, et al. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279:9298–9305. doi: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- 18.Goodchild SC, et al. Oxidation promotes insertion of the CLIC1 chloride intracellular channel into the membrane. Eur Biophys J. 2009;39:129–138. doi: 10.1007/s00249-009-0450-0. [DOI] [PubMed] [Google Scholar]
- 19.Morimoto K, et al. The asymmetric IscA homodimer with an exposed [2Fe-2S] cluster suggests the structural basis of the Fe-S cluster biosynthetic scaffold. J Mol Biol. 2006;360:117–132. doi: 10.1016/j.jmb.2006.04.067. [DOI] [PubMed] [Google Scholar]
- 20.Jain V, et al. Structure of Prolyl-tRNA synthetase-halofuginone complex provides basis for development of drugs against malaria and toxoplasmosis. Structure. 2015;23:819–829. doi: 10.1016/j.str.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 23.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc Natl Acad Sci USA. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambroggio XI, Kuhlman B. Computational design of a single amino acid sequence that can switch between two distinct protein folds. J Am Chem Soc. 2006;128:1154–1161. doi: 10.1021/ja054718w. [DOI] [PubMed] [Google Scholar]
- 25.Porter LL, He Y, Chen Y, Orban J, Bryan PN. Subdomain interactions foster the design of two protein pairs with ∼80% sequence identity but different folds. Biophys J. 2015;108:154–162. doi: 10.1016/j.bpj.2014.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuinstra RL, et al. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter LL, Rose GD. A thermodynamic definition of protein domains. Proc Natl Acad Sci USA. 2012;109:9420–9425. doi: 10.1073/pnas.1202604109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann MT, Tischer A, Whitten ST, Auton M. Structural origins of misfolding propensity in the platelet adhesive von Willebrand factor A1 domain. Biophys J. 2015;109:398–406. doi: 10.1016/j.bpj.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 30.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E, Pielak GJ. Quinary structure modulates protein stability in cells. Proc Natl Acad Sci USA. 2015;112:1739–1742. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mylona A, et al. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science. 2016;354:233–237. doi: 10.1126/science.aad1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung N, et al. Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate. Proc Natl Acad Sci USA. 2016;113:2952–2957. doi: 10.1073/pnas.1516167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain V, Kikuchi H, Oshima Y, Sharma A, Yogavel M. Structural and functional analysis of the anti-malarial drug target prolyl-tRNA synthetase. J Struct Funct Genomics. 2014;15:181–190. doi: 10.1007/s10969-014-9186-x. [DOI] [PubMed] [Google Scholar]
- 34.Takala H, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heffernan R, et al. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci Rep. 2015;5:11476. doi: 10.1038/srep11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cock PJ, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrödinger LLC. 2017. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 2.0.
- 39.Yang Y, et al. SPIDER2: A package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. Methods Mol Biol. 2017;1484:55–63. doi: 10.1007/978-1-4939-6406-2_6. [DOI] [PubMed] [Google Scholar]
- 40.Fitzkee NC, Fleming PJ, Rose GD. The protein coil library: A structural database of nonhelix, nonstrand fragments derived from the PDB. Proteins. 2005;58:852–854. doi: 10.1002/prot.20394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.