Significance
The biological species concept (BSC) has served as the basis for defining species for over 75 years. Members of a biological species are defined by their ability to exchange genetic material, and it was originally thought that asexual lineages were not amenable to species-level classification based on the BSC since clonal individuals are reproductively isolated from one another. In this study, we demonstrate that the rates and patterns of gene exchange in acellular organisms (viruses and bacteriophages) allow the assignment of true biological species, an essential step to organizing the tree of life. Our results show that a universal species definition, based on the BSC, can be used to define biological species in all major lifeforms.
Keywords: speciation, recombination, biological species concept, gene flow, asexuality
Abstract
Due to their dependence on cellular organisms for metabolism and replication, viruses are typically named and assigned to species according to their genome structure and the original host that they infect. But because viruses often infect multiple hosts and the numbers of distinct lineages within a host can be vast, their delineation into species is often dictated by arbitrary sequence thresholds, which are highly inconsistent across lineages. Here we apply an approach to determine the boundaries of viral species based on the detection of gene flow within populations, thereby defining viral species according to the biological species concept (BSC). Despite the potential for gene transfer between highly divergent genomes, viruses, like the cellular organisms they infect, assort into reproductively isolated groups and can be organized into biological species. This approach revealed that BSC-defined viral species are often congruent with the taxonomic partitioning based on shared gene contents and host tropism, and that bacteriophages can similarly be classified in biological species. These results open the possibility to use a single, universal definition of species that is applicable across cellular and acellular lifeforms.
The delineation of species, and the assignment of individuals to species, is notoriously problematic for asexual organisms (1). Like other asexual organisms, viruses reproduce clonally; but unlike cellular organisms, viruses do not possess the universally distributed genes, such as ribosomal genes and replication-related proteins, that are typically used to reconstruct molecular relationships. Also, due to their relatively small genome sizes, and their high rates and diverse modes of evolution, it has been difficult to classify viruses by a unified framework.
At the most fundamental level, viruses are grouped into broad classes according to their form of genetic material and replication mechanism (e.g., the Baltimore classification) (2); and below these classes are refinements into taxonomic ranks based on virion morphology, genome contents, host range, antigenicity, and sequence identity (3–6). Although generally conforming to a Linnaean taxonomic hierarchy (www.ictvonline.org), viral classification below the level of subfamily is fraught with inconsistencies (7). There are guidelines for classifying viruses to a species—currently defined by the International Committee on Taxonomy of Viruses (ICTV) as a “monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria” (8)—but because these defining properties are not uniformly applied, species designations can be contrived rather than having a consistent biological basis (7). Moreover, this definition of a viral species, although stipulating monophyly, remains arbitrary about the benchmark by which clades attain species status, since multiple clades at various phylogenetic depths can be extracted from a single tree.
A more pragmatic approach has been to classify viral species according to host tropism; however, this implies genetic isolation between conspecifics that infect different host species. Moreover, viral sequences extracted from metagenomic datasets often lack information about hosts or about characteristics of the virion particle, making classification based on these characteristics unfeasible. Due to these limitations, it has been advised that a genome-based classification scheme would be useful for viral taxonomy in the metagenomic era (7). Species delineation based on genomic sequence information has become standard practice for bacteria and archaea (9), and similar taxonomic schemes can be applied to viruses, although different criteria might be needed for dsDNA, ssDNA, and RNA viruses to account for their diverse genome structures and rates of evolution.
To circumvent the demands of phylogenetic analysis and the frequent lack of species-defining characteristics, many species of viruses have been delineated using sequence identity thresholds, an approach that is commonly applied to asexual lifeforms but is often criticized on account of the arbitrary nature of such thresholds (10–12). Because viruses, as a whole, span a wide range of mutation rates, it is not possible to specify a particular sequence identity threshold that might correspond to species, as has been suggested for bacteria (13). Current classifications of viral species based on sequence similarity are varied and rely on clade-specific thresholds (11), preventing the emergence of a unified species definition in viruses. However, if viruses engage in homologous gene exchange, either by encoding their own recombination system or by recruiting their host’s (14, 15), it is possible that they form true biological species analogous to those defined in sexual cellular organisms.
Despite their asexual mode of reproduction, several microorganisms engage in sufficient levels of gene flow (i.e., homologous recombination) to distinguish biological species (16–21). We recently introduced a methodology that uses genome sequences to delineate reproductively isolated groups within bacteria and archaea based on their barriers of gene exchange, thereby offering a single framework to define biological species for any set of recombining organisms (21). Similarly, viruses do not rely on sexual reproduction; however, recent analyses of homologous recombination and network reconstructions suggest that species delineation based on gene flow would be possible for bacteriophages infecting cyanobacteria (22–26). Moreover, an experimental evolution study has shown that the mechanisms of speciation occurring in sexual organisms might also operate in viruses (27).
Although the routes and rates of homologous recombination remain poorly described for most viruses, we show that viruses with similar gene contents—typically those classified as members of the same genus—frequently recombine and that gene exchange occurs readily among highly divergent orthologs. Because so few viral taxa were found to evolve clonally, viruses and bacteriophages, like cellular organisms, can be classified into true biological species in accordance with the Biological Species Concept, and that their species limits are largely defined by their host tropism and overall genome contents.
Results
Defining True Biological Species.
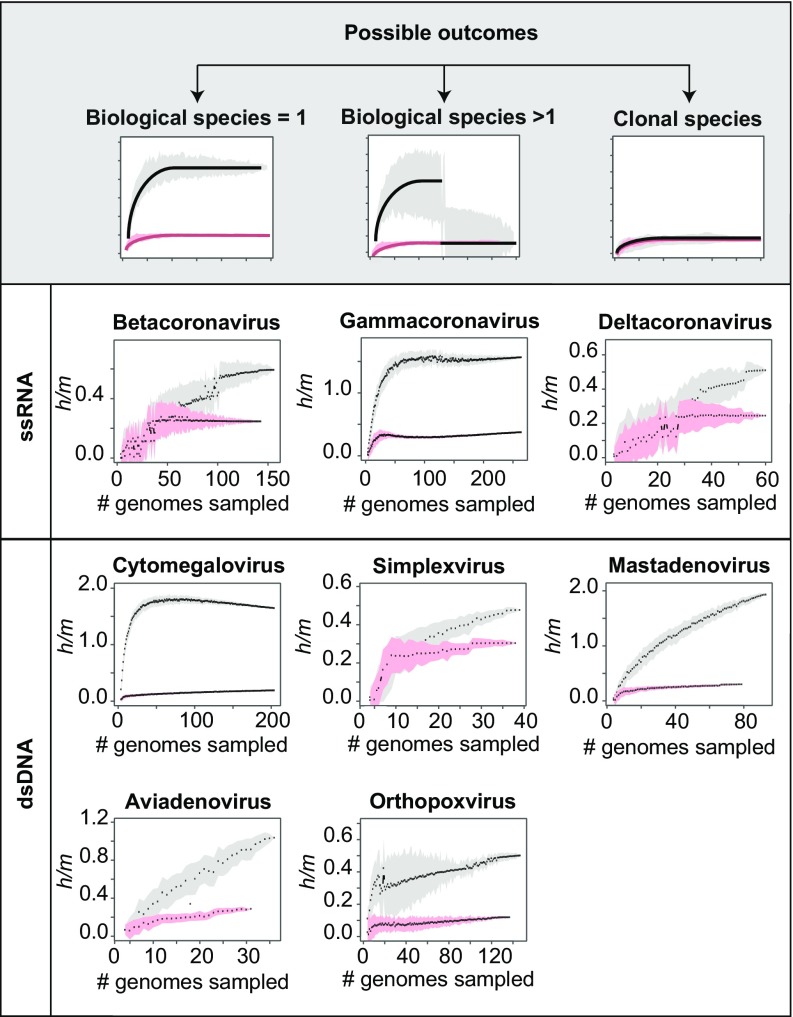
Because members of biological species are characterized by their capacity for gene exchange, we assessed the degree of recombination within eight genera of animal viruses for which there were both large numbers of fully sequenced genomes (n ≥ 15) and core genomes of sufficient size (≥20 kb) to accurately determine whether polymorphic sites arose by mutation or recombination. Our analyses identify gene flow between homologous sequences (i.e., recombination, homologous gene exchange) but does not infer events of horizontal gene transfer (i.e., gene acquisition), which need not involve homologous exchange. In some instances, genomes from a particular genus were excluded from the analysis due to their low amounts of shared gene content with other members of the genus (Methods and SI Appendix, Fig. S1). To estimate the prevalence of gene flow (i.e., homologous recombination) within these redefined genera, we estimated the ratio of homoplasic (h) to nonhomoplasic (m) polymorphisms along the core genome of each genus. Homoplasies are polymorphisms that are not compatible with vertical inheritance from a single ancestral mutation and likely result from the exchange of alleles through homologous recombination. High h/m ratios are indicative of a substantial signal of gene flow, and low h/m ratios are indicative of clonal or nearly clonal evolution (Fig. 1, Top). Sharp variations in the h/m ratios computed across different combinations of genomes—as detected by the exclusion criterion (Methods)—can indicate the presence of one or more genomes that do not engage in gene flow with the rest of the population. In all cases, viruses classified to the same genus showed a signal of gene flow (Fig. 1), and there was no statistical evidence that some viruses are “sexually” isolated from other members of the genus. These results suggest that many ICTV-defined viral genera are actually true biological species, as defined by the BSC.
Fig. 1.
Recognizing biological species. In each set of genomes comprising n strains, nonredundant combinations of i strains (ranging from 4 to n − 2 strains) were subsampled 100 times for each value of i. At each iteration of subsampling, the h/m ratio—the ratio of polymorphisms attributable to homoplasy relative to those attributable to mutation—was calculated for the concatenated alignment of genes common to all strains. Within the bivariate plots, black dots are medians and the gray-shaded region is the SD of the indicated number of subsampled combinations of strains. Red dots and pink-shaded regions denote median h/m values and SD for simulations in which all homoplasies are introduced by convergent mutations, as described in the text. Differences between the distributions of observed and simulated h/m values indicate the extent to which homoplasies are introduced by recombination. Top shows the three possible outcomes of these analyses: when there are no barriers to gene flow among strains (Left); when a discontinuity is produced by inclusion of a strain that does not participate in gene exchange (Center); and when there is clonal evolution or an absence of gene flow (Right). Lower displays results obtained for dsDNA and ssRNA viruses.
We extended this analysis to bacteriophages infecting Mycobacterium smegmatis, which represent the largest sampling of bacteriophage genomes infecting a single host and which have been classified into multiple clusters and subclusters based on gene content (28, 29). By applying the same procedures, we observed that all but one of the 17 bacteriophage clusters (cluster C1) are compatible with a BSC-like definition of species (SI Appendix, Fig. S2). Within cluster C1, we determined that one bacteriophage (Tonenili) is not recombining with the other members of the cluster, but after excluding this genome from our analysis, we retrieved a signal of homogeneous gene flow within the cluster (SI Appendix, Fig. S3). It should be noted that the bacteriophages in our dataset have diverse origins and analyzing viruses from similar geographic and ecological locations would likely result in tighter networks of gene flow, as suggested by previous studies (22–26).
These results suggest that gene flow significantly impacts viral microevolution, as has been reported for cyanophages (22–26). To further visualize the impact of gene flow on viral and bacteriophage evolution, we built dendrograms with SplitsTree, which depicts events of homologous recombination on a phylogenetic network (SI Appendix, Figs. S4 and S5). As predicted by the h/m ratios estimated from the core genomes of these taxa, most phylogenetic networks show high levels of reticulate evolution, indicative of a gene flow among members of a taxon.
Effects of Sampling and Evolutionary Rates on Defining Viral Species.
A key challenge for defining viral species based on gene flow is that viruses typically evolve at very fast rates (especially RNA and ssDNA viruses) (30), which could affect the inference of recombinant sites. It is possible for homoplasies to be introduced by independent, convergent mutations, which would result in an overestimation in the number of polymorphic sites that are introduced by recombination. To test for the effects of convergent mutations on the generation of homoplasies, we simulated sequences that mimicked the features of each viral dataset with respect to level of polymorphisms, nucleotide composition, and tree topology.
The large majority of viral clades and bacteriophage clusters that we defined as species displayed higher h/m ratios than the simulated sequences, indicating that convergent mutations represent only a small fraction of the homoplasies. However, three viral genera (Simplexvirus, Betacoronavirus, and Deltacoronavirus) and three bacteriophage clusters (A6, A9, and N) did not diverge substantially from random expectation. Such patterns can be caused by the lack of recombination but can arise also from the inclusion of multiple sexually isolated subclades, which would reduce the overall signal of recombination (21).
To test whether the three viral genera recognized as clonal (Simplexviruses, Betacoronaviruses, and Deltacoronaviruses) were actually composed of multiple recombining subclades, we partitioned the members of each genus into subclades based on their phylogenetic relationships. These subclades were built by progressively eliminating external taxa, and we then evaluated the pattern of recombination among the members within each of the newly assembled subclades. After decomposing these genera into smaller ensembles, we recovered clear signals of gene flow for Simplexvirus and Deltacoronavirus (SI Appendix, Fig. S6), indicating that these two genera do not evolve clonally, but rather, each contains multiple biological species that do not recombine with one another. Only a single viral genus, the Betacoronaviruses, appeared evolving in a clonal—or nearly clonal—manner. These results show that having similar gene contents is not the only condition for viruses to engage in gene flow and indicate that other forces—such as ecological factors—might restrict the ability of viruses to recombine.
Since recombination between viruses can only take place within coinfected cells, we reasoned that the degree of host tropism might affect the delineation of viral species, such that biological species might comprise viruses that infect a single host species. Despite infecting a wide range of mammalian and bird species, Mastadenoviruses, Aviadenoviruses, and Orthopoxviruses, were each determined to be a single biological species, suggesting that they switch hosts frequently enough to allow opportunities for recombination or that any interruption of gene flow is too recent to be detected.
In contrast, two viral genera that were originally viewed as clonal but later redefined as containing multiple biological species (Simplexviruses and Deltacoronaviruses) initially displayed a wide host tropism: the analyzed strains of Simplexviruses were isolated from humans, monkeys, bats, and rabbits; and Deltacoronaviruses from pigs and several species of birds. After redefining biological species in both of these genera, we observed that each of the newly defined species is confined to a single host species. The redefined biological species of Simplexviruses is confined to strains infecting human hosts, and the redefined biological species of Deltacoronaviruses is confined to strains infecting pigs. In contrast, the Betacoronaviruses, the only genus for which we were unable to define a single biological species, comprise viruses isolated from humans and camels, indicating that this genus might include two biological species of different host tropisms; but its sample size was too small to test this hypothesis.
Recombining Viruses Are Highly Promiscuous.
We analyzed the degree of genomic divergence among members in each of the species that we delineated and observed that despite potential sampling biases, several species contain highly divergent members (Fig. 2). For example, Mastadenovirus contains members that share only 58% sequence identity for genes constituting their core, despite the clear signal of gene flow among members. Such cases suggest that viruses are able—directly or indirectly—to engage in homologous recombination despite high levels of sequence divergence.
Fig. 2.
Maximum sequence divergence between members of viral and bacteriophage species. Shown are average nucleotide sequence identity values for orthologous genes shared by the two maximally divergent strains within each biological species.
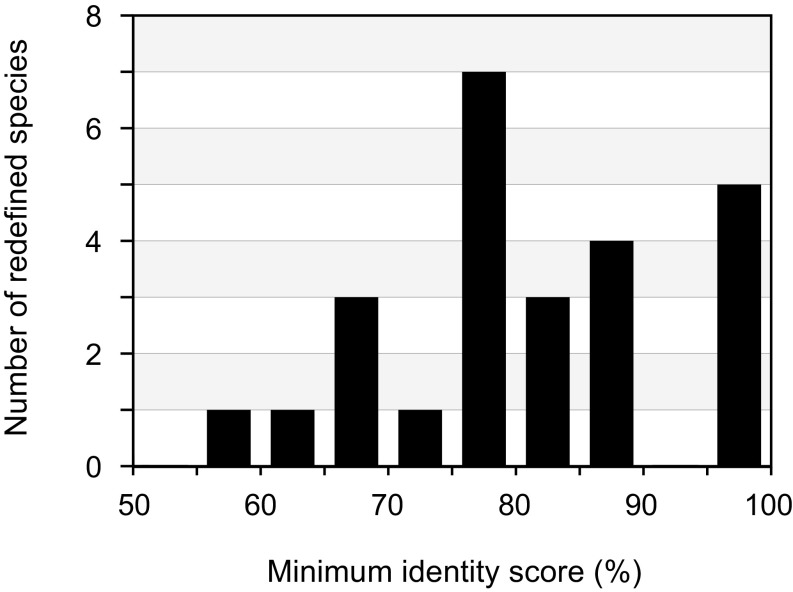
To compare the boundaries of viral species defined by the BSC to those designated by an alternative genome-based method, we estimated the average nucleotide identity (ANI) along the core genome for members of the same biological species and then calculated the number of groupings (ANI species) at different sequence-identity thresholds. (This analysis did not assume a single reference genome and it grouped together any pair of genomes that had a higher ANI value than the given threshold.) At sequence-identity thresholds that are commonly employed, there are often large numbers of ANI species within a single BSC-defined species (SI Appendix, Fig. S7), owing to the fact that divergent genomes retain the ability for homologous exchange. Naturally, the number of ANI species depends on the selected threshold; but importantly, each viral and bacteriophage taxon is impacted differently when analyzed at alternate sequence thresholds. For example, bacteriophage cluster E forms a single ANI species regardless of the chosen threshold, whereas cluster A2 constitutes one, or up to 30, ANI species depending on the threshold (SI Appendix, Fig. S7). Similarly, Cytomegalovirus would constitute a single ANI species, whereas Mastadenovirus would comprise ≥20 species if applying the same ANI threshold (SI Appendix, Fig. S7). Therefore, sequence thresholds do not define cohesive populations and—in many cases—distinct ANI species engage in gene flow.
The ability of divergent viruses to recombine can be associated with the presence of their own recombination systems, which are often more permissive than those of the host (31). A previous study identified recombinase genes across a large set of bacteriophages (14), and three of these genes, Bxz1 gp201, 244 gp117, and PMC gp61, were identified in bacteriophage species C1, E, and F1, respectively. These are, in fact, the three species that displayed the highest rates of recombination among the bacteriophage clusters we tested (SI Appendix, Fig. S2), supporting the view that phage-encoded recombinase genes promote high rates of genomic diversification through permissive homologous recombination.
In light of the potential for recombinational exchange between highly divergent sequences, we also explored whether members of different bacteriophage clusters exchange genes and actually constitute a single biological species. By examining those bacteriophage clusters that share high numbers of homologous genes (e.g., clusters A2, A6, and A9, and clusters A3 and A4), we observed that certain clusters engage in gene flow with one another (SI Appendix, Fig. S8). However, the pattern of gene flow among some of these bacteriophage clusters is asymmetric: members of cluster A2 are able to capture DNA from members of A6 and A9, but members of clusters A6 and A9 do not recombine with one another. This situation is analogous to so-called “ring species” in animals (32): clusters A2, A6, and A9 could be viewed as a single biological species, although some subpopulations (i.e., A6 and A9) are sexually isolated from one another. It is also possible that this asymmetry in gene flow is affected by sampling biases, since these bacteriophages were isolated from different locations.
Discussion
By analyzing patterns of recombination among members of named viral genera and clusters of bacteriophages, we show that the Biological Species Concept, i.e., the delineation of species based on the ability for gene flow among members, can be extended to include viruses and bacteriophages. Despite the limited number of viral clades for which there is sufficient genomic information for analysis, our results show that taxonomic boundaries based on gene flow can be established in viruses, as previously reported for bacteriophages infecting cyanobacteria (22–26) and in bacteriophage lambda that have coevolved with their host (27), contrasting traditional views that viruses, being asexual, evolve clonally. Although homologous recombination has been reported for many viruses, including the genera analyzed in this study (33–39), its frequency and significance on species delineation has been underestimated. In contrast to bacteria, viruses and bacteriophages have high rates of mutation, genetic reassortment and exchange, and gene uptake (especially for RNA and dsDNA viruses) (30), which can generate substantial genetic and genomic diversity. However, viral recombination is not indiscriminate but is confined largely to entities with similar gene contents, such that several of previous groupings based on shared gene contents actually constitute biological species.
Viruses can be organized into a hierarchical classification scheme based on genome contents and overall sequence similarity (6, 26, 40–45); however, genomic features are not the only attributes that dictate species membership. Because homologous exchange requires access to conspecifics, the boundaries of certain viral and bacteriophage species are also imposed by host range or tropism. Therefore, it is necessary to integrate species-level classifications based on gene flow with network-based approaches for higher taxonomic ranks to produce an accurate and consistent classification scheme for viruses and bacteriophages.
Our delineation of viral species is based on recognizing gene flow between viruses, here defined as exchange between gene homologs. However, viruses and bacteriophages engage in additional and more complex exchange processes in which genes are gained or lost through events of horizontal transfer. Acquisition of new genetic material can lead to genome mosaicism, with the result that very divergent viruses might share modules of highly similar contents and sequence (46). Notoriously, such genome mosaicism led to taxonomic conflict in lambdoid bacteriophages, since some possess gene modules of nearly identical sequence (and potentially engage in gene flow) while others lack these sequences (47).
Genome mosaicism has mainly hampered the taxonomy of temperate bacteriophages (48), which are thought to be prone to events of horizontal transfer due to their stable existence in the host chromosome and exposure to other infecting bacteriophages. Although high levels of gene acquisition will disrupt hierarchical schemes of classification (49), horizontal transfer seems to affect relatively few genes in cellular organisms and in most viruses (26, 48), such that a set of core genes can be evaluated for homologous exchange. And in fact, gene-sharing networks suggest that temperate bacteriophages can be ordered into a hierarchal taxonomic framework despite their mosaic structure (26).
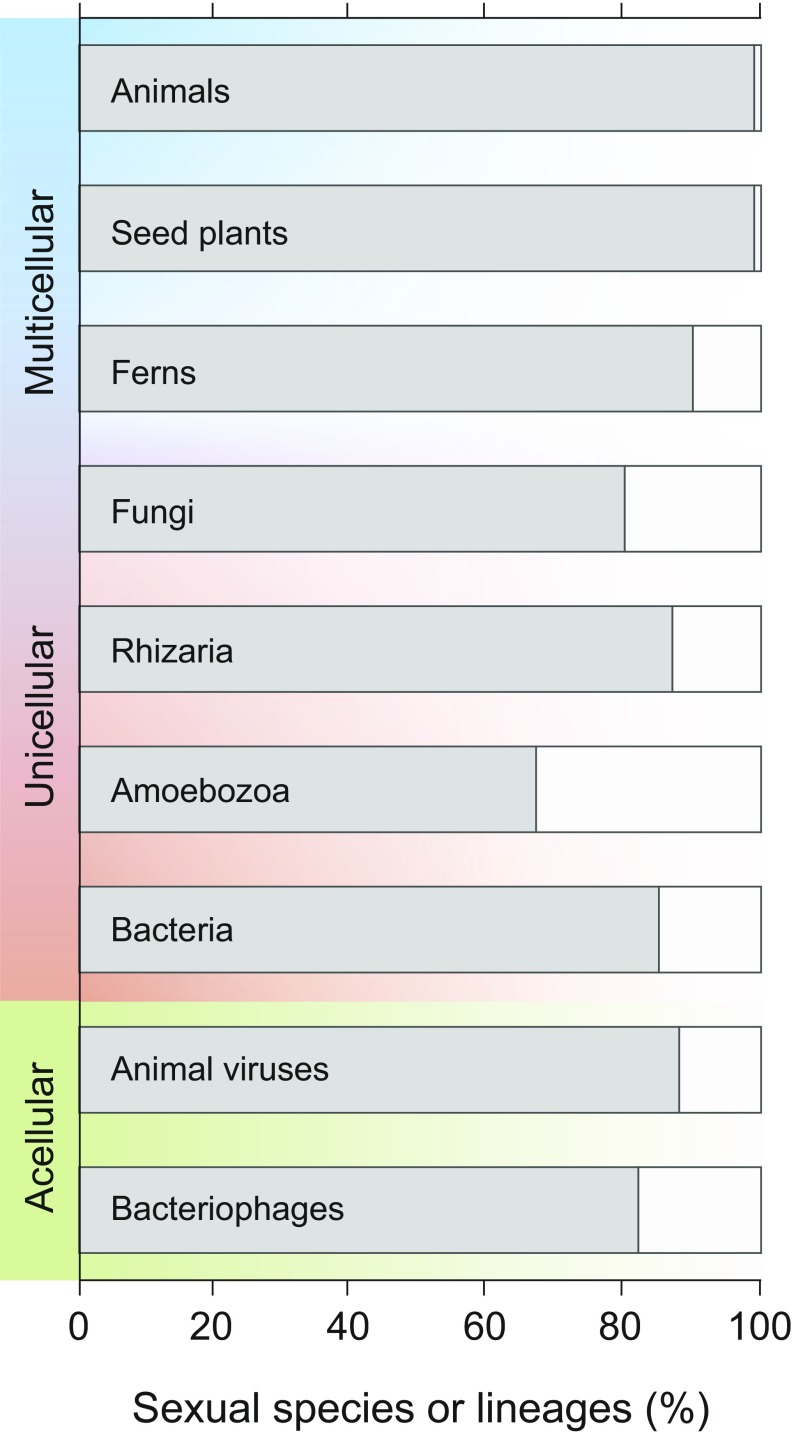
That viruses and bacteriophages can be classified into species based on a biological process—and on the same biological process used for cellular organisms—allows the mechanics of cladogenesis to be compared across all lifeforms. First, we can evaluate the adequacy and applicability of the BSC for species-level classification within each of the major groups of organisms. One shortcoming of the BSC has been its inability to delineate species in asexual organisms, which are ubiquitous among prokaryotes but usually considered to be rare in animals, plants, and fungi (50). Despite differences in cellularity and reproductive processes, the overwhelming majority of lineages in every group can be classified into species based on the BSC (Fig. 3). The BSC was originally formulated for animals, in which parthenogenesis and self-compatibility are rare, and its application to plants was somewhat more limited because it was originally estimated that upwards of 20% of plants reproduced solely by selfing (51, 52). However, reevaluation of the botanical literature suggests that outcrossing is as common in seed plants as in animals and that only about 10% of ferns do not regularly outcross (50). The ability to reproduce clonally is more common in protists (50), but evidence suggests that the extent of sexual reproduction—or some related mechanisms—remains largely underestimated in these lineages (53–55). Although bacteria are asexual sensu stricto, only about 15% are not amenable to classification by the BSC (21), and similarly, less than 20% of bacteriophage and viral groups do not engage in sufficient genetic exchange to be classified as biological species.
Fig. 3.
Prevalence of sex and related mechanisms in cellular and acellular taxa. Frequency of sexual species was compiled by ref. 50 for animals, seed plants, ferns, and fungi and by ref. 21 for bacteria (n = 91) and in the present study for animal viruses (n = 8) and bacteriophages (n = 17). Values reported for Rhizaria (n = 15) and Amoebozoa (n = 15) refer to the frequency of lineages containing sexual species reported in ref. 55.
The boundaries of viral and bacteriophage species are much broader than those in other groups. Whereas the maximal divergence between members of animal and plant species is rarely generally close to 1%, and rarely greater than 5% [as tabulated for neutral sites (56–58)], in every viral species that we examined, there are conspecifics whose homologs differ in sequence by over 20% (Fig. 2). In general, viral species accommodate much higher levels of sequence divergence than bacteriophage species, and all but three of the bacteriophage species (B2, B3, and E) show less than 5% sequence difference between their maximally divergent members (but note that all analyzed bacteriophages infect the same host: M. smegmatis). This ability to recombine with very distant homologs even exceeds what has been reported for bacteria, in which 30% of BSC-defined species have a maximal divergence among conspecifics greater than 5% (21).
The ability for highly divergent viruses to engage in gene flow likely results from their high rates of mutation coupled with action of diversifying selection, since hosts typically impose strong selective pressures that promote diversification (59, 60). Compared with cellular organisms, viruses are capable of recombining more highly mismatched sequences (31); and consistent with this hypothesis, we note that bacteriophages encoding their own recombination systems have the highest rates of gene flow. That viruses can be classified into species based on gene exchange is somewhat paradoxical, since recombination is only rarely required to accomplish viral lifecycles (15, 61), and the majority does not encode for recombination enzymes or rely on this mechanism to propagate. In such cases, recombination can be achieved through using the host recombinase or through “copy choice” processes, in which hybrid genomes are generated by template disassociation and reassociation of the replication complex in coinfected cells (62–64).
Finding that viruses and bacteriophages are amenable to species-level classification based on the Biological Species Concept implies that they can be fully organized in the Linnaean hierarchy established for cellular organisms. We find that biological species of viruses and bacteriophages can be associated with host tropism; however, the high incidence of host switching and uncertainties about primary host reservoirs, combined with the potential for recombination between very divergent genomes, contravenes the attachment of viral species’ names to a particular host (65, 66). Moreover, linking viruses to hosts has become particularly problematic in metagenomic studies, since sequences often originate from sources unrelated to host infection and many of the traits typically used for viral classification remain inaccessible (67, 68). Although our analyses are currently restricted to fully sequenced genomes, there has been substantial progress in the recovery of identification and assembly of viral sequences from metagenomic datasets (69), thereby extending the applicability of our approach. By defining the fundamental units of viral taxonomy, our approach constitutes a step toward the consensus reached by the ICTV (7), stating that a sequence-based approach, centered on a uniform and universally relevant criterion, represents the optimal tool for classifying viruses and bacteriophages into a coherent structure.
Methods
Viral Datasets and Defining Core Genomes.
We retrieved 3,107 genomes of animal viruses representing 15 named genera from the RefSeq database (https://www.ncbi.nlm.nih.gov/) and the entire collection of 1,154 bacteriophage genomes infecting M. smegmatis (phagesdb.org) in May 2017. Animal viruses were selected based on two criteria: (i) the availability of a large number (n ≥ 15) of genomes classified under the same genus name and (ii) an average genome size exceeding 20 kb for members of a genus (SI Appendix, Fig. S1, step 1). Bacteriophages were annotated in GLIMMER v3.02 (70) and grouped taxonomically into clusters based on gene content, as defined in refs. 28 and 29. For each viral genus and bacteriophage cluster, we obtained the set of orthologous proteins shared by each pair of genomes with USEACH Global v5.2 with 70% identity and 80% length conservation (71) and then defined a “core” genome as the set of single-copy orthologous genes shared by at least 85% of its members.
In cases where there were large differences in genome contents among members of a genus, we systematically redefined the genus borders as follows: For each pair of genomes, we defined the ratio of shared orthologs (S) between genomes A and B as SAB = OAB/min(A,B), where OAB represents the number of orthologs shared by genomes A and B, and min(A,B) represent the total number of genes of the smaller genome (72). From the matrix of scores S, we grouped genomes in MCL v14-137 (73) with an inflation parameter of 1.2 (i.e., the minimal value for obtaining large clusters) and redefined each genus as the genomes included in the largest cluster. Following this procedure, a total of four genera were redefined, and their corresponding core genomes were based on each of the redefined sets of genomes (SI Appendix, Fig. S1, step 2). All cases in which the size of the core genome was less than 20% of the average genome size were excluded, leaving a total of the following eight viral genera: Cytomegalovirus, Simplexvirus, Mastadenovirus, Aviadenovirus, Orthopoxvirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (SI Appendix, Fig. S1, step 3). None of the bacteriophage clusters (A1–A6, A9, B1–B3, C1, E, F1, J, K1, L2, N) was subdivided, since each was characterized by a sufficiently large core genome. The lists of analyzed viruses and bacteriophages are available in Datasets S1 and S2, respectively.
For each viral genus or bacteriophage cluster, the protein sequences of each gene in the core genome were aligned with MAFFT v7.271 (74), reverse translated into their corresponding nucleotide sequences, and merged into a single concatenate. Additionally, we attempted to build additional core genomes for pairs of viral genera and of bacteriophage clusters that had similar genome contents, with the result that four pairs of bacteriophage clusters (A2–A6, A2–A9, A6–A9, and A3–A4) possessed relatively well-conserved joint core genomes consisting of 16 or more shared orthologs.
Analysis of Gene Flow.
For each viral genus and bacteriophage cluster, estimates of recombination were based on homoplasies as in ref. 21. First, a matrix of core genome distances D was built for each genus or cluster using RAxML v8.2.7 (75) under a generalized time-reversible (GTR) model, and redundant genomes—those with no or little divergence (D ≤ 0.00005) to another genome—were randomly removed (SI Appendix, Fig. S1, step 4). For each core genome, polymorphic sites were inferred as homoplasic when max(D11) > min(D10), where max(D11) represents the distance between the genomes harboring the minor allele, and min(D01) represents the minimal distance between genomes harboring a minor allele and a major allele. We then computed the ratio h/m, defined as the ratio of homoplasic (h) alleles to nonhomoplasic (m) alleles for multiple combinations of genomes, such that groups of genomes with higher h/m ratios have more polymorphisms attributable to recombination. For each viral genus or bacteriophage cluster, we randomly sampled 100 nonredundant combinations of genomes for different numbers of genomes (from 4 to n – 2, with n the total number of genomes in the genus or cluster being analyzed). Within each viral genus or bacteriophage cluster, we identified genomes that led to a sharp reduction of the h/m ratio relative to other genomes by applying an exclusion criterion as in ref. 21. Such genomes, as they do not recombine with other members of the population, are not considered members of the same biological species (SI Appendix, Fig. S1, step 5). We also applied our exclusion criterion to those pairs of bacteriophage clusters (A2–A6, A2–A9, A6–A9, and A3–A4) with highly similar core genomes. In these cases, those pairs of clusters that did not display a significant decrease in gene flow based on our exclusion criterion were considered to be the same biological species (SI Appendix, Fig. S1, step 6).
Simulations.
We assessed the expected number of homoplasies that might be introduced by convergent mutations in each of the datasets through simulations as in ref. 21. We built a maximum likelihood tree for the core genomes in each genus or cluster with RAxML v8.2.7 (75) under a GTR model. Then, using SeqGen v1.3.3 (76), the resulting tree was applied to generate an alignment that maintained the nucleotide composition, the number of genomes, and the length of the alignment. Because phylogenetic inference considers recombination events as multiple independent mutation events (thereby overestimating the number of mutations that accumulated in the simulated alignments), we rescaled the length of the branches of the trees uniformly to match the level of polymorphisms in the simulated alignments and the real data. Each simulated alignment was then subjected to the same resampling strategy to detect homoplasies.
Phylogenetic Networks.
For each viral genus and bacteriophage cluster, phylogenetic networks were built with SplitsTree v4 (77) with default parameters.
Supplementary Material
Acknowledgments
We thank Kim Hammond for help with preparation of the figures. This work was supported by the National Institutes of Health award R35GM118038 (to H.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717593115/-/DCSupplemental.
References
- 1.Shapiro BJ, Polz MF. Microbial speciation. Cold Spring Harb Perspect Biol. 2015;7:a018143. doi: 10.1101/cshperspect.a018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abedon ST, Calendar RL. The Bacteriophages. 2nd Ed. Oxford Univ Press; Oxford: 2005. p. 768. [Google Scholar]
- 4.Krupovič M, Bamford DH. Order to the viral universe. J Virol. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy: VIIIth Report of the International Committee on Tanoxomy of Viruses. Elsevier Academic; London: 2005. [Google Scholar]
- 6.Rohwer F, Edwards R. The phage proteomic tree: A genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmonds P, et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 8.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. Recently agreed changes to the International Code of Virus Classification and Nomenclature. Arch Virol. 2013;158:2633–2639. doi: 10.1007/s00705-013-1749-9. [DOI] [PubMed] [Google Scholar]
- 9.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson AT. Defining viral species: Making taxonomy useful. Virol J. 2014;11:131. doi: 10.1186/1743-422X-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmonds P. Methods for virus classification and the challenge of incorporating metagenomic sequence data. J Gen Virol. 2015;96:1193–1206. doi: 10.1099/jgv.0.000016. [DOI] [PubMed] [Google Scholar]
- 12.Krause DJ, Whitaker RJ. Inferring speciation processes from patterns of natural variation in microbial genomes. Syst Biol. 2015;64:926–935. doi: 10.1093/sysbio/syv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–3962. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobay LM, Touchon M, Rocha EP. Manipulating or superseding host recombination functions: A dilemma that shapes phage evolvability. PLoS Genet. 2013;9:e1003825. doi: 10.1371/journal.pgen.1003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 17.Simmons SL, et al. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadillo-Quiroz H, et al. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 2012;10:e1001265. doi: 10.1371/journal.pbio.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol. 2014;12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro BJ, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bobay LM, Ochman H. Biological species are universal across life’s domains. Genome Biol Evol. 2017;9:491–501. doi: 10.1093/gbe/evx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marston MF, Amrich CG. Recombination and microdiversity in coastal marine cyanophages. Environ Microbiol. 2009;11:2893–2903. doi: 10.1111/j.1462-2920.2009.02037.x. [DOI] [PubMed] [Google Scholar]
- 23.Gregory AC, et al. Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genomics. 2016;17:930. doi: 10.1186/s12864-016-3286-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marston MF, Martiny JB. Genomic diversification of marine cyanophages into stable ecotypes. Environ Microbiol. 2016;18:4240–4253. doi: 10.1111/1462-2920.13556. [DOI] [PubMed] [Google Scholar]
- 25.Cordero OX. Endemic cyanophages and the puzzle of phage-bacteria coevolution. Environ Microbiol. 2017;19:420–422. doi: 10.1111/1462-2920.13674. [DOI] [PubMed] [Google Scholar]
- 26.Bolduc B, et al. vConTACT: An iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ. 2017;5:e3243. doi: 10.7717/peerj.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer JR, et al. Ecological speciation of bacteriophage lambda in allopatry and sympatry. Science. 2016;354:1301–1304. doi: 10.1126/science.aai8446. [DOI] [PubMed] [Google Scholar]
- 28.Hatfull GF, et al. Comparative genomic analysis of 60 mycobacteriophage genomes: Genome clustering, gene acquisition, and gene size. J Mol Biol. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pope WH, et al. Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science; Phage Hunters Integrating Research and Education; Mycobacterial Genetics Course Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife. 2015;4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duchêne S, Holmes EC. Estimating evolutionary rates in giant viruses using ancient genomes. Virus Evol. 2018;4:vey006. doi: 10.1093/ve/vey006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinsohn JT, Radman M, Petit MA. The lambda red proteins promote efficient recombination between diverged sequences: Implications for bacteriophage genome mosaicism. PLoS Genet. 2008;4:e1000065. doi: 10.1371/journal.pgen.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cain AJ. Animal Species and Their Evolution. Hutchinson House; London: 1954. [Google Scholar]
- 33.Liao CL, Lai MM. RNA recombination in a coronavirus: Recombination between viral genomic RNA and transfected RNA fragments. J Virol. 1992;66:6117–6124. doi: 10.1128/jvi.66.10.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faure-Della Corte M, et al. Variability and recombination of clinical human cytomegalovirus strains from transplantation recipients. J Clin Virol. 2010;47:161–169. doi: 10.1016/j.jcv.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Sijmons S, Van Ranst M, Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses. 2014;6:1049–1072. doi: 10.3390/v6031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 37.Nagy M, Nagy E, Tuboly T. Sequence analysis of porcine adenovirus serotype 5 fibre gene: Evidence for recombination. Virus Genes. 2002;24:181–185. doi: 10.1023/a:1014580802250. [DOI] [PubMed] [Google Scholar]
- 38.Benko M, Harrach B, Russell WC. Family Adenoviridae. In: Regenmortel MHVv, et al., editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Academic; San Diego: 2000. [Google Scholar]
- 39.Moss B. Poxviridae: The viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields’ Virology. 3rd Ed. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 2637–2671. [Google Scholar]
- 40.Lima-Mendez G, Toussaint A, Leplae R. Analysis of the phage sequence space: The benefit of structured information. Virology. 2007;365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 41.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 42.Iranzo J, Koonin EV, Prangishvili D, Krupovic M. Bipartite network analysis of the archaeal virosphere: Evolutionary connections between viruses and capsidless mobile elements. J Virol. 2016;90:11043–11055. doi: 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corel E, Lopez P, Méheust R, Bapteste E. Network-thinking: Graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016;24:224–237. doi: 10.1016/j.tim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iranzo J, Krupovic M, Koonin EV. A network perspective on the virus world. Commun Integr Biol. 2017;10:e1296614. doi: 10.1080/19420889.2017.1296614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiewsakun P, Simmonds P. The genomic underpinnings of eukaryotic virus taxonomy: Creating a sequence-based framework for family-level virus classification. Microbiome. 2018;6:38. doi: 10.1186/s40168-018-0422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juhala RJ, et al. Genomic sequences of bacteriophages HK97 and HK022: Pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 47.Casjens SR. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res Microbiol. 2008;159:340–348. doi: 10.1016/j.resmic.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mavrich TN, Hatfull GF. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol. 2017;2:17112. doi: 10.1038/nmicrobiol.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doolittle WF, Bapteste E. Pattern pluralism and the tree of life hypothesis. Proc Natl Acad Sci USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burt A. Perspective: Sex, recombination, and the efficacy of selection–Was Weismann right? Evolution. 2000;54:337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 51.Stebbins GL. Perspectives. I. Animal species and evolution by Ernst Mayr, a review. Am Sci. 1963;51:362–370. [Google Scholar]
- 52.White MJ. Modes of Speciation. Freeman; San Francisco: 1978. p. 456. [Google Scholar]
- 53.Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One. 2007;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E. The chastity of amoebae: Re-evaluating evidence for sex in amoeboid organisms. Proc Biol Sci. 2011;278:2081–2090. doi: 10.1098/rspb.2011.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cutter AD, Jovelin R, Dey A. Molecular hyperdiversity and evolution in very large populations. Mol Ecol. 2013;22:2074–2095. doi: 10.1111/mec.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romiguier J, et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature. 2014;515:261–263. doi: 10.1038/nature13685. [DOI] [PubMed] [Google Scholar]
- 58.Roux C, et al. Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 2016;14:e2000234. doi: 10.1371/journal.pbio.2000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H. Phage-host interaction: An ecological perspective. J Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 61.Smith GR. General recombination. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1983. pp. 175–210. [Google Scholar]
- 62.Coffin JM. Structure, replication, and recombination of retrovirus genomes: Some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 63.Kim MJ, Kao C. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: Implications for RNA-RNA recombination. Proc Natl Acad Sci USA. 2001;98:4972–4977. doi: 10.1073/pnas.081077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc Natl Acad Sci USA. 2011;108:238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geoghegan JL, Duchêne S, Holmes EC. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi M, Zhang Y-Z, Holmes EC. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018;243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 72.Bobay LM, Rocha EP, Touchon M. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol. 2013;30:737–751. doi: 10.1093/molbev/mss279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 76.Rambaut A, Grassly NC. Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 77.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.